Abstract
Naringin exhibited various pharmacological activities. However, its biological function and underlying mechanism in regulating macrophage polarization remain elusive. This study aimed to investigate the regulatory network between naringin and macrophage polarization in sepsis-induced intestinal injury. Cecal ligation and puncture (CLP) was used to establish the animal model of sepsis. Chromatin immunoprecipitation and a luciferase reporter assay were used to determine the interplay between peroxisome proliferator-activated receptor γ (PPARγ) and miR-21 promoter, as well as miR-21 and its target genes. Naringin enhanced the overall survival of septic mice and alleviated the CLP-induced inflammatory response and intestinal damage. This was accompanied by the increased expression of PPARγ in the intestines and the stimulation of ileal macrophages toward the M2 phenotype. Furthermore, in lipopolysaccharide-stimulated bone marrow-derived macrophages, naringin stimulated M2 polarization. Mechanistically, PPARγ inhibition attenuated the promotion of M2 polarization caused by naringin, and the naringin/PPARγ regulatory work was compromised by miR-21 inhibition. The present study suggested that naringin promoted M2 polarization via the PPARγ/miR-21 axis, thus relieving sepsis-induced intestinal injury. This study provides novel insights into the mechanism by which naringin alleviated sepsis-induced intestinal injury through regulation of macrophage polarization.
Keywords: naringin, PPARγ, miR-21, M1/M2 polarization, sepsis-induced intestinal injury
Graphical abstract
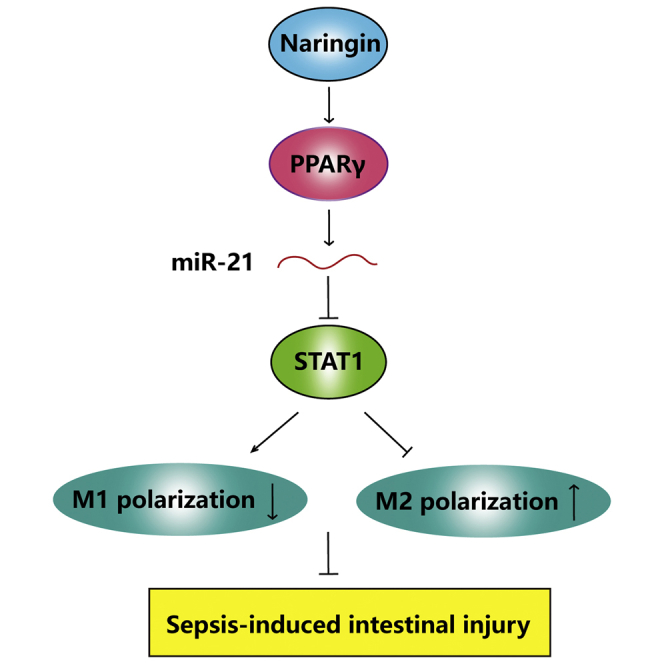
The present study first demonstrates that naringin alleviated sepsis-induced intestinal injury by modulating macrophage polarization. PPARγ regulated transcription of miR-21, and STAT1 was identified as a direct target of miR-21. Naringin promoted M2 macrophage polarization via regulating the PPARγ/miR-21/STAT1 axis, thus relieving sepsis-induced intestinal injury.
Introduction
Sepsis is a deleterious clinical condition caused by dysregulation of the inflammatory response to infection and has a very high mortality rate.1,2 The uncontrolled inflammation response caused by pathogens, the consequent immune disorder, and immunosuppression are the underlying mechanisms.3 Among these processes, the dynamics and polarization of macrophages play important roles, especially in sepsis-induced intestinal injury.1,4
As indispensable components of innate immunity, macrophages contribute significantly to the inflammatory response. Once activated, they have two distinct phenotypes depending on circumstances: the M1 phenotype that is manifested by the appearance of pro-inflammatory cytokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β; and the M2 phenotype that participates in the secretion of anti-inflammatory cytokines (e.g., IL-4 and IL-10).5, 6, 7 Differential expression of macrophage markers and cell surface receptors are implicated in the phenotypical distinction between M1 and M2 macrophages. CD86 and inducible nitric oxide synthase (iNOS) are associated with M1 markers, while CD206, Arg-1, and IL-10 are well-accepted M2 markers.8,9 These two phenotypes have been well implicated in the progression of sepsis. The large amount of inflammatory cytokines released by the M1 macrophages triggers the cytokine storm syndrome and septic shock, leading to damage of the tissues and even death in serious sepsis.10 In contrast, the anti-inflammatory activities induced by M2 macrophages effectively suppress the occurrence and progression of sepsis.11,12 To the best of our knowledge, only one study has demonstrated that the amelioration of lipopolysaccharide (LPS)-induced intestinal injury was followed by the increased proportion of M2 macrophages.13 The underlying mechanism, however, is complicated and remains to be explored.
Peroxisome proliferation-activated receptor γ (PPARγ) has been increasingly recognized as an important regulator of inflammation, which is also the key to the progression of sepsis.14,15 Activation of PPARγ has been shown to effectively suppress expression of inflammatory cytokines and tissue damages caused by sepsis.16, 17, 18 One possible mechanism is that PPARγ can facilitate macrophage polarization toward the M2 phenotype19,20 through directly regulating the expression of STAT6 and related genes.21 Furthermore, PPARγ has been reported to upregulate the expressions of miR-23 and miR-223, which in turn promotes M2 macrophage polarization.22,23 As one of the most abundant microRNAs (miRNAs) in human tissues, miR-21 has been implicated in regulating the polarization of bone marrow-derived macrophages (BMDMs).24 A previous study further revealed that miR-21 inhibits M1 polarization and promotes M2 polarization. In sepsis, increased miR-21 works in concert with decreased miR-128 to induce podocyte injury,25 and STAT3 and C/EBPβ are implicated in the regulation of miR-21 during sepsis.26 Our bioinformatics analysis based on the JASPAR database shows that PPARγ has a binding sequence in the promoter region of miR-21, raising the possibility that PPARγ is also involved in the miR-21-regulated macrophage polarization. In addition, LPS activates nuclear factor κB (NF-κB) and STAT1 pathways to skew macrophages toward the M1 phenotype.27 A previous study reported that pentamethoxyflavanone ameliorates sepsis by regulating macrophage polarization via the STAT1/STAT6 pathway.28 Interestingly, bioinformatics analysis predicts putative binding sites between miR-21 and the STAT1 3′ UTR, suggesting that STAT1 might be a functional target of miR-21 during sepsis.
Despite the importance of macrophage polarization during the inflammation response process, few compounds have been identified that are able to stimulate the transformation of macrophages to the M2 phenotype to improve sepsis.18,28,29 Naringin is a citrus flavonoid generally found in the pericarp of citrus fruits.30,31 Accumulating evidence suggests that naringin possesses a variety of pharmacological activities, including anti-osteoporotic, anti-cancer, anti-apoptotic, and anti-inflammatory effects, as well as effects on metabolic syndrome and central nervous system diseases, including neurological disorders.32, 33, 34 Several reports have confirmed that naringin protects against steroid-induced ischemic necrosis by upregulating PPARγ,35 and naringin can activate the 5’ adenosine monophosphate-activated protein kinase (AMPK) pathway to induce anti-inflammatory activity in sepsis.36 Our previous studies have confirmed that naringin improves sepsis-induced intestinal injury through the regulation of the ROCK1/2 pathway, but the study of macrophage polarization by naringin has not yet been conducted.37 In this study, we investigated the potential of naringin in macrophage polarization and explored the underlying mechanisms. Using a mouse model of sepsis, we first demonstrated the promising anti-inflammatory activity of naringin that led to an amelioration of sepsis-induced intestinal injury. Our mechanistic studies suggested that naringin promoted the transformation of macrophages to the M2 phenotype via the PPARγ/miR-21 axis.
Results
Naringin protected against sepsis-induced intestinal injury in mice
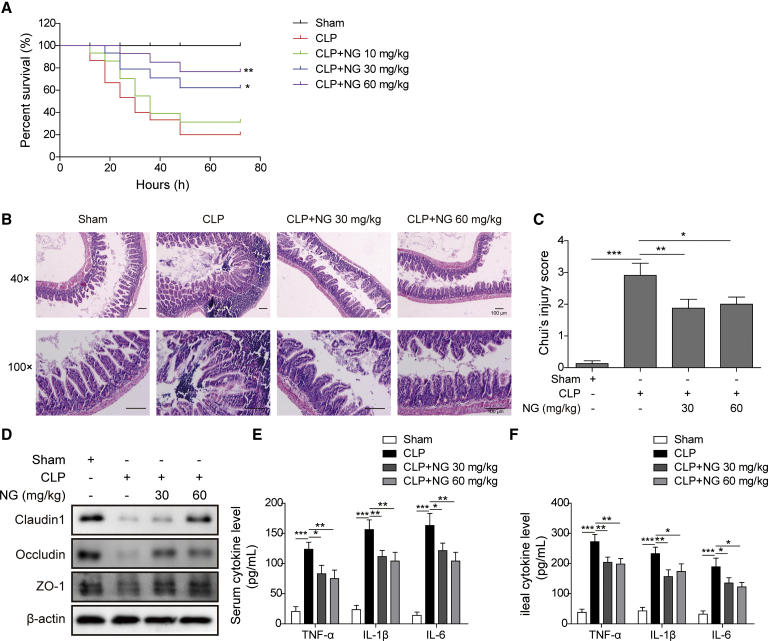
We initially confirmed the protective effects of naringin against sepsis-induced intestinal injury in C57BL/6J mice. Naringin was administrated at doses of 10, 30, and 60 mg/kg intraperitoneally 30 min after challenging by cecal ligation and puncture (CLP). To determine the most effective dose of naringin for the mouse sepsis model, mice were closely monitored for 72 h after CLP challenge. While all mice survived within 72 h in the sham group, the survival rate reduced to 20% in mice with CLP challenge (Figure 1A). Postoperative administration of 10 mg/kg naringin exerted no significant influence on the overall survival of the septic mice. As the dose of naringin increased to 30 or 60 mg/kg, however, the survival rates of the septic mice treated with naringin turned out to be much higher than those for mice in the CLP group (Figure 1A). Given that significant differences were observed by 30 and 60 mg/kg naringin with regard to survival rate, these two doses were selected for the subsequent analyses. Hematoxylin and eosin (H&E) staining showed that CLP caused significant changes in ileal morphology with a markedly increased injury score, while naringin administration reduced the injury score significantly (Figures 1B and 1C). Similar to the ileal morphology, the H&E results showed that CLP-induced histological damage in colon was also alleviated by naringin in a dose-dependent manner (Figure S1). Western blotting was then performed to examine the levels of tight junction proteins. The protein levels of claudin-1, occludin, and ZO-1 markedly decreased in septic mice, whereas administration of naringin in a higher dose (60 mg/kg) restored these proteins to the levels similar to the sham group (Figure 1D). The concentrations of cytokines in ileum (locally) and serum (systemically) were then detected 24 h after surgery to determine the how naringin affects the inflammatory response following CLP. Under the condition of sepsis, inflammatory markers including IL-1β, IL-6, and TNF-α were substantially augmented in ileum and serum (Figures 1E and 1F). The levels of these inflammatory cytokines, however, were reduced substantially after treatment with naringin (Figures 1E and 1F).
Figure 1.
Naringin protected against sepsis-induced intestinal injury in mice
(A) Survival rate of mice after CLP with different doses of naringin. (B) Representative images of ileum stained with H&E. Scale bar: 100 μm. (C) Intestinal injury scores in the four indicated groups. (D) Expression of tight junction proteins was measured by western blotting. (E and F) TNF-α, IL-1β, and IL-6 in serum (E) and ileum (F) were measured by an ELISA assay. Serum and ileum were collected 24 h after CLP. Data represent the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. NG, naringin.
Naringin regulated macrophage polarization in septic mice
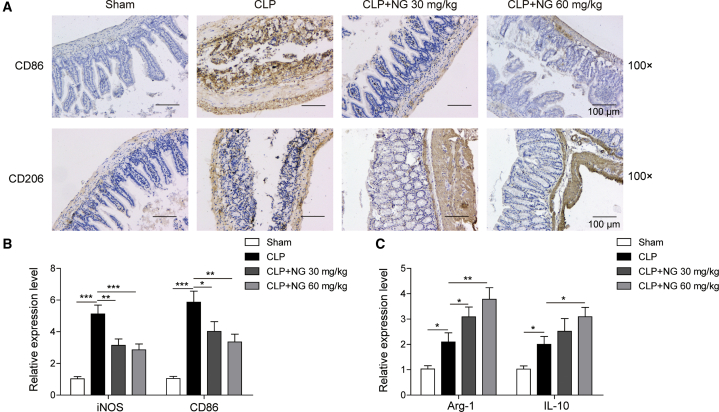
Next, we sought to determine whether naringin regulates macrophage polarization in ileal tissues from septic mice. Immunohistochemistry (IHC) analyses revealed that CLP challenge led to a significant increase in the M1 marker CD86, whereas naringin treatment inhibited M1 macrophage polarization and induced M2 macrophage polarization in which the M2 marker CD206 was increased by naringin (Figure 2A). Quantitative real-time PCR further showed that CLP challenge stimulated the expression of M1 macrophage markers iNOS and CD86, with a slight induction of the expression of M2 macrophage markers Arg-1 and IL-10. Alternatively, administration of naringin dramatically suppressed the CLP-induced M1 marker expression and induced the M2 marker expression (Figures 2B and 2C).
Figure 2.
Naringin regulated macrophage polarization in septic mice
(A) Expression levels of CD86 and CD206 were detected by IHC. Scale bar: 100 μm. (B) Expression levels of iNOS and CD86 were detected by quantitative real-time PCR. (C) Expression levels of Arg-1 and IL-10 were detected by quantitative real-time PCR. Data represent the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Naringin regulated LPS-stimulated macrophage polarization via stimulating PPARγ and its downstream miRNA expression
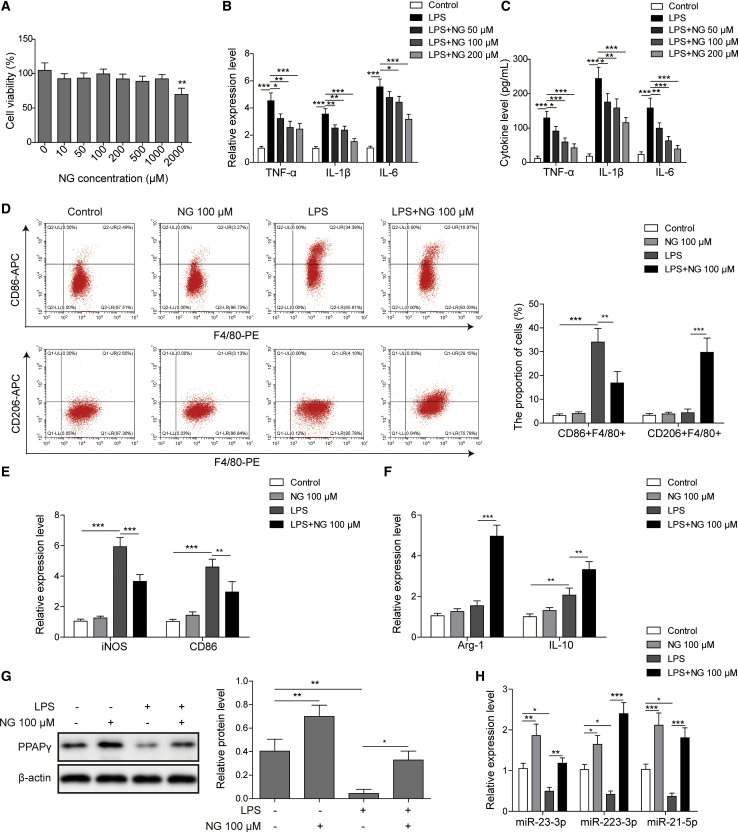
A toxicity test showed that naringin was non-toxic to BMDMs at a concentration of 1,000 μM but exhibited significant cytotoxicity at 2,000 μM (Figure 3A). Quantitative real-time PCR revealed that LPS induced the expression of inflammatory markers IL-1β, IL-6, and TNF-α in BMDMs, whereas naringin at doses of 50, 100, and 200 μM significantly reduced the expression of these cytokines (Figure 3B). For the secreted cytokines, similar results were also observed by an enzyme-linked immunosorbent assay (ELISA) (Figure 3C). 100 μM was thus chosen as the dose to be used in subsequent experiments.
Figure 3.
Naringin regulated LPS-stimulated macrophage polarization via stimulating PPARγ and its downstream miRNA expression
(A) An MTT assay was used to assess the toxicity of naringin with designated doses. (B) The mRNA levels of TNF-α, IL-1β, and IL-6 in BMDMs were measured by quantitative real-time PCR. (C) The secretion levels of TNF-α, IL-1β, and IL-6 in culture medium of BMDMs were measured by ELISA. (D) M1 macrophages (F4/80+CD86+) and M2 macrophages (F4/80+CD206+) in LPS-stimulated BMDMs with or without naringin administration (100 μM) were examined by flow cytometry. (E and F) The mRNA levels of M1 markers iNOS and CD86 (E) and M2 markers Arg-1 and IL-10 (F) were measured by quantitative real-time PCR. (G) The protein level of PPARγ was measured by western blotting with quantitative analysis. (H) The expression levels of miRNAs associated with macrophage polarization were detected by quantitative real-time PCR. Data represent the mean ± SD of three experiments performed in triplicate independently. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Flow cytometry was further performed to investigate the effect of naringin on macrophage polarization in vitro. We found that the proportion of M2 macrophages (CD206+) was unchanged in LPS-stimulated BMDMs compared with BMDMs from the control group (Figure 3D). Accordingly, the numbers of M1 macrophages (CD86+) were enhanced remarkably after LPS stimulation (Figure 3D). Naringin treatment (100 μM) significantly augmented M2 macrophage polarization in LPS-induced BMDMs, while it had no inductive effect in the normal group (Figure 3D). In addition, the LPS-induced number of M1 macrophages (CD86+) was attenuated by naringin (Figure 3D). Additionally, to ascertain the impact of naringin on macrophage polarization, M1 macrophage markers (iNOS and CD86) and M2 macrophage markers (Arg-1 and IL-10) and PPARγ were analyzed (Figures 3E–3G). Naringin downregulated iNOS and CD86, but it upregulated Arg-1, IL-10, and PPARγ in the LPS+naringin group as compared with LPS alone (Figures 3E–3G). It is noteworthy that naringin alone had no effect on iNOS, CD86, Arg-1, and IL-10 levels, but it markedly upregulated PPARγ (Figure 3G).
We finally detected the miRNA levels that have been implicated in macrophage polarization (Figure 3H). We found that LPS significantly decreased the expression of miR-23-3p and miR-223-3p, both of which are regulated by PPARγ,22,23 while naringin treatment reversed the suppressive effects induced by LPS. As expected, similar results were observed for miR-21-5p (miR-21), suggesting miR-21 as the potential target of PPARγ. Taken together, these data demonstrated that naringin stimulated LPS-modulated M2 polarization and upregulated PPARγ and its putative downstream miRNA expression.
Naringin regulated macrophage polarization partly via the PPARγ pathway
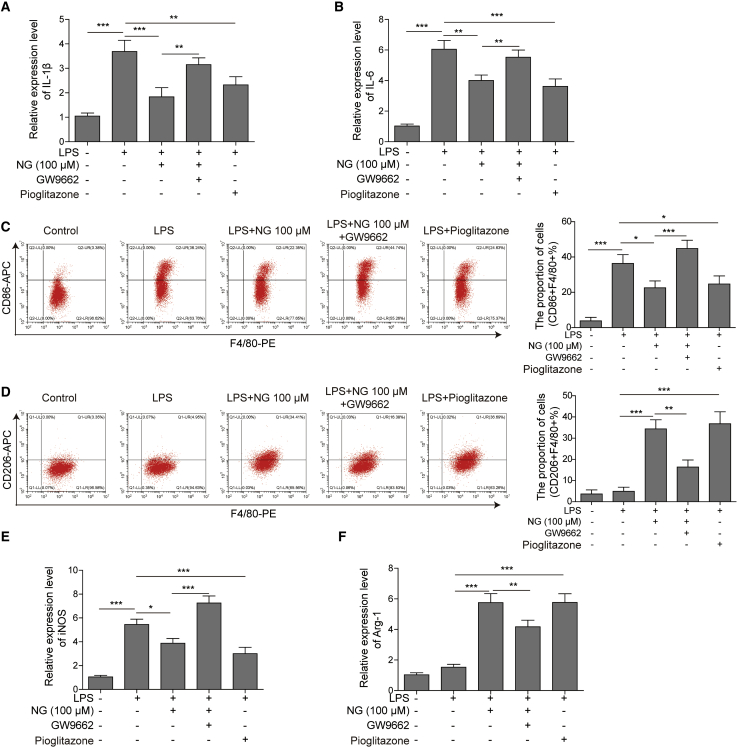
To further explore whether PPARγ mediates naringin-regulated macrophage polarization, we stimulated the mouse BMDMs with LPS challenge and added the following treatments after 30 min: 100 μM naringin, 100 μM naringin plus PPARγ inhibitor (0.1 μM GW9662), or PPARγ agonist (1 μM pioglitazone) as the positive control. We found that inhibition of PPARγ by GW9662 abolished the reduction of inflammatory factors IL-1β and IL-6 induced by naringin on LPS. In contrast, pioglitazone was able to reduce the LPS-induced inflammatory factors (Figures 4A and 4B). Flow cytometry analysis revealed that GW9662 compromised macrophage polarization regulated by naringin in LPS-induced BMDMs, while pioglitazone elevated M2 polarization and inhibited M1 polarization (Figures 4C and 4D). Quantitative real-time PCR detection of M1 macrophage marker iNOS and M2 marker Arg-1 showed that inhibition of PPARγ abolished the reduction of iNOS and the elevation of Arg-1 induced by naringin, while pioglitazone lowered iNOS and increased the expression of Arg-1 (Figures 4E and 4F). Collectively, these findings suggested the involvement of the PPARγ pathway in macrophage polarization regulated by naringin.
Figure 4.
Naringin regulated macrophage polarization partially via the PPARγ pathway
(A and B) The mRNA levels of IL-1β (A) and IL-6 (B) in BMDMs were detected by quantitative real-time PCR. (C and D) M1 macrophages (F4/80+CD86+) and M2 macrophages (F4/80+CD206+) in BMDMs were determined by flow cytometry. (E and F) The mRNA levels of iNOS (E) and Arg-1 (F) in BMDMs were detected by quantitative real-time PCR. Data represent the mean ± SD of three experiments performed in triplicate independently. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
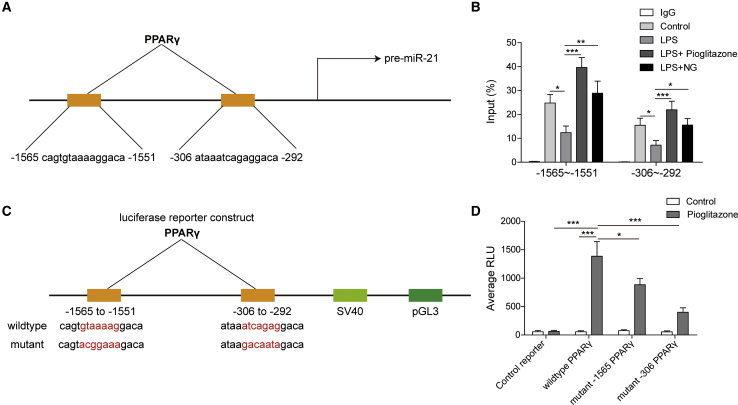
PPARγ directly regulated transcription of miR-21
To determine whether miR-21 is a direct transcriptional target of PPARγ, we first used the JASPAR database to find out the promoter region of miR-21 binding with PPARγ (Figure 5A). Results from chromatin immunoprecipitation (ChIP)-PCR revealed that PPARγ bound to the predicted promoter region of miR-21 (Figure 5B). Moreover, while LPS reduced PPARγ binding, naringin and the PPARγ agonist pioglitazone restored PPARγ binding in the miR-21 promoter region (Figure 5B). We then performed luciferase reporter analysis to validate the binding of PPARγ to miR-21. To this end, we constructed three groups: wild-type (WT) of PPARγ, mutant (MUT) −1565 (mutation of the binding sequence of the −1565 segment), and mutant −306 (mutation of the binding sequence of the −306 segment) (Figure 5C). We found that pioglitazone significantly induced the luciferase activity of the wild-type of PPARγ, while this inductive effect was dramatically decreased in the two mutant types of PPARγ (Figure 5D). Collectively, these results suggested that PPARγ directly regulated transcription of miR-21.
Figure 5.
PPARγ directly regulated transcription of miR-21
(A) The promoter region of pre-miR-21 binding PPARγ from the JASPAR database. (B) The interaction between PPARγ and the miR-21 promoter was detected by ChIP-PCR. (C) Construction of the luciferase reporters. (D) Binding of PPARγ to the predicted sequence of miR-21 validated by a luciferase reporter assay. Data represent the mean ± SD of three experiments performed in triplicate independently. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
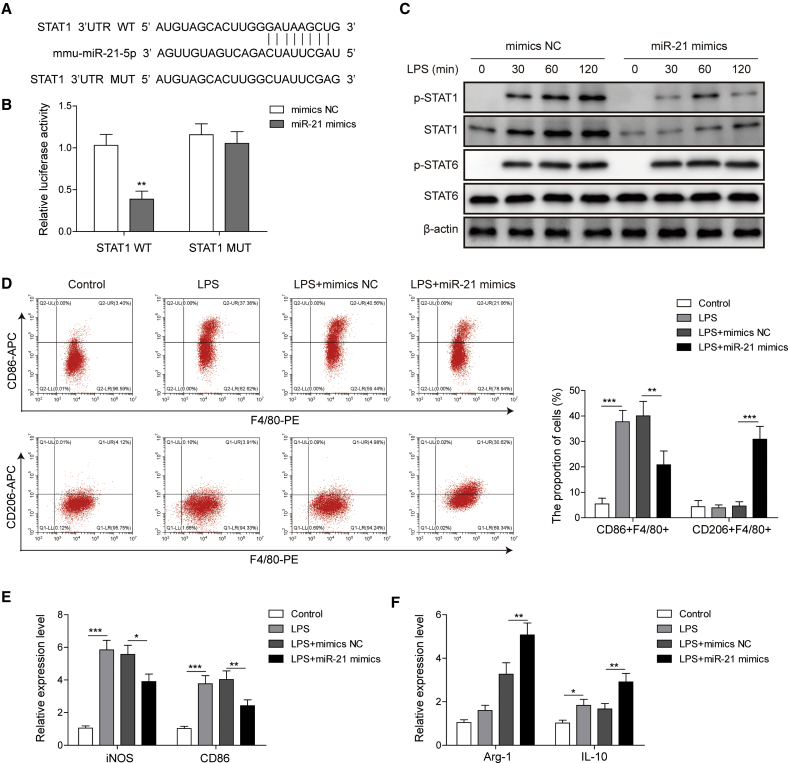
miR-21 targeted STAT1 to regulate macrophage polarization
To identify the molecular mechanism by which miR-21 regulates macrophage polarization, we used a bioinformatics approach to predict the putative binding site between miR-21 and STAT1 (Figure 6A). A luciferase reporter assay was performed to investigate the interplay between STAT1 and miR-21 by constructing the wild-type and mutant type of STAT1. We found that miR-21 decreased the luciferase activity of wild-type rather than the mutant type of STAT1 as compared with the negative control (NC) (Figure 6B). The STAT1/6 pathway is critical to macrophage polarization, and our results showed that miR-21 overexpression significantly inhibited total and phosphorylation of STAT1, while it had no significant effect on STAT6 phosphorylation (Figure 6C). As expected, overexpression of miR-21 significantly promoted M2 polarization whereas it suppressed M1 polarization, as compared with mimic negative control groups in mouse BMDMs challenged with LPS (Figure 6D). Accordingly, miR-21 decreased the expression of M1 markers iNOS and CD86 (Figure 6E) and increased the expression of the M2 markers Arg-1 and IL-10 (Figure 6F). All together, these results indicated that miR-21 targeted STAT1 signaling and regulated macrophage polarization.
Figure 6.
miR-21 targeted STAT1 to regulate macrophage polarization
(A) Binding sites of the STAT1 3′ UTR to miR-21. (B) Luciferase activity of wild-type (WT) and mutant type (MUT) of STAT1 detected by a luciferase reporter assay. (C) The effect of miR-21 on STAT1 and STAT6 was detected by western blotting. (D) The effect of miR-21 on M1 macrophages (F4/80+CD86+) and M2 macrophages (F4/80+CD206+) was detected by flow cytometry. (E and F) The effects of miR-21 on M1 macrophage markers iNOS and CD86 (E) and M2 macrophage markers Arg-1 and IL-10 (F) were detected by quantitative real-time PCR. Data represent the mean ± SD of three experiments performed in triplicate independently. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
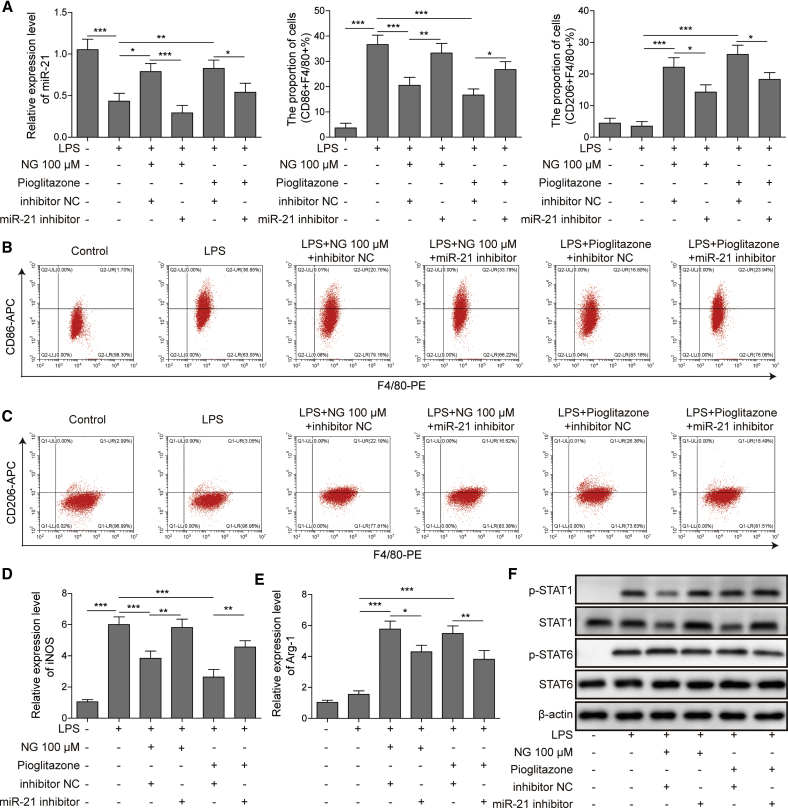
miR-21 partially mediated the regulation of macrophage polarization by the naringin/PPARγ axis
Finally, to investigate the function of miR-21 on naringin/PPARγ axis-regulated macrophage polarization, we used miR-21 inhibitor to inhibit miR-21 expression in BMDMs before naringin or pioglitazone treatment. We validated that miR-21 inhibitor reduced the expression of miR-21 induced by naringin or pioglitazone (Figure 7A). In addition, silencing of miR-21 reversed the increased M2 polarization and decreased M1 polarization induced by naringin or pioglitazone (Figures 7B and 7C). This was also accompanied with the reversion of naringin or pioglitazone-suppressed iNOS and naringin or pioglitazone-induced Arg-1 expression (Figures 7D and 7E). Since PPARγ function is mediated by miR-21, it is likely that phosphorylation of the PPARγ-regulated downstream proteins would thus change. As expected, results from western blotting analysis revealed that silencing of miR-21 reversed naringin or pioglitazone-suppressed STAT1 and phosphorylated (p-)STAT1 (Figure 7F). However, miR-21 inhibition did not change the STAT6 activation elevated by naringin or pioglitazone. Collectively, these data provided strong evidence that miR-21 partially mediated the regulation of macrophage polarization by the naringin/PPARγ axis.
Figure 7.
miR-21 partially mediated the regulation of macrophage polarization by naringin/PPARγ
(A) The level of miR-21 in treated BMDMs was detected by quantitative real-time PCR. (B and C) M1 macrophages (F4/80+CD86+) and M2 macrophages (F4/80+CD206+) in BMDMs were determined by flow cytometry. (D and E) The mRNA levels of M1 marker iNOS (D) and M2 marker Arg-1 (E) in treated BMDMs were detected by quantitative real-time PCR. (F) The protein levels of indicated proteins in treated BMDMs were detected by western blotting. Data represent the mean ± SD of three experiments performed in triplicate independently. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Macrophage polarization has important implications in the incidence and progression of sepsis, with sepsis-induced intestinal injury being the most frequent one, accounting for approximately 70% of cases.38 To develop diagnostic and therapeutic strategies for sepsis-induced intestinal injury, a good understanding of the molecular mechanism behind this severe disease is needed. Using a well- recognized model of CLP-induced acute intestinal injury, we demonstrated that naringin ameliorated intestinal injuries, and this might be achieved by the promotion of anti-inflammatory M2 macrophage polarization in vivo and in vitro. Importantly, our data suggested the possible involvement of PPARγ/miR-21/STAT1 signaling in the promoting effects of naringin on M2 macrophage polarization. It is noteworthy that (1) although a recent study simply mentioned the macrophage polarization in sepsis-induced intestinal injury,13 the detailed underlying mechanism remains largely unknown. We first and systematically indicated M2 macrophage polarization transformation in improving sepsis-induced intestinal injury. (2) We first demonstrated that PPARγ and miR-21 acted as key players in sepsis-induced intestinal injury. Despite that the role of STAT1 in sepsis had been illustrated, we first focused on its biological function in sepsis-induced intestinal injury. (3) We first demonstrated the naringin effects on inducing M2 macrophage polarization. Our findings thus advance the current knowledge of the immunomodulatory potential of naringin on macrophage polarization and provide new insights into optimizing the therapeutic modalities of naringin in sepsis.
Naringin has plenty of biological and pharmacological properties, including anti-inflammatory, anti-oxidant, anti-atherosclerosis, and anti-hypertensive effects.39 Therapeutic potentials of naringin were demonstrated in diverse disorders including atherosclerosis, diabetes mellitus, cardiovascular disorder, and cancers. In terms of its effects on sepsis, naringin was reported to improve intestinal mucosa barrier dysfunction caused by sepsis, possibly via the RhoA/ROCK/NF-κB/MLCK/MLC signaling pathway. Naringin also ameliorated LPS-induced acute lung injury.40 Naringin inhibited the expression levels of pro-inflammatory cytokines and the consequent acute lung injury by suppressing neutrophil migration into the lung. Moreover, through decreasing TNF-α and HMGB1 derived from LPS-induced macrophages, naringin was reported to decrease sepsis-induced mortality and alleviate pathological changes in lung. Our study, for the first time, demonstrated that naringin protected against sepsis-induced intestinal injury by regulating M2 macrophage polarization. Further investigations will focus on how naringin upregulates PPARγ.
As effector cells key to innate immunity, macrophages are of critical importance to sepsis resolution and play key roles in the aggravation of disease severity. Distinctive effector molecules have been found for M1 and M2 macrophages, including Arg-1, iNOS, CD206, and TNF-α, which exert different influences on outcomes of inflammation.41,42 In our study, naringin treatment significantly suppressed M1 macrophage polarization and stimulated M2 macrophage polarization. We further discovered that the regulation of naringin on macrophage polarization is dependent on the upregulation of PPARγ, an important molecule that regulates macrophage polarization.19,29 Based on previous findings, PPARγ regulated macrophage polarization by regulating the transcription of miR-223, miR-23, and other miRNAs.2,23 Our study demonstrated for the first time that PPARγ played this role by regulating miR-21 transcription. As one of the most common miRNAs, the biological function of miR-21 in modulating necroptosis and apoptosis and its involvement in the inflammatory progression were well recognized.43 We speculated that there might be the binding site of miR-21 on PPARγ according to our bioinformatics analysis. As expected, we found that miR-21 was dramatically downregulated in LPS-induced BMDMs, and PPARγ bound to miR-21 directly in BMDMs. In our subsequent loss-of-function experiments, we further identified the regulatory interplay between PPARγ and miR-21. Given that genetic defect of miR-21 augmented STAT1 signaling, and that in the existence of tumor cells, induced the macrophage polarization toward M1 phenotype both in vivo and in vitro. We postulated that miR-21 might also exert its effect on macrophage polarization through regulating STAT1 expression. To determine whether STAT1 functions as a sponge for miR-21 in BMDMs, bioinformatics analysis was used to seek out the potential miRNA binding sites in the STAT1 3′ UTR. A luciferase reporter assay and quantitative real-time PCR demonstrated that miR-21 positively regulated STAT1 expression in mouse BMDMs. Thus, we have demonstrated for the first time that miR-21 protected against sepsis-induced intestinal injury by regulating macrophages via suppressing STAT1.
Several transcriptional factors have been identified to play key roles in regulating the differentiation and polarization of macrophages with regard to the intracellular mechanism. In particular, STAT1 and STAT6 have been demonstrated to be important regulators of macrophage polarization. Previous studies indicated that the STAT1/STAT6 pathway regulated PPARγ, and that PPARγ was involved in STAT6-induced M2 macrophage polarization. Our study found that the PPARγ activator pioglitazone can upregulate miR-21 to regulate STAT1 and STAT6 to a certain degree, suggesting that PPARγ might be a feedback regulator to upstream signaling. Further studies are needed to investigate the exact mechanism. In addition, note that miR-21 had no regulative effect on STAT6 while the naringin/PPARγ axis showed significant influence. This difference suggested that there might be another downstream mechanism of the naringin/PPARγ axis to regulate STAT6 signaling.
In conclusion, the present study demonstrates that naringin promotes M2 macrophage polarization to alleviate sepsis-induced intestinal injury via the PPARγ/miR-21 axis. These results might provide a mechanistic insight for naringin as a potential therapeutic agent for sepsis in the future.
Materials and methods
Reagents
Naringin (catalog number [Cat. no.] HY-N0153), PPARγ agonist pioglitazone (Cat. no. HY-13956), and PPARγ antagonist GW9662 (Cat. no. HY-16578) were all purchased from MedChemExpress (Princeton, NJ, USA). LPS (Cat. no. L2654) was ordered from Sigma-Aldrich (St. Louis, MO, USA). Phycoerythrin (PE)-anti-F4/80 (Cat. no. 123109; 1:100), allophycocyanin (APC)-anti-CD86 (Cat. no. 105011; 1:100), and APC-anti-CD206 (Cat. no. 141708; 1:100) were purchased from BioLegend (San Diego, CA, USA).
Sepsis animal model using CLP
Animal experiments received approval from the Animal Care and Use Committee of The Third Xiangya Hospital, Central South University (Changsha, Hunan, China). Male C57BL/6 mice (6 weeks old, 18–22 g) were obtained from Hunan SJA Laboratory Animal Co. (Changsha, Hunan, China) and raised in the Experimental Animal Center of The Third Xiangya Hospital. Mice were monitored in a 12-h light/12-h dark cycle within groups of up to five in each cage, accommodated with full access to food and water and an appropriate temperature (23°C ± 2°C).
Mice were allocated to the following five groups to explore the influence of naringin on the sepsis-induced intestinal injury: sham group (n = 20), CLP plus vehicle group (n = 40), and CLP plus naringin at 10 mg/kg (n = 40), 30 mg/kg (n = 40), and 60 mg/kg (n = 40) groups. To establish the mouse septic model induced by CLP, 2%–4% isoflurane inhalation was used to anesthetize the mice. The disinfected abdomen was incised at a longitudinal skin midline and the cecum was exposed. The distal thirds of the cecum were subsequently ligated with Perma-Hand silk (Ethicon), punctured with a 30G needle once a time, and the fecal matter was extruded. After that, the cecum was placed back to the abdomen and sutured in two layers. To provide postoperative fluid resuscitation, PBS (1 mL) was given subcutaneously following the surgical process. In addition, bupivacaine (Hospira) was given at the incision site, and flunixin meglumine (Phoenix) was given for analgesia postoperatively. The above operations generated a septic status that was featured by a loss of body weight and appetite, ruffled hair, or periorbital exudates. For the sham group, the cecum was only exposed, without ligation or puncture, and then returned to the peritoneal cavity. An equivalent volume of vehicle was given to CLP and the sham groups and other mice were given naringin at doses of 10, 30, and 60 mg/kg intraperitoneally 30 min after challenging by CLP. To evaluate the survival rates, mice (n = 10 in sham group, and n = 20 in other groups) were counted every 5 h for 3 days. The survival curves were calculated by using the Kaplan-Meier method, and the difference between groups was analyzed by a log-rank test. The ileum, colon, and serum in other mice (n = 10 in sham group, and n = 20 in other groups) were collected to perform histological assessment, quantitative real-time PCR, western blotting, and an ELISA.
Histological assessment for intestinal injury
Mice in each group were anesthetized 24 h after CLP. The ileum and colon tissues were fixed with 4% paraformaldehyde and embedded in paraffin. Sections (5 μm) were then stained with H&E. The Chui’s scoring system that represents the averaged findings of two investigators who independently read the H&E-stained slide in a blinded manner44 was used to evaluate the organ injury score. For IHC analysis, the paraffin-embedded samples were subjected to deparaffinization, rehydration, and antigen retrieval. The sections were incubated with 3% hydrogen peroxide to block endogenous peroxidase activity, followed by the incubation with primary antibodies: anti-CD86 (Cat. no. 19589; 1:100, Cell Signaling Technology, Danvers, MA, USA) and anti-CD206 (Cat. no. 24595; 1:200, Cell Signaling Technology). The sections were then incubated with MaxVision horseradish peroxidase (HRP) polymer immunoglobulin G (IgG) secondary antibody and counterstained with hematoxylin. The images were acquired using an inverted light microscope (Olympus, Tokyo, Japan).
Isolation and differentiation of BMDMs
C57BL/6 mice were sacrificed by cervical dislocation and subsequently sterilized with 75% ethanol. This was followed by the incision of the skin at the root of hind legs and the removal of the muscle tissues from the bones with scissors. The bones cut off from both ends were then flushed with medium by a 5-mL syringe. The bone marrow cells were suspended by pipetting up and down. To obtain BMDMs, bone marrow cells were harvested in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and 60 ng/mL macrophage colony-stimulating factor (M-CSF) for 4 days. For LPS stimulation, BMDMs were subjected to 2 μg/mL LPS for 36 h. BMDMs were assigned to corresponding groups 30 min after the LPS challenge.
Cell culture
HEK293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were inoculated in DMEM supplemented with 10% FBS at room temperature supplied with 5% CO2 and 95% atmosphere. Cells were harvested in 24-well or 6-well plates for further experiments when reaching 80% confluence.
Cell viability assay
BMDMs were harvested in 96-well plates (6 × 103 cells per well) followed by treatment with a series of concentrations of naringin for 24 h. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was then used to test the cell viability. MTT (50 μL) was added into each well. After incubation for 6 h, medium consisting of the unreacted MTT was carefully eliminated, and MTT formazan crystals were dissolved by adding DMSO (150 μL) to each well. The absorbance (Abs.) was measured by using a microplate reader (M680, Bio-Rad, Hercules, CA, USA) at 490 nm after 1 h. To calculate the cell viability, the following equation was used: Cell viability (%) = (mean of Abs. of treatment group/mean of Abs. of control group) × 100%.
Total RNA extraction and quantitative real-time PCR
Total RNA was extracted from BMDMs or ileum tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was generated by using random primers and SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time PCR was conducted using the QuantiTect RT-PCR kit (QIAGEN), and U6 served as an internal control for miRNA. Reverse transcription reaction was performed to detect mRNAs by using a PrimeScript RT reagent kit (Takara Bio, Shiga, Japan). β-Actin was used as an internal control. The PCR reaction was run in triplicate using SYBR Premix Ex Taq II (Takara Bio) with a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The amplification included a 10-min denaturation at 95°C, followed by 60 cycles of denaturation at 95°C for 10 s, annealing at 50°C for 40 s, and extension at 60°C for 2 min. The 2−ΔΔCt method was employed to determine the relative expression levels of target miRNAs or genes. Table 1 presents the primer sequences used for quantitative real-time PCR.
Table 1.
Quantitative real-time PCR primer sequences
| Primers | Sequences |
|---|---|
| mmu-miR-23b-3p | 5′-GCCATCACATTGCCAGGG-3′ (forward) |
| 5′-GTGCAGGGTCCGAGGT-3′ (reverse) | |
| mmu-miR-223-3p | 5′-GCCGCTGTCAGTTTGTCAAAT-3′ (forward) |
| 5′-GTGCAGGGTCCGAGGT-3′ (reverse) | |
| mmu-miR-21-5p | 5′-CGCGCTAGCTTATCAGACTGA-3′ (forward) |
| 5′-GTGCAGGGTCCGAGGT-3′ (reverse) | |
| CD206 | 5′-ATGCCAAGTGGGAAAATCTG-3′ (forward) |
| 5′-TGTAGCAGTGGCCTGCATAG-3′ (reverse) | |
| Arg-1 | 5′-AACACTCCCCTGACAACCAG-3′ (forward) |
| 5′-GCAAGCCAATGTACACGATG-3′ (reverse) | |
| TNF-α | 5′-CCGATGGGTTGTACCTTGTC-3′ (forward) |
| 5′-TGGAAGACTCCTCCCAGGTA-3′ (reverse) | |
| IL-6 | 5′-TGCAAGAGACTTCCATCCAG-3′ (forward) |
| 5′-TCCACGATTTCCCAGAGAAC CA-3′ (reverse) | |
| PPARγ | 5′-AAGAGCTGACCCAATGGTTG-3′ (forward) |
| 5′-ACCCTTGCATCCTTCACAAG-3′ (reverse) | |
| iNOS | 5′-AAGCCCCGCTACTACTCCAT-3′ (forward) |
| 5′-AGCTGGAAGCCACTGACACT-3′ (reverse) | |
| CD86 | 5′-ATCAAGGACATGGGCTCGTA-3′ (forward) |
| 5′-TTAGGTTTCGGGTGACCTTG-3′ (reverse) | |
| IL-1β | 5′-GCCACCTTTTGACAGTGATGAG-3′ (forward) |
| 5′-AAGGTCCACGGGAAAGACAC-3′ (reverse) | |
| IL-10 | 5′-GGTTGCCAAGCCTTATCGGA-3′ (forward) |
| 5′-TTCAGCTTCTCACCCAGGGA-3′ (reverse) | |
| β-Actin | 5′-GGCTGTATTCCCCTCCATCG-3′ (forward) |
| 5′-CCAGTTGGTAACAATGCCATGT-3′ (reverse) |
Flow cytometry
BMDMs were harvested 24 h after LPS stimulation, followed by rinsing with cold PBS and subsequent permeabilization. The collected cells were then incubated with either PE-anti-F4/80 and APC-anti-CD86, or APC-anti-CD206 at 4°C in darkness for 1 h. After washing twice by centrifugation with fluorescence-activated cell sorting (FACS) buffer, cells were resuspended in 300 μL of PBS. The data were obtained using a Beckman Coulter flow cytometer (Beckman Coulter, Brea, CA, USA) and analyzed by using FlowJo software (Tree Star, Ashland, OR, USA).
ChIP assay
A ChIP assay kit (Millipore, Billerica, MA, USA) was used to carry out the ChIP assay according to the manufacturer’s instructions. Immunoprecipitation was performed using an antibody against PPARγ (Cat. no. 2435; 1:100, Cell Signaling Technology). In addition, purified DNA was further analyzed by qPCR. Agarose gel electrophoresis was used to verify the PCR products.
Oligonucleotides and transfections
The mimics and inhibitor of miR-21, as well as the corresponding negative controls, were obtained from GenePharma (Shanghai, China). Transfections were carried out by using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions.
Western blotting analysis
BMDMs or ileal tissues were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA). A bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) was used to determine the protein concentration. Approximate 30 μg of protein was separated by 10% SDS-PAGE and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 1% BSA/Tris-buffered saline (TBS), the blots were then incubated with primary antibodies at 4°C overnight. The following primary antibodies obtained from Cell Signaling Technology were used in this study: anti-β-actin (Cat. no. 3700; 1:1,000), anti-PPARγ (Cat. no. 2435; 1:1,000), anti-claudin-1 (Cat. no. 13255; 1:2,000), anti-occludin (Cat. no. 91131; 1:1,500), anti-ZO-1 (Cat. no. 13663; 1:1,000), anti-STAT1 (Cat. no. 14994; 1: 3,000), anti-p-STAT1 (Tyr701, Cat. no. 9167; 1:1,000), anti-STAT6 (Cat. no. 5397; 1: 2,000), and anti-p-STAT6 (Tyr641, Cat. no. 9361; 1:1,500). The membranes were incubated with corresponding HRP-conjugated secondary antibody at room temperature for 30 min. The signals were visualized by Pico Plus enhanced chemiluminescence (ECL) substrate (Thermo Fisher Scientific) and analyzed by Quantity One software (Bio-Rad, CA, USA). β-Actin served as the loading control.
Dual-luciferase reporter assay
A luciferase reporter assay was carried out using a Dual-Luciferase reporter assay kit (Promega, Madison, WI, USA). A DNA fragment containing the upstream region of precursor (pre-)miR-21 was cloned into pGL3-control reporter vector (Promega), with the purpose of verifying that PPARγ binds to the promoter region of pre-miR-21. To confirm the interaction between miR-21 and the STAT 3′ UTR, the sequence of the STAT1 3′ UTR containing the putative miR-21 binding site was cloned into psiCHECK-2 vector (Promega). The miR-21 mimics and its negative control were subsequently co-transfected with the reporter constructs into HEK293T cells. Renilla luciferase activity was used as an internal control.
ELISA
Secretion of TNF-α, IL-1β, and IL-6 in culture medium collected from BMDMs or serum was determined by commercial ELISA kits (Dakewe, Shenzhen, China) according to the manufacturer’s instructions. In brief, cell culture medium and serum were collected and centrifuged at 1,400 rpm for 1 min, and the supernatant was subjected to subsequent analysis.
Statistical analysis
Data were presented as the mean ± standard deviation (SD). The difference in means between two groups was compared by using the Student’s t test. Multiple comparisons for more than two groups were done by one-way analysis of variance (ANOVA) with a Tukey post hoc test. All analyses was performed by GraphPad Prism 6, and p < 0.05 was considered statistically significant.
Acknowledgments
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Author contributions
Z.-L.L., B.-C.Y., and M.G. contributed to the conception of the study and wrote the paper. Z.-L.L., B.-C.Y., and M.G. conducted the experiments and collected data. M.G., X.-F.X., S.-P.Z., and Z.-L.L. analyzed the data. All authors reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.005.
Supplemental information
References
- 1.Stearns-Kurosawa D.J., Osuchowski M.F., Valentine C., Kurosawa S., Remick D.G. The pathogenesis of sepsis. Annu. Rev. Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dellinger R.P., Levy M.M., Rhodes A., Annane D., Gerlach H., Opal S.M., Sevransky J.E., Sprung C.L., Douglas I.S., Jaeschke R., Surviving Sepsis Campaign Guidelines Committee including The Pediatric Subgroup Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent J.L., Opal S.M., Marshall J.C., Tracey K.J. Sepsis definitions: Time for change. Lancet. 2013;381:774–775. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda M., Surewaard B.G.J., Watanabe M., Hedrick C.C., Lee W.-Y., Brown K., McCoy K.D., Kubes P. Perivascular localization of macrophages in the intestinal mucosa is regulated by Nr4a1 and the microbiome. Nat. Commun. 2020;11:1329. doi: 10.1038/s41467-020-15068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sica A., Mantovani A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David S., Kroner A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci. 2011;12:388–399. doi: 10.1038/nrn3053. [DOI] [PubMed] [Google Scholar]
- 7.Mantovani A., Biswas S.K., Galdiero M.R., Sica A., Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013;229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Martinez F.O., Sica A., Mantovani A., Locati M. Macrophage activation and polarization. Front. Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 10.Mehta A., Brewington R., Chatterji M., Zoubine M., Kinasewitz G.T., Peer G.T., Chang A.C., Taylor F.B., Jr., Shnyra A. Infection-induced modulation of m1 and m2 phenotypes in circulating monocytes: Role in immune monitoring and early prognosis of sepsis. Shock. 2004;22:423–430. doi: 10.1097/01.shk.0000142184.49976.0c. [DOI] [PubMed] [Google Scholar]
- 11.Grailer J.J., Haggadone M.D., Sarma J.V., Zetoune F.S., Ward P.A. Induction of M2 regulatory macrophages through the β2-adrenergic receptor with protection during endotoxemia and acute lung injury. J. Innate Immun. 2014;6:607–618. doi: 10.1159/000358524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kambara K., Ohashi W., Tomita K., Takashina M., Fujisaka S., Hayashi R., Mori H., Tobe K., Hattori Y. In vivo depletion of CD206+ M2 macrophages exaggerates lung injury in endotoxemic mice. Am. J. Pathol. 2015;185:162–171. doi: 10.1016/j.ajpath.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Guo L., Meng M., Wei Y., Lin F., Jiang Y., Cui X., Wang G., Wang C., Guo X. Protective effects of live combined B. subtilis and E. faecium in polymicrobial sepsis through modulating activation and transformation of macrophages and mast cells. Front. Pharmacol. 2019;9:1506. doi: 10.3389/fphar.2018.01506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Knethen A., Soller M., Brüne B. Peroxisome proliferator-activated receptor γ (PPARγ) and sepsis. Arch. Immunol. Ther. Exp. (Warsz.) 2007;55:19–25. doi: 10.1007/s00005-007-0005-y. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y., Zhang Y., Hong K., Luo F., Gu Q., Lu N., Bai A. AMPK inhibition blocks ROS-NFκB signaling and attenuates endotoxemia-induced liver injury. PLoS ONE. 2014;9:e86881. doi: 10.1371/journal.pone.0086881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croasdell A., Duffney P.F., Kim N., Lacy S.H., Sime P.J., Phipps R.P. PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015;2015:549691. doi: 10.1155/2015/549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D., Shi L., Xin W., Xu J., Xu J., Li Q., Xu Z., Wang J., Wang G., Yao W. Activation of PPARγ inhibits pro-inflammatory cytokines production by upregulation of miR-124 in vitro and in vivo. Biochem. Biophys. Res. Commun. 2017;486:726–731. doi: 10.1016/j.bbrc.2017.03.106. [DOI] [PubMed] [Google Scholar]
- 18.Xia H., Chen L., Liu H., Sun Z., Yang W., Yang Y., Cui S., Li S., Wang Y., Song L. Protectin DX increases survival in a mouse model of sepsis by ameliorating inflammation and modulating macrophage phenotype. Sci. Rep. 2017;7:99. doi: 10.1038/s41598-017-00103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A.W., Chawla A. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Huang Y., Xu Y., Ruan W., Wang H., Zhang Y., Saavedra J.M., Zhang L., Huang Z., Pang T. A dual AMPK/Nrf2 activator reduces brain inflammation after stroke by enhancing microglia M2 polarization. Antioxid. Redox Signal. 2018;28:141–163. doi: 10.1089/ars.2017.7003. [DOI] [PubMed] [Google Scholar]
- 21.Kim M.J., Lee Y.J., Yoon Y.S., Kim M., Choi J.H., Kim H.S., Kang J.L. Apoptotic cells trigger the ABCA1/STAT6 pathway leading to PPAR-γ expression and activation in macrophages. J. Leukoc. Biol. 2018;103:885–895. doi: 10.1002/JLB.2A0817-341RR. [DOI] [PubMed] [Google Scholar]
- 22.Ying W., Tseng A., Chang R.C., Morin A., Brehm T., Triff K., Nair V., Zhuang G., Song H., Kanameni S. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z., Yuan P., Sun X., Tang K., Liu H., Han S., Ye T., Liu X., Yang X., Zeng J. Pioglitazone decreased renal calcium oxalate crystal formation by suppressing M1 macrophage polarization via the PPAR-γ-miR-23 axis. Am. J. Physiol. Renal Physiol. 2019;317:F137–F151. doi: 10.1152/ajprenal.00047.2019. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Brandt S., Medeiros A., Wang S., Wu H., Dent A., Serezani C.H. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS ONE. 2015;10:e0115855. doi: 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Wang J., Zhang Z., Miao H. Decreased miR-128 and increased miR-21 synergistically cause podocyte injury in sepsis. J. Nephrol. 2017;30:543–550. doi: 10.1007/s40620-017-0405-y. [DOI] [PubMed] [Google Scholar]
- 26.McClure C., McPeak M.B., Youssef D., Yao Z.Q., McCall C.E., El Gazzar M. Stat3 and C/EBPβ synergize to induce miR-21 and miR-181b expression during sepsis. Immunol. Cell Biol. 2017;95:42–55. doi: 10.1038/icb.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 28.Feng L., Song P., Zhou H., Li A., Ma Y., Zhang X., Liu H., Xu G., Zhou Y., Wu X. Pentamethoxyflavanone regulates macrophage polarization and ameliorates sepsis in mice. Biochem. Pharmacol. 2014;89:109–118. doi: 10.1016/j.bcp.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Xu Y., Zhang P., Ruan W., Zhang L., Yuan S., Pang T., Jia A.Q. Smiglaside A ameliorates LPS-induced acute lung injury by modulating macrophage polarization via AMPK-PPARγ pathway. Biochem. Pharmacol. 2018;156:385–395. doi: 10.1016/j.bcp.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Pang W.Y., Wang X.L., Mok S.K., Lai W.P., Chow H.K., Leung P.C., Yao X.S., Wong M.S. Naringin improves bone properties in ovariectomized mice and exerts oestrogen-like activities in rat osteoblast-like (UMR-106) cells. Br. J. Pharmacol. 2010;159:1693–1703. doi: 10.1111/j.1476-5381.2010.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yow T.T., Pera E., Absalom N., Heblinski M., Johnston G.A., Hanrahan J.R., Chebib M. Naringin directly activates inwardly rectifying potassium channels at an overlapping binding site to tertiapin-Q. Br. J. Pharmacol. 2011;163:1017–1033. doi: 10.1111/j.1476-5381.2011.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R., Qi Q.L., Wang M.T., Li Q.Y. Therapeutic potential of naringin: An overview. Pharm. Biol. 2016;54:3203–3210. doi: 10.1080/13880209.2016.1216131. [DOI] [PubMed] [Google Scholar]
- 33.Deenonpoe R., Prayong P., Thippamom N., Meephansan J., Na-Bangchang K. Anti-inflammatory effect of naringin and sericin combination on human peripheral blood mononuclear cells (hPBMCs) from patient with psoriasis. BMC Complement. Altern. Med. 2019;19:168. doi: 10.1186/s12906-019-2535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed S., Khan H., Aschner M., Hasan M.M., Hassan S.T.S. Therapeutic potential of naringin in neurological disorders. Food Chem. Toxicol. 2019;132:110646. doi: 10.1016/j.fct.2019.110646. [DOI] [PubMed] [Google Scholar]
- 35.Huang D., Li Z., Chen B., Fang G., Sun X., Li F., Xu H., Chen Y., Ding W. Naringin protects against steroid-induced avascular necrosis of the femoral head through upregulation of PPARγ and activation of the Notch signaling pathway. Mol. Med. Rep. 2018;17:3328–3335. doi: 10.3892/mmr.2017.8247. [DOI] [PubMed] [Google Scholar]
- 36.Gil M., Kim Y.K., Hong S.B., Lee K.J. Naringin decreases TNF-α and HMGB1 release from LPS-stimulated macrophages and improves survival in a CLP-induced sepsis mice. PLoS ONE. 2016;11:e0164186. doi: 10.1371/journal.pone.0164186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Gao M., Yang B., Zhang H., Wang K., Liu Z., Xiao X., Yang M. Naringin attenuates MLC phosphorylation and NF-κB activation to protect sepsis-induced intestinal injury via RhoA/ROCK pathway. Biomed. Pharmacother. 2018;103:50–58. doi: 10.1016/j.biopha.2018.03.163. [DOI] [PubMed] [Google Scholar]
- 38.Haussner F., Chakraborty S., Halbgebauer R., Huber-Lang M. Challenge to the intestinal mucosa during sepsis. Front. Immunol. 2019;10:891. doi: 10.3389/fimmu.2019.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bharti S., Rani N., Krishnamurthy B., Arya D.S. Preclinical evidence for the pharmacological actions of naringin: A review. Planta Med. 2014;80:437–451. doi: 10.1055/s-0034-1368351. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Wu H., Nie Y.C., Chen J.L., Su W.W., Li P.B. Naringin attenuates acute lung injury in LPS-treated mice by inhibiting NF-κB pathway. Int. Immunopharmacol. 2011;11:1606–1612. doi: 10.1016/j.intimp.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Arora S., Olszewski M.A., Tsang T.M., McDonald R.A., Toews G.B., Huffnagle G.B. Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection with Cryptococcus neoformans. Infect. Immun. 2011;79:1915–1926. doi: 10.1128/IAI.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Ming X.F. Functions of arginase isoforms in macrophage inflammatory responses: Impact on cardiovascular diseases and metabolic disorders. Front. Immunol. 2014;5:533. doi: 10.3389/fimmu.2014.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buscaglia L.E., Li Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer. 2011;30:371–380. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiu C.J., McArdle A.H., Brown R., Scott H.J., Gurd F.N. Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal. Arch. Surg. 1970;101:478–483. doi: 10.1001/archsurg.1970.01340280030009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.