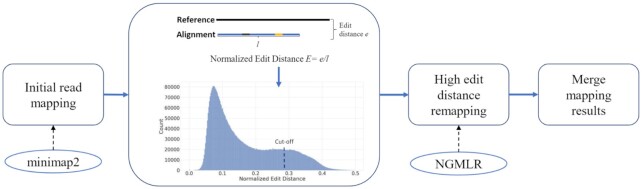
Figure 1:
Overview of Vulcan: As step 1, Vulcan takes raw ONT or PacBio reads as input, then uses minimap2 to map them to the provided reference genome. Subsequently, in step 2, Vulcan performs a normalized edit distance calculation (see Methods) to identify the reads with the highest normalized edit distances. In step 3, Vulcan realigns the high edit distance reads with NGMLR. Finally, in step 4 Vulcan merges the minimap2 and NGMLR remapped reads to create a new bam file.