Abstract
Transcriptional silencing in Saccharomyces cerevisiae occurs at specific loci and is mediated by a multiprotein complex that includes Rap1p and the Sir proteins. We studied the function of a recently identified gene, DOT4, that disrupts silencing when overexpressed. DOT4 encodes an ubiquitin processing protease (hydrolase) that is primarily located in the nucleus. By two-hybrid analysis, the amino-terminal third of Dot4p interacts with the silencing protein Sir4p. Cells lacking DOT4 exhibited reduced silencing and a corresponding decrease in the level of Sir4p. Together, these findings suggest that Dot4p regulates silencing by acting on Sir4p. In strains with several auxotrophic markers, loss of DOT4 ubiquitin hydrolase activity also results in a slow-growth defect. The defect can be partially suppressed by mutations in a subunit of the 26S proteasome, suggesting that Dot4p has the ability to prevent ubiquitin-mediated degradation. Furthermore, wild-type SIR2, SIR3, and SIR4 are required for full manifestation of the growth defect in a dot4 strain, indicating that the growth defect is caused in part by a silencing-related mechanism. We propose that Dot4p helps to restrict the location of silencing proteins to a limited set of genomic loci.
Transcriptional silencing in Saccharomyces cerevisiae is caused by specialized chromatin structures that repress gene expression. Such silencing has been detected at the silent mating-type loci HMR and HML (56), within RDN1, which encodes the ribosomal RNA genes (13, 23, 79), and adjacent to telomeres (27). Transcriptional silencing must be tightly controlled, since excessive silencing can repress loci whose expression is necessary for normal growth and survival (45, 69). The restriction of silencing activity normally begins with recruitment of the silencing complex to cis-acting chromosomal silencer elements (reviewed in reference 60).
Silencing of a locus requires the assembly on the DNA of a multicomponent protein complex (60). Although the complexes that function at the different silent loci are not identical, they do share many factors. For example, silencing at telomeres and the HM loci requires Sir2p, Sir3p, Sir4p, Rap1p, and histones H3 and H4 (3, 33, 70, 74). At the RDN1 locus, the repression of recombination, as well as of RNA polymerase II-transcribed genes inserted within the locus, requires Sir2p (13, 23, 26, 79). A consequence of sharing a common set of limiting components is that the silent loci must achieve a balance with one another in recruiting the silencing proteins (18, 25, 80).
The complex nature of silencing results in a system that is sensitive to the relative concentrations of the silencing factors. For example, overexpressing SIR3 results in increased spreading of silent chromatin from the telomere inward along the chromosome (38, 69) and the suppression of weak defects in HM silencing (62). In contrast, overexpressing SIR4 disrupts silencing at telomeric, RDN1, and HM loci (19, 48, 62, 78, 80). Deletion of SIR4 prevents the recruitment of Sir3p to the telomeres (24), and eliminates telomeric and HM silencing, but enhances RDN1 silencing, apparently through the redistribution of Sir2p to the nucleolus (25, 79, 80). Consistent with this idea, overexpressing SIR2 increases RDN1 silencing and suppresses the silencing defects in RDN1 caused by SIR4 overexpression (23, 80).
To identify additional components and regulators of silencing, we took advantage of the sensitive stoichiometric balance normally required among the silencing components and performed a screen for genes that when overexpressed disrupted telomeric silencing (77, 78). One of the genes identified was DOT4. Overexpression as well as deletion of DOT4 resulted in a partial loss of silencing at telomeres and the HM loci, with very minor changes at RDN1 (78). The DOT4 sequence contained homology to a family of genes encoding ubiquitin-specific processing proteases (Ubps), also referred to as ubiquitin hydrolases or deubiquitinating enzymes (90).
Covalent attachment of the 76-amino-acid (aa) ubiquitin polypeptide to proteins is a well-studied posttranslational modification (35). It can change the activity of a protein or act as a signal for protein degradation by the 26S proteasome (reviewed in reference 44). Some ubiquitin hydrolases (such as Doa4p and Ubp14p in S. cerevisiae) are important for promoting degradation, by clearing the proteasome of ubiquitinated peptide remnants and facilitating ubiquitin recycling (2, 39, 54, 67, 91). Consequently, the absence of these degradation-promoting activities results in pleiotropic phenotypes such as increased sensitivity to temperature and chemical stress conditions (2, 67), reduced ubiquitin-dependent plasma membrane protein turnover (59), and DNA replication defects (76). These phenotypes reflect a general reduction in the rate of ubiquitin-dependent degradation of proteins in the mutants (2, 67).
S. cerevisiae contains 17 genes that potentially encode ubiquitin hydrolases, raising the possibility that some of these enzymes are involved in regulating ubiquitination to prevent, rather than promote, degradation (44, 84, 90). However, deletion of most ubiquitin hydrolase genes in yeast results in little or no genetic phenotype, making further analysis difficult (44). Our characterization of DOT4 and its involvement in silencing is consistent with such a regulatory role for this class of genes.
The involvement of the ubiquitin system in regulating transcriptional silencing has been suggested by studies in yeast (46, 63) and Drosophila (41). In one case, a ubiquitin hydrolase, Ubp3p, which can biochemically interact with Sir4p, was found to interfere with silencing, since deletion of the UBP3 gene resulted in hyperrepression of telomeric genes (63). Our results extend the association between the silencing and ubiquitin-dependent processes, showing that Dot4p is an important regulator of SIR-dependent silencing and raising the possibility that Dot4p acts to help restrict transcriptional repression to normally silenced loci.
MATERIALS AND METHODS
Plasmid manipulations.
DOT4 was originally isolated as an amino-terminally truncated allele (78). A 4-kb ApaI/PvuI fragment containing the full-length DOT4 gene, was subcloned from ATCC lambda clone 3256 into the SmaI site of pVZ1 (42) by blunt-end ligation to produce pVZDOT4(G).
pUC9-DOT4 was made by subcloning the BamHI fragment from pVZDOT4(G) into the BamHI site of a pUC9 derivative in which the PstI site was eliminated by digestion, blunting, and religation.
pRS314-DOT4 and pRS424-DOT4 were made by subcloning the SalI/NotI fragment from pVZDOT4(G) into the SalI and NotI sites of pRS314 or pRS424, respectively.
pRS424-dot4-5 was made in multiple steps. A SacI/BamHI fragment from pVZDOT4(G) was cloned into the SphI site of pVZ1 by blunt-end ligation such that the SacI end was oriented toward the HindIII site of pVZ1 to produce pVZ1-DOT4(3′). This resulted in a 5′ truncation of the DOT4 gene. pVZDOT4(G) was then digested with HindIII, and the resulting fragment, containing the DOT4 promoter region and nucleotides 1 to 279 of the open reading frame (ORF), was ligated in frame into the HindIII site of pVZ1-DOT4(3′) to produce pVZ1-dot4-5, encoding a Dot4 mutant protein missing aa 94 to 250. Finally, a BamHI fragment from pVZ1-dot4-5 containing the dot4-5 allele was subcloned into the BamHI site of pRS424.
The dot4-1 allele was constructed by PCR using pVZDOT4(G) as the template and primers 1 (5′-GTG CTA TGG AAA AAG AGC TCC CTG AAG) and 2 (5′-AGC CTG TAC AGC AGC ATT TGT GTA ACT AGT AAC ACC). Primer 2 contained a single base substitution (underlined) that would generate the cysteine-to-serine mutation. The PCR product was digested with SacI and BsrGI and ligated into the SacI/BsrGI sites of pUC9-DOT4. Since the mutation generates an SpeI site, transformants were screened by SpeI digestion, and putative clones were confirmed by sequencing.
The dot4-6 allele was also constructed by PCR as described above. Primer 1 was as given above, and the primer 2 sequence was 5′-AGC CTG TAC AGC AGC ATT TGT GTA AGC AGT AAC ACC. The PCR product was digested with SacI and BsrGI and cloned into the SacI/BsrGI sites of pUC9-DOT4.
pRS424-dot4-1 was made by cloning the BamHI fragment from pUC9-dot4-1 into the BamHI site of pRS424.
The dot4Δ allele was constructed by digesting pUC9-DOT4 with PstI and HpaI, generating blunt ends using T4 DNA polymerase, and religating to produce pUC9-dot4Δ. This dot4Δ allele lacks most of the DOT4 ORF.
DOT4 fusions were constructed in multiple steps. First, a DOT4 allele was constructed by PCR that would allow in-frame fusions at both the 5′ and 3′ ends of the ORF. An EcoRV site was introduced at the 5′ end and a SmaI site was introduced at the 3′ end of the DOT4 ORF by PCR.
The SmaI site was introduced at the 3′ end of the DOT4 ORF by a two-step PCR technique. Primers SmaIanti3′ (5′-TTT TTT CAC CCG GGG AAC TTC CTT TTT TTA TTT TTT TTC CAT TTT TTT CTG) and DOT4TAG-B (5′-TTC AGG TCA CTA CAT TGC) were used to amplify a fragment beginning near the 3′ end of the DOT4 ORF and ending with the SmaI site placed just before the stop codon. Primers SmaI3′sense (5′-AAA AAA AAT AAA AAA AGG AAG TTC CCC GGG TGA AAA AAC TCG ATA TTC C) and DOT4TAG-D (5′-GGG GAA TTC GAA TTT AAT GCA AGA TCA GC) were used to amplify a fragment beginning with a 3′ DOT4 sequence and a SmaI site and ending with sequences downstream of the DOT4 ORF. The PCR products from these two reactions were mixed and amplified by using primers DOT4TAG-B and DOT4TAG-D. The resulting fragment was digested with HpaI and EcoRI and cloned by blunt-end ligation into pUC9-DOT4 that was digested with HpaI and SmaI. This produced pUC9-DOT4(SmaI).
Primers EcoRVsens2 (5′-GAT ATC ATG ACC ACT CAA GAA TCG ATC AAA CC) and H3anti (5′-GTA TAC AAC AAT AAA GCT TCA GCC) were used to produce a fragment beginning with an EcoRV site and ending with the 5′ sequence of the DOT4 ORF. Primers DOT4TATA+ (5′-CTT ATT TTT ATA TAG TGC CAC CAT CG) and EcoRVanti1 (5′-TTT GAT CGA TTC TTG AGT GGT CAT GAT ATC AGT CTG TGA TTG TGA TAT GAC AAT AGG) were used to produce a fragment beginning with DOT4 upstream sequence and ending with an EcoRV-containing DOT4 ORF sequence. The PCR products from these two reactions were mixed and amplified by PCR by using primers DOT4TATA+ and H3anti. pUC9-DOT4(SmaI) was partially digested with HindIII as described previously (5), using a final HindIII concentration of 0.037 U/μl, incubating for 15 min, and quenching with an EDTA-Sarkosyl solution. The final PCR product containing EcoRV was digested with SphI and HindIII and ligated into the partially digested pUC9-DOT4(SmaI). This produced pDOT4-PO (for “pop-out”), in which the DOT4 ORF is flanked by EcoRV at its 5′ end and by SmaI at its 3′ end. Correct clones were confirmed by sequencing. The DOT4-PO allele was cloned as a BamHI/EcoRI fragment into the BamHI/EcoRI sites of pRS314 or pRS424. The resulting plasmids were used to test for complementation of a dot4Δ allele by DOT4-PO or for the ability of DOT4-PO overexpression to cause telomeric derepression. DOT4-PO was found to behave in a wild-type manner (data not shown).
A pop-out version of dot4-1 (pdot4-1-PO) was made by cloning a SacI/NcoI fragment from pdot4-1 into the SacI/NcoI sites of pDOT4-PO. A pop-out version of dot4-5 (pdot4-5-PO) was made by cloning a HindIII/NcoI fragment from pVZ1-dot4-5 into the HindIII/NcoI sites of pDOT4-PO.
To fuse Dot4p to a six-Myc epitope tag, pBS/KS+MT6 (a generous gift from Mark Roth, Fred Hutchinson Cancer Research Center) was digested with DraI and PstI and cloned in frame into the SmaI site of pDOT4-PO by blunt-end ligation. Correct clones were confirmed by sequencing. pDOT4-MT6 was then used to make pdot4-1-MT6 by replacing the NcoI/SfuI fragment in pdot4-1-PO with a NcoI/SfuI fragment from pDOT4-MT6. Likewise, the NcoI/SfuI fragment in pdot4-5-PO was replaced with a NcoI/SfuI fragment from pDOT4-MT6 to yield pdot4-5-MT6.
pRS306-dot4Δ was made by cloning the BamHI fragment from pUC9-dot4Δ into the BamHI site of pRS306. pRS306-dot4-1-MT6 and pRS306-dot4-5-MT6 were made by cloning BamHI/SfuI fragments from pdot4-1-MT6 and pdot4-5-MT6, respectively, by blunt-end ligation into pRS306 that was digested with BamHI and HindIII.
The GAL4 binding domain was fused to the amino terminus of DOT4 alleles by cloning an EcoRV/EcoRI fragment of pDOT4-PO (or pdot4-1-PO or pdot4-5-PO) in frame into the NcoI site of pAS2 (Clontech, Palo Alto, Calif.) by blunt-end ligation. pAS2-dot4-2 was made by cloning an EcoRV/SacI fragment from pDOT4-PO in frame into the NcoI site of pAS2 by blunt-end ligation. Correct clones were confirmed by sequencing.
Standard molecular genetic manipulations were used as previously described (27, 71). Details of some standard protocols may also be found online (27a).
Yeast strain construction.
All strains were grown in standard culture media, and standard yeast genetic methods were used (1, 34). Genotypes of strains used in this study are presented in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype | Parent | Reference |
|---|---|---|---|
| UCC3505 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ppr1::HIS3 adh4::URA3-TEL-VIIL ADE2-TEL-VR | 77 | |
| UCC4599 | MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 adh4::URA3-TEL-VIIL ADE2-TEL-VR ppr1::LYS2 dot4::HIS3 | UCC4573 | 78 |
| UCC4604 | MATα ura3-52 ade2-101ochre | YNN215 | 75 |
| UCC4605 | MATα ura3-52 | UCC4604 | |
| UCC4606 | MATa/α ura3-52/ura3-52 lys2-801amber/+ ade2-101ochre/+ | UCC4605x | 75 |
| trp1-Δ63/+ his3-Δ200/+ leu2-Δ1/+ | YPH499 | ||
| UCC4608 | MATa/α ura3-52/ura3-52 lys2-801amber/+ ade2-101ochre/+ trp1-Δ63/+ his3-Δ200/+ leu2-Δ1/+ DOT4-URA3-dot4Δ/+ | UCC4606 | |
| UCC4610 | MATa/α ura3-52/ura3-52 lys2-801amber/+ ade2-101ochre/+ trp1-Δ63/+ his3-Δ200/+ leu2-Δ1/+ dot4Δ/+ | UCC4608 | |
| UCC4639 | MATα ura3-52 | UCC4610 | |
| UCC4687 | MATa ura3-52 his3-Δ200 trp1-Δ63 lys2-801 ade2-101 dot4Δ | UCC4610 | |
| UCC4693 | MATα his3-Δ200 trp1-Δ63 ura3-52 | UCC4610 | |
| UCC4711 | MATα ura3-52 his3-Δ200 trp1-Δ63 lys2-801 ade2-101 | UCC4610 | |
| UCC4717 | MATα his3-Δ200 ura3-52 | UCC4610 | |
| UCC4773 | MATa trp1-Δ1 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ sir2::URA3 | PJ69-4a | 49 |
| UCC4774 | MATa trp1-Δ1 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ sir3::URA3 | PJ69-4a | 49 |
| UCC4775 | MATa trp1-Δ1 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ sir4::URA3 | PJ69-4a | 49 |
| UCC4776 | MATa ura3-52 trp1 lys2-801 leu2-Δ1 his3-200 pep4::HIS3 prb1-Δ1.6R can1 GAL 6His-HA3-SIR4 | DM428 | 64 |
| UCC4786 | MATα | UCC4639 | |
| UCC4794 | MATα dot4::KanMX | UCC4786 | |
| UCC4799 | MATa ura3-52 trp1 lys2-801 leu2-Δ1 his3-200 pep4::HIS3 prb1-Δ1.6R can1 GAL 6His-HA3-SIR4 dot4::KanMX | UCC4776 | |
| UCC4817 | MATa his3-Δ200 trp1-Δ63 ura3-Δ0 adh4::URA3-TEL-VIIL | BY4728 | 11 |
| UCC4818 | MATa/α ura3-52/ura3-52 lys2-801amber/+ ade2-101ochre/+ trp1-Δ63/+ his3-Δ200/+ leu2-Δ1/+ DOT4-GFP-KanMX/+ | UCC4606 | |
| UCC4825 | MATa ade2Δ::hisG ura3-Δ0 ADE2-TEL-VR | BY4725 | 11 |
| UCC4826 | MATa his3-Δ200 trp1-Δ63 ura3-Δ0 ppr1::TRP1 adh4::URA3-TEL-VIIL | UCC4817 | |
| UCC4857 | MATa ade2Δ::hisG ura3-Δ0 ADE2-TEL-VR dot4::KanMX | UCC4825 | |
| UCC4865 | MATa ade2Δ::hisG ura3-Δ0 ADE2-TEL-VR dot4-1-MT6-URA3-DOT4 | UCC4825 | |
| UCC4870 | MATa ade2Δ::hisG ura3-Δ0 ADE2-TEL-VR dot4::dot4-1-MT6 | UCC4865 | |
| UCC4875 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa3-Δ1::HIS3 (YCp50DOA3) dot4::KanMX | MHY784 | 16 |
| UCC4876 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa3-Δ1::HIS3 (YCplac22doa3-1) dot4::KanMX | MHY792 | 16 |
| UCC4877 | MATa URA3-TEL-VIIL ppr1::TRP1 ura3Δ0 trp1-Δ63 | UCC4826 | |
| UCC4879 | MATa URA3-TEL-VIIL ppr1::TRP1 ura3Δ0 trp1-Δ63 dot4::KanMX | UCC4877 | |
| UCC4881 | MATα ade2Δ::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 dot4::KanMX | BY4705 | 11 |
| UCC4884 | MATa leu2Δ0 dot4::KanMX | BY4712 | 11 |
| UCC4887 | MATα trp1-Δ63 ura3-52 | UCC4693 | |
| UCC4888 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir2::HIS3 | UCC4693 | |
| UCC4889 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir3::HIS3 | UCC4693 | |
| UCC4890 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir4::HIS3 | UCC4693 | |
| UCC4891 | MATα trp1-Δ63 ura3-52 dot4::KanMX | UCC4887 | |
| UCC4892 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir2::HIS3 dot4::KanMX | UCC4888 | |
| UCC4893 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir3::HIS3 dot4::KanMX | UCC4889 | |
| UCC4894 | MATα his3-Δ200 trp1-Δ63 ura3-52 sir4::HIS3 dot4::KanMX | UCC4890 | |
| UCC4895 | MATa ade2Δ::hisG ura3Δ0 ADE2-TEL-VR dot4-5-MT6-URA3-DOT4 | UCC4825 | |
| UCC4896 | MATa ade2Δ::hisG ura3Δ0 ADE2-TEL-VR dot4-5-MT6 | UCC4895 |
URA3 was placed near the left arm of chromosome VII as described previously (27).
DOT4 was replaced with KanMX by PCR-mediated gene disruption as described elsewhere (7, 11). The reaction mixture contained pRS400 as the template and primers that contained homology to the sequences flanking the DOT4 locus [dot4 RS(+) (TCC AGG AAT ATC GAG TTT TTT CAT TTG GTG AAC CTG TGC GGT ATT TCA CAC CG) and dot4 RS(−) (TCC AGG AAT ATC GAG TTT TTT CAT TTG GTG AAC CTG TGC GGT ATT TCA CAC CG)].
DOT4 was replaced with a deletion allele by two-step gene replacement (34). pRS306-str4Δ was digested with SphI and used to transform cells. Transformants were first selected for growth on media lacking uracil, followed by selection on fluoro-orotic acid (5-FOA) (8) to produce the desired genotype. Similarly, DOT4 was replaced with the dot4-1 or dot4-5 allele by using pRS306-dot4-1-MT6 or pRS306-dot4-5-MT6, respectively, that was digested with SphI prior to transformation.
PPR1 was disrupted with TRP1 by using pPPR1::TRP1-1 that was digested with EcoRI prior to transformation (81).
ADE2 was placed near the right arm of chromosome V by transformation with pHR10-6 that was digested with EcoRI as previously described (69).
UCC4776 was made from DM428 (64), which is a protease-deficient strain based on BJ5459 (51). DM428 carries a 5′-tagged SIR4 allele and the URA3 gene at the SIR4 locus, along with a truncated duplication of SIR4. To remove the truncated allele, recombination events were selected on 5-FOA. Correct recombinants that retained the tagged allele but lost URA3 and the truncated allele were named UCC4776.
GFP (green fluoresent protein)-S65T (40) was fused to the carboxy terminus of Dot4p via a one-step PCR-based technique (89) by amplifying a GFP-KanMX cassette from pFA6a-GFP S65T-KanMX6, using primers DOT4GFP(+) (CAG AAA AAA ATG GAA AAA AAA TAA AAA AAG GAA GTT CAC CAA AAG TAA AGG AGA AGA ACT TTT CAC TGG) and DOT4GFP(−) (ATG CCT ATG AAA AGA GGA AAA TCC AGG AAT ATC GAG TTT TTT GGA TGG CGG CGT TAG TAT CGA ATC G).
SIR2, SIR3, and SIR4 were deleted by PCR (7) using the appropriate pRS30x plasmid.
UCC4606 was made by mating YPH499 with a modified version of YNN215 in which lys2-801amber and ade2-101ochre were converted to wild-type alleles by transformation with PCR-amplified and gel-isolated wild-type genes and selection for Lys+ and Ade+ phenotypes. Genes were amplified from pRS317 (LYS2) or pRS402 (ADE2), using standard primers to pRS series shuttle vectors (7).
Serial dilution and ubiquitin hydrolase assays.
Telomeric URA3 silencing was determined by a serial dilution assay as previously described (27). Similarly, plating of serial dilutions was used to qualitatively assess strain growth. Briefly, colonies were grown for 3 to 4 days on appropriate media (selective media if plasmid selection was required; complete media if otherwise), and colonies were suspended in water. The suspension was serially diluted, plated on appropriate media (as indicated), and allowed to grow for 3 (silencing assay) and up to 5 (growth rate assay) days.
Bacterial ubiquitin hydrolase assays were performed by assaying β-galactosidase activity on standard 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) color indicator Luria-Bertani plates containing ampicillin and chloramphenicol (5). Strains containing different plasmid constructs were all compared on the same plate. Blue colony color was scored as positive for β-galactosidase activity, while white colony color was scored as negative for β-galactosidase activity. Myc-tagged alleles of DOT4 and dot4-1 were used in this assay to ensure equivalent expression levels (data not shown and reference 52).
Calculation of growth rates and FACS analysis.
The doubling time of yeast strains was measured by using a hemocytometer to count cells that were growing in rich medium at 30°C. Simultaneously, exponentially growing cells were harvested for fixation and staining by the method of Nash et al. (66). Briefly, cells were collected, fixed in ethanol, and resuspended in 50 mM sodium citrate (pH 7). Following sonication and treatment with RNase A (0.25 mg/ml), cells were resuspended in sodium citrate containing propidium iodide (16 μg/ml) and analyzed by fluorescence-assisted cell sorting (FACS). MultiCycler (Phoenix Flow Systems) was used to calculate the percentage of cells in G1 and G2 phases of the cell cycle.
Western analysis.
Total yeast protein extracts were made by harvesting exponentially growing cells, resuspending them in sodium dodecyl sulfate (SDS)–β-mercaptoethanol protein loading buffer containing benzamidine and phenylmethylsulfonyl fluoride, adding glass beads, and vortexing for a few minutes with intermittent incubations on ice. The cells were then incubated at 95°C for 10 min. On occasion, cells were frozen in liquid nitrogen before resuspension in protein loading buffer.
SDS-polyacrylamide gel electrophoresis and Western detection were performed as previously described (5). Resolving gels contained 0.325 M Tris (pH 8.8).
Monoclonal antibodies 9E10 and HA.11 (BAbCo, Richmond, Calif.) against Myc and hemagglutinin (HA) tags were used to detect tagged proteins. Goat polyclonal antibodies against Sir2p (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used to detect Sir2p. Antibodies against Rap1p were the generous gift of Judith Berman (University of Minnesota). Antibodies against Sir3p were the generous gift of Danesh Moazed (Harvard Medical School).
Antibodies 9E10 and HA.11 were detected with horseradish peroxidase (HRP)-coupled anti-immunoglobulin G1 subtype-specific secondary antibodies (Boehringer Mannheim, Indianapolis, Ind.). Anti-Sir2p antibodies were detected with HRP-coupled donkey anti-goat secondary antibodies (Santa Cruz Biotechnology). Detection of the HRP signal was performed with ECL (enhanced chemiluminescence) or ECLplus reagents (Amersham-Life Sciences).
Monoclonal antibodies to ubiquitin were made by a modification of the protocol of Haas and Bright (36). Briefly, antigen was prepared in the following manner. Bovine ubiquitin (Sigma) was cross-linked using glutaraldehyde to mouse immunoglobulin G2b at an 18:1 molar ratio and dialyzed against phosphate-buffered saline. Samples were denatured by adding SDS (1%, final concentration) and boiling. These antibodies are now available from BAbCo and Santa Cruz Biotechnology.
Extracts from strain MHY840 (carrying a ubp14Δ allele; the generous gift of M. Hochstrasser, The University of Chicago), which accumulates free polyubiquitin chains (2), were used to help determine the molecular masses of ubiquitin chains in Fig. 7D.
FIG. 7.
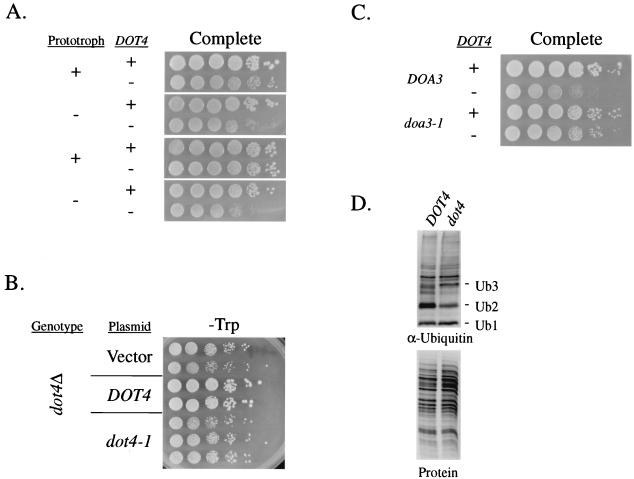
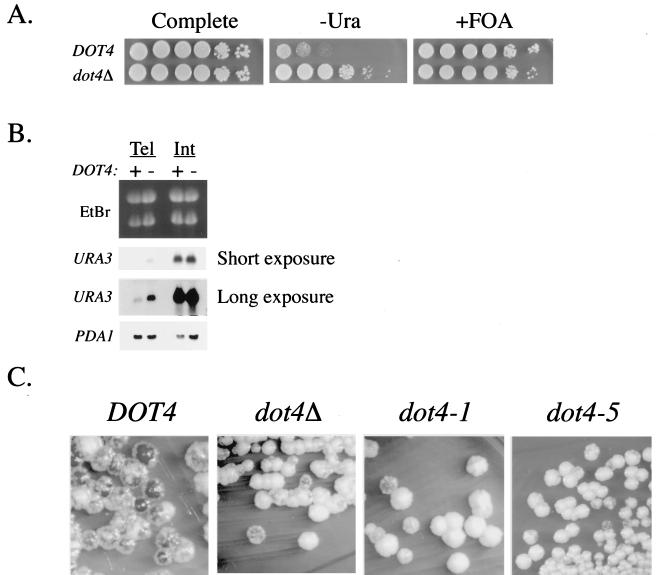
Deleting DOT4 in auxotrophic strains causes a slow-growth defect that is dependent on 26S proteasome activity. (A) DOT4 wild-type and mutant strain pairs containing few (prototroph +) or multiple (prototroph −) auxotrophic markers were assayed for growth on nutritionally complete medium. From the top down, the strain pairs tested were UCC4786 plus UCC4794 (no markers), UCC4711 plus UCC4687 (ura3-52 his3-Δ200 trp1-Δ63 lys2-801 ade2-101), UCC4825 plus UCC4857 (ura3-Δ0), and BY4705 plus UCC4881 (ade2Δ::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0). In all cases, DOT4 was deleted without altering the auxotrophy of the strain. + in the prototroph column represents relative prototrophy. (B) Growth of strains carrying DOT4, dot4-1, or dot4Δ. Strain UCC4599 (dot4::HIS3 ura3-52 lys2-801 amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 adh4::URA3-TEL ADE2-TEL ppr1::LYS2) carried a single-copy TRP1/CEN plasmid containing wild-type DOT4, the enzymatically inactive dot4-1, or empty vector. Multiple independent transformants were grown on medium lacking tryptophan but containing all other required nutritional supplements. (C) A deletion of DOT4 was combined with a mutation in DOA3, encoding a proteasome subunit, and growth was tested on the same plate at the permissive temperature (23°C) on nutritionally complete medium. Strain pairs, containing a DOT4 and dot4Δ::KanMX allele, respectively, were (from the top down) MHY784 plus UCC4875 (his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa3-Δ1::HIS3 YCp50DOA3::URA3) and MHY792 plus UCC4876 (his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa3-Δ1::HIS3 YCplac22doa3-1::TRP1). (D) Levels of free ubiquitin were compared in DOT4 wild-type and mutant strains. Ub1, monoubiquitin; Ub2, diubiquitin; Ub3, triubiquitin. Antiubiquitin antibodies were used on extracts from UCC4786 (DOT4) and UCC4794 (dot4Δ). In a parallel loading experiment, proteins were stained with Coomassie blue to control for differences in sample loading.
Northern analysis.
RNA was isolated by a phenol-freeze protocol (72). Northern analysis was performed as previously described (4), and detection was performed with digoxigenin-labeled riboprobes (Boehringer Mannheim). Riboprobes were made by transcribing a PCR product in the antisense orientation with T7 RNA polymerase in the presence of digoxigenin-labeled UTP as instructed by the supplier (Boehringer Mannheim).
A probe for SIR4 transcripts was made by amplifying a fragment of SIR4 from pKAN59 (48), using primers midSIR4+ (5′-GAT TAC TCT AAA GAG ATT CTA GG) and T7SIR4(−) (5′-ATC GAT AAT ACG ACT CAC TAT AGG GAG GTG ACT TTA AGA TTT CCA TCC).
A probe for URA3 was made as described above, using pRS306 as the template and primers URA3N+ (5′-TCG AAA GCT ACA TAT AAG GAA CG) and T7_URA3(−) (5′-ATC GAT AAT ACG ACT CAC TAT AGG GAG TAC CCT TAG TAT ATT CTC C). A probe for PDA1 was made by using pSD183 (containing the PDA1 gene; a generous gift of S. Diede) as the template and primers T7PDA_P1 (5′-ATC GAT AAT ACG ACT CAC TAT AGG GTG TTC GTC AAC GTA TTT TCT AGC GG) and T7PDA_P2 (5′-TGG TTC CAT GCA CCT TTA CGC TCC AGG).
Fluorescence microscopy.
Cells were grown at room temperature to exponential growth phase and fixed with 3.7% formaldehyde as described elsewhere (1). Fluorescence microscopy (Nikon Eclipse E800) was performed with a progressive charge-coupled device camera (Sony) and Metamorph software (Universal Imaging Corporation, West Chester, Pa.) for image capture and analysis. Nikon 4′,6-diamidino-2-phenylindole (DAPI) and fluorescein isothiocyanate filters were used for DAPI and GFP detection, respectively.
RESULTS
Dot4p is a deubiquitinating enzyme.
The predicted protein sequence of Dot4p contains regions of homology (especially at the highly conserved Cys box [Fig. 1A]) to the family of deubiquitinating enzymes known as Ubps (44, 90). To determine if Dot4p had deubiquitinating activity, we used a bacterial assay that makes use of the N-end rule (6, 88). A chimeric protein made of ubiquitin fused to the N terminus of β-galactosidase is stable in Escherichia coli and has β-galactosidase activity. However, when a ubiquitin-specific protease is expressed in the same cells (E. coli does not normally have deubiquitinating activity), ubiquitin is cleaved from the fusion polypeptide, leaving a new amino terminus on β-galactosidase. According to the N-end rule, residues at the amino terminus of a protein regulate the protein’s rate of proteolysis. Hence, if the new amino terminus of the β-galactosidase is a methionine, the protein remains stable. However, if the amino-terminal residue following deubiquitination is leucine, the β-galactosidase will be very unstable.
FIG. 1.
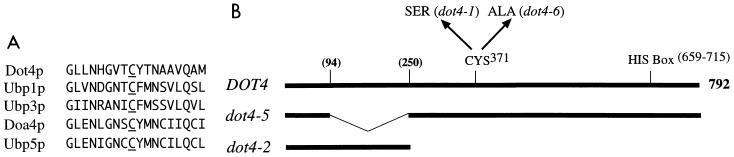
Dot4p mutant map and Cys box sequence. (A) Dot4p contains sequence homology to the active site Cys box of other yeast deubiquitinating enzymes. (B) Schematic map of the Dot4 protein and various engineered alleles.
We used such fusion proteins to test the ubiquitin hydrolase activity of Dot4p, as measured by the presence or absence of β-galactosidase activity on an X-Gal plate (Table 2). E. coli cells that expressed the ubiquitin–Leu–β-galactosidase (Ub-Leu-βgal) fusion and no DOT4 exhibited strong β-galactosidase activity, but when DOT4 was coexpressed in these cells, the β-galactosidase activity was virtually eliminated (Table 2, vector versus DOT4), suggesting that Dot4p was indeed a ubiquitin hydrolase. Dot4p did not appear to have any other proteolytic activities that reduced the β-galactosidase activity, because when leucine was replaced by the stabilizing methionine at the N terminus of β-galactosidase, activity was stable even in the presence of DOT4 (Table 2, Ub-Met-βgal).
TABLE 2.
Dot4p is a ubiquitin hydrolase
| DOT4 allele | Ubiquitin substrate | β-Galactosidase activitya |
|---|---|---|
| Vector | Ub-Leu-βgal | + |
| DOT4 | Ub-Leu-βgal | − |
| DOT4 | Ub-Met-βgal | + |
| dot4-1 | Ub-Leu-βgal | + |
| dot4-6 | Ub-Leu-βgal | + |
| dot4-5 | Ub-Leu-βgal | − |
| dot4-2 | Ub-Leu-βgal | + |
| dot4Δ | Ub-Leu-βgal | + |
Dot4p enzymatic activity was assayed on X-Gal color indicator plates by coexpression with a ubiquitin–β-galactosidase fusion. All tests were performed on the same plate. When a Ub-Leu-βgal substrate is used, the presence of β-galactosidase activity correlates with the lack of ubiquitin hydrolase activity. Conversely, the absence of β-galactosidase activity indicates the proteolytic separation of ubiquitin from Leu-βgal and subsequent degradation of the Leu-βgal peptide. The Ub-Met-βgal substrate serves as a control for the presence of nonspecific peptidase activity.
The putative active site of ubiquitin proteases contains a conserved and essential cysteine that forms a thiol-ester bond with ubiquitin during the process of deubiquitination (Fig. 1A) (44, 90). When a mutant allele of DOT4 containing a mutation of cysteine 371 to serine (dot4-1) or alanine (dot4-6 [Fig. 1B]) was expressed in E. coli, or when the DOT4 coding region was deleted from the clone (dot4Δ), deubiquitinating activity was abolished (Table 2). The lack of hydrolase activity in cells expressing dot4-1 was not the result of reduced Dot4 protein levels, as determined by Western analysis (data not shown). Thus, the conserved cysteine residue of Dot4p was required for its activity, consistent with DOT4 encoding a ubiquitin-specific protease.
Yeast ubiquitin-specific proteases are composed of two general regions; one contains the conserved domains with the putative hydrolase active site, and the other is highly divergent among the 17 ubiquitin hydrolases and thought to be required for specific protein interactions (44, 90). In Dot4p, the amino-terminal 46% (residues 1 to 362) contains the novel sequence region. As expected, this region was not required for Dot4p activity in the bacterial assay. Deletion of residues 94 to 250 of the bacterially expressed Dot4p (Fig. 1B) still produced enzymatic activity (Table 2, dot4-5). Conversely, a clone expressing only the amino-terminal 250 residues (dot4-2) had no detectable deubiquitinating activity (Table 2).
Overexpression of the amino-terminal region of Dot4p disrupts telomeric silencing.
The overexpression of DOT4 in S. cerevisiae disrupted silencing at telomeres and the silent mating-type loci (Fig. 2A) (78). This phenotype may result from excess ubiquitin hydrolase activity or, alternatively, from Dot4p’s interactions with another molecule that prevents the latter from functioning properly. To distinguish between these two possibilities, the dot4-1 allele, which lacks hydrolase activity, was overexpressed in a yeast strain containing a telomeric URA3 gene. Overexpression of dot4-1 caused derepression of the telomeric URA3 (Fig. 2A), suggesting that the ubiquitin hydrolase activity of Dot4p was not necessary for this effect.
FIG. 2.
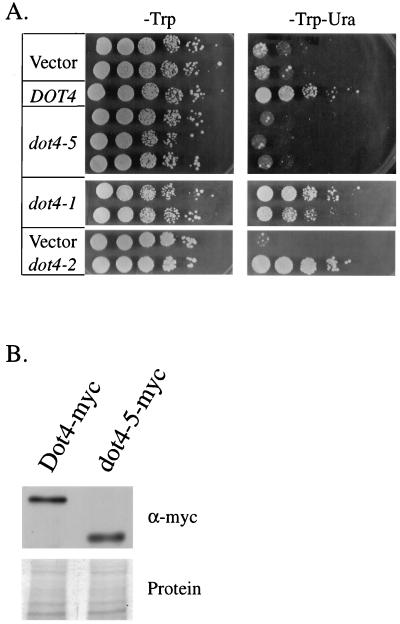
Overexpression of the amino terminus of Dot4p disrupts silencing. (A) A wild-type DOT4 strain (UCC3505) containing URA3 near a telomere was transformed with a high-copy TRP1 plasmid carrying various alleles of DOT4 under the control of the DOT4 promoter. Telomeric silencing was measured by testing for growth on media lacking uracil in a serial dilution plating assay. In the case of dot4-2, control of transcription occurred through the GAL1 promoter, and silencing was assayed on medium containing galactose as the sole carbon source. (B) The extent of dot4-5 overexpression in panel A was determined by comparison with wild-type DOT4 overexpression, using Western analysis of total yeast protein extracts. Proteins were tagged with a six-Myc epitope, and blots were probed with anti-Myc antibodies (α-myc). Prestaining of the blot with India ink showed equivalent sample loading.
Next, the amino-terminal region of Dot4p, which contains the divergent, putative protein interaction domain, was tested for its overexpression phenotype. When aa 94 to 250 were deleted from Dot4p, the resulting mutant protein, dot4-5p, maintained its enzymatic activity (Fig. 1B), but overexpression in yeast did not affect silencing of a telomeric URA3 (Fig. 2A). Expression levels of dot4-5p and full-length Dot4p were equivalent (Fig. 2B). Conversely, when a peptide containing only the amino-terminal 250 aa of Dot4p was overexpressed (dot4-2), strong derepression of the telomeric URA3 was observed (Fig. 2A). Thus, the amino-terminal region of Dot4p, which does not contain ubiquitin hydrolase activity, is necessary and sufficient for the overexpression phenotype of full-length Dot4p.
Dot4p can interact with Sir4p.
Since many of the key factors required for transcriptional silencing in yeast have been identified, we used the two-hybrid assay (17) to test whether Dot4p interacted with Sir3p or Sir4p, both of which are known structural components of telomeric silent chromatin. As shown in Fig. 3A, Dot4p was found to interact, directly or indirectly, with two different but overlapping regions of Sir4p. These regions are known to be involved in forming complexes between Sir4p molecules as well as between Sir4p and the telomeric silencing factors Sir2p, Sir3p, Rap1p, and Ku70 (17, 30, 64, 65, 86).
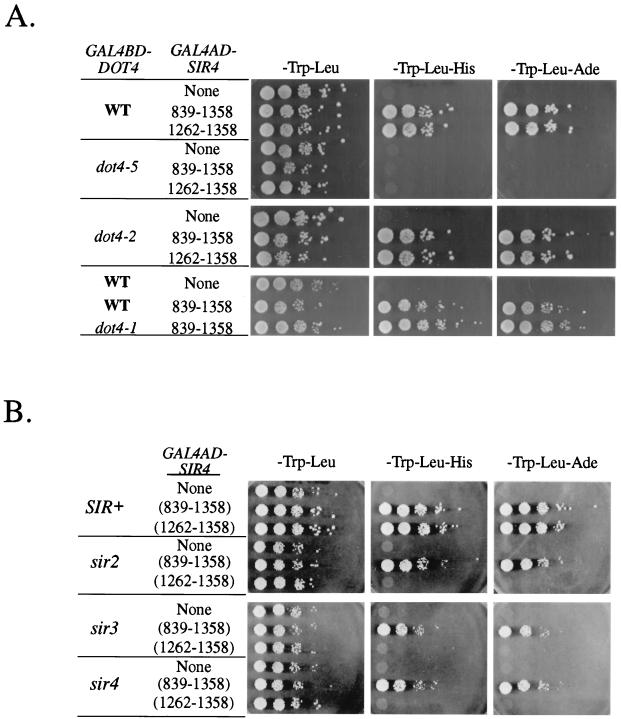
FIG. 3.
Dot4p interacts with Sir4p in a two-hybrid assay. (A) A two-hybrid assay was used to test the interaction between a Dot4p bait and Sir4p C-terminal preys. GAL4BD-DOT4 fusions (bait) on a TRP1 plasmid and GAL4AD-SIR4 fusions (prey) on a LEU2 plasmid were transformed into PJ69-4a, which contains HIS3 and ADE2 under the control of synthetic GAL promoters. The ability of cells to grow in the absence of histidine or adenine was tested in a serial dilution plating assay. Activation of HIS3 and ADE2 indicates a positive interaction between the two protein fusions. WT, wild type. (B) Dot4p-Sir4p two-hybrid interaction was tested in a PJ69 background in which SIR2 (UCC4773), SIR3 (UCC4774), or SIR4 (UCC4775) was deleted.
The interaction between Dot4p and Sir4p could explain the silencing defect of DOT4 overexpression because silencing is particularly sensitive to Sir4p dosage (19, 20, 48, 62, 64, 78). Therefore, the alleles of DOT4 used to define its overexpression phenotype (Fig. 1B) were tested for the ability to interact with SIR4 in the two-hybrid assay. The enzymatically inactive dot4-1p, which disrupted silencing when overexpressed (Fig. 2A), could still interact with a Sir4p peptide (Fig. 3A). An interaction was also detected with the amino-terminal region of Dot4p (dot4-2p [Fig. 3A]) that, when overexpressed, was necessary and sufficient for disrupting silencing (Fig. 2A). In contrast, dot4-5p, which did not disrupt silencing when overexpressed (Fig. 2A), showed no interaction with Sir4p by the two-hybrid assay (Fig. 3A). The interactions with Sir4p appeared to be specific, since neither Sir3p nor Snf4p, an unrelated protein, interacted with Dot4p (52). Thus, a direct correlation was found between the ability of overexpressed Dot4p derivatives to disrupt silencing and their ability to interact with Sir4p in the two-hybrid assay.
As previously mentioned, Sir4p interacts with a number of other proteins and can be found in a large silencing complex that includes Sir2p, Sir3p, and additional Sir4p molecules (reviewed in reference 60). Therefore, we tested whether the Dot4p-Sir4p interaction was maintained in sir2, sir3, or sir4 mutant strains (Fig. 3B). In the absence of a wild-type Sir protein complex, the interaction between Dot4p and the carboxy-terminal tail of Sir4p was abolished, while interaction with the larger Sir4p fusion remained intact. These results suggest that the interaction between Dot4p and the Sir4p carboxy-terminal peptide (aa 1262 to 1358) is the result of this peptide’s ability to recruit a silencing complex that included full-length Sir4p. Furthermore, the middle region of Sir4p (aa 839 to 1261) likely mediates the interaction with Dot4p.
Dot4p is localized to the nucleus.
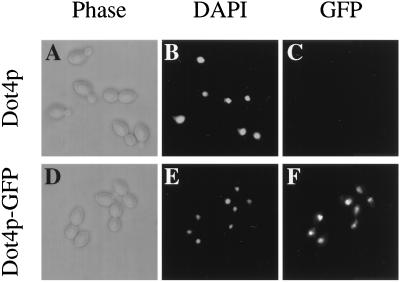
Given that Sir4p is a nuclear chromatin-associated protein, we determined whether Dot4p was also normally located in the nucleus. The genomically encoded Dot4p was carboxy-terminally tagged with the GFP from Aequorea victoria (15, 40) to produce a fully functional genomic DOT4-GFP allele that retained full transcriptional control by the DOT4 promoter and fully complemented a dot4 deletion mutant (data not shown). As shown in Fig. 4, the Dot4-GFP fusion preferentially colocalized with DAPI staining of the nucleus, consistent with its involvement in silent chromatin.
FIG. 4.
Dot4p is localized primarily to the nucleus. Dot4p localization was determined by using a DOT4-GFP-S65T gene fusion integrated at the DOT4 genomic locus of a diploid yeast strain (UCC4606). The strain (UCC4818) is heterozygous for the fusion gene. By fluorescence microscopy, the GFP signal was compared to a DAPI signal in cells that were fixed during exponential growth phase in 3.7% formaldehyde. (A to C) Control experiment using parental strain UCC4606 which lacks DOT4-GFP fusion; (D to F) experiment in which strain UCC4818 was stained with DAPI and tested for localization of Dot4-GFP.
The enzymatic activity of Dot4p is necessary for its role in silencing.
Consistent with our previous report (78), deletion of DOT4 caused a defect in transcriptional silencing (Fig. 5A). Two pathways could cause the increase in expression of a telomeric marker gene: crippling of the silencing mechanism or up-regulating the marker gene’s transcription. To distinguish between these two possibilities, we performed Northern analysis of strains containing URA3 either at its normal chromosomal locus or at a telomere. Deleting DOT4 increased expression of the telomeric URA3 but had no effect on transcription at the normal URA3 locus (Fig. 5B). This result further supports the notion that DOT4 acts on transcriptional silencing.
FIG. 5.
Dot4p and its deubiquitinating activity are important for transcriptional silencing. (A) Silencing was measured in DOT4 (UCC4877) and dot4Δ (UCC4879) strains grown on complete, uracil-lacking, and 5-FOA-supplemented media. These strains are fully prototrophic except that the URA3 gene was placed near a telomere and PPR1 was deleted. Growth on medium lacking uracil indicates a disruption of telomeric silencing. (B) Northern analysis of URA3 expression at a telomeric (Tel; UCC4877 [DOT4] and UCC4879 [dot4Δ::KanMX]) or internal (Int; BY4712 [DOT4] and UCC4884 [dot4Δ::KanMX]) locus. Northern analysis of PDA1 mRNA was used as a loading control in a parallel loading experiment. (C) A strain containing ADE2 near a telomere (UCC4825) was used to test the effects of two DOT4 alleles on silencing. DOT4 was deleted (UCC4857) or replaced in the genome with the dot4-1 (UCC4870) or dot4-5 (UCC4896) alleles, and telomeric silencing of ADE2 was observed following growth on rich medium. ADE2 expression results in a white colony color, whereas ADE2 repression results in a red colony color.
We next asked whether Dot4p’s function in silencing required its enzymatic activity, which was dispensable for the overexpression phenotypes of DOT4 (Fig. 2 and 3). This was examined by replacing the genomic copy of DOT4 with the dot4-1 allele, which is enzymatically inactive (Table 2), in a strain where the ADE2 gene was placed near a telomere. Normally, yeast strains expressing ADE2 produce white colonies, whereas an ade2 mutation results in red colonies. By placing ADE2 near a telomere, it is possible to visualize changes in telomeric silencing by simply observing colony color. A strain in which ADE2 is placed near a telomere produces predominantly red colonies with white sectors; the red sectors reflect large populations of cells in which the ADE2 gene is silenced by the telomere (27). In the dot4-1 strain, however, the colonies were predominantly white, indicating that the mutation in DOT4 compromised telomeric silencing (Fig. 5C). Therefore, the enzymatic activity of Dot4p is necessary for its role in silencing.
As mentioned above, the amino-terminal 250 aa of Dot4p interacted with Sir4p (Fig. 3A), and this domain was necessary for the overexpression phenotype of DOT4 (Fig. 2A). Therefore, we tested whether the amino-terminal domain was needed for silencing by replacing the genomic copy of DOT4 with the dot4-5 allele. This allele, which has a deletion of residues 94 to 250 (Fig. 1B), showed no interaction with Sir4p in the two-hybrid assay (Fig. 3A) but was otherwise enzymatically active (Table 2). The dot4-5 mutation caused a modest derepression of a telomeric ADE2 gene (Fig. 5C). We interpret these results to mean that dot4-5p possesses intermediate functionality in telomeric silencing, with the decreased functionality of the enzyme possibly due to its decreased interaction with Sir4p.
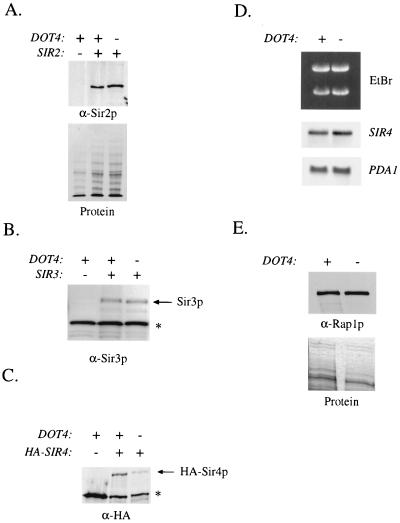
Deleting DOT4 causes a reduction in Sir4 protein levels.
Using Western analysis, we examined the levels of silencing proteins in DOT4 wild-type and mutant strain backgrounds. We found that Sir2p and Sir3p levels did not change in a DOT4 deletion strain (Fig. 6A and B), nor was there a significant change in Rap1p levels (Fig. 6E). However, Western analysis of an HA-tagged Sir4p revealed a significant reduction of Sir4p levels in the dot4 strain (Fig. 6C). Equivalent results were obtained with anti-Sir4p antibodies (data not shown). Northern analysis determined that the changes in Sir4p levels were not the result of reduced SIR4 transcription (Fig. 6D). The reduction in Sir4p levels associated with a dot4Δ mutation suggested that Dot4p may be important in maintaining normal Sir4p levels in vivo.
FIG. 6.
Sir4p levels decrease in a dot4Δ strain. (A) To detect Sir2p, Western analysis was performed on total protein extracts from strains carrying wild-type DOT4 (UCC4786) or dot4Δ (UCC4794). A SIR2 deletion strain (UCC4888) is included as a negative control. Sir2p was detected with an anti-Sir2p antibody (α-Sir2p). A parallel loading experiment in which proteins were stained with Coomassie blue shows equivalent loading of proteins. (B) Anti-Sir3p antibody was used for Western analysis of protein extracts from strains UCC4825 (DOT4 SIR3), UCC4857 (dot4Δ SIR3), and UCC4889 (DOT4 sir3Δ). The presence of a cross-reacting band (asterisk) serves as a control for protein loading. (C) As for panel A except that anti-HA antibody was used to detect Sir4p in extracts of UCC4776 (DOT4 HA-SIR4), UCC4799 (dot4Δ HA-SIR4) and BJ5459 (DOT4 SIR4). The cross-reacting band (asterisk) serves as a control. (D) Northern analysis was used to test the effect of deleting DOT4 on SIR4 expression. In parallel loading experiments, SIR4 and PDA1 expression was analyzed by using RNA from exponentially growing DOT4 and dot4Δ cells. EtBr, ethidium bromide. (E) As for panel A except that anti-Rap1p antibody was used on extracts from UCC4786 (DOT4) and UCC4794 (dot4Δ). The nitrocellulose blot was stained with India ink to control for loading.
Loss of Dot4p function can cause a growth defect that requires full proteasome activity.
Deleting DOT4 led to another interesting discovery: in certain strain backgrounds, the absence of Dot4p caused a growth defect (Fig. 7A) (78). Since simply disrupting silencing does not itself lead to slow growth (data not shown) (3), we decided to investigate the relationship between this slow-growth phenotype and the silencing functions of Dot4p.
When DOT4 was deleted in strains that contained several auxotrophic markers (leading to deficiencies in amino acid biosynthesis), growth defects were observed (especially when his3, leu2, lys2, and trp1 mutations were combined), even though the cells were grown in nutritionally complete media (Fig. 7A and data not shown). This auxotrophic-dependent slow-growth phenotype was the result of eliminating the Dot4p deubiquitination activity, since the same defect was detected in strains containing the dot4-1 allele (Fig. 7B) (52).
The conjugation of ubiquitin is typically thought of as a signal for protein degradation by the 26S proteasome, a large multisubunit enzyme (44). As a deubiquitinating enzyme, Dot4p may remove conjugated ubiquitin and thus prevent protein degradation. If proteins necessary for normal growth were degraded too rapidly when Dot4p was absent, then crippling ubiquitin-dependent degradation might suppress the slow-growth defect (47). Therefore, a mutation in DOA3, which encodes a subunit of the 26S proteasome, was introduced into strains to slow the degradation rate of ubiquitinated proteins (16). As shown in Fig. 7C, the doa3-1 allele imparted significant suppression of the dot4Δ growth defect. This result was consistent with a role for Dot4p in preventing degradation. It was not possible to determine whether a doa3 mutation could suppress the silencing defect of dot4 cells, because the doa3-1 allele alone severely crippled telomeric silencing (data not shown).
Deleting ubiquitin hydrolase genes that are required for ubiquitin recycling results in a dramatic reduction of free ubiquitin levels (67). Therefore, we examined whether DOT4 was important in recycling ubiquitin from degraded peptides by Western analysis of cellular ubiquitin. The level of free monoubiquitin was essentially the same in a dot4Δ strain as in a wild-type strain (Fig. 7D). Thus, Dot4p appears to be involved not in ubiquitin recycling but rather in removing ubiquitin from specific protein substrates. However, the difference in the pattern of ubiquitinated proteins between wild-type and dot4 cells seen in Fig. 7D suggests that Dot4p may act on a number of proteins.
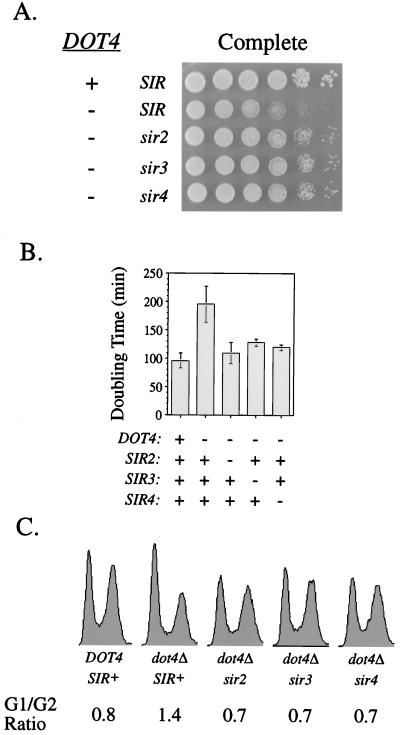
The full growth defect of a dot4 mutant requires a complete silencing complex.
A stoichiometric imbalance of silencing proteins, such as when Sir2p, Sir3p, and Rap1p are overexpressed, results in growth defects (22, 45). In some cases it appears that a fully functional silencing complex is needed for such a growth defect. For example, when SIR3 is overexpressed, wild-type SIR4 is required to detect a defect (45). In dot4Δ mutants, an imbalance also occurred since Sir4p was underrepresented relative to the other silencing proteins (Fig. 6). Therefore, we tested whether functional Sir proteins were required for the growth defect of a dot4 mutant strain. When a DOT4 deletion was combined with a deletion of SIR2, SIR3, or SIR4, the growth defect was partially suppressed (Fig. 8A and B). Cell cycle analysis revealed that dot4 mutant strains spent more time in the G1 phase of the cell cycle, consistent with a metabolic growth defect (Fig. 8C). Suppression of the growth defect by mutations in the SIR genes was correlated with a return to a nearly wild-type cell cycle profile (Fig. 8C). Therefore, an intact silencing complex was necessary for the full manifestation of the growth defect in an auxotrophic dot4Δ strain.
FIG. 8.
The growth defect of dot4Δ mutants requires functional SIR genes. (A) SIR2, SIR3, or SIR4 was deleted in combination with a deletion in DOT4, and growth was assayed on nutritionally complete growth medium. The strains assayed were, from the top to bottom, UCC4887, UCC4891, UCC4892, UCC4893, and UCC4894. (B) The growth rates of strains from panel A were quantified. Doubling times were calculated by regression analysis for exponential equations. The bars represent the mean of three independent experiments, and the error bars represent standard deviations. (C) FACS analysis was used to determine the cell cycle profiles of strains from panel A. The proportions of cells in G1 and G2 phases of the cell cycle are presented.
A loss of silencing at the HM loci, as when a SIR gene is deleted, causes both a and α information to be expressed, leading to the program of diploid gene expression in a haploid cell. To test whether this diploid program was responsible for suppression of the growth defect in dot4Δ strains, the above set of experiments was repeated in an auxotrophic strain with an HMLa MATa HMRa genotype (only a information can be expressed). Even in this strain, the dot4Δ growth defect was suppressible by mutations in the SIR genes (data not shown) (52). Taken together, these results suggest that in dot4 strains, the SIR silencing complex inappropriately repressed loci required for normal cellular growth.
DISCUSSION
In this study, we present an initial characterization of the DOT4 gene, which was originally identified by its ability to disrupt telomeric silencing when overexpressed (78). The DOT4 gene product has ubiquitin hydrolase activity (Table 2), consistent with its predicted protein structure. This activity is required for cells to attain a high level of telomeric silencing. Dot4p appears to mediate this effect through interactions with silencing proteins, in particular Sir4p.
DOT4 interacts with SIR4.
There are several lines of evidence that support this idea: (i) Dot4p interacts with Sir4p by the two-hybrid assay (Fig. 3); (ii) this interaction requires Dot4p sequences necessary and sufficient for the overexpression phenotype of DOT4, while other sequences are dispensable (Fig. 2 and 3); (iii) the two-hybrid interaction is stabilized by functional SIR genes (Fig. 3); (iv) a dot4 mutant which cannot interact with Sir4p by the two-hybrid assay also cannot support full silencing (Fig. 5); (v) mutations in the SIR genes can suppress a dot4 mutant phenotype (Fig. 8); and (vi) Sir4p levels are sensitive to DOT4 deletion (Fig. 6).
Our results add Dot4p to an extensive list of proteins that interact directly or indirectly with Sir4p. This list includes Sir4p itself, as well as Sir1p, Sir2p, Sir3p, Rap1p, Dis1p, Hdf1p (Ku70), Sif2p, Ubp3p, and histones H3 and H4 (17, 20, 37, 38, 63 to 65, 82, 85, 86, 92). The sequence-specific DNA binding protein Rap1p is the central factor in recruiting Sir4p to silent loci. Rap1p apparently collaborates with Sir1p to localize Sir4p to the HM loci (14, 18, 65) or with Ku70 (Hdf1p) to localize Sir4p to telomeres (10, 19, 30, 68). Once it is in proximity to the chromosome, Sir4p may act as a scaffold for recruiting other members of the silencing complex, Sir2p and Sir3p, to telomeres or HM loci (19, 24, 82). While Sir4p can also interact with histones H3 and H4, it is not clear whether this interaction helps to stabilize the Sir4p complex or whether it is an interaction that directly causes silencing (37).
Other Sir4p-interacting proteins seem to antagonize silencing. Dis1p has sequence similarity to the SWI2/SNF2 family of DNA-dependent ATPases and has been proposed to make the HM loci more accessible for recombination during mating-type switching (92). Deletion of another gene, SIF2, increases the level of silencing at telomeres, suggesting that it too normally antagonizes Sir4p action at telomeres (20).
Ubp3p, like Dot4p, is a ubiquitin hydrolase that interacts with Sir4p (63). Yet these related enzymes appear to act in different ways on telomeric silencing. Loss of UBP3 causes increased silencing (63), whereas loss of DOT4 decreases silencing (Fig. 5). It has been suggested that Ubp3p stabilizes antagonists of Sir4p-mediated silencing at telomeres (e.g., transcriptional activators) (63). In contrast, Dot4p activity appears to positively regulate silencing at telomeres. Although we have not succeeded in detecting ubiquitinated Sir4 protein, the interaction between Sir4p and Dot4p (Fig. 3) and the decrease in Sir4p levels as a result of deleting DOT4 (Fig. 6) support the idea that silencing is regulated by ubiquitin-dependent mechanisms, possibly through the regulation of Sir4p degradation. Hence, our results support and extend previously published observations in yeast (63) and Drosophila (41) that suggested a connection between the ubiquitin system and chromatin-dependent silencing.
Regulation of silencing by Sir4p and involvement of Dot4p.
Some of the many protein interactions that Sir4p participates in suggest that it plays a key regulatory role in partitioning silencing components to specific loci in the S. cerevisiae genome. For example, Sir4p appears to help target silent chromatin to telomeres and the HM loci. Specific mutations in the SIR4 gene cause Sir2p and Sir3p to become localized to the nucleolus, at the expense of the HM loci and telomeres (25, 53, 79, 80, 82). The mechanism of this nuclear partitioning is unclear but may involve differential binding between the Sir proteins. For instance, a complex of Sir2p with Sir4p prevents Sir3p from interacting with Sir4p in vitro (64, 82).
Further support for the regulatory importance of Sir4p comes from two other types of studies. First, Sir3p and Sir4p associate with silent chromatin in different ways. While Sir3p is bound to telomeric chromatin all along the silenced structure, Sir4p is found only relatively close to the end of the chromosome (82). Thus, Sir4p may not be an integral structural component of the repressive chromatin structure but rather may act as a tether between Sir3p and the telomeric DNA binding proteins (e.g., Rap1p and Hdf1p). Second, deleting SIR4, reducing it to a single copy in a diploid cell, or increasing SIR4 dosage can differentially affect the amount of silencing at telomeres, RDN1 and the HM loci (3, 48, 62, 70, 78–80, 83). Thus, Sir4p levels are regulated to maintain proper chromatin function: too little or too much Sir4p affects normal silencing.
Our data suggest a model by which Dot4p affects silencing by regulating Sir4p. We propose that the reason for the decrease in silencing in strains with either DOT4 loss of function alleles or overexpressed DOT4, was a reduction in the functional level of Sir4p. However important distinctions exist between the two cases. In the case of overexpressed DOT4, the higher level of Dot4p likely engages Sir4p in a futile complex that prevents it from efficiently participating in the formation of silent chromatin. This conclusion is supported by the correlation between the ability of DOT4 alleles to disrupt silencing and to interact with Sir4p by the two-hybrid assay (Fig. 2 and 3).
In the case when Dot4p activity is absent, the cellular levels of Sir4p were reduced with little effect on Sir2p and Sir3p levels (Fig. 6). The simplest explanation for the reduced level of Sir4p is that without Dot4p, a ubiquitin-mediated proteolysis pathway more readily degrades Sir4p. An alternative explanation for Dot4p activity in silencing draws on evidence that ubiquitination may alter protein activity rather than protein stability per se (e.g., histone ubiquitination) (12, 21, 43, 57). In this instance, a ubiquitinated Sir4p may be incapable of assembling into silent chromatin at specific loci. Sir4p molecules that are not engaged in a silent chromatin may then be degraded at a higher rate. Thus, the decreased level of Sir4p in dot4Δ strains may be an indirect effect. Last, we cannot rule out the possibility that ubiquitin-related peptides, such as Rub1p and Smt3p (50, 55, 58, 73), are attached to Sir4p and hence involved in the Dot4p regulation of Sir4p.
DOT4 may help to restrict Sir protein activity.
The slow-growth phenotype of dot4 mutants implicates Dot4p in the regulation of molecules other than Sir4p. Dot4p is likely to counteract the ubiquitin-mediated degradation of these molecules because a defect in the 26S proteasome significantly suppressed the slow-growth phenotype (Fig. 7C), while ubiquitin remains abundant in dot4 mutants (Fig. 7D). In addition, there is an important connection between the silencing and slow-growth phenotypes of dot4Δ: deleting SIR2, SIR3, or SIR4 significantly suppressed the growth defect (Fig. 8). Since deleting the SIR genes abolishes SIR-dependent silencing (60), this suppression may be the result of derepressing a silent locus that influences growth in a dot4 strain. Our experiments with a strain carrying a information at HML, MAT, and HMR suggest that the mating-type loci are not involved in the growth defect (52). Nevertheless, it is possible that another, yet undiscovered silent locus must be expressed in order to compensate for the growth defect caused by the absence of Dot4p.
Alternatively, the Sir proteins may act promiscuously in dot4Δ strains to repress genes not normally silenced, including metabolic genes required for normal growth rates. The well-known problem of silencing promiscuity stems from observations that the Sir protein complexes do not interact directly with DNA. Instead, they interact with DNA-binding proteins such as histones and Rap1p (60). Since histones and Rap1p also bind DNA throughout the genome in nonsilencing capacities (31, 74, 87), mechanisms must exist for restricting Sir-dependent silencing activity to proper sites. There is strong evidence that histone acetylation plays a crucial role in restricting silencing activity (32, 33, 60). Yet, overexpressing Sir3p can extend silencing until cell death ensues (28, 38, 69). Likewise, inappropriate targeting of Rap1p onto the chromosome can result in the extension of Sir-dependent silencing to these sites (65). Therefore, in addition to histone acetylation, other mechanisms likely contribute to the restriction of silencing under normal conditions.
The finding that dot4Δ mutants exhibit SIR-dependent slow growth suggests that Dot4p’s activity in the cell may be to help restrict silencing to proper loci. Based on our results, we suggest that redirection of the Sir proteins in a dot4Δ strain may occur via Rap1p, which binds and regulates numerous promoters throughout the genome, including genes coding for ribosomal proteins and regulators of metabolism (29, 74). Rap1p activity depends on the genetic context of its binding site (9, 74). Normally, Rap1p binding at silent loci leads to a repressed chromatin state, while binding to promoters leads to activation of transcription. When bound to a heterologous promoter, Rap1p fails to properly activate transcription because it inappropriately recruits the Sir protein complex (65). However, upon deletion of one of the required SIR genes, Rap1p regains its transcriptional activating ability (65). Furthermore, when Sir3p is artificially tethered to a genomic site, it too can bypass normal regulation of silencing and repress transcription (61).
We propose that Dot4p restricts Sir proteins to the normal silent loci, possibly by regulating Sir4p levels or by altering Sir4p binding specificity (see above). In the absence of Dot4p, Sir activity becomes promiscuous and results in the formation of Rap1p-Sir complexes at inappropriate loci. This leads to reduced transcription of Rap1p-regulated genes and a decrease in metabolic competence. This model would also explain the decrease of silencing at telomeres and the HM loci in dot4 mutants (Fig. 5) (78), because the silencing factors (i.e., Sir2p and Sir3p) may be redirected to different genomic sites.
ACKNOWLEDGMENTS
We thank J. Berman, M. Hochstrasser, P. James, D. Moazed, P. Philippsen, M. Roth, and R. Sternglanz for generously providing plasmids, yeast strains, and/or antibodies, and we thank Liz Wayner for her expertise and help in generating antiubiquitin antibodies. We also thank our friends at the FHCRC and The University of Chicago for helpful discussions and/or critical reading of the manuscript.
A.K. gratefully acknowledges support from a Glenn Foundation/AFAR Award for Aging Research and Medical Scientist National Research Service Award 5T32 GM07281. This work was supported by a Pew Charitable Trust Biomedical Scholars Fellowship, a Cancer Research Foundation Fletcher Scholarship, and National Institutes of Health grant GM43893 (D.E.G.).
REFERENCES
- 1.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Amerik A, Swaminathan S, Krantz B A, Wilkinson K D, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D O, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1998. [Google Scholar]
- 6.Baker R T, Tobias J W, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 9.Boscheron C, Maillet L, Marcand S, Tsai-Pflugfelder M, Gasser S M, Gilson E. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- 10.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 13.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 14.Buck S W, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 15.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 19.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cockell M, Renauld H, Watt P, Gasser S M. Sif2p interacts with the Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr Biol. 1998;8:787–790. doi: 10.1016/s0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- 21.Davie J R, Murphy L C. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry. 1990;29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 22.Freeman K, Gwadz M, Shore D. Molecular and genetic analysis of the toxic effect of RAP1 overexpression in yeast. Genetics. 1995;141:1253–1262. doi: 10.1093/genetics/141.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritze C E, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B K, Grunstein M, Gasser S M. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb S, Esposito R E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 27.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 27a.Gottschling Lab Home Page. 27 July 1999, revision date. [Online.] http://www.fhcrc.org/∼gottschling/homepage.html. [27 July 1999, last date accessed.]
- 28.Gottschling laboratory. Unpublished observations.
- 29.Graham I R, Chambers A. Use of a selection technique to identify the diversity of binding sites for the yeast RAP1 transcription factor. Nucleic Acids Res. 1994;22:124–130. doi: 10.1093/nar/22.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 31.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 32.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 33.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 34.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 35.Haas A L. Introduction: evolving roles for ubiquitin in cellular regulation. FASEB J. 1997;11:1053–1054. doi: 10.1096/fasebj.11.13.9367340. [DOI] [PubMed] [Google Scholar]
- 36.Haas A L, Bright P M. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- 37.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 38.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 39.Hegde A N, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain D G, Martin K C, Kandel E R, Schwartz J H. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 40.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 41.Henchoz S, De Rubertis F, Pauli D, Spierer P. The dose of a putative ubiquitin-specific protease affects position-effect variegation in Drosophila melanogaster. Mol Cell Biol. 1996;16:5717–5725. doi: 10.1128/mcb.16.10.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Eghtedarzadeh M K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987;117:711–25. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 44.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 45.Holmes S G, Rose A B, Steuerle K, Saez E, Sayegh S, Lee Y M, Broach J R. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H, Kahana A, Gottschling D E, Prakash L, Liebman S W. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6693–6699. doi: 10.1128/mcb.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Baker R T, Fischer-Vize J A. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 48.Ivy J M, Klar A J S, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson E S, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 51.Jones E W. The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu Rev Genet. 1984;18:233–270. doi: 10.1146/annurev.ge.18.120184.001313. [DOI] [PubMed] [Google Scholar]
- 52.Kahana A. University of Chicago. 1998. Chicago, Ill. [Google Scholar]
- 53.Kennedy B K, Gotta M, Sinclair D A, Mills K, McNabb D S, Murthy M, Pak S M, Laroche T, Gasser S M, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 54.Lam Y A, Xu W, DeMartino G N, Cohen R E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 55.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Nagaraja S, Delcuve G P, Hendzel M J, Davie J R. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem J. 1993;296:737–744. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lustig A J. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- 61.Lustig A J, Liu C, Zhang C, Hanish J P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall M, Mahoney D, Rose A, Hicks J B, Broach J R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 64.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 66.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papa F, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 68.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 70.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 72.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 75.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singer J D, Manning B M, Formosa T. Coordinating DNA replication to produce one copy of the genome requires genes that act in ubiquitin metabolism. Mol Cell Biol. 1996;16:1356–1366. doi: 10.1128/mcb.16.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 78.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Identification of high copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith J S, Boeke J D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 80.Smith J S, Brachmann C B, Pillus L, Boeke J D. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stevenson J, Gottschling D E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 83.Sussel L, Vannier D, Shore D. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood S A, Mattick J S, Kaibuchi K. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 86.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 87.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 88.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 90.Wilkinson K D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 91.Wilkinson K D, Tashayev V L, O’Connor L B, Larsen C N, Kasperek E, Pickart C M. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Z, Buchman A R. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]