Abstract
Small interfering RNAs (siRNAs) are widely studied for their highly specific gene silencing activity. However, obstacles remain to the clinical application of siRNAs. Attaching conjugates to siRNAs can improve their stability and broaden their application, and most functional conjugates of siRNAs locate at the 3′-terminus of the sense or antisense strand. In this work, we found that conjugating a group at the 5′-terminus of the antisense strand via phosphodiester was practicable, especially when the group was a flexible moiety such as an alkyl linker. When conjugating a bulky ligand, such as cRGD, the length of the 5′-phosphodiester linker between the ligand and the 5′-terminus of the antisense strand was the key in terms of RNA interference (RNAi). With a relative longer linker, the conjugates showed potency similar to siRNA. A highly efficient transfection system composed of a neutral cytidinyl lipid (DNCA) and a gemini-like cationic lipid (CLD) was employed to deliver siRNAs or their conjugates. The cRGD conjugates showed superior targeting delivery and antitumor efficacy in vivo and also selective cellular uptake in vitro. This unity of encapsulation and conjugation strategy may provide potential strategies for siRNA-based gene therapy.
Keywords: siRNA, 5′-cRGD-conjugated siRNAs, DNCA, CLD, phosphodiester linkage, targeted drug delivery
Graphical abstract
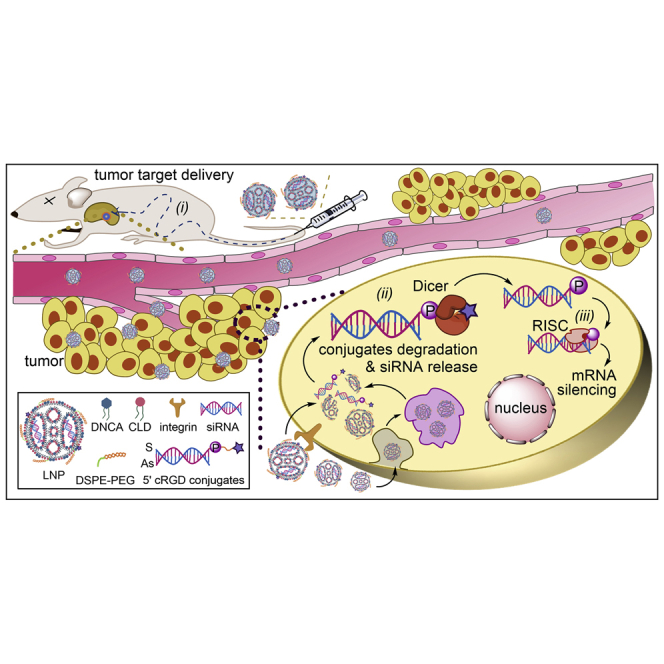
cRGD conjugation at the 5′-terminus of the antisense strand of siRNA by phosphodiester linkage as a tumor targeting moiety could be hydrolyzed to release siRNA, which provided efficient gene silence after the intracellular transportation.
Introduction
Small interfering RNAs (siRNAs) are important gene silencing tools that are widely studied for use in the treatment of rare diseases, metabolic diseases, and tumors, because of their sequence specificity. Natural siRNA is a 21–25-mer RNA, processed from long duplex RNA by Dicer enzyme, with 5′-phosphate and 3′-hydroxyl group.1 The 5′-hydroxyl of synthetic siRNAs can be rapidly phosphorylated by cellular kinase hClp1.2 The 5′-phosphorylation is essential for siRNA incorporation into a RNA-induced silencing complex (RISC) and is a mediating target of mRNA cleavage.3,4 Chen et al.5 used small groups of antisense strand 5′-O-methyl-siRNA to show that 5′-phosphorylation status within duplex siRNA plays an important role in strand incorporation into RISC. The stable phosphate mimic 5′-E-vinylphosphonate (E-VP) siRNA can also incorporate into human Argonaute-2 (hAGO2) protein, with improved activity through a better fitted stereochemistry.6,7 Moreover, other 5′-modifications at the 5′-terminus, such as altritol-nucleotide (ANA), could increase 5′-exonuclease resistance but failed to silence the target mRNA in most cases.8
It was previously supposed that 5′-conjugates could block RNA interference (RNAi), and thus most conjugated ligands to date are at the 3′-terminus of the sense strand.9 The second, third, and fourth siRNA drugs, GIVLAARI, approved by the US Food and Drug Administration (FDA) in 2019, OXLUMO, approved in 2020, and Leqvio, approved by the European Medicines Agency (EMA) in 2020, are siRNA conjugates that contain a GalNAc (triantennary N-acetylgalactosamine carbohydrates) moiety at the 3′-terminus of the sense strand.10,11 Most siRNA drugs now in phase 3 clinical trials are GalNAc conjugates.12,13 Thus, these GalNAc conjugates have acted as the treatment for liver-related disorders.14 Conjugating various ligands, such as lipids, aptamers, peptides, polymers, and other small molecules, to siRNA is widely used in clinical and preclinical studies.15 These conjugation strategies significantly improve the stability, biological half-life, and targeting abilities of siRNA drugs and also provide potential methods for extrahepatic siRNA delivery.16
Introducing a large steric group at the 5′-terminus of the antisense strand via a cleavable linkage based on a phosphodiester bond can control the function of the siRNA, since a bulky moiety blocks the siRNA from loading into a RISC.17 Jain et al.18 designed light-sensitive modifications (caging groups) of antisense strands at the 5′-terminus via a phosphodiester linker to block interaction of the duplex with the cellular machinery responsible for RNAi. These caging groups allowed RNAi modulation in space, time, and degree through light-activated control. Ji et al.19 introduced both photolabile caging groups and biology ligands onto the 5′-terminus of antisense strands to block siRNA activities by increasing the steric hindrance between ligands and their corresponding binding receptor. However, recent research indicates that caging groups only partially block the silencing activities of siRNAs before irradiation,20 even when stability is increased by incorporating phosphorothioate groups at the terminus of siRNA strands.21 These results suggest that either the conjugated group does not completely prevent binding between the 5′-terminus and RISC or it can be cleaved by nuclease in cells. This has led us to consider the tolerance of gene silencing when bulky group conjugated at the 5′-terminus of antisense strands with a flexible phosphodiester linker.
The cyclo(Arg-Gly-Asp-D-Phe-Lys) peptide (cRGD) acts as a tumor-target group by recognizing the αv class of integrins upregulated in tumors,22 usually covalently conjugating to siRNA,23 and is also a bulky group that can block RNAi when conjugated at the 5′-terminus of the antisense strand.24 In this study, cRGD-siRNA conjugates at the 5′-terminus antisense strand via phosphodiester linkage have been reported, which were encapsulated with neutral cytidinyl lipid DNCA and the gemini-like cationic lipid CLD, both developed by our group (Figure S1).25, 26, 27, 28, 29, 30 DNCA could provide hydrogen bond and π-π stacking with siRNA.31 Overall, with a relative longer flexible linker, the siRNA conjugates effectively silenced the target mRNA in vitro and inhibited tumor proliferation in vivo.
Results
Design, synthesis, and characterization of different linker lengths between the target moiety and 5′-terminus of antisense strand of siMB3
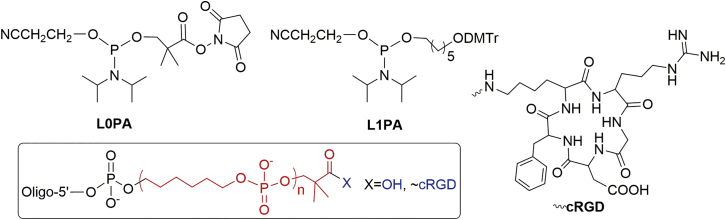
Experiments were carried out using the siRNA siMB3, which targets the mRNA of mutant BRAF (v-raf murine sarcoma viral oncogene homolog B1) protein. Modified siMB3s were synthesized by inserting two alkyl linkers (L0PA and L1PA; Supplemental information, Experimental section) and the cRGD peptide onto the 5′-terminus of the siMB3 antisense strand with a solid-phase synthesizer. These two alkyl linkers were synthesized as previously reported.20 In brief, L0PA was designed for the conjugation with the amino group of the cRGD. L1PA was prepared for the cRGD 5′-conjugates to increase the linker amounts between cRGD and the terminus of the antisense strand. Thus, the cRGD-conjugated-siMB3s with different linker lengths (RLnAs, where n = 0–4, Figure 1) were created by inserting L0PA and different numbers of L1PA phosphoramidites. To evaluate the effect of cRGD modification on RNAi, seven RNA oligonucleotide strands were synthesized for further investigation (Table S1).
Figure 1.
The structural outline of linker building blocks and 5′-siRNA conjugates
Sizes and zeta potentials of DNCA/CLD-encapsulated cRGD-siMB3 nanocomplexes
Nanoparticles were prepared using siMB3 or siMB3 conjugates with DNCA/CLD (8/3 mol/mol) encapsulation; the nanoparticles ranged in size from ∼90- to 300-nm diameter (Table 1). Because of the usage of CLD with a cationic charge, oligonucleotides with an anionic charge, and DNCA with nearly neutral charge, the zeta potentials of nanoparticles ranged from approximately −7 to 33 mV. The zeta potentials of DNCA/CLD-encapsulated RL2As/S, RL3As/S, and RL4As/S were opposite to those of DNCA/CLD-encapsulated As/S, RL0As/S, and RL1As/S because cRGD has a certain amount of cationic charge and this decreases the exposure of anionic oligonucleotides.
Table 1.
The characteristics of siRNA/DNCA/CLD nanocomplexes
| Name | Average particle size (nm) | Polymer dispersity index | Zeta potential (mV) | |
|---|---|---|---|---|
| 1 | DNCA | 246 ± 15.7 | 0.126 | 5.56 ± 0.99 |
| 2 | CLD | 151 ± 10.3 | 0.259 | 33.0 ± 0.72 |
| 3 | DNCA/CLD | 316 ± 70.5 | 0.414 | 14.8 ± 2.98 |
| 4 | DNCA/CLD+As/S | 116 ± 3.71 | 0.462 | −6.39 ± 3.26 |
| 5 | DNCA/CLD+RL0As/S | 195 ± 43.8 | 0.371 | −3.69 ± 0.51 |
| 6 | DNCA/CLD+RL1As/S | 91.6 ± 0.99 | 0.377 | −20.0 ± 2.86 |
| 7 | DNCA/CLD+RL2As/S | 298 ± 9.59 | 0.139 | 1.02 ± 2.48 |
| 8 | DNCA/CLD+RL3As/S | 158 ± 9.49 | 0.205 | 12.8 ± 1.83 |
| 9 | DNCA/CLD+RL4As/S | 154 ± 1.42 | 0.185 | 11.6 ± 0.89 |
Effects of different linker lengths between the target moiety and 5′-terminus of antisense of siRNA on gene silencing efficiency in vitro
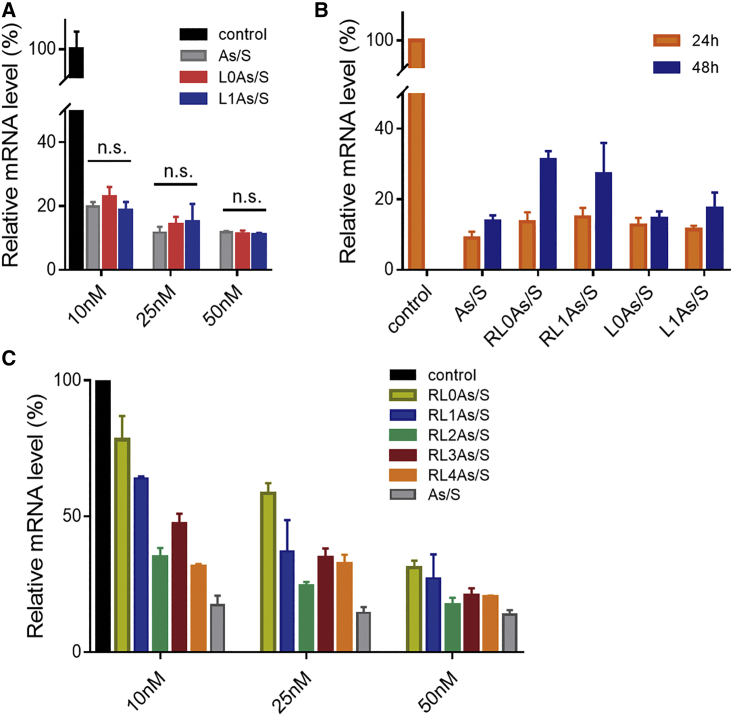
To explore the gene silencing ability of different siMB3 conjugates, qRT-PCR tests were conducted to detect the mRNA levels of the BRAFV600E gene in A375 tumor cells. The effect of 5′-terminus siMB3 conjugates on targeted gene silencing was compared. Cells treated with solvent (GenOpti) provided a control group, and cells treated with unconjugated siMB3 (As/S) encapsulated by DNCA/CLD provided a positive control. First, the gene silencing activities of siMB3 conjugates with different linkers were evaluated. BRAFV600E mRNA levels significantly decreased after treatment with different conjugated and unconjugated siMB3s (Figure 2A). The siMB3 conjugates, L0As/S and L1As/S, showed gene silencing abilities similar to the unconjugated siMB3 (As/S), and the flexible linker modification at the 5′-terminus of the siRNA antisense strand did not block siRNA activity at three concentration conditions.
Figure 2.
The normalization of gene silencing of different siRNA 5′-conjugates
(A) The dose effect of siMB3 conjugates L0As/S and L1As/S on the targeting gene silencing compared with unconjugated siMB3 (As/S) at 10 nM, 25 nM, and 50 nM. (B) The time effect of siMB3 conjugates on the targeting gene silencing at a concentration of 50 nM at 24 and 48 h. (C) The dose effect of different linker lengths of cRGD-conjugated-siMB3s (RLnAs/S, n = 0–4) on the targeting gene silencing. The β-actin housekeeping gene was used as internal control.
Next, A375 cells were treated with control group, As/S, and corresponding conjugates for 24 h and 48 h (Figure 2B). After 24-h incubation of 50 nM conjugates with DNCA/CLD, gene silencing did not differ significantly from the control, whereas after 48-h incubation the gene silencing abilities of 50 nM RL0As/S and RL1As/S were weaker than in the control. The weaker silencing ability is presumably due to the cRGD conjugation on the 5′-terminus of the siMB3s; this large bulky group might inhibit siRNA incorporation into the RISC. However, the silencing activities of the conjugates were not completely blocked. We hypothesize that this is because the cRGD-conjugated part of RL0As/S and RL1As/S may slowly degrade intracellularly, releasing siMB3s with phosphate or hydroxyl groups at the 5′-terminus; these unconjugated siMB3s silenced the expression of mRNA with the conjugates.
We next evaluated the BRAFV600E gene silencing abilities of the same cRGD conjugates with different linker length insertions on the 5′-terminus of the antisense strand of siMB3 (Figure 2C). The silencing efficiency of cRGD conjugates was dose dependent. At the same concentration, the silencing efficiency depended on the distance between the conjugate moieties and the terminus of the antisense strand. The gene silencing of the cRGD conjugates with longer linkers (RL2As/S, RL3As/S, and RL4As/S) had stronger gene knockdown abilities than the others (RL1As/S and RL0As/S).
Selective cellular uptake of integrin targeting with DNCA/CLD/cRGD-siRNA conjugates in vitro
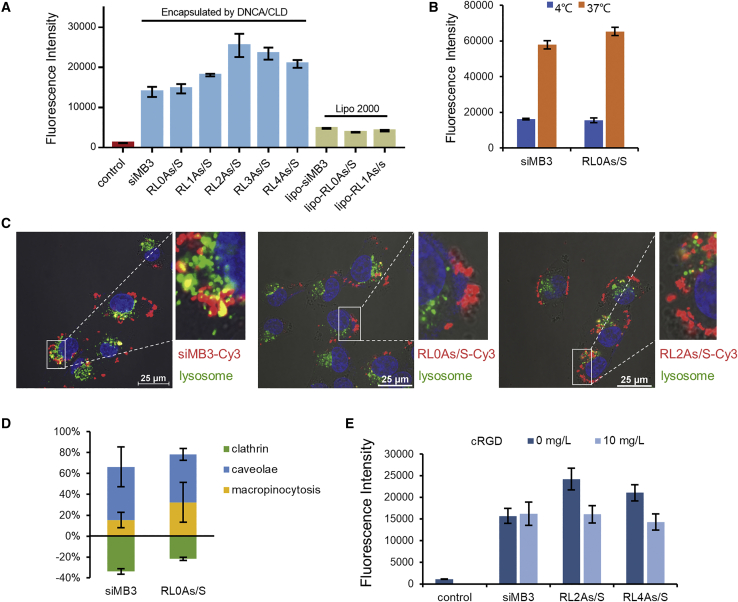
Cellular delivery efficiency of DNCA/CLD/siMB3 nanocomplexes was evaluated in A375 tumor cells, and the difference between cRGD-conjugated and unconjugated siMB3 was compared. As seen in Figure 3A, after incubation for 4 h, flow cytometry of the DNCA/CLD/siMB3 nanocomplexes had higher cellular uptake efficiency than the most-used commercial transfection reagent Lipofectamine 2000 nanocomplexes in the DMEM (10% fetal bovine serum [FBS]). The uptake of DNCA/CLD/siMB3 nanocomplexes was less affected by the presence of serum proteins compared with that of Lipofectamine 2000, even though serum contained small amounts of some potential competitors such as vitronectin and fibronectin. The delivery efficiency of cRGD-conjugated siRNA (RL0As/S-Cy3) was higher than that of unconjugated siRNA (As/S-Cy3) through the DNCA/CLD delivery system, suggesting that some cRGD might leak out of DNCA/CLD nanocomplexes to increase αvβ3-positive A375 cellular uptake. This result matches our previous work showing that cRGD-modified siRNA partially enhanced the αvβ3-positive A375 cellular uptake.24 The effects of cRGD conjugates with different linkers on cellular uptake were also compared (Figure 3A). The cellular uptake abilities of conjugates increased with extended linker lengths until RL2As/S. The cellular uptake of RL3As/S and RL4As/S was less than that of RL2As/S, suggesting that introducing more than 3 phosphodiester linkers between cRGD and the strand terminus may no longer benefit cellular uptake of conjugated siMB3. Confocal laser scanning microscopy (CLSM) showed similar results with flow cytometry (Figure S2). The cellular uptake of As/Cy3-S and RL0As/S-Cy3 delivered by Lipofectamine 2000 was less than that of the DNCA/CLD delivery system with 4-h incubation.
Figure 3.
Cellular uptake of DNCA/CLD/siRNA-conjugate nanocomplexes in A375 cells
(A) The flow cytometry analysis of fluorescence intensity of cells treated with DNCA/CLD/Cy3-siRNA, DNCA/CLD/cRGD-conjugated sense strand 5′-Cy3-siRNA, and Lipo/Cy3-siRNA at 10 nM for 4 h. Lipo 2000, Lipofectamine 2000. (B) The flow cytometry analysis of the cellular uptake mechanism at 4°C of siMB3 and RL0As/S-siMB3 encapsulated by DNCA/CLD for 2 h (siMB3: 50 nM). (C) The flow cytometry analysis of the cellular uptake mechanism of endocytosis of DNCA/CLD-encapsulated siMB3 and RL0As/S-siMB3 for 2 h (siMB3: 10 nM). (D) Confocal analysis of colocalization of Cy3-labeled siMB3 or RL0As/S and lysosome (siMB3: 50 nM), in which nucleus, siRNA, and lysosome are visible by blue, red, and green colors. (E) The flow cytometry analysis of the cellular uptake of siMB3, RL2As/S-siMB3, and RL4As/S-siMB3 encapsulated by DNCA/CLD with or without a pre-incubation of cRGD peptide (10 mg/L); the Cy3-siRNA concentration was 10 nM.
The cellular uptake mechanism of cRGD-conjugated siRNA and unconjugated siRNA encapsulated by DNCA/CLD was evaluated next. Most proteins associated with cellular uptake and metabolism are inhibited at 4°C, and thus cell penetration of both siMB3 and RL0As/S at 4°C was monitored; results showed that the transmembrane transfer efficiency was affected by temperature and energy dependent for both (Figure 3B). As Figure 3C shows, after incubating for 4 h, part of the Cy3-labeled siRNA was located on the cell membrane and part had penetrated and distributed into the cytoplasm. Also, the colocalization of siRNA with lysosome remained at a relatively low level. Furthermore, after conjugated cRGD at the 5′-terminus, the colocalization dropped (RL0As/S or RL2As/S versus As/S, p < 0.05, Figure S3).
Next, the endocytic pathways of siMB3 and its antisense 5′-conjugates were clarified (Figure 3D). The siRNAs encapsulated by DNCA/CLD still entered the cell through non-clathrin-mediated endocytosis as previously reported.31
Since cRGD is the ligand of tumor-related integrin αvβ3 and cRGD was also used as a target moiety when conjugating with DSPE-PEG (bis(1,2-distearoyl-sn-glycero-3-phosphoethanolamine)-N-[(polyethylene glycol)-2000]),32 the selective cellular uptake of siRNA conjugated with the target group cRGD at the 5′-terminus of antisense strand was evaluated (Figure 3E). The A375 cells were first incubated with 10 mg/L cRGD, and then Cy3-labeled siMB3, RL2As/S, and RL4As/S delivered by DNCA/CLD were given. The uptakes of RL2As/S and RL4As/S were competitively inhibited by the cRGD peptide, suggesting that the DNCA/CLD/cRGD-siRNA conjugate nanoparticles have selective cellular uptake of integrin targets. The cRGD groups of siRNA conjugates were on the surface of the nanoparticles, providing tumor-targeting ability for in vivo administration.
The biodistribution of DNCA/CLD/cRGD-conjugated siMB3 in vivo
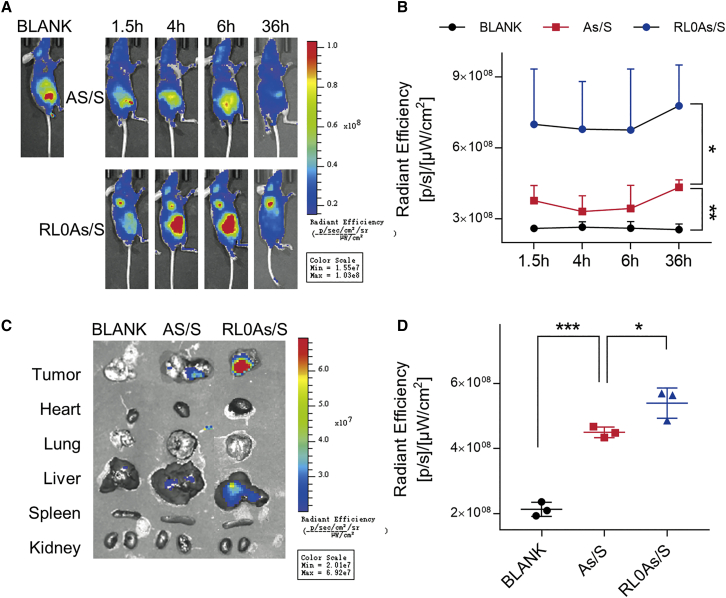
The biodistributions of nanoparticles were monitored in BALB/c-nude mice to verify targeting efficiency. A375 cells were injected into the right shoulder area, and after 10 days tumor volumes reached ∼800 mm3. Subsequently, animals were injected with Cy7-labeled siMB3 (As/S) or conjugates (RL0As/S) encapsulated by DNCA/CLD via the tail vein. The fluorescent signal was measured from 1.5 h to 36 h after injection. Bioimaging of the Cy7 spectrum revealed significant accumulation in tumors of mice injected with cRGD-conjugated siMB3, whereas tumors in mice injected with unconjugated siMB3 had only weak fluorescent signals (Figure 4A). However, the accumulation in liver and kidneys is difficult to observe because of autofluorescence of food in the gastrointestinal tract, which was not associated with the tumor signal. The intratumor fluorescent signal was measured (Figure 4B), and tumor accumulation of cRGD-conjugated siMB3 was ∼90% higher than that of unconjugated siMB3 (p < 0.05). At 36 h, tumor tissues and organs were isolated and photographed. The Cy7-labeled siMB3 was mostly contained in tumor and liver tissue, and the fluorescent signal of RL0As/S was much stronger than that of As/S in the tumor (Figure 4C). Quantitative results corresponded with in vivo distribution (Figure 4D), with more cRGD conjugates accumulating in the tumor (p < 0.05). These results demonstrate that siMB3 with cRGD target group encapsulated by DNCA/CLD and administered via the tail vein persists longer than unencapsulated siMB3. As the conjugated siRNA nanoparticles entered into cell through the receptor-mediated pathway, the cRGD moiety can act as a target group when applied in vivo. The combined conjugation and delivery strategies achieved tumor-targeted siRNA therapy by systemic delivery.
Figure 4.
The biodistribution of sense strand 5′-Cy7-labeled siRNA nanoparticles
(A) The nude mice treated with GenOpti (Blank), mix/siRNA (As/S), and mix/cRGD-conjugated siRNA (RL0As/S) by tail intravenous injection. Images were taken at 1.5, 4, 6, and 36 h after the injection by an in vivo imaging system. (B) The corresponding total radiant efficiency at tumor area of different treatments. (C) The representative images of dissected tumor tissues and organs 36 h after injection. (D) The corresponding total radiant efficiency of isolated tumor tissues. The results are presented as mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
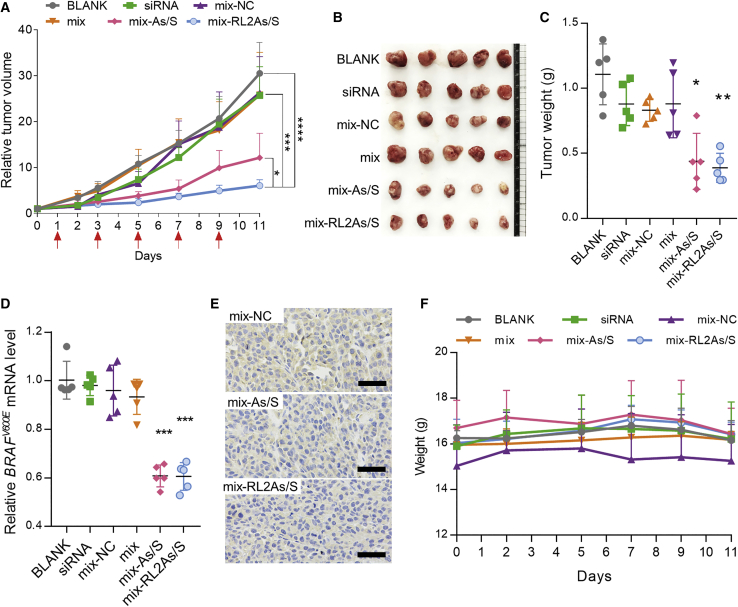
The antitumor and gene inhibition efficiency of DNCA/CLD/cRGD-conjugated siMB3 in vivo
Given the similar gene silencing activities of siMB3 and RL2As/S-siMB3 in vitro, and the significantly enhanced tumor accumulation with cRGD in vivo, antitumor and gene inhibition efficiency were evaluated. A375 tumor cells were subcutaneously injected into female BALB/c-nude mice, and mice were separated into six groups when tumor sizes reached ≤50 mm3 at the 7th day after injection. Treatments, given via tail vein every 2 days, included (1) solvent (BLANK); (2) naked siMB3 (siRNA); (3) nanoparticles of DNCA/CLD/PEG-DSPE (mix); (4) encapsulated scrambled siRNA (mix-NC); (5) encapsulated siMB3 (mix-As/S), and (6) encapsulated cRGD-conjugated-siMB3 (mix-RL2As/S). After five administrations, the relative tumor growth of siMB3 treatments had slowed compared with these blank and negative controlled groups (BLANK, naked siRNA, mix-NC, Figure 5A). The volume of tumor tissues in negative groups increased >25-fold over the 10 days, whereas the mix-As/S treatment group increased only 12-fold, suggesting that the siMB3 could inhibit the tumor growth under the encapsulation of DNCA/CLD/PEG-DSPE (mix).
Figure 5.
In vivo antitumor efficiency and BRAFV600E gene silencing activity of siRNA and antisense 5′-conjugates in A375 tumor-bearing BALB/c-nude mice
(A) Relative tumor volume of A375 xenograft tumor-bearing nude mice treated with siMB3 or RL2As/s-siMB3 delivered by DNCA/CLD/PEG-DSPE (represented as mix). (B) Images of harvested A375 tumors at the end of the experiment. (C) Each tumor was weighed after being photographed (mix-As/S compared with BLANK, siRNA, mix-NC, and mix, ∗p < 0.05; mix-RL2As/S compared with BLANK, siRNA, mix-NC, and mix, ∗∗p < 0.01). (D) The expression of BRAFV600E mRNA in tumors (mix-As/S or mix-RL2As/S compared with BLANK, siRNA, mix-NC, and mix, ∗∗∗p < 0.001). (E) Immunohistochemistry of BRAFV600E expression in the tumor after different treatments to reveal the gene silencing abilities of siRNA. The scale bars represent 50 μm. (F) The body weight changes during the experiment. The results are presented as mean ± SD (n = 5). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Importantly, the tumor growth rate of the mix-RL2As/S group was much slower than that of the mix-As/S group (p < 0.05), with the tumor volume increasing only ∼6-fold. The 5′-terminus conjugated siMB3 was 50% more effective than unconjugated siMB3 in reduction of tumor growth. Tumor tissues were isolated and photographed 48 h after the last injection. As shown in Figures 5B and 5C, the change of ex vivo tumor sizes and weights corresponded to measurement done in vivo. The average isolated tumor weight of mix-RL2As/S (0.39 ± 0.1 g) was smaller than mix-siMB3 (0.44 ± 0.2 g). Furthermore, the gene silencing activities of conjugated and unconjugated siMB3 were evaluated by qRT-PCR and immunohistochemistry. BRAFV600E mRNA levels in tumors revealed that both the mix-siMB3 and mix-RL2As/S groups effectively reduced expression of target mRNA by 40%, a significant difference from negative control (siRNA, mix, mix-NC) and blank groups (p < 0.001, Figure 5D). Immunolocalization of BRAF-mutated V600E on paraffin-embedded sections of tumors matched qRT-PCR results. The yellow area represents the expression of BRAFV600E protein, and different color shades reflect different protein levels. Compared to NC, conjugated siMB3 and unconjugated siMB3 both downregulated BRAFV600E, and the silencing efficiency of RL2As/S equaled that of As/S (Figure 5E).
This superior antitumor ability of RL2As/S was due to the combination of increased tumor accumulation and siRNA activity recovery, since the average tumor volume of RL0As/S treatment was higher than in unconjugated siMB3 treatment at end of the in vivo experiment (Figure S4), although they did not differ significantly (p = 0.25). Moreover, the steady change of body weight and survival of all mice during the experiment suggests that the treatments were safe enough (Figure 5F).
Discussion
The siRNA drugs can efficiently and specifically silence mRNA in vivo and in vitro. This so-called RNAi is a gene therapy strategy that has been extensively studied in the past two decades. At present, the FDA and the EMA have approved four liver-targeted siRNA drugs.33,34 However, research on the delivery system of siRNA lipid complexes based on cationic lipids has stalled; the interest in siRNA drug development now focuses on the siRNA conjugates, especially the 3′-GalNAc conjugates.35 Previous studies have demonstrated that the siRNA-conjugated strategy is being realized as the dominant technology in clinical research. However, the 5′-conjugates at the antisense strand are little investigated.
Our former reports used CLD and DNCA to encapsulate siRNA, showing superior antitumor and gene silencing activity both in vitro and in vivo.31 This mixed CLD and DNCA delivery system has also been used in the 3′,3″-bis-peptide siRNA and antisense oligonucleotides (ASOs).29,32
In this study, several conjugated siRNAs at 5′-antisense terminus, LnAs/S or RLnAs/S (n = 0–4, Figure 1), have been successfully synthesized. The mRNA silencing activity of LnAs/S reveals that a flexible group alkyl linker with appropriate length at the 5′-terminus of the antisense strand does not affect siRNA activity. When a large bulky part, such as cRGD, is close to the 5′-terminus, the siRNA activity is impaired, meaning that conjugating cRGD with shorter linker chains (RL0As/S and RL1As/S) blocked siRNA activity. However, the leakage of gene silencing exists, as these blocked siRNA conjugates still maintain 20%–30% silencing abilities at the concentration of 10 nM. These results correspond with previous studies in which insertion of a flexible long linker (19 atoms) between cRGD and the 5′-terminus of the antisense strands20 or a single small photo-sensitive group (NPE) incompletely blocked siRNA activity.36
Interestingly, the silencing activity was gradually recovered when the distance between cRGD and the 5′-terminus increased (RL2As/S, RL3As/S, and RL4As/S), because of its flexible structures and decreased steric hindrance for enzymatic decomposition, or incorporating siRNA conjugates into RISC. As a previous study reports, modification on the 5′-terminus only blocked one of the charges on the phosphate and left another oxygen unmodified, which just like the phosphodiester caused less effect of the binding to RISC. In addition, conjugates with the longer alkyl linkage may undergo biodegradation in the cell, and the conjugated moieties can be recognized by enzymes and cleaved. Then, with the large bulky cRGD group dropped, the 5′-phosphate antisense strands are exposed. During these processes, a longer linker provides a flexible cleavage position.
The vehicle used is a mixture of neutral cytidinyl/cationic lipids, which are developed by our laboratory, to encapsulate siRNA and its cRGD conjugates and then transfect it into cells in vitro or target deliver it into tumor by tail vein administration in vivo. This carrier shows superior efficiency over the commercial transfection reagent Lipofectamine 2000. The cellular uptake mechanism is also underevaluated. We find that the conjugate moiety enhanced the interaction between siRNA and DNCA/CLD lipids. Thus, the siRNA conjugate nanoparticles show superior cellular uptake ability. And RLnAs/Ss encapsulated by DNCA/CLD enters into cell mainly through the caveolae-mediated endocytosis and macropinocytosis, which can avoid clathrin-mediated endocytosis, reducing degradation in the lysosome. The confocal results also show few colocalizations between the lysosome and Cy3-labeled siRNA conjugates (RL0As/S), supporting the non-lysosome cellular uptake. The results are consistent with our former reports, which means that the existence of the conjugating moiety does not affect the uptake mechanism. Also, the siRNA conjugates/DNCA/CLD nanoparticles still enter into the cell by the αvβ3-mediated uptake, suggesting that the cRGD moiety still acts as targeting group even after encapsulating.
Moreover, with the tumor targeting group cRGD, the antisense 5′-terminus conjugates show improved gene silencing and antitumor activity in vivo. The cRGD-increased accumulation of siRNA nanoparticles in tumor area and the extended distance between cRGD and the 5′-terminus with a longer linker (RL2 compared with RL0) are the key points for the superior antitumor efficiency of 5ʹ-conjugates at antisense strands. This strategy of targeting group conjugation with cationic/cytidinyl lipid encapsulation could provide potential therapeutic applications of siRNA in future.
In conclusion, by combining an efficient neutral cationic/cytidinyl lipid delivery system and 5′-conjugation, the druggability of siRNA could be improved, also providing a new strategy for the in-depth development of siRNA drugs.
Materials and methods
Materials
The gemini-like cationic lipids (CLD) and cytidinyl lipid (DNCA) were synthesized by our group (Figure S1),28,37 and the synthesis method for RNA samples was described in previous studies.20 In brief, siMB3s were obtained by an ABI 394 DNA/RNA synthesizer and purified via Waters 1525 HPLC. The column used was the XBridge Oligonucleotide BEH C18 OBD Prep Column (2.5 μm, 4.6 mm × 50 mm, 1/pkg). Their identities were confirmed by electrospray ionization mass spectrometry. Finally, the corresponding siRNAs were formed by annealing antisense or sense strands. All studies were done with siMB3 (As, 5′-AUC GAG AUU UCU CUG UAG Cdtdt; S, 5′-GCU ACA GAG AAA UCU CGA Udtdt), which was targeting to BRAFV600E mRNA.38 The detailed synthesis methods of siRNAs and their conjugates are shown in the Supplemental information, and Figures S5–S20.
Cell culture
The A375 cell line was bought from KeyGen Biotech (China), and cells were cultured in DMEM supplemented with 10% v/v FBS at 37°C with 5% CO2.
The preparation of nanoparticles
In brief, the CLD and DNCA were dissolved in ethanol at a final concentration of 10 mmol/L and then dropped into siRNA solution (GenOpti, M&C, China). For in vitro studies, the molar ratio of different parts was 84/31.5/1 (DNCA/CLD/siRNAs). For in vivo studies, the molar ratio of different parts was 21/31.5/0.4 (DNCA/CLD/DSPE-PEG2000, mix); the molar ratio of all these lipids to siRNA was 52.9/1. After centrifugation briefly at 3,000 rpm, the mixture was sonicated at 70°C for 10 min.
Size and zeta potential assay
The particle sizes and zeta potentials were measured at 25°C with DLS (dynamic light scattering, Malvern Zetasizer Nano ZS, Malvern, UK). The siRNA and siRNA conjugate concentrations were 50 nM.
Cellular uptake efficiency assay
A375 cells were seeded in 24-well plates (1 × 105 per well) and grown to 70%–80% confluence after 24-h proliferation. Then, nanocomplexes containing sense strand 5′-Cy3-labeled siRNA (cRGD conjugated or unconjugated) at a final concentration of 10 nM with DNCA/CLD were exposed to the cells and incubated for an additional 4 h at 37°C. After incubation, the cells were washed three times with pre-cooled fresh DMEM and then harvested by 0.25% trypsin. After centrifugation, cells were resuspended by DMEM, and the uptake of Cy3-labeled siRNA was immediately detected with a FACSCalibur flow cytometer (Becton Dickinson, USA).
Lysosome escape assay
A375 cells (6 × 104 cells/well) were seeded in 14-mm confocal dishes (Solarbio, China) and grown to 70%–80% confluence after 24-h proliferation. After that, nanocomplexes containing Cy3-labeled siRNA (cRGD conjugated or unconjugated) at a final concentration of 50 nM with DNCA/CLD were added to the cells and incubated for an additional 4 h at 37°C. The cells were washed three times with pre-cooled fresh DMEM. Then LysoBrite NIR (AAT Bioquest, USA) and Hoechst 33342 (Solarbio, China) were added to the medium and incubated with the cells for 30 min at 37°C for lysosome and nucleus labeling. Next, the cells were washed with fresh DMEM three times. Intracellular distribution of Cy3-labeled siRNA and lysosome colocalization were observed with an A1Rsi confocal microscope (Nikon, Japan).
In vitro gene silencing assay
A375 cells were seeded in 12-well plates (2 × 105 per well) and grown to 70%–80% confluence after 24-h proliferation. Then, nanocomplexes containing siRNAs (cRGD conjugated or unconjugated) at final concentrations of 10, 25, and 50 nM were exposed to the cells and incubated for an additional 48 h at 37°C. After incubation, the cells were washed three times with pre-cooled fresh DMEM, and total RNAs were extracted with the TRIzol reagent (Invitrogen, USA) and then reversed to cDNA by Reverse Transcription System A3500 (Promega). cDNA was then mixed with forward and reverse primers of BRAFV600E and housekeeping gene β-actin and GoTaq qPCR Master Mix (A6002, Promega, USA) and then analyzed by a real-time PCR amplifier (MX3005P; Stratagene, USA) The threshold cycles (Ct) of each sample were normalized to the β-actin gene, and the inhibition of gene silencing is represented as the percentage of BRAFV600E expression.
Animals
All the animal studies were approved by the Animal Care and Use Committee of Peking University (No. LA2017194), and all the operations about animals conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978).
In vivo biodistribution assay
The female nude mice were bearing A375 xenograft tumors on their right flank. After tumor sizes grew to ∼800 mm3, mice were divided into three groups, with three mice in each group. The siRNAs used in biodistribution assay were sense strand 5ʹ-Cy7 labeled on their sense strands, and the siRNAs encapsulated by mix were given by intravenous injection at 1.0 mg/kg. The fluorescent signal was detected with an in vivo imaging system at 1.5, 4, 6, and 36 h. At the end time point, the nude mice were euthanized, and their tumor tissues and organs were isolated and examined.
In vivo antitumor efficiency
The A375 cells (2 × 106) were inoculated subcutaneously on the right flank of female nude mice. When the tumor volume reached 30–80 mm3 (length × width2/2), mice were divided into six groups, with five mice in each group. Different formulations were given by intravenous injection at days 1, 3, 5, 7, and 9. The mice were treated with different formulations at a siRNA dosage of 1.48 mg/kg. After 48 h of final injection, the mice were euthanized, and tumor tissues were isolated, weighted, and photographed. Then parts of isolated tumors were homogenated by homogenizer in TRIzol (Thermo Fisher, USA), and the total RNA was extracted and analyzed. The other parts of isolated tumors were immersed in 4% paraformaldehyde overnight at 4°C. Then the tumors were embedded and cut into slices to analyze immunohistochemistry. The BRAFV600E protein was stained as brown color, and nuclei were stained by hematoxylin as blue color.
Statistical analysis
For statistical analysis between two groups, Student’s t test for independent means was applied. For multiple comparison, one-way ANOVA was applied. A p value < 0.05 was considered statistically significant. Each value is expressed as mean ± SD. Statistical analysis was performed with SPSS software (version 16.0; SPSS, Chicago, IL, USA).
Statement of animal rights
All animal experiments were approved by the Committee for Animal Research of Peking University (No. LA2017194), and all the operations about animals conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978). The specific pathogen free (SPF)-grade female nude mice (3–4 weeks) were obtained from Wantonglihua (China) and kept in the Department of Laboratory Animal Science, Peking University Health Science Center.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (Grant No. 2017ZX09303013), the National Natural Science Foundation of China (Grant Nos. 21778006, 21907021), the Innovation Fund for Outstanding Doctoral Candidates of Peking University Health Science Center (X.Z., Grant No. BMU2020BSS001), and the Open-End Funds from the State Key Laboratory of Natural and Biomimetic Drugs, Peking University (L.Y.).
Author contributions
Z.Y. conceived the project, proposed constructive discussions, and supervised the research. X.Z. and L.Y. designed and performed the experiment, analyzed the data, and wrote the manuscript. Y.P., L.Y., and H.L. provided siRNA synthesis support. Z.L. provided experimental support and helped edit the manuscript. J.W., and Z.G. provided experimental support. All authors contributed to the general discussion.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.004.
Supplemental information
References
- 1.Kandasamy S.K., Fukunaga R. Phosphate-binding pocket in Dicer-2 PAZ domain for high-fidelity siRNA production. Proc. Natl. Acad. Sci. USA. 2016;113:14031–14036. doi: 10.1073/pnas.1612393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weitzer S., Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 3.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 4.Nykänen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 5.Chen P.Y., Weinmann L., Gaidatzis D., Pei Y., Zavolan M., Tuschl T., Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parmar R., Willoughby J.L.S., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of siRNA-GalNAc Conjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 7.Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L. siRNA carrying an (E)-vinylphosphonate moiety at the 5 end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2017;45:3528–3536. doi: 10.1093/nar/gkw1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P., Degaonkar R., Guenther D.C., Abramov M., Schepers G., Capobianco M., Jiang Y., Harp J., Kaittanis C., Janas M.M. Chimeric siRNAs with chemically modified pentofuranose and hexopyranose nucleotides: altritol-nucleotide (ANA) containing GalNAc-siRNA conjugates: in vitro and in vivo RNAi activity and resistance to 5′-exonuclease. Nucleic Acids Res. 2020;48:4028–4040. doi: 10.1093/nar/gkaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai W. Current aspects of siRNA bioconjugate for in vitro and in vivo delivery. Molecules. 2019;24:2211. doi: 10.3390/molecules24122211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Paula Brandão P.R., Titze-de-Almeida S.S., Titze-de-Almeida R. Leading RNA interference therapeutics part 2: Silencing delta-aminolevulinic acid synthase 1, with a focus on Givosiran. Mol. Diagn. Ther. 2020;24:61–68. doi: 10.1007/s40291-019-00438-6. [DOI] [PubMed] [Google Scholar]
- 11.Alnylam Pharmaceuticals I. 2020. Alnylam announces U.S. Food and Drug Administration (FDA) approval of OXLUMO™ (lumasiran), the first and only treatment approved for primary hyperoxaluria type 1 to lower urinary oxalate levels in pediatric and adult patients.https://www.businesswire.com/news/home/20201124005407/en/ [Google Scholar]
- 12.Huang Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol. Ther. Nucleic Acids. 2017;6:116–132. doi: 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng Y., Xiao H., Zhang J., Liang X.J., Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Weingärtner A., Bethge L., Weiss L., Sternberger M., Lindholm M.W. Less is more: Novel hepatocyte-targeted siRNA conjugates for treatment of liver-related disorders. Mol. Ther. Nucleic Acids. 2020;21:242–250. doi: 10.1016/j.omtn.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig K., Abrams M., Amiji M. Recent preclinical and clinical advances in oligonucleotide conjugates. Expert Opin. Drug Deliv. 2018;15:629–640. doi: 10.1080/17425247.2018.1473375. [DOI] [PubMed] [Google Scholar]
- 16.Klein D., Goldberg S., Theile C.S., Dambra R., Haskell K., Kuhar E., Lin T., Parmar R., Manoharan M., Richter M. Centyrin ligands for extrahepatic delivery of siRNA. Mol. Ther. 2021;29:2053–2066. doi: 10.1016/j.ymthe.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debart F., Dupouy C., Vasseur J.J. Stimuli-responsive oligonucleotides in prodrug-based approaches for gene silencing. Beilstein J. Org. Chem. 2018;14:436–469. doi: 10.3762/bjoc.14.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain P.K., Shah S., Friedman S.H. Patterning of gene expression using new photolabile groups applied to light activated RNAi. J. Am. Chem. Soc. 2011;133:440–446. doi: 10.1021/ja107226e. [DOI] [PubMed] [Google Scholar]
- 19.Ji Y., Yang J., Wu L., Yu L., Tang X. Photochemical Regulation of Gene Expression Using Caged siRNAs with Single Terminal Vitamin E Modification. Angew. Chem. Int. Ed. Engl. 2016;55:2152–2156. doi: 10.1002/anie.201510921. [DOI] [PubMed] [Google Scholar]
- 20.Yu L., Liang D., Chen C., Tang X. Caged siRNAs with single cRGD modification for photoregulation of exogenous and endogenous gene expression in cells and mice. Biomacromolecules. 2018;19:2526–2534. doi: 10.1021/acs.biomac.8b00159. [DOI] [PubMed] [Google Scholar]
- 21.Kala A., Friedman S.H. Enhanced light-activated RNA interference using phosphorothioate-based dsRNA precursors of siRNA. Pharm. Res. 2011;28:3050–3057. doi: 10.1007/s11095-011-0529-z. [DOI] [PubMed] [Google Scholar]
- 22.Svensen N., Walton J.G.A., Bradley M. Peptides for cell-selective drug delivery. Trends Pharmacol. Sci. 2012;33:186–192. doi: 10.1016/j.tips.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Cen B., Wei Y., Huang W., Teng M., He S., Li J., Wang W., He G., Bai X., Liu X. An efficient bivalent cyclic RGD-PIK3CB siRNA conjugate for specific targeted therapy against glioblastoma In vitro and In vivo. Mol. Ther. Nucleic Acids. 2018;13:220–232. doi: 10.1016/j.omtn.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z., Liu S., Zhang Y., Yang X., Ma Y., Guan Z., Wu Y., Zhang L., Yang Z. Reductive nanocomplex encapsulation of cRGD-siRNA conjugates for enhanced targeting to cancer cells. Int. J. Nanomedicine. 2017;12:7255–7272. doi: 10.2147/IJN.S136726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J., Qiu C., Diao Y.P., Wei W., Jin H.W., Zheng Y., Wang J.C., Zhang L.H., Yang Z.J. Delivery Pathway Regulation of 3′,3″-Bis-Peptide-siRNA Conjugate via Nanocarrier Architecture Engineering. Mol. Ther. Nucleic Acids. 2018;10:75–90. doi: 10.1016/j.omtn.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L., Huang Y., Wang X., Ma Y., Liu Y., Guan Z., Zhang L., Yang Z. Serum stability enhancement of siRNA caused by peptide conjugation at 3′-terminus of sense strand. J. Chin. Pharm. Sci. 2014;23:215–219. [Google Scholar]
- 27.Yang M.Y., Sun J., Wang C., Zhang Y.F., Zhang L.H., Yang Z.J. Transfection of 3′,3′’-bis-peptide-siRNA conjugate by cationic lipoplexes mixed with a neutral cytosin-1-yl-lipid. J. Chin. Pharm. Sci. 2017;26:719–726. [Google Scholar]
- 28.Ma Y., Zhu Y., Wang C., Pan D., Liu S., Yang M., Xiao Z., Yang X., Zhao W., Zhou X. Annealing novel nucleobase-lipids with oligonucleotides or plasmid DNA based on H-bonding or π-π interaction: Assemblies and transfections. Biomaterials. 2018;178:147–157. doi: 10.1016/j.biomaterials.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Ma Y., Zhao W., Li Y., Pan Y., Wang S., Zhu Y., Kong L., Guan Z., Wang J., Zhang L., Yang Z. Structural optimization and additional targets identification of antisense oligonucleotide G3139 encapsulated in a neutral cytidinyl-lipid combined with a cationic lipid in vitro and in vivo. Biomaterials. 2019;197:182–193. doi: 10.1016/j.biomaterials.2018.12.033. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X., Wang S., Zhu Y., Pan Y., Zhang L., Yang Z. Overcoming the delivery barrier of oligonucleotide drugs and enhancing nucleoside drug efficiency: The use of nucleolipids. Med. Res. Rev. 2020;40:1178–1199. doi: 10.1002/med.21652. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X.Y., Pan Y.F., Li Z., Li H.T., Wu J., Ma Y., Guan Z., Yang Z.J. siRNA packaged with neutral cytidinyl/cationic/PEG lipids for enhanced antitumor efficiency and safety in vitro and in vivo. ACS Appl. Bio Mater. 2020;3:6297–6309. doi: 10.1021/acsabm.0c00775. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y.F., Li S.X., Zhou X.Y., Sun J., Fan X.M., Guan Z., Zhang L.H., Yang Z.J. Construction of a Targeting Nanoparticle of 3′,3″-Bis-Peptide-siRNA Conjugate/Mixed Lipid with Postinserted DSPE-PEG2000-cRGD. Mol. Pharm. 2019;16:4920–4928. doi: 10.1021/acs.molpharmaceut.9b00800. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M.M., Bahal R., Rasmussen T.P., Manautou J.E., Zhong X.B. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021;189:114432. doi: 10.1016/j.bcp.2021.114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb Y.N. Inclisiran: First Approval. Drugs. 2021;81:389–395. doi: 10.1007/s40265-021-01473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L., Pei F., Zhang J., Wu J., Feng M., Wang Y., Jin H., Zhang L., Tang X. Synthesis of site-specifically phosphate-caged siRNAs and evaluation of their RNAi activity and stability. Chemistry. 2014;20:12114–12122. doi: 10.1002/chem.201403430. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y., Guo Y.J., Li Y.T., Wu Y., Zhang L.H., Yang Z.J. A novel gemini-like cationic lipid for the efficient delivery of siRNA. New J. Chem. 2014;38:4952–4962. [Google Scholar]
- 38.Fisher M., Abramov M., Van Aerschot A., Rozenski J., Dixit V., Juliano R.L., Herdewijn P. Biological effects of hexitol and altritol-modified siRNAs targeting B-Raf. Eur. J. Pharmacol. 2009;606:38–44. doi: 10.1016/j.ejphar.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.