Abstract
Mutant p53 (mutp53) commonly loses its DNA binding affinity to p53 response elements (p53REs) and fails to induce apoptosis fully. However, the p53 mutation does not predict chemoresistance in all subtypes of breast cancers, and the critical determinants remain to be identified. In this study, mutp53 was found to mediate chemotherapy-induced long intergenic noncoding RNA-p21 (lincRNA-p21) expression by targeting the G-quadruplex structure rather than the p53RE on its promoter to promote chemosensitivity. However, estrogen receptor alpha (ERα) suppressed mutp53-mediated lincRNA-p21 expression by hijacking mutp53 to upregulate damaged DNA binding protein 2 (DDB2) transcription for subsequent DNA repair and chemoresistance. Levels of lincRNA-p21 positively correlated with the clinical responses of breast cancer patients to neoadjuvant chemotherapy and had an inverse correlation with the ER status and DDB2 level. In contrast, the carboplatin-induced DDB2 expression was higher in ER-positive breast tumor tissues. These results demonstrated that ER status determines the oncogenic function of mutp53 in chemoresistance by switching its target gene preference from lincRNA-p21 to DDB2 and suggest that induction of lincRNA-p21 and targeting DDB2 would be effective strategies to increase the chemosensitivity of mutp53 breast cancer patients.
Keywords: mutant p53, estrogen receptor, lincRNA-p21, DDB2, G-quadruplex, chemotherapy
Graphical abstract

ER status determines the oncogenic function of mutp53 in chemoresistance by switching its target gene preference from lincRNA-p21 to DDB2. The co-treatment of chemotherapy with ER antagonists in a neoadjuvant setting may benefit luminal A/B patients through upregulation of lincRNA-p21 to enhance chemotherapy efficacy in mutp53-expressing breast cancers.
Introduction
In response to DNA damage by chemotherapeutic agents, activated p53 acts as a decision-making transcription factor that selectively mediates gene expressions involved in inducing cell-cycle arrest, DNA repair, and apoptosis.1 When DNA damage exists, p53 halts the cell cycle at the G1 phase by transcriptionally inducing the expression of cyclin-dependent kinase (CDK) inhibitor p21, allowing the time for the DNA repair machinery to remove the lesions before cell proliferation resumes.2 In addition to its well-defined role in regulating the cell cycle, p53 has also been directly implicated in the regulation and orchestration of various DNA-damage-response (DDR) mechanisms in response to chemotherapy,2 including transcriptional upregulation of damaged DNA binding protein 2 (DDB2), which associates with its binding partner DDB1 to recruit XPC protein to the DNA damage lesion in the early step of nucleotide excision repair (NER).2
TP53 gene mutations are found in around 30% of breast cancer cases and are associated with a poor prognosis and chemoresistance.3,4 In particular, TP53 hotspot mutations, which are located mainly within the DNA-binding domain, markedly reduce the ability of wild-type p53 (WTp53) to bind to the promoters of its target genes.5 Mutant p53 (mutp53) can also have a dominant-negative effect over other p53 family members, such as abolishing the apoptotic functions of p63 and p735,6 by co-opting other transcription factors to mediate the transactivation of their target genes involved in tumor progression and drug resistance.5,7 Furthermore, mutp53 has been demonstrated to modulate gene expressions by binding to intronic and intergenic sequences predisposed to form non-B DNA structures that include mismatches, cruciforms, and especially G-quadruplex.8,9 These “gain of function” (GOF) roles suggest that mutp53 has a critical role in conferring chemoresistance.10
Even though basic studies employing genetically engineered cell lines and mutp53 mouse models have demonstrated that the mutp53 GOF role contributes to chemoresistance,11 the role of p53 mutations in clinical response to chemotherapy has not been well defined nor suggested being a clinical prediction marker for chemoresistance. In certain tumors, including esophageal12 and diffuse large B cell lymphoma (DLBCL),13 mutp53 predicts resistance to chemotherapy. Conversely, in bladder cancer14 and chronic lymphocytic leukemia,15 the p53 mutation has not been associated with a chemotherapy response. Intriguingly, mutp53 predicts better chemotherapy responses in early-stage or inflammatory breast cancer but offers no such benefit in metastatic or non-inflammatory breast cancer.16 These inconsistent roles of mutp53 in the response of breast tumors to chemotherapy suggest that certain factors in different subtypes of breast cancer may interfere with mutp53-targeted gene expression, leading to distinctively different predictive patterns. Indeed, the poor prognostic effect of the TP53 mutation in breast cancer is limited to estrogen receptor (ER)-positive disease, especially the luminal B subtype.3 ER has been reported to suppress WTp53-dependent gene transcription and tumor-suppressor functions in breast cancer, whereas disruption of this interaction by ER antagonism restores p53 activity.17 However, the molecular aspects of the interplay between ER and mutp53 in regulating chemosensitivity remain unclear.
Over the past decade, the human genome has been deciphered, and numerous functional long noncoding RNAs (lncRNAs) have been discovered and implicated in many diseases, particularly cancer.18 Emerging evidence has shown that lncRNAs regulate gene expression via diverse mechanisms such as epigenetic regulation, transcriptional control, post-transcriptional regulation, and molecular scaffolding.19 As a crucial sequence-specific transcription factor, p53 regulates the expression of some lncRNAs, and p53-associated lncRNAs have been considered as biomarkers for cancer diagnosis or targets for disease therapy.20 For instance, long intergenic noncoding RNA-p21 (lincRNA-p21; TP53COR1), pint (a p53-induced noncoding transcript), and taurine up-regulated 1 (TUG1) modulate cellular apoptosis and proliferation by directly interacting with heterogeneous nuclear ribonucleoprotein K (hnRNAP-K) and the polycomb-repressive complex 2 (PRC2), respectively.21, 22, 23 Plasmacytoma variant translocation 1 (PVT1) induced by WTp53 was found to inhibit mouse double minute 2 (MDM2) and stabilize p53 protein levels.24 In contrast, p53-induced promoter of CDKN1A antisense DNA damage activated RNA (PANDAR) and regulator of reprogramming (RoR) suppress cellular apoptosis and dictate chemoresistance via negative regulation of p53 in an autoregulatory loop.25,26 Experiments using p53 null cell lines have demonstrated the essential roles of WTp53 in the upregulation of lncRNAs.21,27 However, chemotherapy-induced lncRNA expression is observed in mutp53-expressing head and neck squamous cell carcinoma (HNSCC) and colorectal cancer (CRC).28,29 Furthermore, the expression of lincRNA-p21 in tumor tissue is not associated with TP53 genetic status in either CRC30 or lung cancer.31 Thus, the impact of TP53 mutations on the expression and functions of p53-associated lncRNAs remains unclear.
In this study, our data showed that ERα hijacks mutp53 to enhance DDB2 promoter activity by binding with its ER response elements (EREs) and thereby reduces the availability of mutp53 for lincRNA-p21 transcription in ER-positive cells. Conversely, lincRNA-p21 was found to be upregulated by both WTp53 and mutp53 in lncRNA quantitative polymerase chain reaction (qPCR) array analysis. However, mutp53 induces lincRNA-p21 expression by targeting the non-B structure, rather than the canonical p53 response element (p53RE), on the lincRNA-p21 promoter. These results demonstrated that lincRNA-p21 could serve not only as a predictive marker but also as a sensitizer for a chemotherapeutic response in breast cancer.
Results
ER-positive breast cancer cells were insensitive to chemotherapy
To characterize the chemosensitivity of different breast cancer subtypes, we first examined the viability of breast normal cell (MCF-10A), luminal A (MCF7 and T-47D), luminal B (BT-474), human epidermal growth factor receptor 2 (HER2)-enriched (SK-BR-3), and triple-negative (MDA-MB-468, MDA-MB-231, and HBL-100) breast cancer cell lines in response to platinum drugs carboplatin (Figure 1A) and cis-diammineplatinum (II) (cisplatin) (Figure 1B) and topoisomerase II inhibitor doxorubicin (Figure 1C). As shown in the heatmap representing cell viability, three ER-positive MCF7, T-47D, and BT-474 breast cancer cells were more resistant to carboplatin, cisplatin, and doxorubicin. In contrast, four ER-negative SK-BR-3, MDA-MB-468, MDA-MB-231, and HBL-100 breast cancer cells were relatively sensitive to carboplatin, cisplatin, or doxorubicin regardless of TP53 genetic status (Figures 1A−1C; Table S1). Thus, mutp53-expressing breast cancer cell lines were used in the rest of the experiments to examine the role of ER in chemosensitivity. In parallel, carboplatin induced apoptosis in ER-negative SK-BR-3 and MDA-MB-231 but not in ER-positive T-47D and BT-474 breast cancer cell lines (Figure 1D). DNA repair is one of the important reasons to avoid chemotherapy-induced DNA damage and cell apoptosis.32 Therefore, we also examined the impact of ER on chemotherapy-induced DNA damage in comet assays. Indeed, the results showed that chemo-sensitive ER-negative, but not chemo-resistant ER-positive, breast cancer cell lines exhibited a dramatic tail moment and length index in response to carboplatin (Figure 1E), raising the possibility that ER may possess the anti-apoptotic function through enhancing DNA repair in response to chemotherapy.
Figure 1.
The cellular sensitivity of various subtypes of breast cancer cell lines
(A−C) The viability of breast cancer cell lines in response to various concentrations of carboplatin, cisplatin, and doxorubicin was determined by using the MTT assay and visualized as a heatmap by using Morpheus (https://software.broadinstitute.org/morpheus). (D) Apoptotic cell death in different breast cancer cell lines in response to carboplatin (50 μM) in a time-dependent manner was examined by using flow cytometry analysis. (E) The comet assay was performed to examine the effect of carboplatin (50 μM) on DNA damage in T-47D and MDA-MB-231 cancer cell lines. The tail moment and tail-length index were calculated using Comet Assay III software. Data in all panels were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
ERα contributes to DNA repair in mutp53-expressing breast cancer cells
To further demonstrate the essential role of ER in conferring to the chemoresistance of mutp53-expressing breast cancer cells, the effect of ERα silencing by two independent short hairpin RNAs (shRNAs) on chemotherapy-induced cell death of T-47D cancer cells was examined. The heatmap data presented that the inhibition of cell viability by platinum drugs was enhanced while ERα was silenced (Figure 2A). The synergistic effects of the combination of ER partial antagonist tamoxifen (4-OHT) on the chemotherapy-induced cytotoxicity of T-47D and BT-474 cancer cells were next found in MTT assays (Figure 2B). In addition, the carboplatin-induced apoptosis, as indicated as caspase-3 cleavage, of BT-474 cancer cells was also enhanced by co-treatments with tamoxifen (Figure 2C). Furthermore, the silencing of ERα also enhanced the level of damaged DNA in BT-474 cancer cells in response to carboplatin, as evidenced by the more extended tail length in the comet assays (Figure 2D). Taken together, ERα may function as a critical regulator of DNA repair in conferring the chemoresistance of mutp53-expressing breast cancer cells.
Figure 2.
Inhibition of ERα increases chemosensitivity in ER-positive breast cancer cells
(A) The effect of ERα silenced by two independent shRNAs on the viability of T-47D cancer cells in response to platinum drugs was examined in the MTT assay, and the result was visualized as a heatmap. (B) The inhibitory effects of chemo drugs (carboplatin, cisplatin, and doxorubicin) with or without 10 μM tamoxifen on the viability of T-47D and BT-474 cancer cell lines were examined in the MTT assay. (C) The effect of 10 μM tamoxifen on carboplatin (50 μM)-induced apoptotic marker expression in BT-474 cancer cells was examined in a western blot assay. (D) BT-474 cancer cells were treated with carboplatin (50 μM) in a time-dependent manner following ERα gene silencing and were then subjected to the comet assay to detect the extent of DNA damage. The tail-length index was calculated from images by Comet Assay III analysis. Data in (A), (B), and (D) were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
ERα transcriptionally activates DDB2 gene expression for DNA repair in mutp53-expressing breast cancer cells
Previous studies revealed that ERα reduced chemotherapy-induced cell death by repressing the apoptosis pathway or activating the survival pathway via interacting with transcription factors, such as p53, Myc, and SP1.33, 34, 35, 36, 37, 38, 39, 40, 41 However, its roles in regulating DNA repair remain unclear. Gene Set Enrichment Analysis (GSEA) analysis showed the positive correlations of the ER level with the expression of gene sets involved in DNA repair or the p53-dependent pathway in four databases (Gene Expression Omnibus [GEO]: GSE50948, GSE5847, GSE23988, and GSE44272) (Figure S1A). Furthermore, DDB2, a downstream gene of WTp53 and a well-known component of NER, was highly associated with ER-positive expression in both DNA repair and p53 pathway gene sets (Figure S1A), implying the involvement of DDB2 in mediating ER-dependent chemoresistance and DNA repair.
Basal levels of DDB2 mRNA (Figure 3A) and protein (Figure 3B) were higher in ER-positive than in ER-negative breast cancer cell lines, regardless of TP53 genetic status. The positive correlation of DDB2 expression with ER/progesterone receptor (PR) status was also observed in human breast tumor tissues (Tables S2 and S3). In support of the upregulation of DDB2 expression by ERα, silencing ERα gene expression repressed the protein (Figures 3C and S2A) and mRNA (Figure 3C) levels of DDB2 in ER-positive breast cancer cell lines. Three predicted EREs and one validated p53RE exist on the DDB2 promoter; we constructed three luciferase reporter plasmids containing these different elements as illustrated in Figure 3D. As compared to control (pGL4.22-Luc), the basal DDB2 promoter#1, containing three EREs and one p53RE (from −1,091 to −7), was found to dramatically stimulate luciferase activity. This phenomenon was minimally affected by the deletion of three EREs (DDB2 promoter#2 from −299 to −7; containing one p53RE) but was dramatically reduced by the deletion of p53RE (DDB2 promoter#3 from −930 to −281; two EREs) (Figure 3E). Interestingly, treatment with carboplatin further enhanced the activity of only DDB2 promoter#1, but not #2 or #3 (Figure 3E). Furthermore, silencing of ERα reduced the carboplatin-induced luciferase activity driven by DDB2 promoter#1 in mutp53-expressing breast cancer cells (Figure 3F). These results suggest that ERα may cooperate with mutp53 to mediate chemotherapy-induced DDB2 expression.
Figure 3.
ERα mediates chemotherapy-induced DDB2 gene transcription
(A and B) Basal mRNA (A) and protein (B) levels of DDB2 were higher in ER-positive than ER-negative breast cancer cell lines, regardless of their TP53 genetic status. (C) Silencing of ERα by two independent shRNAs suppressed DDB2 protein and mRNA expression. Reduction of both S118-phosphorylated and total ERα revealed the inhibition of ERα activity. (D) ER response elements (EREs) and p53 RE (p53RE) motifs on the DDB2 promoter and the luciferase reporter constructs driven by different lengths of the DDB2 promoter were illustrated (vermilion, red, and pink: EREs; orange: p53RE). (E) Treatment with carboplatin (50 μM) increased the luciferase activity driven by the DDB2 promoter containing both p53RE and EREs in mutp53-expressing T-47D cancer cells. Firefly luciferase activity was normalized with β-gal activity. (F) Silencing of ERα decreased the carboplatin (50 μM)-induced DDB2 promoter activity in T-47D cancer cells, and firefly luciferase activity was normalized with β-gal activity. Data in (A), (C), (E), and (F) were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
ERα interacts with mutp53 to enhance DDB2 gene expression in response to DNA damage
To demonstrate the cooperation between mutp53 and ERα in mediating DDB2 transcription, the effect of mutp53 silencing by using shRNAs on DDB2 promoter luciferase activity was examined. Similar to the results with ERα silencing, carboplatin-induced luciferase activity driven by DDB2 promoter#1 was slightly reduced, whereas mutp53 was silenced (Figure 4A). Moreover, mutation of these EREs but not p53RE significantly repressed the carboplatin-induced DDB2 promoter activity (Figures 4B and S2B). Findings from chromatin immunoprecipitation (ChIP) assays further revealed that ERα and mutp53 were preferentially recruited to the DDB2 promoter on ERE#1 in response to carboplatin and doxorubicin treatment (Figure 4C), suggesting that ERα and mutp53 may collaboratively mediate DDB2 gene transcription through binding to ERE#1. Interestingly, the acetylation of mutp53 at K382, a transcriptional activating marker of p53, was also inhibited by silencing ERα expression (Figure 4D). The protein-protein interaction between ERα and mutp53 was induced by carboplatin and doxorubicin (Figure 4E). ERα enhanced WTp53- and mutp53-driven DDB2 promoter activity (Figure S2C) as well as DDB2 protein expression (Figure 4F). However, the reduction of DDB2 expression by silencing ERα was not accompanied by the downregulation of WTp53 K382 acetylation in MCF7 cancer cells, suggesting the distinct regulations by ERα in WTp53- and mutp53-mediated DDB2 expressions (Figure S2D). Furthermore, silencing of DDB2 enhanced the level of damaged DNA in BT-474 cancer cells in response to carboplatin, as evidenced by the longer tail length index in the comet assays (Figure 4G). Thus, these results demonstrated that mutp53 is an important co-factor to interact with ERα and targets EREs to transcriptionally upregulate DDB2 gene expression for DNA repair.
Figure 4.
ERα cooperates with mutp53 to upregulate DDB2 gene transcription and mediates chemoresistance in ER-positive breast cancer cells
(A) Silencing of p53 decreased the carboplatin (50 μM)-induced DDB2 promoter activity in T-47D cancer cells. (B) Deletions of EREs, but not p53RE, reduced carboplatin (50 μM)-induced DDB2 promoter activity in T-47D cancer cells. Firefly luciferase activity was normalized with β-gal activity in (A) and (B). (C) Carboplatin (50 μM) induced chromatin-binding affinity of both ERα and mutp53 preferentially on ERE#1 of the DDB2 promoter in T-47D cancer cells in the ChIP assay. (D) Silencing of ERα reduced p53 activity, as evidenced by the acetylation at K382 in T-47D and BT-474 cancer cells. (E) Chemotherapy (50 μM carboplatin or 0.5 μM doxorubicin) induced the protein interaction between p53 and ERα in the coimmunoprecipitation (coIP) assay. (F) Ectopic co-expression of ERα and p53 mutants (R280K and R273H) synergistically enhanced DDB2 expression in HEK293T cells. (G) Silencing of DDB2 by two independent shRNAs enhanced carboplatin (50 μM)-induced DNA damage in the comet assay. The tail moment and tail-length index were calculated from images by Comet Assay III analysis. Data in (A−C) and (G) were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
ERα reduces chemosensitivity by repressing the mutp53-upregulated lincRNA-p21 expression
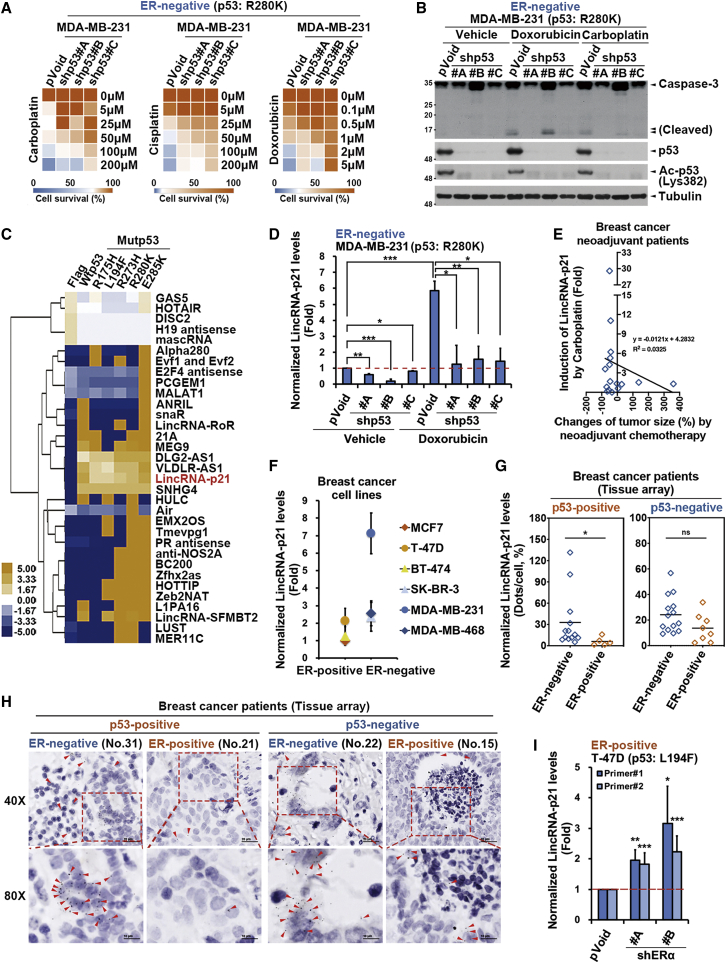
The mutp53 GOF plays an oncogenic role in suppressing apoptosis6,16 but still did not fully predict the chemoresistance in several cancer types.12, 13, 14, 15 Interestingly, mutp53-expressing breast cancer cells remain sensitive to chemotherapeutic agents in the absence of ERα expression (Table S1), leading us to address whether mutp53 mediates the chemosensitivity in ER-negative breast cancer cells. As shown in the result of the heatmap analysis, the silence of the endogenous p53R280K reduced the sensitivity of ER-negative MDA-MB-231 cancer cells to carboplatin, cisplatin, and doxorubicin (Figure 5A) as well as the chemotherapy-induced cleavage of pro-caspase-3 (Figure 5B), suggesting that mutp53 remains essential for the chemotherapy-induced apoptosis in ER-negative breast cancer cells.
Figure 5.
ERα represses the mutp53-dependent lincRNA-p21 expression to confer chemoresistance
(A) Silencing p53 by the independent shRNAs reduced the cell viability of ER-negative/p53R280K MDA-MB-231 cancer cells in response to chemo drugs, as presented in the heatmap. (B) Silencing p53 by the independent shRNAs reduced chemotherapy (0.5 μM doxorubicin and 50 μM carboplatin)-induced apoptotic marker expression in ER-negative/p53R280K MDA-MB-231 cancer cells. (C) Heatmap results from the lncRNA qPCR array showed the regulation of lncRNA expression by WTp53 and mutp53 in HEK293T cells. (D) Silencing of the endogenous p53R280K mutant suppressed doxorubicin (0.5 μM)-induced lincRNA-p21 expression in MDA-MB-231 cancer cells. (E) The ex vivo induction of lincRNA-p21 expression by carboplatin (50 μM) in primary human breast cancer tumors positively correlated with tumor shrinkage in response to neoadjuvant chemotherapy (n = 16). (F) lincRNA-p21 expression was higher in ER-negative than in ER-positive breast cancer cell lines in quantitative real-time PCR analysis. (G and H) lincRNA-p21 expression examined in ISH assay was higher in human ER-negative/p53-positive (n = 13) than ER-positive/p53-positive (n = 5) breast cancer tumors, but the lincRNA-p21 level in ER-negative/p53-negative (n = 14) and ER-positive/p53-negative (n = 8) breast cancer tumors did not show significant difference. The red arrows indicated the signals of lincRNA-p21 expression, which was calculated by the average number of dots per nucleus. Welch two-sample t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (I) Silencing of ERα by two independent shRNAs increased lincRNA-p21 expression in ER-positive/p53L194F T-47D cancer cells in quantitative real-time PCR analysis. Data in (A), (D), (F), and (I) were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
LncRNAs are abundantly expressed and involved in gene regulation in cancer cells.18,19,42 To understand the impact of p53 mutations on the expression of lncRNAs that may interfere with the sensitivity of breast cancer to chemotherapy, lncRNA expression in the WTp53- and mutp53-expressing human embryonic kidney 293 transformed with a large T antigen (HEK293T) cells were analyzed and compared using lncRNA qPCR array analysis (Figure 5C). Overexpression of WTp53 or various mutp53 proteins (R175H, L194F, R273H, R280K, and E285K) affected the expression of around one-third (36%) of the lncRNAs, whereas four lncRNAs (DLG2-AS1, VLDLR-AS1, lincRNA-p21, and SNHG4) were upregulated by both WTp53 and p53 mutants (Figure 5C). Interestingly, among these four lncRNAs, lincRNA-p21 is the only lncRNA that is known to be a direct target of WTp53 for mediating p53-induced apoptosis, but its regulation by mutp53 remains unclear. The upregulating effect of mutp53 on lincRNAp21 expression was further confirmed in quantitative real-time PCR analysis with two different primer sets (Figure S3A, upper), but these p53 mutants failed to enhance the protein expression of p21, a canonical target gene of WTp53 (Figure S3A, lower). Silencing of endogenous p53R280K mutant expression also repressed basal and doxorubicin-induced lincRNA-p21 levels in ER-negative MDA-MB-231 cancer cells (Figure 5D). The induction of lincRNA-p21 levels in response to ex vivo treatment with carboplatin was also positively correlated with tumor shrinkage in response to chemotherapy in the neoadjuvant settings (Figure 5E). Silencing of lincRNA-p21 suppressed carboplatin-induced DNA damage in ER-negative and mutp53-expressing MDA-MB-231 cancer cells (Figure S3B), indicating the essential role of mutp53-induced lincRNA-p21 in chemo-sensitization. Interestingly, ER-negative breast cancer cell lines express higher basal lincRNA-p21 levels than ER-positive breast cancer cell lines did regardless of TP53 genetic status (Figure 5F). Consistently, the level of lincRNA-p21 was negatively associated with ER status in human breast cancer tissue (Tables S4 and S5), and this association was only found in p53-positive breast cancer tissue (Figures 5G and 5H). Notably, lincRNA-p21 levels were increased by silencing ERα expression in ER-positive/p53L194F T-47D cancer cells (Figure 5I). These results indicated that mutp53 could upregulate lincRNA-p21 expression and thereby mediate chemosensitivity in ER-negative cancer cells. In contrast, in ER-positive cancer cells, ERα confers chemoresistance by suppressing mutp53-dependent lincRNA-p21 expression.
Mutp53 mediates lincRNA-p21 expression by targeting the non-B DNA structures
It has been reported that WTp53 binds to not only the sequence-based p53RE in linear B-DNA but also the structure-based local non-B DNA motifs such as cruciform DNA (CF), G-quadruplex DNA (GQ), and triplex DNA.9 Although mutp53 proteins do not bind or bind only weakly to the p53RE on the promoter of canonical target genes such as p21 (CDKN1A),7 they still possess the ability to interact with local non-B DNA structures, especially GQ,43 in intergenic sequences that globally modulate transcription.8 An analysis of the non-B DB version v2.0 database (https://nonb-abcc.ncifcrf.gov/apps/site/default)44 predicted several non-B structures, including a GQ motif and four CF motifs, on the promoter of lincRNA-p21 (Figure 6A; Table S6), but these were not conserved in the mouse lincRNA-p21 promoter (Figure S4A). The promoter-reporter assays revealed that p53 mutants only enhanced the activity of the reporter gene constructed with lincRNA-p21 promoter regions containing non-B DNA motifs plus p53RE but not that containing p53RE alone (Figure 6B). Mutations of GQ but not p53RE reduced the effects of chemotherapy- (Figure 6C) and mutp53-induced (Figure S4B) lincRNA-p21 promoter activity. In ChIP assays, endogenous p53R280K bound to some of these non-B structures but not the canonical p53RE in MDA-MB-231 cancer cells in response to carboplatin and doxorubicin (Figure 6D). Moreover, p53R280K from carboplatin-treated MDA-MB-231 cancer cells showed a binding affinity with biotin-labeled oligonucleotides corresponding to the GQ motif of the lincRNA-p21 promoter in vitro (Figure 6E). In support of these findings, N-methyl mesoporphyrin IX (NMM) and auramine, two stabilizers for the GQ structure,45,46 enhanced the binding of p53R280K with the biotinylated GQ motif (Figure 6F) and the expression of lincRNA-p21 in p53R280K-expressing MDA-MB-231 (Figure 6G) and p53R175H-expressing SK-BR-3 cancer cells (Figure S4C). These results demonstrated that mutp53 remains able to mediate lincRNA-p21 expression by targeting the non-B DNA structures on its promoter, primarily through the GQ structure.
Figure 6.
ERα hijacks mutp53 from the G-quadruplex DNA (GQ) structure of the lincRNA-p21 promoter to EREs of the DDB2 promoter
(A) Illustration of the predicted non-B structure and p53RE on the lincRNA-p21 promoter and two luciferase-reporter constructs driven by lincRNA-p21 promoters containing different motifs. (B and C) GQ motif is required for mutp53-increased lincRNA-p21 promoter activity, and firefly luciferase activity was normalized with β-gal activity. (D) The binding efficacy of endogenous mutp53 on different motifs of the lincRNA-p21 promoter in response to chemotherapies (50 μM carboplatin and 0.5 μM doxorubicin) in the ChIP assay. (E and F) Carboplatin (50 μM) and two GQ stabilizers (1 μM NMM and 5 μM auramine) increased the binding of mutp53 to the GQ motif of the 5′-biotinylated lincRNA-p21 promoters in the in vitro pull-down assay. (G) GQ stabilizers increased lincRNA-p21 expression in MDA-MB-231 cancer cells. (H) Silencing of ERα switched the chromatin-binding activity of mutp53 from EREs of the DDB2 promoter (left) to the non-B DNA motifs of the lincRNA-p21 promoter (right) in ER-positive/p53L194F T-47D cancer cells. (I) The inversed correlation between the ex vivo induction of lincRNA-p21 and DDB2 expression by carboplatin (50 μM) treatments in human primary breast cancer tissues in an ER status-dependent manner. Data in (B−D), (G), and (H) were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test.
Since both DDB2 and lincRNA-p21 transcription involves mutp53 but is enhanced and suppressed by ERα, respectively, we examined the role of ERα in influencing the mutp53-binding preference to these two target genes. In response to carboplatin treatment, silencing of ERα reduced the DNA binding of mutp53 to EREs on the DDB2 promoter but enhanced its binding to non-B DNA structures on the lincRNA-p21 promoter in ER-positive/p53L194F T-47D cancer cells (Figure 6H). Interestingly, the inverse correlation between carboplatin-induced DDB2 and lincRNA-p21 expression was also observed in human primary breast tumor tissue (Figures 6I and S5A). Similar to the inhibition of DDB2 promoter activity in mutp53-expressing breast cancer cells (Figure 3F), the silence of ERα also repressed the carboplatin-induced DDB2 promoter activity in WTp53-expressing MCF7 cancer cells (Figure S6A). Unlike the induction of lincRNA-p21 expression in mutp53-expressing breast cancer cells (Figure 5I), however, the silencing of ERα did not restore the expression of lincRNA-p21 significantly (Figure S6B) and also has no change in carboplatin-induced lincRNA-p21 promoter activity in WTp53-expressing MCF7 cancer cells (Figure S6A). In contrast to the enhancing effect on DDB2 promoter activity (Figure S2C), ERα overexpression repressed the WTp53- and mutp53-induced lincRNA-p21 promoter activity (Figure S6C). These findings suggest that ERα cooperates with both WTp53 and mutp53 for the upregulation of DDB2 expression and downregulation of lincRNA-p21 expression in distinct manners, but suppression of ERα only can rescue the mutp53-mediated lincRNA-p21 expression.
Induction of lincRNA-p21 reduces the ERα-mediated chemoresistance
To examine the chemo-sensitizing effect of lincRNA-p21 in vivo, we established the tetracycline-inducible lincRNA-p21 expressing system in ER-positive T-47D cancer cells (T-47D#Tet-On-LincRNA-p21), which were then injected into the mammary fat pad of mice to study tumor growth. We first validated the expression of lincRNA-p21 in the T-47D#Tet-On-LincRNA-p21 stable clone, and the data showed that the expression of lincRNA-p21 was indeed upregulated, accompanied with the downregulation of DDB2 expression upon the tetracycline treatment (Figure 7A). The treatment timeline of the xenograft mouse model was illustrated in Figure 7B. We first subcutaneously implanted 17β-estradiol and 1 × 107 cells of the T-47D#Tet-On-LincRNA-p21 clone into non-obese diabetic (NOD)/severe combined immunodeficiency (SCID) mice, which were then treated with saline, tetracycline, doxorubicin, or a tetracycline/doxorubicin combination after 1 month of tumor inoculation. The combination treatment with tetracycline/doxorubicin showed a synergistic effect to minimize the growth rate (Figure 7C) and the tumor size at the end point (Figure 7D) of chemo-resistant T-47D xenograft tumors. In situ hybridization (ISH) and immunohistochemistry staining were then performed to validate the molecular alterations in the xenograft tumor tissues in response to these treatments. The results showed that the combination treatment of tetracycline/doxorubicin decreased the expressions of DDB2 and Ki67 and slightly increased the cleavage of caspase-3 accompanied by the lincRNA-p21 induction (Figure 7E). These results indicated that the tetracycline-induced lincRNA-p21 reduced the tumor growth and also increased the chemosensitivity of ER-positive breast cancer cells to doxorubicin.
Figure 7.
lincRNA-p21 reduces the growth and chemoresistance of the ER-positive tumor in vivo
(A) The T-47D#Tet-On-LincRNA-p21 stable clone was treated with or without tetracycline (10 μg/mL), and the expressions of lincRNA-p21 and DDB2 were detected by quantitative real-time PCR analysis and western blot assay, respectively. Data were representative of three experiments and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test. (B) Illustration of the treatment timeline in the tumor-xenograft mouse model (yellow arrow: the starting point for tetracycline administration [0.2 mg/mL]; red arrow: the points for the intraperitoneal injection with doxorubicin [2.5 mg/kg]). (C and D) The tumor growth rate (C) and size at the end point (D) in four groups of these mice. Data were representative of n = 3 in every group and were shown as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus the control group, Student’s t test. (E) The expressions of lincRNA-p21, DDB2, Ki67, and cleaved caspase-3 in the tumor tissues were examined in the in situ hybridization and immunohistochemistry assays.
In summary, these findings revealed the involvement of a non-B DNA structure in mutp53-mediated lincRNA-p21 expression and the inverse correlation between lincRNA-p21 and DDB2 expression. The expression of ERα increases DDB2 expression and suppresses lincRNA-p21 transcription by switching the promoter-binding affinity of mutp53, thereby resulting in DNA repair and chemoresistance (Figure 8).
Figure 8.
The proposed model of ERα/mutp53 mediated DDB2 and lincRNA-p21 in contributing to chemoresistance
The proposed model illustrates how ERα determined chemoresistance of breast cancer by disrupting the balance between mutp53-dependent DDB2 and lincRNA-p21 transcriptions. In the ER-negative cancer cells (left), lincRNA-p21, transcribed by mutp53 in a G-quadruplex of non-B structure-dependent fashion, mediate apoptosis for chemosensitivity. In the ER-positive cancer cells (right), however, ERα switches mutp53 to preferentially mediate DDB2 transcription via targeting its EREs and thereby reduces lincRNA-p21 expression, conferring chemoresistance (p53∗, mutp53).
Discussion
TP53 is one of the most frequently mutated genes in breast cancer.3,4 Several retrospective studies have investigated the predictive role of the p53 mutation in breast cancer chemotherapy, with controversial results.16 Thus, the p53 mutation has not yet served as a biomarker for predicting clinical responses to breast cancer chemotherapy.47 Although many published studies have shown that the presence of the p53 mutation is associated with poor outcomes in breast cancers,16 this observation is probably confined to patients with specific molecular subtypes.47 A follow-up analysis revealed that high levels of p53 protein due to protein stabilization by p53 mutations were associated with worse disease-free survival in ER-positive patients but correlated with good outcomes in ER-negative patients.48 However, the advantage of concurrent neoadjuvant chemotherapy and hormone therapy in ER-positive breast cancer remains controversial.49, 50, 51 In light of our study, we discovered that ERα renders chemoresistance by regulating p53-dependent gene transcription from lincRNA-p21 to DDB2 for DNA repair in mutp53-expressing breast cancer cells, supporting that the addition of the ER inhibitor to neoadjuvant chemotherapy provides the improvement of the clinical response in patients with ER-positive breast cancer, especially for those individuals with p53 mutations.
In reflection of its diverse functions in regulating the p53 pathway, lincRNA-p21 is upregulated by WTp53 to trigger apoptosis and cell-cycle arrest by acting as a transcriptional repressor,21 co-activator,52 or translation suppressor53 for different gene-expression patterns. In this study, we further discovered that lincRNA-p21 also could be upregulated by mutp53 through the direct binding on non-B DNA structures of its promoter. Moreover, lincRNA-p21 expression was inversely correlated to DDB2 in human breast cancer tissue (Figure 6I) and had an enhancing role in chemosensitivity, showing that lincRNA-21 may thereby participate in the modulating function of mutp53 in NER and chemoresistance.
WTp53 directly targets p53RE within exon 1 of lincRNA-p21 to drive its expression in mouse embryonic fibroblasts,21 whereas a mutation of a single p53 allele represses ultraviolet-induced mouse lincRNA-p21 expression in mouse keratinocytes.54 However, basal levels of human lincRNA-p21 are not associated with p53 mutations in human CRC30 or lung31 tumor tissue. Several noncoding RNAs were approved or are being tested in clinical trials as predictive biomarkers for diagnosis and prognosis or were used in the oligonucleotide format of locked nucleic acids (LNA) for therapy in cancers.55, 56, 57 The tumor-suppressive roles of lincRNA-p21 have been found in several cancer types, including stomach, prostate, glioma, hepatocarcinoma, and lung cancers but not in breast cancer, according to the analysis in LncRNADisease database (http://www.rnanut.net/lncrnadisease/)58 (Table S7). Our data showed that mutp53 effectively mediates chemotherapy-induced human lincRNA-p21 in breast cancer cells, consistent with findings in HNSCC,28 by targeting the non-B DNA structures (especially GQ) within the proximal promoter region rather than binding to the canonical p53RE. This mechanism may explain the lack of association between the p53 mutation and lincRNA-p21 expression in human tumor tissue.
Mutp53 DNA binding to non-B DNA has been demonstrated to be solely dependent on its stereo-specific configuration rather than the nucleotide sequence, and the binding affinity varies among different mutp53 proteins.59 The binding regions enriched for mutp53 are from 1 kb upstream to 1 kb downstream of the transcription start sites (TSSs).43 Mutp53 preferentially and autonomously binds to G/C-rich DNA, prone to form G-quadruplex structures around TSSs of many genes.43 Consistent with this finding, our data revealed that some p53 mutants increase lincRNA-p21 expression and preferentially bind to GQ near the TSS of lincRNA-p21. Interestingly, the GQ sequence was not found in the proximal region of mouse lincRNA-p21 promoters, which only share 25% homology with human lincRNA-p21 promoters (Figure S4A), which explains why mutp53 did not induce mouse lincRNA-p21 expression.54 Although mutp53 mediates lincRNA-p21 expression by targeting non-B DNA structures, our data also revealed that the presence of ERα repressed mutp53-dependent lincRNA-p21 expression by hijacking mutp53 to mediate DDB2 gene expression for chemoresistance. It accounts, in part, for the more significant responses to neoadjuvant chemotherapy in ER-negative breast cancer patients rather than in ER-positive breast cancer patients35 and the roles of hormone receptors in DNA repair34 and DDB2-dependent proliferation.60 Consistent with our findings, DDB2 expression was not associated with the somatic mutations of p53 in the genotypic mutation analysis (muTarget; https://www.mutarget.com).61 The lower expression of DDB2 was also found in the responder of ER-positive breast cancer patients who received treatments with chemotherapy, indicating low DDB2 expression as a potential biomarker predicting chemotherapy efficacy (area under the curve [AUC]: 0.582) in the receiver operating characteristic (ROC) curve analysis (ROC Plotter; http://www.rocplot.org)62 (Figure S7A).
ERα has been reported to mediate gene regulation through several co-factors to enhance transcription; activate activator protein 1 (AP-1), nuclear factor κB (NF-κB); and form complexes with CBP/p300.33 ERα has also been reported to regulate several DNA repair pathways through interactions with DNA-dependent protein kinase (DNA-PK), BRCA1 DNA repair associated (BRCA1), and WTp53.34 In addition to the reciprocal regulation of ERα and p53 expression at the transcriptional level,17 the physical interaction between ERα and WTp53 has also been found to protect breast cancer from apoptosis by inhibiting the p53-dependent transcriptional repression of anti-apoptotic genes and preventing the accumulation of DNA damage.17,40,63, 64, 65 Disruption of the interaction between ERα and WTp53 by ER antagonists, fulvestrant and tamoxifen, can restore the WTp53 activity to upregulate doxorubicin-induced apoptosis regulatory genes, ATF3, BTG2, and TRAF4.63 Ionizing radiation was also demonstrated to disrupt ERα-WTp53 interaction and restores the expression of p21 (CDKN1A), a CDK inhibitor, and downregulates the gene of the inhibitor of apoptosis (IAP) family, Survivin.64 However, the regulation between ERα and mutp53 is not clear. In our study, the interaction between ERα and mutp53 was found to repress mutp53-dependent lincRNA-p21 expression but to upregulate DDB2 gene expression by binding to EREs within the promoter.
In the results of in situ hybridization staining, lincRNA-p21 had lower expression in tumor tissues of p53-positive/ER-positive breast cancer patients, suggesting that both WTp53 and mutp53 can upregulate lincRNA-p21 expression, and this effect was repressed while ER exists. However, lincRNA-p21 expression in p53-negative tissues was not affected by ER status (Figures 5G and 5H). In addition to p53, hypoxia-inducible factor 1-alpha (HIF-1α) was found to induce lincRNA-p21 expression rapidly in response to hypoxia,66 and inhibitor of growth 1 (ING1b) was demonstrated to promote apoptosis by regulating lincRNA-p21 transcription in both p53 null and p53-expressing cells.27 These findings suggest that ER-regulated lincRNA-p21 expression is p53 specific.
In conclusion, our data indicate the involvement of a non-B DNA structure in mutp53-mediated lincRNA-p21 expression and that the switch of mutp53-mediated gene transcriptions from lincRNAp21 to DDB2 expression by ERα confers chemoresistance (Figure 8). Thus, these findings suggest that the co-treatment of chemotherapy with ER antagonists in a neoadjuvant setting to enhance the chemosensitivity by inducing the lincRNA-p21 level may benefit luminal A/B patients. However, this enhancing effect of ER antagonists on lincRNA-p21 expression may be limited to patients with mutp53-expressing breast cancers. Other strategies to enhance the level of lincRNA-p21 in both mutp53- and WTp53-expressing cancer for chemo-sensitization warrant further investigations.
Materials and methods
lncRNA qPCR microarray and dataset collection
A LncProfiler qPCR Array (RA900A-1) was purchased from SBI (System Biosciences, Mountain View, CA, USA) to determine the expression of 90 human lncRNAs in WTp53- and mutp53-overexpressing cells. The datasets produced in this study are available in the following database: lncRNA qPCR microarray: GEO: GSE159185 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159185).
Microarray datasets from breast cancer patients (GEO: GSE50948, GSE5847, GSE23988, and GSE44272) were subjected to GSEA (https://www.broadinstitute.org/gsea/; Research Resource Identification [RRID]: SCR_003199) to determine the relationship between ER status and enrichment genes in the function of DNA repair and the p53 pathway.
Clinical specimens and primary culture treatments
G∗Power software (RRID: SCR_013726) was used to perform power analysis, and an α error probability of 0.05 and a power level of 0.8 were employed to decide the collection sample size. A total of 61 residual breast tumor tissue specimens with written, informed consents were collected from patients who received surgery for different breast cancer subtypes in Chung Shan Medical University Hospital (Taichung, Taiwan) following the protocol (CS2-18150) approved by the Institute of Research Board Committee. The patient inclusion criteria are as follows: patients diagnosed between 2018 and 2019 at a single tertiary care institution, histological confirmed invasive ductal carcinoma breast cancer, randomly collected and non-selected samples from each molecular subtype, and tumor grade. Only one male patient was enrolled in the collection of breast cancer patients, and thus patient sex is not considered as a biological variable. All tissues were homogenized and cultured with or without carboplatin for 5 days. Following treatment, total RNA and protein lysates were prepared with Trizol Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The correlations among multiple variables, including gender, age, molecular subtypes, tumor types, tumor grade, and Ki67 status, with the expressions of lincRNA-p21 and DDB2 were analyzed.
Tissue microarray and in situ hybridization
Breast cancer tissue microarrays were purchased from SuperBioChips Laboratories (Seoul, Korea) and used to detect lincRNA-p21 for in situ hybridization experiments. Tissue microarray specimens included different breast cancer subtypes from 40 patients. The RNAscope lincRNA-p21 (TP53COR1) probe for use in the in situ hybridization assay was designed and purchased from Advanced Cell Diagnostics (Newark, CA, USA). We used the RNAscope 2.5 HD Detection Kit-BROWN according to the manufacturer’s protocol to screen for lincRNA-p21 signaling in breast cancer tissues. Signals for lincRNA-p21 expression were quantified with Fiji ImageJ (RRID: SCR_002285) and normalized with nuclei to calculate the areas and percentages of probe numbers.
Cell culture
All cell lines were purchased from the American Type Culture Collection (ATCC) and were tested by non-contamination from mycoplasma by the MycoAlert Mycoplasma Detection Kit (LT07-318; Thermo Fisher Scientific, Waltham, MA, USA). Except for the MCF-10A cell line (RRID: CVCL_0598), cell lines HEK293T (RRID: CVCL_0063), HBL-100 (RRID: CVCL_4362), MCF7 (RRID: CVCL_0031), T-47D (RRID: CVCL_0553), BT-474 (RRID: CVCL_0179), SK-BR-3 (RRID: CVCL_0033), MDA-MB-231 (RRID: CVCL_0062), and MDA-MB-468 (RRID: CVCL_0419) were cultured in Dulbecco’s modified Eagle’s medium:nutrient mixture F-12 (DMEM/F12; HyClone, Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and HyClone penicillin-streptomycin solution. The medium for MCF-10A consisted of DMEM/F12 supplemented with 5% horse serum (Gibco), HyClone penicillin-streptomycin solution, and other additives, including 1.05 mM calcium chloride anhydrous (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), 0.1 μg/mL cholera toxin, 10 μg/mL insulin (Sigma-Aldrich), 20 ng/mL human EGF (Sigma-Aldrich), and 0.5 μg/mL hydrocortisone (Sigma-Aldrich). All cells were incubated at 37°C in a humidified incubator containing 5% CO2.
Antibodies
Antibody against p21WAF1 (Calbiochem; OP64, RRID: AB_2335868) was purchased from Merck KGaA (Darmstadt, Germany). Antibodies against p-HER2 (Tyr1221/1222, #2243, RRID: AB_490899), p-ERα (Ser118, #2511, RRID: AB_331289), Ac-p53 (Lys382, #2525S, RRID: AB_330083), DDB2 (#5416, RRID: AB_10694497), PARP (#9542, RRID: AB_2160739), and histone H3 (#9715, RRID: AB_331563) were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies against HER2 (Neu, sc-393712, RRID: AB_2810840), ERα (sc-8002, RRID: AB_627558), p53 (sc-126, RRID: AB_628082), and DDB2 (sc-81246, RRID: AB_2261381) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The antibody against caspase-3 (Imgenex; IMG-144A, RRID: AB_316677) was purchased from Novus Biologicals (Centennial, CO, USA). Antibodies against FLAG M2 (F1804, RRID: AB_262044), α-tubulin (T5168, RRID: AB_477579), and β-actin (A2228, RRID: AB_476697) were purchased from Merck KGaA (Darmstadt, Germany).
Inhibitors and reagents
Carboplatin (41575-94-4), (Z)-4-hydroxy tamoxifen (68047-06-3), NMM (142234-85-3), and tetracycline (64-75-5) were all purchased from Cayman Chemical (Ann Arbor, MI, USA). Cisplatin (P4394), doxorubicin hydrochloride (Sigma-Aldrich; D1515), and auramine (Sigma-Aldrich; 492-80-8) were all purchased from Merck KGaA (Darmstadt, Germany). Clarity, Amersham, or Millipore enhanced chemiluminescence (ECL) was purchased from Bio-Rad Laboratories (Hercules, CA, USA), GE Healthcare Life Sciences (Pittsburgh, PA, USA), or Merck KGaA (Darmstadt, Germany), respectively.
Western blot analysis
Total protein lysate concentration was determined using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA, USA), whereby 30–50 μg of protein lysate was heated at 95°C in the sample buffer for 5 min. Denatured proteins were separated in SDS-PAGE with a running buffer consisting of 25 mM Tris-HCl, 192 mM glycine, and 0.1% SDS, before being transferred to polyvinylidene fluoride (PVDF) membranes (0.45 μM; Millipore, Merck KGaA, Darmstadt, Germany) or NC membranes (0.22 μM, Amersham, GE Healthcare Life Sciences, Pittsburgh, PA, USA) with transfer buffer (700 mL of ddH2O, 200 mL of methanol, and 100 mL of 10× running buffer [250 mM Tris-HCl and 1.92 M glycine]). The transferred membrane was blocked with 5% milk or BSA in Tris-buffered saline-Tween (10 mL of 2 μM Tris-HCl, pH 7.4, 100 mL of 5 M NaCl, 0.5 mL of 100% Tween 20, and 890 mL of ddH2O) and stained with the indicated primary antibodies at 4°C overnight, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. ECL signaling was detected using a ChemiDoc Touch Imaging System (Bio-Rad).67
Plasmids and site-directed mutations
WTp53 was inserted into the pcDNA6A-FLAG plasmid for ectopic protein overexpression, and different p53 mutants were acquired using a site-directed mutation assay (KAPA HiFi HotStart PCR Kit; Kapa Biosystems, Wilmington, MA, USA). To examine the promoter activities of lincRNA-p21 and DDB2, we constructed the promoter regions of both genes into the pGL4.22 (luc2CP/Puro) vector, which was purchased from Promega (Madison, WI, USA). The non-B elements, p53RE, and ER elements were further mutated in the pGL4.22-lincRNA-p21 or pGL4.22-DDB2 promoters for transcriptional activity analysis. ERα genes were constructed into pcDNA3 for overexpression (for details of all site-directed mutation primer sets, see Table S8).
RNA extraction and reverse transcriptase PCR (RT-PCR)
After the indicated treatments, cells were washed three times with ice-cold PBS and lysed with Trizol Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was isolated by adding 0.2 mL of chloroform per 1 mL of Trizol Reagent, followed by centrifugation at 12,000 × g for 15 min to separate the aqueous, interphase, and organic phases. Next, RNA from the aqueous phase was precipitated by mixing with 0.25–0.5 mL of isopropanol, followed by centrifugation at 12,000 × g for 15 min. After removal of the supernatant, the gel-like pellet was washed twice with 1 mL of 75% ethanol, air dried, and then dissolved in DEPC-treated water. The RT-PCR was performed with 1 μg total RNA, Invitrogen M-MLV RT (Thermo Fisher Scientific, Waltham, MA, USA), random hexamer, deoxyribonucleotide triphosphate (dNTP), 5× M-MLV buffer, and DTT.
Quantitative real-time PCR
For quantitative real-time PCR, the KAPA SYBR FAST qPCR Master Mix (2×) Kit (Kapa Biosystems, Wilmington, MA, USA) was used to detect the expression of target genes with specific primers. The threshold cycle (Ct) value was analyzed using the LightCycler 480 Real-Time PCR System (Roche Molecular Systems, Pleasanton, CA, USA) or Applied Biosystems QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). ddCt (RRID: SCR_003396) was calculated with normalization to housekeeping genes as the reference (for details of all primer sets, see Table S8).
Luciferase reporter assay protocol
Cells were seeded into 12-well plates (2 × 105 cells/well) and co-transfected with a reporter gene containing target gene promoters and the β-gal gene, using TransIT-X2 (Mirus Bio, Madison, WI, USA) and Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) transfection reagents for breast cancer cells and HEK293T, respectively. After 6 h of incubation, cells were cultured in a complete medium and treated with the indicated drugs for 48 h. Lysates were harvested, and promoter activity was determined using the Luciferase Assay System (Promega, Madison, WI, USA), whereas firefly luciferase activity was normalized against the β-gal activity.68
ChIP assay protocol
After the indicated treatments, cells were harvested and subjected to the ChIP assay, according to the manufacturer’s instructions (EZ-magna ChIP A/G Chromatin Immunoprecipitation Kit or the HighCell# ChIP Kit). Chromatin fragmentation was performed using micrococcal nuclease (#88216; Thermo Fisher Scientific, Waltham, MA, USA) or chromatin shearing sonicator Bioruptor, while the binding elements for transcription factors were analyzed by quantitative real-time PCR using specific primer sets (see Table S8).
Preparation of biotinylated probes for the promoter region of lincRNA-p21 and biotin pull-down assay
To examine the direct binding of mutp53 to the non-B structure of the lincRNA-p21 promoter, the biotinylated probes containing non-B and p53RE sequences were synthesized by GeneDireX (Taipei, Taiwan). For the in vitro pull-down assay, 3 μg of biotin-labeled DNA was heated to 90°C for 2 min and restructured in structure buffer (10 mM Tris-HCl to pH 7.0, 0.1 M KCl, and 10 mM MgCl2) at room temperature for 20 min. Cells (2 × 107) were treated with or without chemotherapy treatments and then resuspended in nuclear isolation buffer (1.28 M sucrose, 40 mM Tris-HCl, pH 7.5, 20 mM MgCl2, and 4% Triton X-100). The nuclei pellets were hybridized to the folded DNA in RIP buffer (150 mM KCl, 25 mM Tris-HCl, pH 7.4, 0.5 mM DTT, 0.5% NP-40, 1 mM PMSF, and cOmplete Protease Inhibitor Cocktail [1 tablet contains protease inhibitors sufficient for a 10-mL cell extract]; Roche Molecular Systems, Pleasanton, CA, USA) for 1 h; then DNA-protein complexes were pulled down using Novagen streptavidin agarose beads (Novagen, San Diego, CA, USA) and analyzed by western blot (for details of all biotinylated probes, see Table S8).
Cell viability assay protocol
The MTT assay was used to detect cell viability. Cells (5 × 103) grown in 96-well plates were treated with different concentrations of the indicated chemotherapy treatments for 48 or 72 h. The culture medium was then changed to a serum-free medium containing 5× Sigma-Aldrich MTT solution (Merck KGaA, Darmstadt, Germany) and incubated for 3 h. The cells were then lysed with DMSO, and the optical density (OD) at 550 nm was detected by an ELISA reader.69
Comet assay
1% NMA gel (Invitrogen; 15510-027), purchased from Thermo Fisher Scientific (Waltham, MA, USA), was used as the basal layer. 1.5% LMAP gel (J.T.Baker; A426-05), purchased from Capitol Scientific (Austin, TX, USA), was mixed with chemotherapy-treated cells (1 × 105) at a 2:1 ratio as the second layer and then was covered with 1.5% LMAP gel mixed with PBS. Each layer was placed on ice to solidify into a gelatinous mass. The slide was sequentially treated with 50 μM H2O2/PBS for 5 min and freshly prepared lysing solution (2.5 M NaCl, 200 mM NaOH, 100 mM EDTA, 10 mM Tris-HCl, pH 10.0, 1% Triton X-100, and 10% DMSO) overnight at 4°C in darkness to expose DNA. The slides were then soaked in electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 12.0) for 20 min, and the cells migrated in electrophoresis (25 V, 300 mA, for 20 min) to unwind DNA. Slides were subjected to wash with neutralization buffer (0.4 M Tris-HCl) three times to fix double-strand DNA after electrophoresis. DNA was further stained with ethidium bromide (EtBr) and visualized under fluorescence microscopy to observe damaged cells, and the tail moment was calculated using Comet Assay III software.
Flow cytometry protocol
The different stages of apoptotic cells were examined using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Kit (BioVision, Milpitas, CA, USA). Early and late stages of apoptotic cells were stained with Annexin V, and necrotic cells were stained with propidium iodide and then analyzed using the FACSVerse flow cytometer (BD Biosciences, San Jose, CA, USA).
Tet-on system and xenograft mouse model
The cDNA of lincRNA-p21 was constructed into the pAS4.1w.Ppuro-aOn vector and was used for the selection of the T-47D#Tet-On-LincRNA-p21 clone of ER-positive T-47D cancer cells with puromycin treatment. 5-week-old female NOD/SCID mice, purchased from BioLASCO Taiwan (Taipei, Taiwan), were subcutaneously implanted with 60-day release pellets containing 1.7-mg 17β-estradiol (Innovative Research of America) 2 days before the subcutaneous inoculation with the mixture of 0.1 mL 50% PBS plus 50% Matrigel (Corning Life Sciences, Corning, NY, USA) containing 1 × 107 T-47D#Tet-On-LincRNA-p21 cancer cells in the mammary gland. After 1 month of tumor growth, mice were classified into four groups (n = 3): saline, tetracycline, doxorubicin, and tetracycline/doxorubicin combination. Tetracycline (0.2 mg/mL) was prepared in the drinking water for mice, and doxorubicin (2.5 mg/kg) was administered intraperitoneally three times to reach the total dose of 7.5 mg/kg. Tumor diameters were serially measured with calipers, and tumors volume was calculated by the formula: volume = length × width2/2. Mice were sacrificed in a CO2 chamber, and in situ hybridization as well as immunohistochemistry staining were performed to compare the molecular status in these four groups in the end.
Immunohistochemistry assay
Tumor tissues from the T-47D#Tet-On-LincRNA-p21 NOD/SCID mouse model were dewaxed by xylene and rehydrated by 100%, 95%, 80%, and, 75% ethanol and by distilled water. The tissue sections were incubated with DDB2 (1:1,000 dilution; Abcam; ab77765, RRID: AB_1951708), Ki67 (1:200 dilution; Thermo Fisher Scientific; MA5-14520, RRID: AB_10979488), or cleaved caspase-3 (1:300 dilution; Cell Signaling Technology; #9661) antibodies overnight, followed by the staining with a polymer HRP-conjugated secondary antibody for 10 min according to the protocol of the UltraVision Quanto detection system (TL-060-QHD; Thermo Fisher Scientific, Waltham, MA, USA). Finally, these tissue slides were stained with hematoxylin for 5 min and dehydrated.70
Statistical analysis
The difference between two categorical variables was analyzed using the Student’s t test or Welch’s two-sample t test, whereas differences among more than two category variables were analyzed by one-way ANOVA. The results are presented as the mean ± SD, n ≥ 3. The p value was calculated with the two-tailed test, and a statistically significant difference was defined as p < 0.05. Pearson’s chi-square test was used to analyze characteristics of breast cancer patients, and unconditional logistic regression was used to obtain the odds ratio. All statistical analysis was performed using SigmaPlot 10.0 (RRID: SCR_003210), GraphPad Prism 8 (RRID: SCR_002798), or SPSS 21 software (RRID: SCR_002865).
Acknowledgments
We would like to thank Iona J. MacDonald from the China Medical University for her English language revision of this manuscript. Experiments and data analysis were performed, in part, through the use of the Medical Research Core Facilities, Office of Research & Development, at the China Medical University, Taichung, Taiwan. This work was supported by grants from the Ministry of Science and Technology of Taiwan (grant number [no.] MOST 105-2314-B-040-011-MY3 to M.-H.Y. and grant nos. MOST 105-2320-B-039-056-MY3 and MOST 108-2314-B-039-032 to W.-C.H.), from China Medical University (grant no. CMU106-ASIA-18 to W.-C.H.), and from China Medical University Hospital (grant nos. DMR-109-021, DMR-109-022, and DMR-109-213 to W.-C.H.). The following was funding for the open access charge: China Medical University. This work was also financially supported by the “Drug Development Center, China Medical University” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author contributions
Y.-H.H. conceived the study, designed and performed the experiments, analyzed data, interpreted results, and wrote the manuscript. M.-H.Y., L.-C.L., and C.-H.H. provided human breast cancer tissue specimens and clinical information. H.-F.C. supervised experimental designs and performed experiments. T.-S.W., R.-H.W., Y.C., and J.Y. provided the protocol, materials, microscope, and software for the comet assay and tissue slide scanner. Y.-L.W., T.K.H., D.-W.H., F.-J.C., J.-Y.C., and S.-W.H. performed experiments. F.-J.C., W.-C.C., and P.-C.S. carried out the GSEA computational analysis. R.-H.W. and C.-C.H. provided statistical concepts for the clinical analysis. Y.-J.C. revised the manuscript. W.-C.H. conceived and supervised the entire project, designed experiments, interpreted results, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.022.
Supplemental information
References
- 1.Hafner A., Bulyk M.L., Jambhekar A., Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 2.Ali Syeda Z., Langden S.S.S., Munkhzul C., Lee M., Song S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020;21:1723. doi: 10.3390/ijms21051723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silwal-Pandit L., Vollan H.K., Chin S.F., Rueda O.M., McKinney S., Osako T., Quigley D.A., Kristensen V.N., Aparicio S., Børresen-Dale A.L. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. 2014;20:3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 4.Hientz K., Mohr A., Bhakta-Guha D., Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2017;8:8921–8946. doi: 10.18632/oncotarget.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosh R., Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 7.Pfister N.T., Prives C. Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53. Cold Spring Harb. Perspect. Med. 2017;7:a026054. doi: 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brázdová M., Quante T., Tögel L., Walter K., Loscher C., Tichý V., Cincárová L., Deppert W., Tolstonog G.V. Modulation of gene expression in U251 glioblastoma cells by binding of mutant p53 R273H to intronic and intergenic sequences. Nucleic Acids Res. 2009;37:1486–1500. doi: 10.1093/nar/gkn1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brázda V., Coufal J. Recognition of Local DNA Structures by p53 Protein. Int. J. Mol. Sci. 2017;18:375. doi: 10.3390/ijms18020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz-Heddergott R., Moll U.M. Gain-of-Function (GOF) Mutant p53 as Actionable Therapeutic Target. Cancers (Basel) 2018;10:188. doi: 10.3390/cancers10060188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donehower L.A. Insights into wild-type and mutant p53 functions provided by genetically engineered mice. Hum. Mutat. 2014;35:715–727. doi: 10.1002/humu.22507. [DOI] [PubMed] [Google Scholar]
- 12.Kandioler D., Schoppmann S.F., Zwrtek R., Kappel S., Wolf B., Mittlböck M., Kührer I., Hejna M., Pluschnig U., Ba-Ssalamah A. The biomarker TP53 divides patients with neoadjuvantly treated esophageal cancer into 2 subgroups with markedly different outcomes. A p53 Research Group study. J. Thorac. Cardiovasc. Surg. 2014;148:2280–2286. doi: 10.1016/j.jtcvs.2014.06.079. [DOI] [PubMed] [Google Scholar]
- 13.Young K.H., Leroy K., Møller M.B., Colleoni G.W., Sánchez-Beato M., Kerbauy F.R., Haioun C., Eickhoff J.C., Young A.H., Gaulard P. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood. 2008;112:3088–3098. doi: 10.1182/blood-2008-01-129783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plimack E.R., Hoffman-Censits J.H., Viterbo R., Trabulsi E.J., Ross E.A., Greenberg R.E., Chen D.Y., Lallas C.D., Wong Y.N., Lin J. Accelerated methotrexate, vinblastine, doxorubicin, and cisplatin is safe, effective, and efficient neoadjuvant treatment for muscle-invasive bladder cancer: results of a multicenter phase II study with molecular correlates of response and toxicity. J. Clin. Oncol. 2014;32:1895–1901. doi: 10.1200/JCO.2013.53.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez D., Martinez P., Wade R., Hockley S., Oscier D., Matutes E., Dearden C.E., Richards S.M., Catovsky D., Morgan G.J. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J. Clin. Oncol. 2011;29:2223–2229. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 16.He C., Li L., Guan X., Xiong L., Miao X. Mutant p53 Gain of Function and Chemoresistance: The Role of Mutant p53 in Response to Clinical Chemotherapy. Chemotherapy. 2017;62:43–53. doi: 10.1159/000446361. [DOI] [PubMed] [Google Scholar]
- 17.Konduri S.D., Medisetty R., Liu W., Kaipparettu B.A., Srivastava P., Brauch H., Fritz P., Swetzig W.M., Gardner A.E., Khan S.A., Das G.M. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. USA. 2010;107:15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 21.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marín-Béjar O., Marchese F.P., Athie A., Sánchez Y., González J., Segura V., Huang L., Moreno I., Navarro A., Monzó M. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barsotti A.M., Beckerman R., Laptenko O., Huppi K., Caplen N.J., Prives C. p53-Dependent induction of PVT1 and miR-1204. J. Biol. Chem. 2012;287:2509–2519. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Fang L., Jiang J., Kuang Y., Wang B., Shang X., Han P., Li Y., Liu M., Zhang Z., Li P. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. 2018;9:1103. doi: 10.1038/s41419-018-1148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang A., Zhou N., Huang J., Liu Q., Fukuda K., Ma D., Lu Z., Bai C., Watabe K., Mo Y.Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran U.M., Rajarajacholan U., Soh J., Kim T.S., Thalappilly S., Sensen C.W., Riabowol K. LincRNA-p21 acts as a mediator of ING1b-induced apoptosis. Cell Death Dis. 2015;6:e1668. doi: 10.1038/cddis.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S., Yang X., Li J., Yang W., Ma H., Zhang Z. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol. Cancer. 2019;18:38. doi: 10.1186/s12943-019-0993-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y., Li Y., Sheng J., Wu F., Li K., Huang R., Wang X., Jiao T., Guan X., Lu Y. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J. Exp. Clin. Cancer Res. 2019;38:379. doi: 10.1186/s13046-019-1375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai H., Fesler A., Schee K., Fodstad O., Flatmark K., Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin. Colorectal Cancer. 2013;12:261–266. doi: 10.1016/j.clcc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Castellano J.J., Navarro A., Viñolas N., Marrades R.M., Moises J., Cordeiro A., Saco A., Muñoz C., Fuster D., Molins L. LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation. J. Thorac. Oncol. 2016;11:2173–2182. doi: 10.1016/j.jtho.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Germano G., Amirouchene-Angelozzi N., Rospo G., Bardelli A. The Clinical Impact of the Genomic Landscape of Mismatch Repair-Deficient Cancers. Cancer Discov. 2018;8:1518–1528. doi: 10.1158/2159-8290.CD-18-0150. [DOI] [PubMed] [Google Scholar]
- 33.Sommer S., Fuqua S.A. Estrogen receptor and breast cancer. Semin. Cancer Biol. 2001;11:339–352. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 34.Caldon C.E. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front. Oncol. 2014;4:106. doi: 10.3389/fonc.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C.Y., Jiang Z.N., Zhou Y., Li J.J., Huang L.M. Estrogen receptor α roles in breast cancer chemoresistance. Asian Pac. J. Cancer Prev. 2013;14:4049–4052. doi: 10.7314/apjcp.2013.14.7.4049. [DOI] [PubMed] [Google Scholar]
- 36.Liu W., Konduri S.D., Bansal S., Nayak B.K., Rajasekaran S.A., Karuppayil S.M., Rajasekaran A.K., Das G.M. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J. Biol. Chem. 2006;281:9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- 37.Tabuchi Y., Matsuoka J., Gunduz M., Imada T., Ono R., Ito M., Motoki T., Yamatsuji T., Shirakawa Y., Takaoka M. Resistance to paclitaxel therapy is related with Bcl-2 expression through an estrogen receptor mediated pathway in breast cancer. Int. J. Oncol. 2009;34:313–319. [PubMed] [Google Scholar]
- 38.Tokuda E., Seino Y., Arakawa A., Saito M., Kasumi F., Hayashi S., Yamaguchi Y. Estrogen receptor-α directly regulates sensitivity to paclitaxel in neoadjuvant chemotherapy for breast cancer. Breast Cancer Res. Treat. 2012;133:427–436. doi: 10.1007/s10549-011-1758-x. [DOI] [PubMed] [Google Scholar]
- 39.Moutsatsou P. The spectrum of phytoestrogens in nature: our knowledge is expanding. Hormones (Athens) 2007;6:173–193. [PubMed] [Google Scholar]
- 40.Sayeed A., Konduri S.D., Liu W., Bansal S., Li F., Das G.M. Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res. 2007;67:7746–7755. doi: 10.1158/0008-5472.CAN-06-3724. [DOI] [PubMed] [Google Scholar]
- 41.Simoncini T., Hafezi-Moghadam A., Brazil D.P., Ley K., Chin W.W., Liao J.K. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu D., Shi Y., Liu J.B., Wu T.M., Jia C.Y., Yang H.Q., Zhang D.D., Yang X.L., Wang H.M., Ma Y.S. Targeting Long Non-coding RNA to Therapeutically Regulate Gene Expression in Cancer. Mol. Ther. Nucleic Acids. 2020;21:712–724. doi: 10.1016/j.omtn.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quante T., Otto B., Brázdová M., Kejnovská I., Deppert W., Tolstonog G.V. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle. 2012;11:3290–3303. doi: 10.4161/cc.21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013;41:D94–D100. doi: 10.1093/nar/gks955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicoludis J.M., Barrett S.P., Mergny J.L., Yatsunyk L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tung J.C., Huang W.C., Yang J.C., Chen G.Y., Fan C.C., Chien Y.C., Lin P.S., Candice Lung S.C., Chang W.C. Auramine O, an incense smoke ingredient, promotes lung cancer malignancy. Environ. Toxicol. 2017;32:2379–2391. doi: 10.1002/tox.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duffy M.J., Synnott N.C., Crown J. Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res. Treat. 2018;170:213–219. doi: 10.1007/s10549-018-4753-7. [DOI] [PubMed] [Google Scholar]
- 48.Coates A.S., Millar E.K., O’Toole S.A., Molloy T.J., Viale G., Goldhirsch A., Regan M.M., Gelber R.D., Sun Z., Castiglione-Gertsch M. Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX. Breast Cancer Res. 2012;14:R143. doi: 10.1186/bcr3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiu K., Iwamoto T., Kelly C.M., Watanabe N., Motoki T., Ito M., Ohtani S., Higaki K., Imada T., Yuasa T. Neoadjuvant Chemotherapy with or without Concurrent Hormone Therapy in Estrogen Receptor-Positive Breast Cancer: NACED-Randomized Multicenter Phase II Trial. Acta Med. Okayama. 2015;69:291–299. doi: 10.18926/AMO/53675. [DOI] [PubMed] [Google Scholar]
- 50.Yu K.D., Wu S.Y., Liu G.Y., Wu J., Di G.H., Hu Z., Hou Y.F., Chen C.M., Fan L., Tang L.C. Concurrent neoadjuvant chemotherapy and estrogen deprivation in patients with estrogen receptor-positive, human epidermal growth factor receptor 2-negative breast cancer (CBCSG-036): A randomized, controlled, multicenter trial. Cancer. 2019;125:2185–2193. doi: 10.1002/cncr.32057. [DOI] [PubMed] [Google Scholar]
- 51.Matsunuma R., Watanabe T., Hozumi Y., Koizumi K., Ito Y., Maruyama S., Ogura H., Goto K., Mori H., Sawai N., Shiiya N. Preoperative concurrent endocrine therapy with chemotherapy in luminal B-like breast cancer. Breast Cancer. 2020;27:819–827. doi: 10.1007/s12282-020-01077-0. [DOI] [PubMed] [Google Scholar]
- 52.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K., Ward A.J., Raj A., Lee J.T., Sharp P.A., Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall J.R., Messenger Z.J., Tam H.W., Phillips S.L., Recio L., Smart R.C. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:e1700. doi: 10.1038/cddis.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blokhin I., Khorkova O., Hsiao J., Wahlestedt C. Developments in lncRNA drug discovery: where are we heading? Expert Opin. Drug Discov. 2018;13:837–849. doi: 10.1080/17460441.2018.1501024. [DOI] [PubMed] [Google Scholar]
- 56.Vicentini C., Galuppini F., Corbo V., Fassan M. Current role of non-coding RNAs in the clinical setting. Noncoding RNA Res. 2019;4:82–85. doi: 10.1016/j.ncrna.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian Y., Shi L., Luo Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. (Lausanne) 2020;7:612393. doi: 10.3389/fmed.2020.612393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47(D1):D1034–D1037. doi: 10.1093/nar/gky905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Göhler T., Jäger S., Warnecke G., Yasuda H., Kim E., Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kattan Z., Marchal S., Brunner E., Ramacci C., Leroux A., Merlin J.L., Domenjoud L., Dauça M., Becuwe P. Damaged DNA binding protein 2 plays a role in breast cancer cell growth. PLoS ONE. 2008;3:e2002. doi: 10.1371/journal.pone.0002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagy Á., Győrffy B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer. 2021;148:502–511. doi: 10.1002/ijc.33283. [DOI] [PubMed] [Google Scholar]
- 62.Fekete J.T., Győrffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer. 2019;145:3140–3151. doi: 10.1002/ijc.32369. [DOI] [PubMed] [Google Scholar]
- 63.Bailey S.T., Shin H., Westerling T., Liu X.S., Brown M. Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:18060–18065. doi: 10.1073/pnas.1018858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu W., Ip M.M., Podgorsak M.B., Das G.M. Disruption of estrogen receptor alpha-p53 interaction in breast tumors: a novel mechanism underlying the anti-tumor effect of radiation therapy. Breast Cancer Res. Treat. 2009;115:43–50. doi: 10.1007/s10549-008-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGowan E.M., Lin Y., Hatoum D. Good Guy or Bad Guy? The Duality of Wild-Type p53 in Hormone-Dependent Breast Cancer Origin, Treatment, and Recurrence. Cancers (Basel) 2018;10:172. doi: 10.3390/cancers10060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 67.Liu S.-C., Tsai C.-H., Wu T.-Y., Tsai C.-H., Tsai F.-J., Chung J.-G., Huang C.-Y., Yang J.-S., Hsu Y.-M., Yin M.-C. Soya-cerebroside reduces IL-1β-induced MMP-1 production in chondrocytes and inhibits cartilage degradation: implications for the treatment of osteoarthritis. Food Agric. Immunol. 2019;30:620–632. [Google Scholar]
- 68.Cheng F.-J., Huynh T.-K., Yang C.-S., Hu D.-W., Shen Y.-C., Tu C.-Y. Hesperidin is a potential inhibitor against SARS-CoV-2 infection. Nutrients. 2021;13:2800. doi: 10.3390/nu13082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee H.-P., Chen P.-C., Wang S.-W., Fong Y.-C., Tsai C.-H., Tsai F.-J., Chung J.-G., Huang C.-Y., Yang J.-S., Hsu Y.-M. Plumbagin suppresses endothelial progenitor cell-related angiogenesis in vitro and in vivo. J. Funct. Foods. 2019;52:537–544. [Google Scholar]
- 70.Lee H.-P., Liu S.-C., Wang Y.-H., Chen B.-C., Chen H.-T., Li T.-M. Cordycerebroside A suppresses VCAM-dependent monocyte adhesion in osteoarthritis synovial fibroblasts by inhibiting MEK/ERK/AP-1 signaling. J. Funct. Foods. 2021;86:104712. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.