Abstract
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide. MicroRNAs (miRNAs) are known to be important regulators of GC. This study aims to investigate the role of miRNA (miR)-497 in GC. We demonstrated that the expression of miR-497 was downregulated in human GC tissues. After N-methyl-N-nitrosourea treatment, the incidence of GC in miR-497 knockout mice was significantly higher than that in wild-type mice. miR-497 overexpression suppressed GC cell proliferation, cell-cycle progression, colony formation, anti-apoptosis ability, and cell migration and invasion capacity. Additionally, miR-497 overexpression decreased the expression levels of cell division cycle 42 (CDC42) and integrin β1 (ITGB1) and inhibited the phosphorylation of focal adhesion kinase (FAK), paxillin (PXN), and serine-threonine protein kinase (AKT). Furthermore, overexpression of miR-497 inhibited the metastasis of GC cells in vivo, which could be counteracted by CDC42 restoration. Furthermore, the focal adhesion of GC cells was found to be regulated by miR-497/CDC42 axis via ITGB1/FAK/PXN/AKT signaling. Collectively, it is concluded that miR-497 plays an important role in the repression of GC tumorigenesis and progression, partly via the CDC42/ITGB1/FAK/PXN/AKT pathway.
Keywords: gastric cancer, miR-497, CDC42, tumorigenesis, focal adhesion kinase, paxillin
Graphical abstract
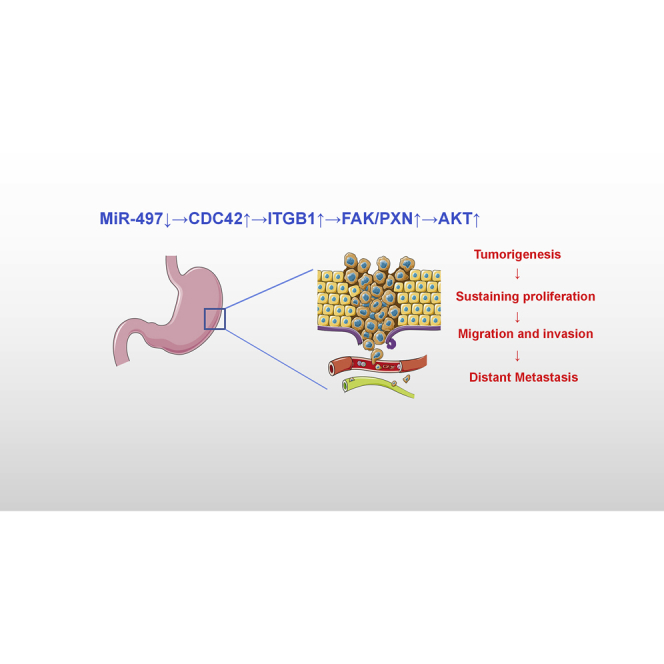
This study reports that the defect in miR-497 promotes the malignant transformation of gastric epithelial cells, and miR-497 is a regulator for the CDC42/ITGB1/FAK/PXN/AKT pathway. These demonstrations provide a novel explanation of the tumorigenesis and progression of gastric cancer and provide clues for the prevention and treatment of this deadly disease.
Introduction
Gastric cancer (GC) is one of the most common and lethal malignancies worldwide with a high morbidity, especially in East Asia population.1,2 With an unsatisfactory 5-year survival rate, the prognosis is worse in GC patients having distant metastasis and relapse after surgery.3 It is crucial to comprehensively understand the underlying molecular mechanism of GC tumorigenesis and progression, which is pivotal to facilitate the development of novel strategies for GC prevention and treatment.
MicroRNAs (miRNAs) are short non-coding RNAs (about 18–30 nucleotides [nt]) that suppress the expressions of target genes at translational level.4 miRNAs have been reported to regulate multiple malignant biological behaviors of cancer cells, including cell proliferation, apoptosis, motility, adhesion, as well as drug resistance.5 miRNA (miR)-497 has been reported to suppress disease progression of many cancers, including non-small cell lung cancer, myeloma, and glioma.6, 7, 8 However, the role, as well as the underlying mechanism of miR-497 in GC, has not been fully elucidated. Interestingly, bioinformatics data suggest that cell division cycle 42 (CDC42) is a candidate target of miR-497.
CDC42, a member of the Rho GTPase family, has been reported to promote cell proliferation by promoting cell-cycle progression.9 Upregulation of CDC42 expression has been found to be associated with crucial biological events during cancer cell metastasis, such as formation of lamellipodium, cytoskeletal rearrangement of migrating cell, and adhesions to the extracellular matrix.10 A previous study has found that CDC42 as an oncogene is overexpressed in GC and that high CDC42 expression is associated with unfavorable prognosis in GC patients.11 However, the mechanisms of CDC42 upregulation remain to be illustrated.
Focal adhesion kinase (FAK), a cytoplasmic protein tyrosine kinase, as well as paxillin (PXN), another key protein in focal adhesion, play a role in promoting cancer progression and metastasis.12 Integrin β1 (ITGB1) is a crucial mediator of signal transduction between extracellular matrix and cells. Intriguingly, CDC42 has been reported to facilitate cancer progression via the activation of ITGB1/FAK/PXN signaling.13 Besides, serine-threonine protein kinase (AKT) has been found to be activated by FAK in the regulation of cell growth.14
Based on the literature so far, we hypothesized that miR-497 might regulate the tumorigenesis and progression of GC via the modulation of CDC42/ITGB1/FAK/PXN/AKT signaling. This study was designed to validate our hypothesis.
Results
miR-497 is downregulated in GC
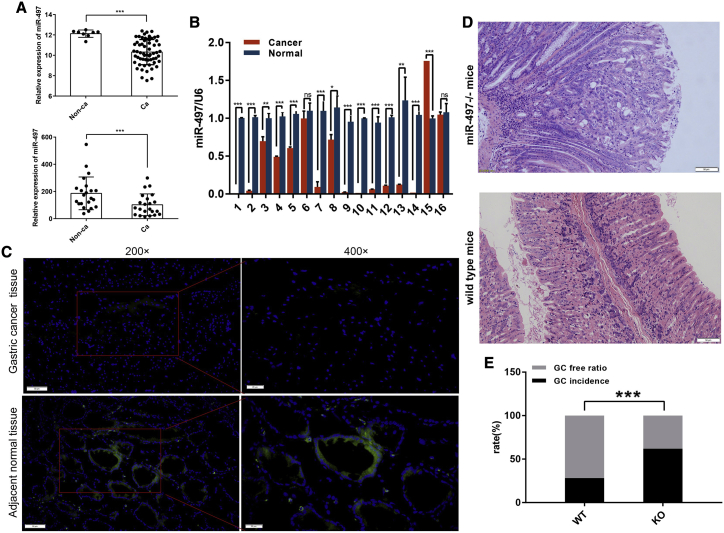
In our previous study, miR-497 has been identified as a tumor suppressor in liver cancer.15 To determine whether miR-497 also plays a tumor-suppressive role in GC, we first analyzed the expression patterns of miR-497 in GC. As shown in Figure 1A, two independent GEO datasets: GSE26595, GSE28700, indicated that miR-497 expression was significantly downregulated in GC tissues compared with that in non-cancerous tissues. Next, quantitative polymerase chain reaction (qPCR) was performed to detect miR-497 expression in 16 pairs of GC tissues/corresponding adjacent tissues. Consistently, miR-497 expression levels were found to be significantly decreased in most GC tissues (Figure 1B). To verify our results, fluorescence in situ hybridization (FISH) was conducted to explore miR-497 expression in GC tissues. In agreement with our qPCR data, the miR-497 signal was hardly detected in GC tissues (Figure 1C).
Figure 1.
miR-497 is downregulated in clinical GC samples, and its knockout (miR-497−/−) promotes GC formation in mice
(A) Comparison of miR-497 expression in GC tissues and para-carcinoma tissues using the data from database GEO Database: GSE26595 (upper) and GEO Database: GSE28700 (lower). (B) qPCR was used to detect miR-497 expression in GC tissues and para-carcinoma tissues (16 pairs). (C) FISH was used to detect the expression of miR-497 in GC tissues (upper) and para-carcinoma tissues (lower) (blue, DAPI; green, hsa-miR-497) (×200 scale bars, 50 μm; ×400 scale bars, 20 μm). (D) Representative images of H&E staining section of the gastric tissues of miR-497 KO mice (n = 21) and wild-type (WT) mice (n = 39) (scale bars, 50 μm). (E) Quantification of GC occurrence rate in miR-497 KO mice (n = 21) and WT mice (n = 39). ns, no significance; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
miR-497 inhibits GC tumorigenesis in vivo
Although many previous studies have reported that miR-497 expression is dysregulated in cancers and that miR-497 regulates malignant biological behaviors of cancer cells,6, 7, 8,15 the association between miR-497 expression dysregulation and malignant transformation of normal cells is unknown. In this study, miR-497 knockout (miR-497−/− [KO]) mice were created by CRISPR-Cas9-mediated genome editing (Figure S1A). The successful establishment of KO mice was verified by both qPCR and Southern blot (Figures S1B and S1C). After 40 weeks of treatment with water containing N-methyl-N-nitrosourea (MNU), the rate of tumorigenesis was significantly higher in mice of the miR-497 KO group (13 of 21) than that in mice of the wild-type (WT) group (11 of 39; 61.9% versus 28.2%, p = 0.0145) (Figures 1D and 1E), suggesting that malignant transformation of normal gastric epithelial cells was promoted in the absence of miR-497.
miR-497 regulates the growth and motility of GC cells
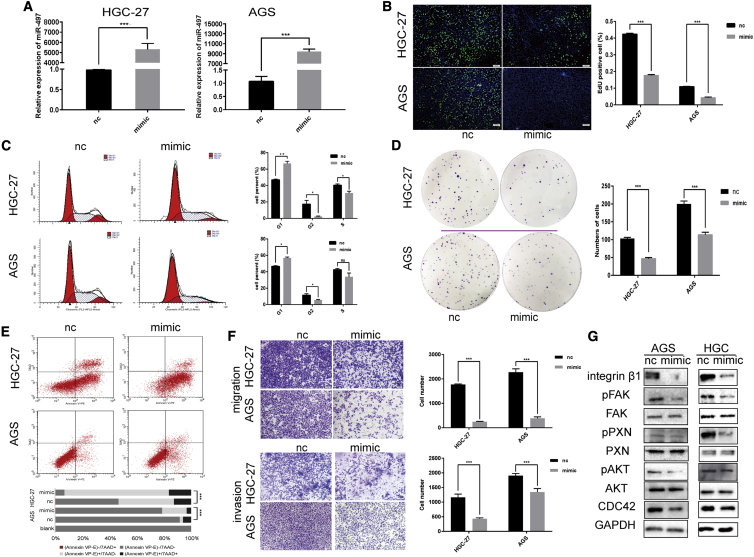
Next, functional experiments were performed in GC cells lines (HGC-27 and AGS) transfected with miR-497 mimics or control miRNAs (Figure 2A). An 5-ethynyl-20-deoxyuridine (EdU) assay indicated that miR-497 overexpression led to significantly decreased levels of CG cell proliferation (Figure 2B). Additionally, miR-497 overexpression induced cell-cycle progression arrest and inhibited the colony-formation ability of GC cells (Figures 2C and 2D). Furthermore, in both GC cell lines, miR-497 overexpression induced apoptosis (p < 0.001; Figure 2E) and suppressed the migration and invasion capacity of GC cells (Figure 2F). ITGB1/FAK/PXN/AKT signaling has been shown as an important pathway in the mediation of cancer cell motility, migration, and invasion.14,16 Therefore, we investigated the expression patterns of the ITGB1/FAK/PXN/AKT signal pathway upon miR-497 overexpression. As shown in Figure 2G and Figure S2A, the expression levels of CDC42, ITGB1, pAKT (phospho-FAK) (Tyr397), phospho-paxillin (pPXN) (Tyr118), and phospho-serine-threonine protein kinase (pAKT) (Ser473) were prominently downregulated upon miR-497 overexpression.
Figure 2.
miR-497 modulates proliferation, migration, and invasion of GC cells
(A) miR-497 expression was analyzed by qPCR in HGC-27 (NC [nc]) versus HGC-27 (miR-497 mimic) and AGS (nc) versus AGS (miR-497 mimic) cells after transfection. (B−F) After transfection, the proliferation (B), cell cycle (C), colony-formation ability (D), apoptosis (E), and migration and invasion (F) of GC cells were examined by EdU assay, flow cytometry analysis, plate colony-formation assay, flow cytometry analysis, and Transwell assay, respectively. (G) After transfection, western blotting was used to detect the expressions of ITGB1, phospho-focal adhesion kinase (pFAK) (Tyr397), total FAK, pPXN (Tyr118), total PXN, pAKT (Ser473), total AKT, and CDC42. (B) Scale bars, 50 μm; (F) scale bars, 100 μm. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
CDC42 is a direct target of miR-497
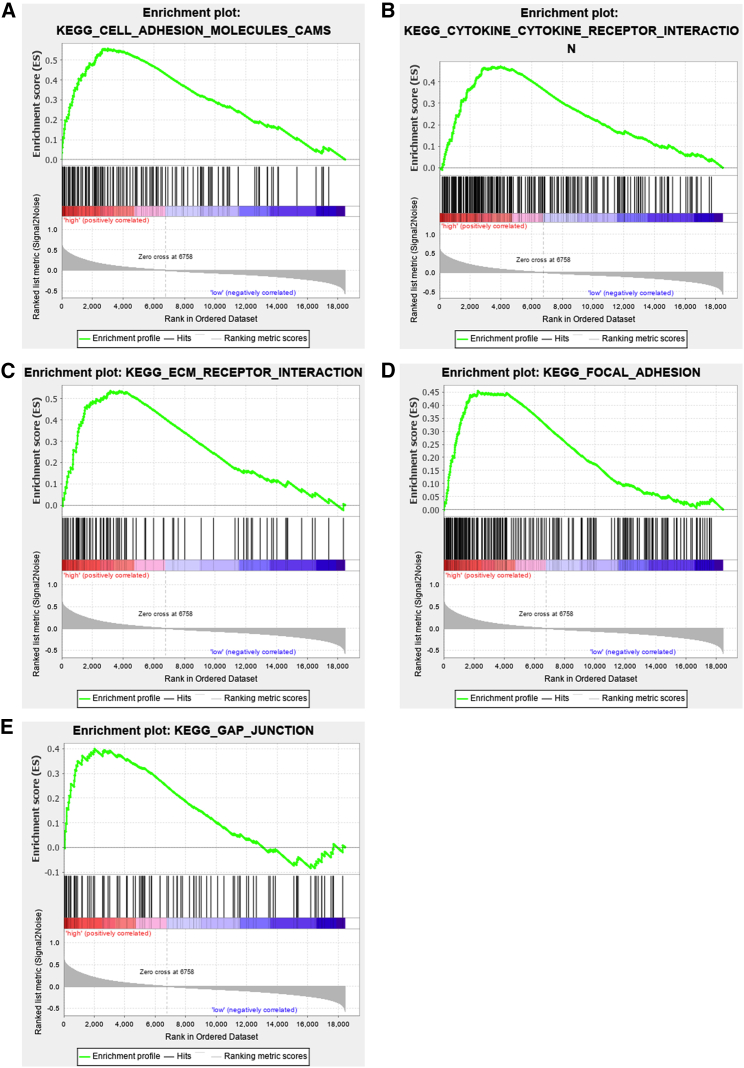
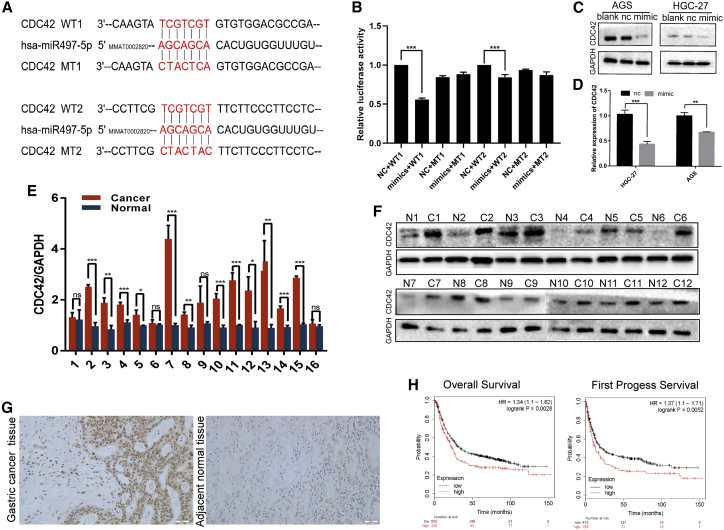
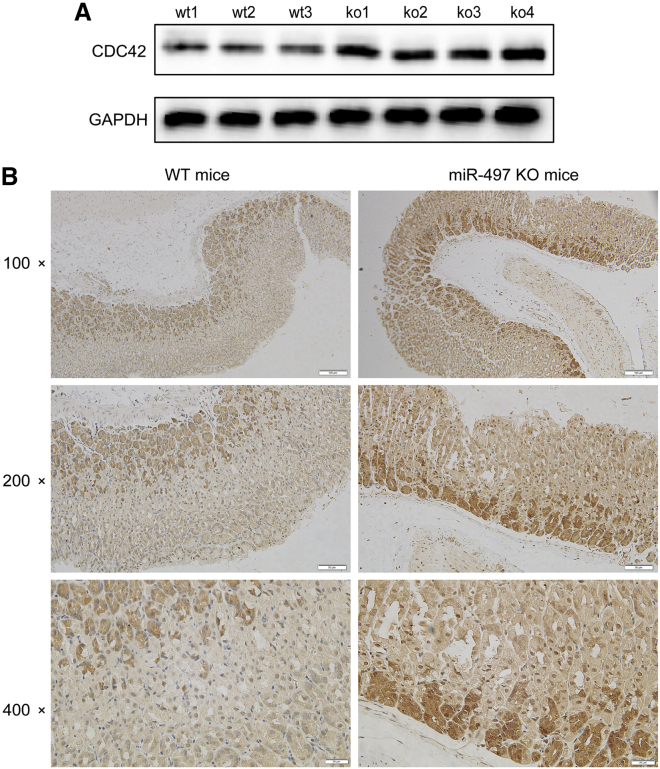
As described above, miR-497 plays a role in regulating the expression/activation of ITGB1, FAK, PXN, and AKT. Consistently, gene set enrichment analysis suggested that miR-497 can probably regulate biological processes related with cell motility, migration, and invasion (Figures 3A−3E). Next, we explored the underlying molecular mechanism in which these biological processes were exerted by miR-497. To predict the target genes of miR-497, miRWalk: http://mirwalk.umm.uni-heidelberg.de/, TargetScan: http://www.targetscan.org/vert_71/, and miRDB: http://mirdb.org/ databases were searched simultaneously. Candidate genes identified by all three databases were considered for further analysis. CDC42, an upstream protein of ITGB1/FAK/PXN signaling,13 was identified with two miR-497 target sites in the 3′ UTR region (Figure 4A). Next, the two potential binding sites identified were validated by a dual luciferase reporter assay (Figure 4B). CDC42 expression in GC cells was observed to be markedly repressed by miR-497 overexpression, as shown by qPCR and western blot analyses (Figures 4C and 4D). CDC42 mRNA and protein expression levels were found to be significantly upregulated in human GC tissues, which were opposite to those of miR-497 (Figures 4E−4G; Figures S2B and S2C). Kaplan-Meier (KM) analysis showed that in GC, increased CDC42 expression was associated with shorter overall survival (OS) time and survival time in patients after the first progression (Figure 4H). Importantly, the expression of CDC42 was detected in the gastric tissues of miR-497 KO and WT mice, respectively. As shown, the expression of CDC42 in miR-497 KO mice was notably higher than that in WT mice (Figures 5A and 5B; Figure S2D), which further validated the regulatory relationship between miR-497 and CDC42.
Figure 3.
Gene set enrichment analysis (GSEA) suggests miR-497 regulates biological processes related with cell motility, migration, and invasion
(A−E) GSEA based on The Cancer Genome Atlas (TCGA) data suggested that in GC, miR-497 was associated with cell adhesion (A), cytokine receptor interaction (B), cell-extracellular matrix interaction (C), focal adhesion (D), and cells’ gap junction (E). Red represents genes that are positively correlated with miR-497; blue represents genes that are negatively correlated with miR-497.
Figure 4.
Identification of CDC42 as a target for miR-497
(A) The target sites of miR-497 in 3′ UTR of CDC42 are shown as a schematic representation. (B) WT or mutant 3′ UTR constructs of CDC42 were cloned into a pmirGLO vector, respectively, and co-transfected with miR-497 mimics into 293T cells. Firefly luciferase activities were normalized to Renilla luciferase activities. (C) CDC42 expression in GC cells was detected by western blotting after the transfection of miR-497. (D) CDC42 expression in GC cells was detected by qPCR after the transfection of miR-497. (E) qPCR was used to detect CDC42 expression in GC tissues and para-carcinoma tissues (16 pairs). (F) Western blotting was used to detect CDC42 expression in GC tissues and para-carcinoma tissues (12 pairs). (G) Representative images of the IHC, which showed that the expression of CDC42 was upregulated in the GC tissues, compared with adjacent gastric tissues (scale bars, 20 μm). (H) Kaplan-Meier survival curve was used to compare the OS and first progress survival (FPS) of GC patients with high or low CDC42 expression. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure 5.
miR-497 KO increases the expression of CDC42 in vivo
(A) Western blot was used to detect CDC42 expression in the gastric tissues of miR-497 KO mice (n = 4) and WT mice (n = 3). (B) Representative images of the IHC, which showed that the expression of CDC42 was upregulated in the gastric epithelium of miR-497 KO mice (×100 scale bars, 100 μm; ×200 scale bars, 50 μm; ×400 scale bars, 20 μm).
Silencing the CDC42 axis inhibits the malignant phenotypes of GC cells
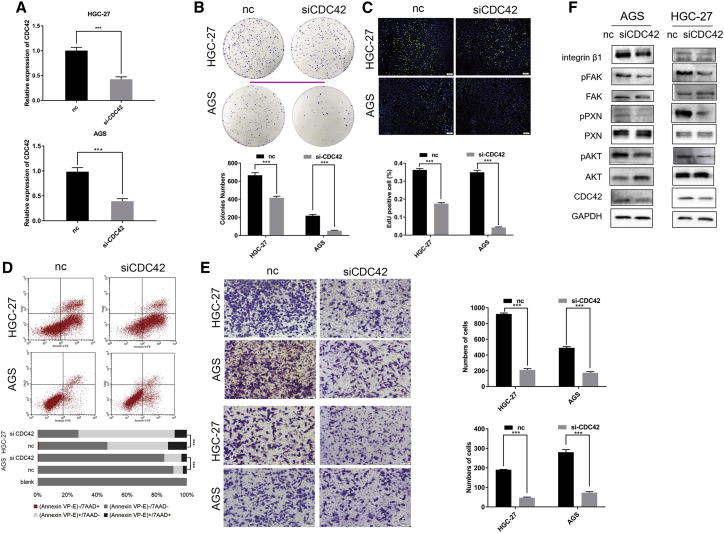
To investigate the biological functions of CDC42 in GC cells, CDC42 was knocked down in GC cells using small interfering RNA (siRNA) (Figure 6A). Functional experiments indicated that the depletion of CDC42 inhibited proliferation (Figures 6B and 6C), induced apoptosis (Figure 6D), and repressed the migration and invasion (Figure 6E) of GC cells. Additionally, the ITGB1/FAK/PXN/AKT pathway was significantly inhibited in GC cells upon CDC42 knockdown (Figure 6F; Figure S2E), which was similar with the effects observed with miR-497 transfection.
Figure 6.
CDC42 knockdown represses the malignant phenotypes of GC cells via regulating ITGB1/FAK/PXN/AKT signaling
(A) The knockdown of CDC42 in HGC-27 (upper) and AGS (lower) cells after CDC42 siRNA transfection was confirmed by qPCR. (B−E) After transfection, the colony-formation ability (B), proliferation (C), apoptosis (D), and migration and invasion (E) of GC cells were examined by plate colony-formation assay, EdU assay, flow cytometry analysis, and Transwell assay, respectively. (F) After transfection, western blotting was used to detect the expressions of ITGB1, phospho-focal adhesion kinase (pFAK) (Tyr397), total FAK, pPXN (Tyr118), total PXN, pAKT (Ser473), total AKT, and CDC42 in GC cells (scale bars, 50 μm). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
The tumor-suppressive effect of miR-497 is partly dependent on CDC42
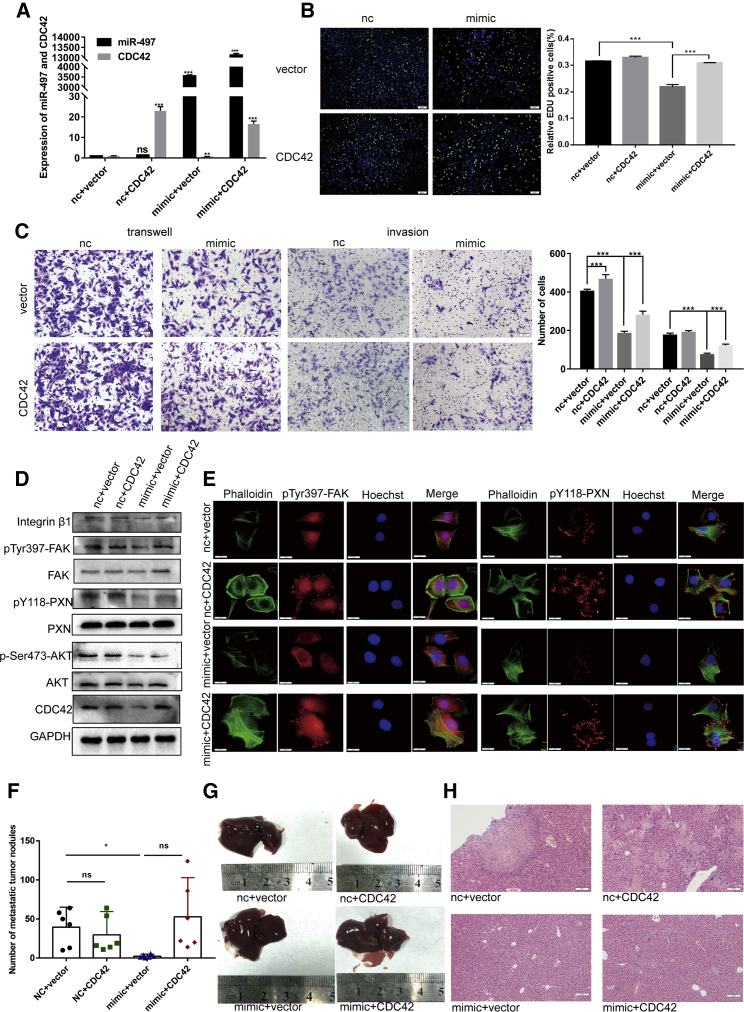
To further explore whether miR-497 can exert its function in GC by regulating CDC42, we conducted rescue assays by co-transfecting HGC-27 cells with the miR-497 mimic and CDC42-overexpressing plasmid. The co-transfection efficiency was validated by analyzing the RNA levels of miR-497 and CDC42 (Figure 7A) before further analyses by EdU assay and Transwell assay (Figures 7B and 7C). As shown in Figures 7B and 7C, whereas miR-497 overexpression decreased the proliferation and migration capacity of HGC-27 cells, CDC42 overexpression partly abolished these effects. Moreover, western blot analysis showed that miR-497 can modulate ITGB1/FAK/PAX/AKT signaling and that the role of miR-497 in ITGB1/FAK/PAX/AKT signaling modulation can be counteracted by CDC42 overexpression (Figure 7D; Figure S2F). Furthermore, immunofluorescence analysis showed that miR-497 inhibited the formation of focal adhesion and the phosphorylation of FAK and PXN and that conversely, CDC42 overexpression promoted these biological events and counteracted the effects of miR-497 (Figure 7E).
Figure 7.
Exogenous CDC42 reversed the effects of miR-497 on GC cells in vitro and in vivo
(A) The expression of miR-497 and CDC42 in GC cells was affirmed by qPCR after transfection. (B and C) The proliferation (B) and migration and invasion (C) of GC cells were detected by the EdU assay and Transwell assay (scale bars, 50 μm). (D) After transfection, western blotting was used to detect the expressions of ITGB1, phospho-focal adhesion kinase (pFAK) (Tyr397), total FAK, pPXN (Tyr118), total PXN, pAKT (Ser473), total AKT, and CDC42 in GC cells. (E) The immunofluorescence assay showed the effect of the miR-497/CDC42 axis on the focal adhesion formation of GC cells (scale bars,10 μm; green, phalloidin; red, pPXN [Tyr118] or pFAK [Tyr397]; blue, Hoechst 33342). (F) The number of metastatic tumor nodules in the liver are compared among the nude mice in different groups. (G) Representative images of metastatic tumor nodules in liver section of nude mice intravenously injected with HGC-27 cells (n = 6 in each group). (H) Representative images of the H&E section of the liver of the nude mice (scale bars, 100 μm). nc + vector, co-transfected with nc mimic and empty vector; nc + CDC42, co-transfected with nc mimic and CDC42 plasmid; mimic + vector, co-transfected with miR-497 mimic and empty vector; mimic + CDC42, co-transfected with miR-497 mimic and CDC42 plasmid. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
miR-497 suppresses the metastasis of GC cells in vivo
We further assessed the effects of miR-497 and CDC42 on the metastasis of GC cells in vivo. HGC-27 cells were injected into nude mice, and the mice were sacrificed after 4 weeks. The metastatic nodules in the lungs and liver surfaces were counted. As shown in Figures 7F and 7G, the number of liver metastasis nodules was decreased in mice of the mimic group but was rescued upon CDC42 co-transfection. No obvious metastasis was observed in the lungs, suggesting that GC was more inclined to metastasize in the liver.17 The nodules on the surfaces of mice livers were confirmed to be metastatic tumors by H&E staining (Figure 7H). These results further validated that the miR-497/CDC42 axis plays a role in the regulation of GC progression.
Discussion
Tumor-suppressive properties of miR-497 have been reported in multiple cancers, including GC; in particular, miR-497 has been shown in the regulation of cancer cell malignant behaviors through multiple downstream genes/pathways, such as Wnt family member 3A (Wnt3a), neurotrophic receptor tyrosine kinase 3 (NTRK3), and mechanistic target of rapamycin kinase (mTOR) signaling.6, 7, 8,15,18, 19, 20, 21, 22, 23 We have previously reported that miR-497 is underexpressed in liver cancer and that miR-497 plays a role in regulating the expression of oncogene structure specific recognition protein 1 (SSRP1).15 A previous study has shown that miR-497 is underexpressed in GC due to DNA methylation and that miR-497 represses GC progression through direct targeting of RAF1.23 However, the role of miR-497 in transforming normal cells into cancer cells remains unexplored. In this study, in agreement with previous reports, we showed that miR-497 is downregulated in GC and that miR-497 suppresses the malignant behaviors of GC cells in vitro and in vivo. Importantly, we also demonstrated that miR-497 KO can increase the susceptibility of mice to carcinogen MNU. Our data suggested that miR-497 is a crucial factor that protects gastric epithelial cells from malignant transformation.
CDC42, a small GTPase with important functions in multiple cancers in humans, has been shown to be involved in the regulation of tumor growth, cell cycle, epithelial-mesenchymal transition, metastasis, angiogenesis, and chemoresistance.24 CDC42 has an upregulated expression or is hyperactivated in cancers.24 In GC, multiple oncoproteins can activate CDC42 in promoting cancer progression. For instance, MICAL-L2 can potentiate epidermal growth factor receptor (EGFR) stability in facilitating GC progression via the activation of CDC42;25 GINS complex subunit 4 (GINS4) can promote GC cell growth and metastasis and suppress apoptosis via the activation of Rac1/CDC42.11 However, the mechanism of CDC24 dysregulation in GC has not been fully clarified. Previous studies have reported that underexpressed miR-133 and miR-137 in GC can contribute to the overexpression of CDC42.26,27 These studies suggest that miRNAs may be the crucial regulators of CDC42. For the first time, in this study, CDC42 has been identified as a target gene of miR-497, and it can be negatively regulated by miR-497.
Integrins are cell-adhesion proteins that mediate the interaction between cytoskeletal elements and the extracellular matrix.28 An increasing number of studies have demonstrated that the upregulation or activation of integrins, including ITGB1, can promote the proliferation, migration, invasion, adhesion, and chemoresistance of cancer cells.28 In GC, it has been reported that ITGB1 can dominate crucial biological events during tumorigenesis and activate multiple downstream oncogenic pathways, including FAK and nuclear factor κB (NF-κB). Specifically, integrins including ITGB1 are dispensable for Helicobacter pylori cytotoxin-associated gene A (CagA) translocation, which is crucial for GC tumorigenesis.29 Helicobacter pylori infection also promotes NF-κB signaling via the activation of ITGB1.30 Additionally, ITGB1 has been shown to activate FAK and PXN in enhancing the malignant phenotypes of GC cells.13,31 Here, we report that the miR-497/CDC42 axis can regulate the activation of ITGB1/FAX/PXN/AKT signaling, which helps clarify the downstream molecular mechanism of miR-497 in the suppression of GC progression.
In summary, in this work, through the establishment of miR-497 KO mice, we demonstrate that miR-497 KO can increase the susceptibility of mice to carcinogen MNU. Additionally, we report that CDC42, a crucial modulator in cancer cell metastasis, is a target gene of miR-497 and that the miR-497/CDC42 axis can regulate the activation of ITGB1/FAK/PXN/AKT signaling in GC. Our data facilitate further understanding of the molecular mechanism of GC tumorigenesis and progression by implying that restoration of miR-497 expression may be a promising strategy in preventing the occurrence and development of GC. In the following studies, the relationship between miR-497 and Helicobacter pylori susceptibility is desirable to be investigated.
Materials and methods
Human specimens
Paired GC (adenocarcinoma) tissues and adjacent noncancerous tissue were collected from the Department of Gastrointestinal Surgery, Renmin Hospital of Wuhan University. This work was supported by the Ethics Committee of Renmin Hospital, Wuhan University. Written, informed consents were obtained from each patient. All procedures were conducted in accordance with the principles of the Declaration of Helsinki.
FISH
Frozen sections of human GC tissues/corresponding adjacent tissues were fixed and hybridized in hybridization buffer with the digoxin-labeled hsa-miR-497 probe (5′-ACAAACCACAGTGTGCTGCTG-3′) (Exonbio, San Diego, CA, USA) at 37°C overnight before signal detection using the FISH Kit (Exonbio, San Diego, CA, USA), according to the manufacturer’s protocol. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime, Shanghai, China). Tissue images were captured by a fluorescence microscope (Olympus, Tokyo, Japan).
Quantitative real-time PCR
Total RNAs in GC tissues and cells were isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA), whereas miRNAs were extracted using the mirPremier miRNA Isolation Kit (Sigma, St. Louis, MO, USA). Reverse transcription was performed using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Rockford, IL, USA), and qPCR was performed on the StepOne Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA) using the Light Cycler Fast Start DNA MasterPlus SYBR Green I Kit (Roche Diagnostics, Burgess Hill, UK). Primer sequences are shown in Table 1.
Table 1.
Sequences used for qRT-PCR
| hsa-miR-497 | F: 5′-CAGCAGCACACTGTGGTTTGTA-3′ |
| R: uni-miR qPCR primer | |
| hsa-CDC42 | F: 5′-CCATCGGAATATGTACCGACTG-3′ |
| R: 5′-CTCAGCGGTCGTAATCTGTCA-3′ | |
| hsa-U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACgAATTTGCGT-3′ | |
| hsa-GAPDH | F: 5′-CCCATCACCATCTTCCAGGAG-3′ |
| R: 5′-CTTCTCCATGGTGGTGAAGACG-3′ |
F, forward; R, reverse; uni-miR, universal-microRNA.
Survival analysis
Survival analysis was performed using the online database KM Plotter (www.kmplot.com).32 GC patients were classified into a high-expression group or low-expression group according to the median gene-expression level. Univariate analysis of survival time was performed using a two-sided log rank test to evaluate the prognosis of patients.
Cell culture and transfection
Human GC cell lines AGS and HGC-27 were purchased from Procell Life Science & Technology (Wuhan, China) and authenticated by short tandem repeats. Cells were cultured at 37°C in a humidified incubator (5% CO2, 37°C) with HyClone DMEM/F12 (Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction.
Cell proliferation assay
Cells were seeded into 96-well plates (5,000 per well). Cell proliferation was measured using an EdU Assay Kit (Beyotime, Shanghai, China).33 Briefly, after incubation with 50 μM of EdU solution for 2 h, cells were fixed in 4% paraformaldehyde and stained with Apollo Dye Solution and Hoechst-33342 staining solution. Images were then acquired using a fluorescence microscope (Olympus, Tokyo, Japan). The percentages of EdU-positive cells were calculated with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Flow cytometry analysis
To evaluate the cell cycle of GC cells, after transfection, GC cells were harvested, then washed with PBS, and then fixed with 70% ethanol. Fixed cells were stained with propidium iodide (PI) staining solution (Beyotime, Shanghai, China) for 15 min at room temperature in the dark. Then, cell suspensions were analyzed using BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Cell cycle was analyzed using FlowJo version (v.)10 software (FlowJo, Ashland, OR, USA). To evaluate GC cell apoptosis, the Annexin V-phycoerythrin (PE)/7-aminoactinomycin D (7-AAD) Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) was used. Briefly, about 1 × 105 GC cells in each group were suspended with 500 μL of binding buffer. Next, 5 μL of Annexin V-PE staining solution and 10 μL of 7-AAD staining solution were added into the cell suspension and mixed thoroughly before the cells were stained in the dark for 30 min. Ultimately, the cells were analyzed by flow cytometry.
Colony-formation assay
GC cells in each group were plated into 10 cm dishes (1 × 103 cells/dish), respectively, and cultured for 14 days. Subsequently, cells were fixed in 4% paraformaldehyde and dyed with crystal violet solution. After a washing step with tap water followed by a drying step, visible colonies were counted with naked eyes.
Transwell assay
For migration assessment, 1 × 105 GC cells (suspended in 200 μL of FBS-free serum) were inoculated in the upper compartment of each Transwell chamber (8 μm pore size; Corning, Corning, NY, USA), whereas 600 μL of complete medium containing 10% FBS was added in each lower compartment. After incubation for 48 h, cells in the upper compartment were cleared by wet cotton swabs, whereas cells on the bottom surface of the membranes were fixed with paraformaldehyde and then dyed for 10 min before staining with 0.4% crystal violet solution. Migrated cells in 6 randomly selected fields were counted utilizing a microscope (Olympus, Tokyo, Japan). For invasion assessment, the same procedure was performed, except that each Transwell chamber (Corning, Corning, NY, USA) was coated with a layer of Matrigel (Clontech, Madison, WI, USA) before cell inoculation.
Western blot
Total proteins from GC tissues and cells were extracted using radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) containing the protease inhibitors and phosphatase inhibitors. Protein samples (20 μL) in each group were separated by SDS-PAGE before being transferred onto polyvinylidene fluoride (PVDF) membranes (Beyotime, Shanghai, China), which were then blocked by 5% non-fat milk followed by incubation overnight at 4°C with primary antibodies against ITGB1 (Cell Signaling Technology, Beverly, MA, USA; #34971), FAK (Cell Signaling Technology, Beverly, MA, USA; #3285), pY397-FAK (Cell Signaling Technology, Beverly, MA, USA; #3281), CDC42 (Abcam, Cambridge, MA, USA; #ab187643), PXN (Abcam, Cambridge, MA, USA; #ab32084), pY118-PXN (Abcam, Cambridge, MA, USA; #ab109547), AKT (Proteintech, Wuhan, China; #10176-2-AP), pS473-AKT (Proteintech, Wuhan, China; #66444-1-IG), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, Dallas, TX, USA; #sc-47724) (Table S4). Subsequently, the membranes were incubated with horseradish peroxidase-labeled secondary antibody (Proteintech, Wuhan, China) at room temperature for 1 h. Protein signals were developed using the Enhanced Chemiluminescence Kit (Beyotime, Shanghai, China).
Dual luciferase reporter assay
The potential target site of miR-497 on 3′ UTR of CDC42 was predicted using miRDB database (http://mirdb.org/). DNA fragments containing a CDC42 WT sequence and CDC42 mutant (MT) sequence were inserted into the pmirGLO basic vector (Promega, Madison, WI, USA), respectively. Then, 293T cells were co-transfected with reporter vectors and miR-negative control (NC) or miR-497 mimics. After 48 h, luciferase activities of the cells were measured using the Dual Luciferase Reporter Gene Assay Kit (Promega, Madison, WI, USA). The luciferase activity of firefly was normalized to that of Renilla.
Immunohistochemistry (IHC)
Following rehydration, deparaffinized tissue sections were subjected to antigen retrieval. Subsequently, the sections were incubated with rabbit polyclonal antibody anti-CDC42 (Proteintech, Wuhan, China; #I0I55-1-AP) at 4°C overnight and then washed three times in PBS. The sections were then incubated with secondary antibody (Beyotime, Shanghai, China) at room temperature for 1 h. Next, the sections were washed three times in PBS, and the signals were detected using the diaminobenzidine (DAB) chromogenic agent (Beyotime, Shanghai, China) by following the manufacturer’s instruction. Images were obtained using an upright microscope (Olympus, Tokyo, Japan) and were then scored by two independent pathologists.
Immunofluorescence
GC cells were inoculated on coverslips placed in 24-well plates and cultured for 24 h before being washed in PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100. Then, cells were incubated with primary antibodies (Table S4) at 4°C overnight, followed by incubation with secondary antibodies labeled with Alexa Fluor 555, Alexa Fluor 488, or phalloidin (fluorescein isothiocyanate labeled), respectively. The antibodies were obtained from Abcam (Cambridge, UK). Hoechst 33258 staining solution (Beyotime, Shanghai, China) was used to stain the nuclei. Stained GC cells were photographed using a fluorescence microscope (Olympus, Tokyo, Japan).
Animal models
All animal experiments in this work were approved by the Institutional Animal Care and Use Committee of the Renmin Hospital of Wuhan University. miR-497 KO C57BL/6 mice (Cyagen, Guangzhou, China) were generated by CRISPR-Cas9 (Supplemental information). WT (n = 39) and KO (n = 21) mice were used to induce GC. All mice were fed under special pathogen-free conditions. At 5 weeks old, mice were given 180 ppm of MNU in drinking water (MNU water) every other week before being sacrificed at week 40.34,35 A metastasis model was established by injecting GC cells into the mice via caudal vein. 4-week-old male nude mice (Charles River, Beijing, China) were fed under specific pathogen-free conditions. After transfection, HGC-27 cells were resuspended in PBS at 1 × 107 cells/mL. Each nude mouse was injected with a total of 1 × 106 cells in 100 μL of PBS before being sacrificed 4 weeks later (n = 6 in each group). The lungs and livers were dissected and embedded in paraffin, followed by pathological examination.
Statistical analyses
All of the experiments were performed in triplicates. Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). Chi-square test, t test, or one-way ANOVA were performed. Data are expressed as mean ± standard error of mean. p < 0.05 was considered to be statistically significant.
Acknowledgments
We thank Hubei Yican Health Industry Co., Ltd. (Wuhan, China), and Editsprings (Wuhan, China) for linguistic assistance during the preparation of this manuscript. The data that support the findings of this study are available from the corresponding author upon reasonable request. We acknowledge the funding support from the Natural Science Foundation of China (nos. 81672387, granted to H.Y.; 81770899, granted to Y.Y.; and 81703030, granted to Q.D.) and the Guided Fund of Renmin Hospital of Wuhan University (no. RMYD2018M65, granted to W.Z.).
Author contributions
H.Y., Q.D., and W.Z. conceived and designed the experiments. H.Y., Q.D., W.Z., and Y.Y. provided the funds and materials. L.Z., L.Y., Z.L., Y.D., D.Y., Z.Z., R.S., M.X., M.Z., and D.C. performed the experiments. B.R. and L.Z. conducted the bioinformatics analysis and statistical analysis. L.Z., L.Y., J.T., and Q.L. drafted this manuscript. H.Y., Y.Y., and Q.D. were responsible for study supervision.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.025.
Contributor Information
Yanning Yang, Email: ophyyn@163.com.
Qianshan Ding, Email: iamdqs@163.com.
Honggang Yu, Email: yuhonggang@whu.edu.cn.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Allemani C., Weir H.K., Carreira H., Harewood R., Spika D., Wang X.S., Bannon F., Ahn J.V., Johnson C.J., Bonaventure A., CONCORD Working Group Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z., Lu Q., Zhu D., Han Y., Zhou X., Ren T. Lnc-SNHG1 may promote the progression of non-small cell lung cancer by acting as a sponge of miR-497. Biochem. Biophys. Res. Commun. 2018;506:632–640. doi: 10.1016/j.bbrc.2018.10.086. [DOI] [PubMed] [Google Scholar]
- 7.Yu T., Xu Z., Zhang X., Men L., Nie H. Long intergenic non-protein coding RNA 152 promotes multiple myeloma progression by negatively regulating microRNA-497. Oncol. Rep. 2018;40:3763–3771. doi: 10.3892/or.2018.6721. [DOI] [PubMed] [Google Scholar]
- 8.Lu F., Ye Y., Zhang H., He X., Sun X., Yao C., Mao H., He X., Qian C., Wang B. miR-497/Wnt3a/c-jun feedback loop regulates growth and epithelial-to-mesenchymal transition phenotype in glioma cells. Int. J. Biol. Macromol. 2018;120:985–991. doi: 10.1016/j.ijbiomac.2018.08.176. [DOI] [PubMed] [Google Scholar]
- 9.Xiao X.H., Lv L.C., Duan J., Wu Y.M., He S.J., Hu Z.Z., Xiong L.X. Regulating Cdc42 and Its Signaling Pathways in Cancer: Small Molecules and MicroRNA as New Treatment Candidates. Molecules. 2018;23:787. doi: 10.3390/molecules23040787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridley A.J. Rho GTPases and cell migration. J. Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z., Yu Z., Rong Z., Luo Z., Zhang J., Qiu Z., Huang C. The novel GINS4 axis promotes gastric cancer growth and progression by activating Rac1 and CDC42. Theranostics. 2019;9:8294–8311. doi: 10.7150/thno.36256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulzmaier F.J., Jean C., Schlaepfer D.D. FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J.W., Lee H.J., Bae G.U., Kang J.S. Promyogenic function of Integrin/FAK signaling is mediated by Cdo, Cdc42 and MyoD. Cell. Signal. 2011;23:1162–1169. doi: 10.1016/j.cellsig.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Balsas P., Palomero J., Eguileor Á., Rodríguez M.L., Vegliante M.C., Planas-Rigol E., Sureda-Gómez M., Cid M.C., Campo E., Amador V. SOX11 promotes tumor protective microenvironment interactions through CXCR4 and FAK regulation in mantle cell lymphoma. Blood. 2017;130:501–513. doi: 10.1182/blood-2017-04-776740. [DOI] [PubMed] [Google Scholar]
- 15.Ding Q., He K., Luo T., Deng Y., Wang H., Liu H., Zhang J., Chen K., Xiao J., Duan X. SSRP1 Contributes to the Malignancy of Hepatocellular Carcinoma and Is Negatively Regulated by miR-497. Mol. Ther. 2016;24:903–914. doi: 10.1038/mt.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J., Yao J.F., Deng X.F., Zheng X.D., Jia M., Wang Y.Q., Huang Y., Zhu J.H. 14, 15-EET induces breast cancer cell EMT and cisplatin resistance by up-regulating integrin αvβ3 and activating FAK/PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018;37:23. doi: 10.1186/s13046-018-0694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marek V., Zahorec R., Palaj J., Durdik S. Gastric cancer with liver metastasis. Bratisl. Lek Listy. 2016;117:59–61. doi: 10.4149/bll_2016_012. [DOI] [PubMed] [Google Scholar]
- 18.Yang H., Wu X.L., Wu K.H., Zhang R., Ju L.L., Ji Y., Zhang Y.W., Xue S.L., Zhang Y.X., Yang Y.F., Yu M.M. MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase. Am. J. Cancer Res. 2016;6:2690–2699. [PMC free article] [PubMed] [Google Scholar]
- 19.Yu T., Zhang X., Zhang L., Wang Y., Pan H., Xu Z., Pang X. MicroRNA-497 suppresses cell proliferation and induces apoptosis through targeting PBX3 in human multiple myeloma. Am. J. Cancer Res. 2016;6:2880–2889. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Bu J.Y., Lv W.Z., Liao Y.F., Xiao X.Y., Lv B.J. Long non-coding RNA LINC00978 promotes cell proliferation and tumorigenesis via regulating microRNA-497/NTRK3 axis in gastric cancer. Int. J. Biol. Macromol. 2019;123:1106–1114. doi: 10.1016/j.ijbiomac.2018.11.162. [DOI] [PubMed] [Google Scholar]
- 21.Guo D., Wang Y., Ren K., Han X. Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression. Exp. Cell Res. 2018;362:172–179. doi: 10.1016/j.yexcr.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Mizrahi A., Barzilai A., Gur-Wahnon D., Ben-Dov I.Z., Glassberg S., Meningher T., Elharar E., Masalha M., Jacob-Hirsch J., Tabibian-Keissar H. Alterations of microRNAs throughout the malignant evolution of cutaneous squamous cell carcinoma: the role of miR-497 in epithelial to mesenchymal transition of keratinocytes. Oncogene. 2018;37:218–230. doi: 10.1038/onc.2017.315. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Li Y., Zou Y., Zhang J., An J., Guo J., Ma M., Dai D. MicroRNA-497 acts as a tumor suppressor in gastric cancer and is downregulated by DNA methylation. Oncol. Rep. 2017;38:497–505. doi: 10.3892/or.2017.5698. [DOI] [PubMed] [Google Scholar]
- 24.Maldonado M.D.M., Dharmawardhane S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018;78:3101–3111. doi: 10.1158/0008-5472.CAN-18-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min P., Zhao S., Liu L., Zhang Y., Ma Y., Zhao X., Wang Y., Song Y., Zhu C., Jiang H. MICAL-L2 potentiates Cdc42-dependent EGFR stability and promotes gastric cancer cell migration. J. Cell. Mol. Med. 2019;23:4475–4488. doi: 10.1111/jcmm.14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Z., Liu F., Wang G., Li Y., Zhang H., Li F. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell. Signal. 2014;26:2667–2673. doi: 10.1016/j.cellsig.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Chen Q., Chen X., Zhang M., Fan Q., Luo S., Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig. Dis. Sci. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- 28.Yang D., Tang Y., Fu H., Xu J., Hu Z., Zhang Y., Cai Q. Integrin β1 promotes gemcitabine resistance in pancreatic cancer through Cdc42 activation of PI3K p110β signaling. Biochem. Biophys. Res. Commun. 2018;505:215–221. doi: 10.1016/j.bbrc.2018.09.061. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q., Busch B., Jiménez-Soto L.F., Ishikawa-Ankerhold H., Massberg S., Terradot L., Fischer W., Haas R. Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation. PLoS Pathog. 2018;14:e1007359. doi: 10.1371/journal.ppat.1007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y., Liu J.P., Li X.Y., Cai Y., He C., Li N.S., Xie C., Xiong Z.J., Ge Z.M., Lu N.H., Zhu Y. Downregulation of tumor suppressor RACK1 by Helicobacter pylori infection promotes gastric carcinogenesis through the integrin β-1/NF-κB signaling pathway. Cancer Lett. 2019;450:144–154. doi: 10.1016/j.canlet.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 31.Yu R., Li Z., Zhang C., Song H., Deng M., Sun L., Xu L., Che X., Hu X., Qu X. Elevated limb-bud and heart development (LBH) expression indicates poor prognosis and promotes gastric cancer cell proliferation and invasion via upregulating Integrin/FAK/Akt pathway. PeerJ. 2019;7:e6885. doi: 10.7717/peerj.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H., Liu Y., Bian Z., Zhang J., Zhang R., Chen X., Huang Y., Wang Y., Zhu J. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol. Cancer. 2018;17:151. doi: 10.1186/s12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humar B., Blair V., Charlton A., More H., Martin I., Guilford P. E-cadherin deficiency initiates gastric signet-ring cell carcinoma in mice and man. Cancer Res. 2009;69:2050–2056. doi: 10.1158/0008-5472.CAN-08-2457. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M., Nomura S., Hosoi A., Nagaoka K., Iino T., Yasuda T., Saito T., Matsushita H., Uchida E., Seto Y. Established gastric cancer cell lines transplantable into C57BL/6 mice show fibroblast growth factor receptor 4 promotion of tumor growth. Cancer Sci. 2018;109:1480–1492. doi: 10.1111/cas.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.