Abstract
The CRISPR-Cas9 technology represents a powerful tool for genome engineering in eukaryotic cells, advancing both fundamental research and therapeutic strategies. Despite the enormous potential of the technology, efficient and direct intracellular delivery of Cas9 ribonucleoprotein (RNP) complexes in target cells poses a significant hurdle, especially in refractive primary cells. In the present work, vapor nanobubble (VNB) photoporation was explored for Cas9 RNP transfection in a variety of cell types. Proof of concept was first demonstrated in H1299-EGFP cells, before proceeding to hard-to-transfect stem cells and T cells. Gene knock-out levels over 80% and up to 60% were obtained for H1299 cells and mesenchymal stem cells, respectively. In these cell types, the unique possibility of VNB photoporation to knock out genes according to user-defined spatial patterns was demonstrated as well. Next, effective targeting of the programmed cell death 1 receptor and Wiskott-Aldrich syndrome gene in primary human T cells was demonstrated, reaching gene knock-out levels of 25% and 34%, respectively. With a throughput of >200,000 T cells per second, VNB photoporation is a scalable and versatile intracellular delivery method that holds great promise for CRISPR-Cas9-mediated ex vivo engineering of cell therapy products.
Keywords: CRISPR-Cas9, gene editing, T cells, stem cells, photoporation, intracellular delivery
Graphical abstract

Gold nanoparticle (AuNP)-sensitized vapor nanobubble (VNB) photoporation represents a promising technique for intracellular delivery of Cas9 RNPs in hard-to-transfect stem cells and human T cells. With advantages such as broad applicability across different cell types, low cytotoxicity, spatial selectivity, and high throughput, the technology holds great potential for research and therapeutic applications.
Introduction
The CRISPR-CRISPR-associated protein (Cas9) technology has immensely expanded our capability to manipulate genes in a precise and targeted manner. Originally discovered as an adaptive immune system in Bacteria and Archaea against viral infections and foreign plasmid DNA,1 the technology has been efficiently repurposed for targeted genome editing in eukaryotic cells.2,3 Highly specific gene editing by the Cas9 endonuclease is obtained in combination with a guide RNA (gRNA), either as CRISPR RNA (crRNA):trans-activating crRNA (tracrRNA) duplex, or single gRNA (sgRNA), which is complementary to the target gene sequence. In turn, double-strand breaks induced by Cas9 can be repaired by two cell-endogenous repair systems: non-homologous end joining (NHEJ), causing insertions or deletions (indels), or homology-directed repair (HDR) in the presence of a homologous donor template. With this, the CRISPR-Cas9 technology has the exciting potential to correct genetic defects or to introduce novel gene functions. Combined with its simplicity and versatility, it has become the tool of choice for a broad variety of applications ranging from basic research to therapeutic cell engineering.4
Successful application of this gene-editing technology relies on the extent to which CRISPR-Cas9 components can be delivered into the cells of interest. Cas9 and gRNA can be introduced as a pre-assembled ribonucleoprotein (RNP) complex, as a plasmid DNA encoding for both components, or as a combination of the gRNA with mRNA encoding for Cas9.4, 5, 6 Of those, the preformed Cas9 RNP complex is generally preferred because of its transient exposure and hence reduced risk for potentially oncogenic off-target DNA cleavages.7 However, efficient delivery of Cas9 RNP complexes still represents a significant challenge, especially in primary difficult-to-transfect cells. Non-viral, physical transfection methods have attracted quite some attention for this, as they offer excellent flexibility in terms of the type of molecules and cell types for which they can be used. However, none of the classical physical approaches offers the combination of (1) high knock-out levels, (2) low cytotoxicity, (3) high throughput, (4) spatial control, and (5) applicability to both adherent and suspension cells. As such, the need remains for innovative transfection technologies that are suitable for fast and safe Cas9 RNP delivery in a broad array of ex vivo cell-engineering applications.6
Vapor nanobubble (VNB) photoporation is one such promising upcoming technique for gentle but efficient intracellular delivery of macromolecules in a broad range of hard-to-transfect cell types.8, 9, 10, 11, 12 It is based on a combination of pulsed laser irradiation and sensitizing nanoparticles, most commonly gold nanoparticles (AuNPs), to induce transient permeabilization of the cell membrane with nanoscale precision. If the laser pulse energy is sufficiently high, then VNBs can emerge from the AuNPs by rapid evaporation of the surrounding liquid as a result of photothermal heating. By rapid expansion and the inevitable collapse of the VNB, local high-pressure shockwaves can induce mechanical poration of the cell membrane.13 Through these transient membrane pores, extracellular biological macromolecules such as Cas9 RNPs can then enter the cell cytoplasm (Figure 1). Conveniently, being an optical technology, it is readily compatible with most traditionally used cell culture recipients as long as they are optically transparent. As such, it can be applied to both adherent and suspension cells with minimal cell manipulations.13, 14, 15 A unique feature is that the technology enables even single-cell transfections or spatially patterned transfections by appropriate scanning of the activating laser beam.8,10,16
Figure 1.
Vapor nanobubble (VNB) photoporation procedure for intracellular delivery of Cas9 ribonucleoproteins (RNPs)
Cells are first incubated with cationic plasmonic gold nanoparticles (AuNPs), after which, unbound AuNPs are washed away, and Cas9 RNPs are supplemented to the cell medium. Upon pulsed laser irradiation, VNBs will be generated from cell-attached AuNPs, leading to the formation of pores in the cell membrane and allowing the extracellular Cas9 RNPs to access the cell’s interior. Once in the cytosol, the nuclear localization sequence (NLS) attached to Cas9 will induce nuclear import of the Cas9 RNP, and target gene-specific editing is initiated.
Capitalizing on recent studies demonstrating safe and efficient intracellular delivery of a wide variety of molecules by VNB photoporation,9,12,17, 18, 19 we here investigate if the technique is suitable for gene editing in hard-to-transfect cell types via intracellular delivery of Cas9 RNPs. As a first proof of concept, EGFP knock-out is demonstrated with >80% efficiency in H1299-EGFP, while maintaining a high level of cell viability. Next, primary hard-to-transfect cells with therapeutic relevance are included, showing effective CRISPR-Cas9 gene editing by photoporation in mesenchymal stem cells (MSCs) and primary T cells. Successful gene editing was achieved in murine MSCs (mMSCs) with up to 60% knock-out efficiency and a cell viability of 87%. In primary human T cells, editing levels up to 25% and 34% were obtained for the clinically relevant Programmed Cell Death 1 (PD-1) immune checkpoint receptor and Wiskott-Aldrich syndrome (WAS) gene, respectively, while maintaining a cell viability >70%. Altogether, this work establishes VNB photoporation as a promising tool for intracellular delivery of Cas9 RNPs in hard-to-transfect cells, such as stem cells and T cells, which is expected to be of benefit to therapeutic cell engineering and research applications.
Results
VNB photoporation of Cas9 RNPs enables highly efficient H1299-EGFP knock-out even at low nanomolar concentrations
In this study, we aim to evaluate VNB photoporation as a promising gentle approach for Cas9 RNP transfection and effective gene editing. For a first proof of concept, a lung epithelial cell line that stably expresses EGFP (H1299-EGFP) was used (Figure 2A). Photoporation requires optimization of the AuNP concentration and laser fluence in order to achieve optimal intracellular delivery for a given cell type with minimal loss in cell viability. Alexa Fluor 647 10 kDa dextran (RD10) was used as a model macromolecule in which its intracellular delivery can be easily verified by confocal fluorescence microscopy (Figure S1A) and quantified by flow cytometry (Figure S1B) and was done as a function of increasing AuNP concentrations (2−32 × 107 AuNP/mL) and laser pulse fluences (λ = 532 nm; E3 = 0.84 J/cm2, E4 = 1.26 J/cm2, E5 = 1.60 J/cm2). Cell viability was assessed in parallel using a CellTiter-Glo assay, which assesses the cell’s metabolic activity (Figure S1C). Both the percentage of RD10-positive cells and the relative mean fluorescence intensity (rMFI) per cell were found to increase for increasing AuNP concentration and laser fluences. This was generally accompanied by a slight decrease in cell viability, as can be expected. The best delivery efficiency was obtained for 16 × 107 AuNPs/mL and laser fluence E5 (=1.60 J/cm2), reaching 96% RD10-positive cells. Since the corresponding cell viability was still above 80%, this condition was selected for further experiments with Cas9 RNPs.
Figure 2.
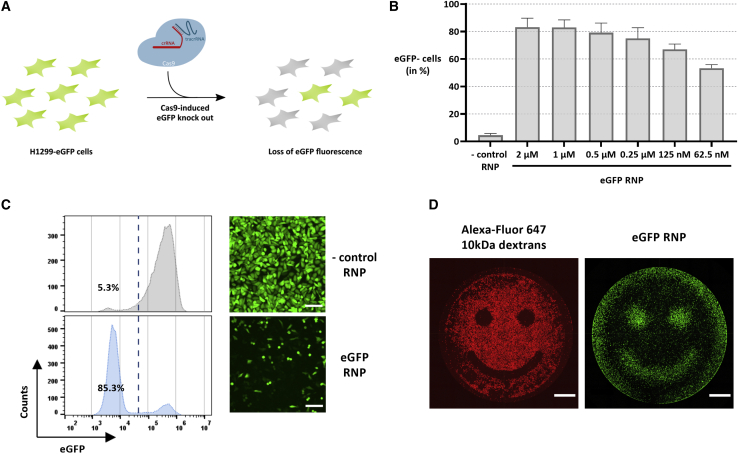
VNB photoporation enables highly efficient and spatial-selective H1299-EGFP knock-out
(A) Schematic depiction of the experimental set-up for Cas9-induced EGFP knock-out. (B) H1299-EGFP knock-out levels after photoporation in the presence of decreasing concentrations of EGFP RNPs (16 × 107 AuNPs/mL, E5; n = 3, mean ± SD). (C) Flow cytometry histograms and confocal microscopy images of a representative experiment on H1299-EGFP cells photoporated in the presence of 1 μM negative control RNPs (non-targeting crRNA) or EGFP-targeting RNPs (16 × 107 AuNPs/mL, E5; scale bars, 150 μm). (D) Spatial-selective photoporation of 0.05 mg/mL Alexa Fluor 647 10 kDa dextran or 1 μM EGFP RNPs (16 × 107 AuNPs/mL, E5). The image with Alexa Fluor 647 10 kDa dextran was recorded 2 h after photoporation, whereas the EGFP image was recorded after 48 h (scale bars, 1,000 μm).
To achieve effective EGFP knock-out, we first screened a set of crRNAs (Table S1), which according to literature, all target the EGFP gene but at different sites (Figure S2A). The screening was performed with commercial transfection reagents RNAiMAX and JetCRISPR with which Cas9 RNP complexes were prepared at a 2.5 molar excess of gRNA to the Cas9 protein (i.e., ratio 2.5:1), similar to earlier studies.20, 21, 22 Knock-out of the EGFP gene was evaluated 48 h after Cas9 RNP transfection by confocal microscopy (Figures S2B and S2C) and flow cytometry (Figure S2D). Three of the tested crRNAs (crRNA_1, crRNA_2, and crRNA_4) showed effective EGFP knock-out, from which crRNA_4 was selected for further experiments, as it returned the best results with 43% EGFP-negative cells in combination with RNAiMAX. Having optimized the photoporation parameters and having selected the optimal gRNA, H1299-EGFP cells were photoporated in the presence of EGFP-targeting Cas9 RNPs. After 48h, EGFP knock-out levels were observed in a RNP concentration-dependent manner (Figures 2B and S3). Representative confocal microscopy images and flow cytometry histograms are shown in Figure 2C for 1 μM RNPs. Whereas knock-out percentages initially increased for increasing RNP concentrations, they remained constant at around 80% from 0.25 μM to 2 μM RNP. To verify EGFP knock-out at the genomic level, we performed a PCR amplification of the EGFP locus and analyzed the obtained PCR amplicons by Sanger sequencing (Figure S4). Note that crRNA_2 was chosen for this specific experiment, as it targets the EGFP gene more central in the PCR amplicon, thereby simplifying sequence analysis of the cleavage site region. By comparison with a PCR amplicon of cells photoporated with a negative control RNP, tracking of indels by decomposition (TIDE) analysis affirmed that an indel frequency of 82.2% could be obtained using 1 μM EGFP-targeting RNPs. Based on these results, to ensure efficient gene editing even in difficult-to-transfect cell types further on, a Cas9 RNP concentration of at least 1 μM was used in all subsequent experiments.
A unique feature of VNB photoporation is the possibility for spatial-selective intracellular delivery in cell subpopulations within a culture, even with single-cell resolution.16 This spatial selectivity is achieved by only irradiating the cells of interest with the photoporation laser beam. An example of this is shown in Figure 2D, where H1299-EGFP cells were photoporated according to a pre-defined “smiley” pattern with Alexa Fluor 647 10 kDa dextran or EGFP-targeting Cas9 RNPs. Whereas the 10-kDa dextran results in increased fluorescence in photoporated cells, Cas9 RNPs result in a loss of EGFP fluorescence at the same places. This demonstrates the possibility to transfect cells according to user-defined patterns, which opens up future possibilities to engineer tissues according to complex pre-defined patterns.16
Effective gene knock-out in mMSCs
In analogy with the experiments on H1299-EGFP cells, mMSCs stably expressing the EGFP gene (mMSC-EGFP) were first photoporated with Alexa Fluor 647 10 kDa dextrans (RD10) to optimize the AuNP concentration (4−32 × 107 AuNPs/mL) and laser fluence settings (E3 = 0.84 J/cm2 and E5 = 1.60 J/cm2). Cell viability was evaluated in parallel using a CellTiter-Glo assay, again showing that higher AuNP concentrations lead to better delivery efficiency at the expense of cell viability (Figure S5A). Multiplication of cell viability with transfection efficiency gives the transfection yield (=percentage of living transfected cells compared to the initial cell population). As shown in Figure S5B, the transfection yield was fairly constant over the tested range of AuNP concentrations and laser fluences, with an apparent maximum for an AuNP concentration of 8 × 107 AuNPs/mL and laser fluence E5. Therefore, we chose to continue with AuNP concentrations 8 × 107 AuNPs/mL and 16 × 107 AuNPs/mL and laser fluence E5 (=1.60 J/cm2). Following photoporation, cells were allowed to grow for 7 days prior to analysis of EGFP knock-out by confocal microscopy and flow cytometry, whereas cell viability was assessed both 24 h and 7 days after transfection (Figure 3A). At both AuNP concentrations around 55%–60%, EGFP-negative cells were obtained with a cell viability of ∼90% (24 h) and ∼98% (7 days) (Figure 3B), confirming the feasibility of Cas9-mediated gene editing of stem cells with photoporation. Exemplary confocal microscopy images and flow cytometry histograms are shown in Figure 3C. Finally, to demonstrate once more the unique ability to perform gene editing in a spatially resolved manner, mMSC-EGFP cells were photoporated according to a square pattern in the presence of Alexa Fluor 647 10 kDa dextran or EGFP-targeting Cas9 RNPs (Figure 3D).
Figure 3.
Effective and spatial-selective murine mesenchymal stem cell stably expressing EGFP (mMSC-EGFP) knock-out by VNB photoporation
mMSC-EGFPs were photoporated with 1 μM EGFP-targeting RNPs (crRNA_4) or negative control RNPs (non-targeting crRNA). (A) Schematic overview of the experimental set-up and timeline. (B) mMSC-EGFP knock-out levels determined 7 days after photoporation by flow cytometry, whereas cell viability was assessed 24 h and 7 days after transfection using a CellTiter-Glo viability assay (n ≥ 2, mean ± SD). (C) Flow cytometry histograms and confocal microscopy images of a representative experiment on mMSC-EGFP cells photoporated in the presence of 1 μM negative control RNPs or EGFP-targeting RNPs (16 × 107 AuNPs/mL, E5; scale bars, 300 μm). (D) Spatial-selective intracellular delivery of Alexa Fluor 647 10 kDa dextran (0.05 mg/mL, readout after 2 h) or EGFP RNPs (1 μM, 16 × 107 AuNPs/mL, E5). The laser-irradiated region is indicated by the white dotted squares (scale bars, 1,000 μm).
Editing of WAS and PD-1 genes in primary human T cells by VNB photoporation
Following the successful experiments on adherent H1299-EGFP and mMSC-EGFP cells, we investigated the potential of VNB photoporation for delivery of Cas9 RNPs and target gene editing in primary human T cells. First, different AuNP concentrations and laser fluences were screened for efficient T cell transfection and minimal cytotoxicity. Effective intracellular delivery was evaluated by flow cytometry using fluorescein isothiocyanate (FITC)-labeled dextran of 250 kDa (FD250) as a model macromolecule, whereas in parallel, cell viability was assessed using a CellTiter-Glo assay (Figure S6A). Again, we found that the delivery efficiency increases with increasing AuNP concentration and laser fluence, at the expense of a gradual decrease in cell viability. Based on the percentage of positive cells and rMFI values (i.e., the average FD250 fluorescence per cell), it was noted that laser fluence E4 (=1.26 J/cm2) performed slightly but systematically better than E3 (=0.84 J/cm2). Increasing the laser fluence further to E5 (=1.60 J/cm2) did not provide further improvement, so that E4 was selected for further experiments on human T cells. To determine what is the most optimal AuNP concentration in combination with this laser fluence, we calculated the transfected cell yield, i.e., the percentage of cells that are both alive and transfected (relative to the initial cell population). From this, it became clear that an optimal AuNP concentration must be situated between 1 × 108 and 2 × 108 AuNPs/mL (Figure S6B). We, therefore, decided to proceed with laser fluence E4 and two AuNP concentrations of 1.1 × 108 and 1.3 × 108 AuNPs/mL, which should provide a good trade-off between delivery efficiency and cytotoxicity.
Having determined suitable photoporation conditions for treatment of human T cells, we next attempted gene editing in primary human T cells, first by targeting the WAS gene. T cells were first collected from donors and culture expanded for 10 days, resulting in a CD3+ T cell purity of 95% (Figure S7). Next, photoporation-mediated intracellular delivery of WAS targeting Cas9 RNPs was performed (Figure 4A). After an additional 11 days of culture to wash out the preformed WAS protein (WASp), the WAS gene-editing efficiency was assessed. Based on intracellular WASp immunostaining and flow cytometry analysis, this resulted in a WAS knock-out level of ∼20% and ∼34% for 1.1 and 1.3 × 108 AuNP/mL, respectively, which remained virtually the same when increasing the RNP concentration from 1.8 to 3.6 μM (Figure 4B). Interestingly, WAS knock-out levels were identical in CD4+ and CD8+ T cells (Figures 4B and S8).
Figure 4.
Wiskott-Aldrich syndrome (WAS) gene editing in primary human T cells by photoporation-mediated Cas9 RNP transfection
(A) Timeline detailing the workflow for induction and detection of WAS gene knock-outs. (B) T cells were photoporated (1.1 × 108 or 1.3 × 108 AuNPs/mL, E4) in the presence of 1.8 μM or 3.6 μM Cas9 RNP targeting the WAS gene or negative control RNPs (1.8 μM, ctrl, non-targeting crRNA). WAS protein (WASp) protein levels were determined 11 days after Cas9 RNP transfection by immunostaining of the intracellular WASp and readout by flow cytometry (n = 3 donors; mean ± SD). Paired Student’s t tests were performed to determine statistical differences between CD4+ and CD8+ T cell subsets (ns, non-significant). (C) Viability of human T cells 2 h and 48 h after WAS Cas9 RNP transfection, as determined by CellTiter-Glo assay. (D) Indel percentages for the tested conditions, as determined by Sanger sequencing and subsequent TIDE analysis (n = 2 donors; mean ± SD).
The cell viability for an AuNP concentration of 1.1 × 108 AuNP/mL was ∼80% after 2 h and 48 h (Figure 4C). For 1.3 × 108 AuNP/mL, cell viability was ∼70%, even though the difference with 1.1 × 108 AuNP/mL was not significant. The results at the protein level were largely confirmed at the genomic level: Sanger sequencing of the targeted exon and subsequent TIDE analysis was performed and showed similar percentages (Figure 4D). The percentages measured at the genomic level nicely correlated with the protein measurements but were generally somewhat lower, possibly due to contaminating DNA originating from dead feeder cells used to expand the transfected T cell population.
Next, we evaluated two sgRNAs (sgRNA_1 and sgRNA_2) targeting the PDCD1 gene, as previously reported by Zhao et al.23 Primary human T cells were expanded for 7 days prior to photoporation-mediated intracellular delivery of PD-1-targeting RNPs (Figure 5A). After photoporation, cells were allowed to rest for 1 day, after which, T cells were restimulated with soluble CD3/CD28 antibody complexes (Immunocult) to induce PD-1 expression. PD-1 expression level was quantified by flow cytometry using PD-1 immunostaining 2 days after stimulation. Effective disruption of PD-1 expression was obtained for both PD-1-specific sgRNAs, reaching PD-1 knock-out levels (=average reduction of PD-1 expression) of, respectively, 15% and 25% for sgRNA_1 and sgRNA_2 (Figure 5B). Importantly, minimal impact on cell viability was observed, with values that remained above 80% both 24 h and 48 h after VNB photoporation (Figure 5C). To put these cell viability values in perspective, a comparison of long-term cell viability and cell proliferation was performed between VNB photoporation (2 × 108 AuNPs/mL, E4) and nucleofection, which is a benchmark technology for non-viral T cell engineering (Figure S9).24 Primary human T cells were treated with two different nucleofection pulse programs, as recommended by the manufacturer, for the transfection of T cells: EO-115 (high-functionality protocol) and FI-115 (high-efficiency protocol). In accordance with previous reports and in contrast with VNB photoporation,9,12,24,25 nucleofection was accompanied by a substantial drop in cell viability to about 40%–50% (dependent on pulse program), which further decreased to ∼5% (FI-115) and ∼25% (EO-115) after 4 days (Figures S9A and S9B). Whereas the electroporation buffer alone did cause some acute toxicity, it did not affect long-term viability, showing that it is really the application of the electric pulses that causes term cell toxicity. T cells treated by VNB photoporation, on the other hand, showed higher initial survival (∼67% cell viability), which did not aggravate over time. Analysis of T cell proliferation furthermore proved that cells treated by VNB photoporation grow similar to the untreated control, whereas nucleofection caused delayed proliferation (EO-115) or even no proliferation at all (FI-115) (Figures S9C and S9D). Together, these data show that VNB photoporation is a more gentle approach toward T cell engineering, supporting rapid T cell proliferation after transfection.
Figure 5.
PD-1 gene editing in primary human T cells by VNB photoporation
(A) Timeline detailing the workflow for induction and detection of PD-1 gene knock-outs. (B) T cells were photoporated in the presence of 1 μM Cas9 RNPs targeting the PD-1 gene or negative control RNPs (non-targeting crRNA; 2 × 108 AuNPs/mL, E4). PD-1 knock-out levels were determined on day 2 by PD-1 immunostaining and readout by flow cytometry (n = 3 donors; mean ± SD). (C) Viability of human T cells 24 h and 48 h after photoporation of PD-1 targeting Cas9 RNPs, as determined by CellTiter-Glo assay (n ≥ 2 donors; mean ± SD).
Discussion
CRISPR-Cas9 genome editing has become an indispensable tool for the next generation of cell-based therapeutics, including adoptive T cell therapy for advanced cancer treatment.26 The effectiveness of this exciting technology strongly depends on the extent to which the CRISPR-Cas9 effector molecules can be delivered in target cells.5 Viral vectors, most often lentiviral and retroviral vectors, prevail for ex vivo engineering of cell therapeutics, given their efficiency to transduce primary cells. However, clinical-scale manufacturing of viral vectors comes with several limitations such as high cost, limited availability, and immunogenicity.27,28 Also, packaging the large Cas9 and sgRNA sequences into these viral vectors, potentially in combination with other transgenes (e.g., chimeric antigen receptors [CARs]), is a bottleneck. To circumvent these problems of viral vectors, non-viral transfection approaches have gained in popularity, of which, electroporation is the oldest and still most-used method for engineering of hard-to-transfect primary cell types.24 Even though high knock-out levels can be obtained using electroporation,22,29, 30, 31 it was repeatedly shown to come with significant loss of cell viability, undesired phenotypic changes, or loss of cell functionality, hence motivating the development of improved non-viral alternative techniques in the last decade.12,25,32,33 In line with these ongoing efforts, we here report on VNB photoporation as a promising technology for intracellular delivery of Cas9 RNPs and gene editing in hard-to-transfect cells, including stem cells and T cells, which is expected to be of benefit to therapeutic cell engineering and research applications.
Our work provides a systematic evaluation and optimization of VNB photoporation for efficacious intracellular Cas9 RNP delivery. Following optimization of the VNB photoporation procedure, we were able to achieve efficient knock-out levels of >80% in H1299-EGFP cells, which even remained >60% when the Cas9 RNP concentration was reduced to 62.5 nM. Encouraged by these results, we next applied VNB photoporation for targeted gene knock-out in MSCs, which are considered notoriously hard to transfect by conventional transfection agents.34,35 CRISPR-Cas9 gene editing of stem cells with RNPs is of interest for stem cell therapeutics36 and regenerative medicine.37,38 Indeed, gene-editing approaches are amply employed to enhance the functionality, responsiveness, and specificity of stem cell derivatives, as well as to provide a way toward allogeneic, “off-the-shelf” stem cell use.36 Editing rates up to ∼60% were obtained in mMSC-EGFP cells with minimal impact on cell viability, whereas at the same time, the possibility was demonstrated to target specific cell populations in a spatial-selective manner. This unique feature of VNB photoporation allows transfection of specific subsets of cells in a culture by proper scanning of the activating laser beam. This unique feature could be useful, for instance, when cell-selective differentiation of stem cell fate is desired, for example, in stem cell co-culture strategies for tissue engineering or to evaluate cell bystander effects.16,39 It is of note that optogenetics also represents a very popular tool for spatiotemporal-controlled redirection of stem cell fate,36,40, 41, 42 although spatial resolution and efficiency are relatively limited. VNB photoporation, which allows versatile and efficient cell transfections of a broad range of effector molecules even down to single-cell resolution,16 therefore has the potential to be of substantial benefit to tissue engineering applications.43,44 As a physical transfection approach, VNB photoporation has been demonstrated to be applicable to diverse cell types.9,11,16,34,35,45 Therefore, it is expected that translation also to human MSCs is readily possible, thereby opening up a wide range of clinical applications.43,44
We additionally demonstrated CRISPR-based knockouts in T cells, which are critical regulators and central effectors of adaptive immunity and cancer immunotherapy.46 VNB photoporation comes with several advantageous characteristics that make the technology attractive for therapeutic T cell engineering and CRISPR-based screening applications, including high efficacy, low cytotoxicity, and high throughput.9,12,24 Although we11,12 and others14,47 have previously demonstrated that VNB photoporation enables safe and efficient delivery of various molecules in human T cells, gene-editing applications have not been reported thus far. One exception is the study by Bošnjak et al.19 who delivered Cas9 RNP in murine CD8+ T cells via photoporation, but negligible knock-out levels of only ∼4% were obtained, even with Cas9 RNP concentrations up to 5 μM. This is likely due to the fact that lower laser pulse fluences were used in this study, which results in cell permeabilization by heating instead of VNB formation. Indeed, we have demonstrated before that intracellular delivery is more efficient by VNB formation instead of heating, especially for larger macromolecules.48 With VNB photoporation, we could selectively knock out the WASp in T cells. Deficiencies in expression of WASp underlie the X-linked primary immunodeficiency disease WAS. T cells of WAS patients respond to antigens with suboptimal proliferation and reduced cytokine production. WAS patients suffer from recurrent infections, thrombocytopenia, and eczema and are treated with allogeneic hematopoietic stem cell transplantation from a healthy donor.49 The CRISPR-Cas9 technology offers a valuable alternative approach not only for improving our understanding of functional consequences of WAS genetic defects but also for restoring physiological WAS gene expression in patient-derived T cells and hematopoietic stem cells. Here, we showed that VNB-mediated Cas9 RNP delivery enables effective WAS knock-out in ∼34% primary human T cells, whereas cell viability remained ∼70%. Moreover, we found that VNB photoporation equally transfects CD4+ and CD8+ T cell subsets, which is an important observation, as T cell therapeutics preferably consist of both subsets.50 Being a physical transfection approach, VNB photoporation enables simultaneous delivery of diverse effector molecules.9,11,24,34 Co-delivery of Cas9 RNPs and a homologous donor template by VNB photoporation CRISPR-based gene knock-in therefore lies within the future realms of possibilities.
Genome-editing strategies, such as CRISPR-Cas9, have also been extensively investigated in recent years to enhance the efficacy of next-generation CAR T cell therapeutics.51,52 Indeed, CAR T cell activity is often insufficient to invoke robust tumor clearance as a result of tumor-induced immunosuppression. Therefore, manipulation of immune-checkpoint blockades by CRISPR-Cas9 gene editing is actively evaluated in multiple clinical trials for treatment of various liquid and solid tumors.46 For instance, activation of the PD-1/PD-1 ligand (PD-L1) immune-checkpoint pathway leads to the inhibition of T cell effector function, so that specific downregulation of PD-1 expression represents an emerging avenue to enhance the anti-tumor efficacy of CAR T cells.51,53 We here evaluated VNB photoporation for Cas9 RNP-mediated knock-out of the PDCD1 gene, which encodes the PD-1 immune-checkpoint receptor. We found that effective disruption of PD-1 expression was obtained in ∼25% of the human T cells, whereas minimal cytotoxicity was induced. Importantly, comparison of our optimized VNB photoporation procedure with nucleofection, i.e., the current non-viral method of choice for T cell engineering, proved that cell proliferation was almost unaffected after VNB photoporation, whereas cells treated by nucleofection showed no or substantially delayed cell proliferation depending on the electroporation program used. These results are in line with previous publications,9,12,25,32,45 hence, motivating the development of gentler non-viral alternatives such as VNB photoporation.24
Apart from high delivery efficiency, cell viability, and functionality, upscaling is another important requirement for intracellular delivery technologies, which are to be used to produce clinically relevant batches of cells. Photoporation performs well in this regard with a current throughput of 200,000 cells per second. By no means is this is a hard limit, as throughput scales linearly with laser power. Exchanging the laser used in this study for a more powerful model would scale up throughput proportionally so that treatment of 106−107 cells per second should be very well feasible. Photoporation is, therefore, a truly scalable intracellular delivery technology suitable for CRISPR-Cas9 gene editing of T cells. Especially in light of our recent finding that AuNPs could be replaced by more biocompatible polymeric nanoparticles,45 it shows that translation of photoporation for the production of therapeutic engineered cells is very well feasible.
Conclusions
In summary, we demonstrated that VNB photoporation enables efficient intracellular delivery of Cas9 RNPs for targeted gene editing in both adherent and suspension cell types. The transfection technology comes with several attractive features that are of interest for both fundamental research as well as therapeutic cell engineering applications: (1) high delivery efficiency, (2) low cytotoxicity, (3) high-throughput, (4) spatial selectivity, and (5) applicability to adherent and suspension cell types. Following optimization of the VNB photoporation procedure, we were able to achieve remarkably efficient knock-out levels >80% in H1299-EGFP cells. Moreover, gene-editing levels up to ∼60% were achieved in hard-to-transfect stem cells with minimal impact on cell viability. At the same time, specific cell populations can be targeted in a spatial-selective manner, which holds promise for spatially controlled stem cell engineering. We have furthermore shown that VNB photoporation can efficiently engineer primary human T cells with minimal cytotoxicity and at a throughput of at least 200,000 cells per second, thereby demonstrating editing efficiencies up to 34% and 25% for the WAS gene and PD-1 gene, respectively. Given these results, VNB photoporation is readily compatible with CRISPR-based T cell gene function studies and has the potential to be used for the production of T cell therapeutics. In conclusion, VNB photoporation constitutes a promising technique for Cas9 RNP-mediated ex vivo engineering in a broad scope of research and therapeutic applications.
Materials and methods
Materials and reagents
Roswell Park Memorial Institute (RPMI)-1640 medium, Dulbecco’s modified Eagle’s medium containing growth factor F-12 (DMEM/F-12), Iscove’s modified Dulbecco’s medium (IMDM), fetal bovine serum (FBS), Opti-MEM (minimal essential medium), L-glutamine, penicillin/streptomycin (P/S) solution (5,000 IU/mL penicillin and 5,000 μg/mL streptomycin), Dulbecco’s phosphate-buffered saline without Ca2+/Mg2+ (DPBS−), geneticin (G418) antibiotic, TO-PRO3 iodide (1 mM), 10 kDa Alexa Fluor 647 dextran, FD250, and RNAiMAX were all obtained Life Technologies (Merelbeke, Belgium). JetCRISPR was received from Polyplus (Illkirch, France). Alt-R S.p. Cas9 Nuclease V3, Alt-R custom crRNAs, Alt-R CRISPR Negative Control crRNA #1, and Alt-R tracrRNA were purchased from Integrated DNA Technologies (Leuven, Belgium). sgRNAs were purchased from either Integrated DNA Technologies (PD-1) or Synthego (WAS). Table S1 lists the different crRNA, sgRNA, and primer sequences used throughout the manuscript.
Cell culture
H1299-EGFP cells (human non-small cell lung cancer cell line) were obtained from the lab of Prof. Foged (Department of Pharmacy, University of Copenhagen, Copenhagen, Denmark). The cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and 500 μg/mL G418. mMSCs stably expressing EGFP,54 were cultured in DMEM/F-12 supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin. All cells were maintained in a humidified atmosphere of 5% CO2 at 37°C, and experiments were performed on cells with a passage number below 25. 1 day prior to transfection, 15 × 103 were seeded in wells of a 96-well plate unless stated differently.
Buffy coats from healthy donors were obtained from the Biobank of the Red Cross Flanders after informed consent and approval and used following the guidelines of the Medical Ethical Committee of Ghent University Hospital in conformance with the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoprep (STEMCELL Technologies, Vancouver, BC, Canada) gradient centrifugation, and the percentage of T cells was determined by flow cytometry. Next, the PBMCs were resuspended in IMDM supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine (complete IMDM [cIMDM]), and 10 ng/mL interleukin (IL)-2 (Roche, Vilvoorde, Belgium) and stimulated by ImmunoCult Human CD3/CD28 T Cell Activator (25 μL/106 T cells; STEMCELL Technologies, Vancouver, BC, Canada). Cultures were split, and fresh cIMDM supplemented with 10 ng/mL IL-2 was added at day 3 and day 7.
Immediately after WAS Cas9 RNP transfection, T cells were rested at 1 × 106 cells/mL in cIMDM supplemented with 5 ng/mL IL-2 in a 48-well plate. The next day, T cells were restimulated on irradiated allogenic feeder cells (mixture of 40 Gy-irradiated PBMC and 50 Gy-irradiated JY cells) in cIMDM supplemented with 1 μg/mL phytohemagglutinin (PHA; Sigma-Aldrich, St. Louis, MO, USA). IL-2 was added on day 5 and day 10.
Following PD-1 Cas9 RNP transfection, T cells were rested in cIMDM supplemented with 5 ng/mL IL-2 for 24 h. Next, the cells were stimulated using ImmunoCult Human CD3/CD28 T Cell Activator (10 μL/106 T cells) to induce PD-1 expression and analyzed 48 h later for PD-1 expression by immunostaining and flow cytometry.
Preparation of Cas9 RNP complexes
crRNA:tracrRNA duplexes were prepared by mixing individual crRNAs in a 1:1 molar ratio with tracrRNA, followed by heating at 95°C for 5 min and annealing at room temperature for 5−10 min. Next, Cas9 RNP complexes were obtained by mixing either crRNA:tracrRNA duplexes or sgRNA in a 2.5:1 molar ratio with Cas9 endonuclease and allowing the complexes to assemble for at least 10 min at room temperature prior to transfection.
VNB photoporation procedure
For transfection by VNB photoporation, cells were first incubated for 30 min with AuNPs diluted in cell culture medium at the specified concentration, followed by a washing step with DPBS− (H1299-EGFP, mMSC-EGFP) or cell culture medium (T cells) to remove unbound AuNPs. Note that an AuNP concentration of 2 × 108 AuNPs/mL was used for transfection of human T cells with PD-1 targeting RNPs, since a new batch of AuNPs was used for these experiments, which provided better results at slightly higher concentrations. Prior to laser treatment, Cas9 RNPs or Alexa Fluor 647 10 kDa dextrans were diluted with Opti-MEM and added to the cells. VNB photoporation was performed with an in-house-developed set-up consisting of a 3-ns-pulsed laser (wavelength = 532 nm), equipped with a Galvano scanner to enable irradiation of the cells in high throughput (±5 s per 96-well plate). Spatial-selective cell transfection of a pre-defined pattern was implemented according to a binary image. Different laser pulse fluences ranging from 0.84 to 1.26 to 1.60 J/cm2, indicated as E3 – E4 – E5, were used in various experiments. After laser treatment, cells were washed twice with DPBS− and supplied with fresh culture medium (Alexa Fluor 647 10 kDa dextran). For cells transfected with Cas9 RNPs, fresh culture medium was immediately supplemented without a washing step. Dependent on the experiment, cells were analyzed after 2 h (Alexa Fluor 647/FITC-dextran), 48 h (H1299-EGFP; T cells, PD-1), 7 days (mMSC-EGFP), or 11 days (T cells, WAS).
Cas9 RNP transfection in H1299-EGFP by RNAiMAX and JetCRISPR
H1299-EGFP cells were seeded at a density of 104 cells/well of a 96-well plate 24 h prior to transfection. On the day of transfection, Cas9 RNPs were prepared as described above. 50 pmol of the Cas9 RNP was first mixed with 0.25 μL of the transfection reagent and allowed to complex for 15 min. RNP-RNAiMAX and RNP-JetCRISPR transfection complexes were diluted in Opti-MEM, added to the cells, and incubated for 4 h at 37°C, 5% CO2. Next, the cells were washed once with DPBS−, supplied with new culture medium, and further incubated at 37°C, 5% CO2, prior to analysis of EGFP knock-out by confocal microscopy or flow cytometry.
Nucleofection
Primary human T cells were electroporated with the 4D-Nucleofector system (Lonza, Breda, the Netherlands) according to the manufacturer’s recommendations with the P3 4D-Nucleofector kit (V4XP-3032). First, 1 × 106 T cells were resuspended in 20 μL P3 primary cell-line solution and transferred to a 16-well Nucleocuvette strip. The cells were transfected using Pulse Program EO-115 (high functionality program) or FI-115 (high efficiency program) and immediately supplemented with 80 μL-preheated culture medium. Eventually, 50 μL of that cell suspension was transferred to a 96-well plate containing 150 μL-preheated cell culture medium. Completely untreated T cells and T cells incubated in nucleofection buffer solution during the time of the procedure were taken along as controls.
Confocal microscopy
Visualization of H1299-EGFP cells after transfection with EGFP-targeting Cas9 RNPs or Alexa Fluor 647 10 kDa dextran was performed by confocal laser-scanning microscopy (C2 or A1R; Nikon Benelux, Brussels, Belgium) and a 10× (Plan Apo, NA 0.45; Nikon Benelux, Brussels, Belgium) or 20× (Plan Apo VC, NA 0.75; Nikon Benelux, Brussels, Belgium) objective lens. The EGFP fluorophore was excited either with a 488-nm argon-ion laser (CVI Melles Griot, Albuquerque, NM, USA) or 488-nm laser diode (LU-N4 LASER UNIT 405/488/561/640; Nikon Benelux, Brussels Belgium) and, respectively, detected using 525/50 nm (ET525/50M) or 525/50 nm (MHE57030, A1 Filter Cube 525/50) detection filters, depending on the specific confocal microscope. Alexa Fluor 647 10 kDa dextrans were excited using a 640-nm laser diode (LU-N4 LASER UNIT 405/488/561/640; Nikon Benelux, Brussels Belgium) or 640 nm laser source (56ICS/S2669; Melles Griot, Bensheim, Germany) and, respectively, detected using 700/75 nm (MHE57070, A1 Filter Cube 585/65 700/75) or 660 nm LP (ZET 420-470 660 LP) detection filters.
Flow cytometry and immunostaining
To evaluate the delivery or EGFP knock-out efficiency, H1299-EGFP or mMSC-EGFP cells were detached by trypsin/EDTA (0.25%), centrifuged (500 × g, 5 min), and resuspended in flow buffer (DPBS, 0.1% sodium azide, 1% bovine serum albumin) with TO-PRO3 iodide (1,000×) as a cell-viability dye. Flow cytometry was performed using a CytoFLEX (Beckman Coulter, Suarlée, Belgium) flow cytometer, and data analysis was carried out using the FlowJo software (Tree Star).
Immunostaining of surface markers on primary human T cells were performed in DPBS− with 1% FCS and using antibody concentrations as recommended by the manufacturer. Intracellular WASp staining was performed using the BD Cytofix/Cytoperm (BD Biosciences, Erebodegem, Belgium), according to the manufacturer’s instructions. The following anti-human monoclonal antibodies were used in various experiments: phycoerythrin (PE)-PD-1 (Miltenyi Biotec, Leiden, the Netherlands); PE-Cy7-conjugated CD3, peridinin chlorophyll protein complex (PerCP)-cyanin (Cy)5.5-conjugated CD4, and allophycocyanin (APC)-Fire 750-conjugated CD8 (BioLegend, San Diego, CA, USA); and Alexa Fluor 700-conjugated WASp (Santa Cruz Biotechnology, Santa Cruz, CA, USA). All cell populations were analyzed after doublet and dead cell exclusion by Fixable Viability Dye eFluor 506 staining (eBioscience, Vienna, Austria) or TO-PRO3 iodide. Flow cytometry was performed using a BD LSR II (BD Biosciences) or CytoFLEX flow cytometer, and FlowJo software (Tree Star) was used for data analysis. For calculation of the PD-1 knock-out level, the MFI of each sample (MFIsample) was subtracted with the MFI of the unstimulated control (MFIunstimulated control) and expressed relative to the MFI of a control sample transfected with a negative control RNP (MFInegative control RNP) under identical experimental conditions, according to the following equation:
| (Equation 1) |
CellTiter-Glo cell viability assay
Cell viability after VNB photoporation was determined using the CellTiter-Glo luminescence cell viability assay (Promega, Leiden, the Netherlands), according to the manufacturer’s instructions. In brief, cells in culture medium were supplemented with an equal volume of CellTiter-Glo reagent and shaken for 10 min using an orbital shaker at 120 rpm. Next, the content of the wells was transferred to an opaque 96-well plate, and the luminescence was allowed to stabilize for at least 10 min. The luminescent signal in each well was measured using a Glomax luminometer (Promega, Leiden, the Netherlands), and cell viability was determined relative to the untreated control.
Evaluation of cell viability and cell proliferation by trypan blue cell counting
Primary human T cell density was assessed at different time points (6 h, 24 h, 48 h, 72 h, 96 h) following VNB photoporation or nucleofection using a using a Bürker counting chamber (Brand, Wertheim, Germany) and trypan blue exclusion (0.4%; Sigma-Aldrich, Overijse, Belgium). Cell viability of the different conditions was calculated relative to their respective untreated control. For calculation of normalized cell growth, cell proliferation was normalized against the respective 6-h samples.
Quantification of gene-editing efficiency by Sanger sequencing
Genomic DNA of H1299-EGFP or primary human T cells was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Chatsworth, CA, USA), according to the manufacturer’s instructions. Next, the DNA target sites of interest were amplified by PCR using KAPA HiFi HotStart ReadyMix (Roche Diagnostics Belgium, Diegem, Belgium). Amplified PCR products were purified by the QIAquick PCR purification kit (QIAGEN, Chatsworth, CA, USA), following the manufacturer’s protocol. Next, PCR amplicons were sequenced by the GATC lightrun service (Eurofins Genomics, Ebersberg, Germany), after which the sequencing data were analyzed using TIDE (http://shinyapps.datacurators.nl/tide/).55
Statistical analysis
All data are shown as mean ± standard deviation (SD), unless specified differently. Statistical differences were analyzed using GraphPad Prism 8 software (GraphPad Software, La Jolla, CA, USA). The statistical tests used in each figure are mentioned in the figure captions. Statistical differences with a p value < 0.05 were considered significant.
Acknowledgments
We acknowledge the Centre for Advanced Light Microscopy at Ghent University (Belgium) for use of its microscopes and support for the microscopy experiments. The authors would like to acknowledge funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements number (no.) 810685 (DelNam project) and no. 648124. S.S. acknowledges the support of a VLAIO grant (grant number: HBC.2017.0542). M.P. (FWO grant G036727N and FWO-SB grant 1S14318N), J.V.H. (FWO-SB grant 1S62519N), and J.C.F. (FWO grant 1210120N) thank the financial support by the Flemish Research Foundation.
Author contributions
Conceptualization, L.R., K.R., and K.B.; investigation, L.R., M.P., A.H., G.G., and J.V.H.; project administration, L.R.; writing – original draft, L.R.; writing – review & editing, L.R., M.P., G.G., J.V.H., S.S., J.C.F., T.B., O.G.d.J., R.M.-B., E.M., P.V., S.C.D.S., B.V., K.R., and K.B.; funding acquisition, M.P., J.V.H., S.S., J.C.F., and K.B.; resources, G.G., O.G.d.J., R.M.-B., E.M., and P.V.; supervision, S.C.D.S., B.V., K.R., and K.B.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.014.
Supplemental information
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oude Blenke E., Evers M.J.W., Mastrobattista E., van der Oost J. CRISPR-Cas9 gene editing: Delivery aspects and therapeutic potential. J. Control. Release. 2016;244(Pt B):139–148. doi: 10.1016/j.jconrel.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Glass Z., Lee M., Li Y., Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fajrial A.K., He Q.Q., Wirusanti N.I., Slansky J.E., Ding X. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics. 2020;10:5532–5549. doi: 10.7150/thno.43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S., Kim D., Cho S.W., Kim J., Kim J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong R., Samal S.K., Demeester J., Skirtach A., De Smedt S., Braeckmans K. Laser-assisted photoporation : fundamentals, technological advances and applications. Adv. Phys.-X. 2016;1:596–620. [Google Scholar]
- 9.Wayteck L., Xiong R., Braeckmans K., De Smedt S.C., Raemdonck K. Comparing photoporation and nucleofection for delivery of small interfering RNA to cytotoxic T cells. J. Control. Release. 2017;267:154–162. doi: 10.1016/j.jconrel.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Xiong R., Verstraelen P., Demeester J., Skirtach A.G., Timmermans J.-P., De Smedt S.C., De Vos W.H., Braeckmans K. Selective Labeling of Individual Neurons in Dense Cultured Networks With Nanoparticle-Enhanced Photoporation. Front. Cell. Neurosci. 2018;12:80. doi: 10.3389/fncel.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raes L., Van Hecke C., Michiels J., Stremersch S., Fraire J.C., Brans T., Xiong R., De Smedt S., Vandekerckhove L., Raemdonck K., Braeckmans K. Gold Nanoparticle-Mediated Photoporation Enables Delivery of Macromolecules over a Wide Range of Molecular Weights in Human CD4+ T Cells. Crystals. 2019;9:411. [Google Scholar]
- 12.Raes L., Stremersch S., Fraire J.C., Brans T., Goetgeluk G., De Munter S., Van Hoecke L., Verbeke R., Van Hoeck J., Xiong R. Intracellular Delivery of mRNA in Adherent and Suspension Cells by Vapor Nanobubble Photoporation. Nano-Micro Lett. 2020;12:185. doi: 10.1007/s40820-020-00523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukianova-Hleb E., Hu Y., Latterini L., Tarpani L., Lee S., Drezek R.A., Hafner J.H., Lapotko D.O. Plasmonic nanobubbles as transient vapor nanobubbles generated around plasmonic nanoparticles. ACS Nano. 2010;4:2109–2123. doi: 10.1021/nn1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukianova-Hleb E.Y., Wagner D.S., Brenner M.K., Lapotko D.O. Cell-specific transmembrane injection of molecular cargo with gold nanoparticle-generated transient plasmonic nanobubbles. Biomaterials. 2012;33:5441–5450. doi: 10.1016/j.biomaterials.2012.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgart J., Humbert L., Boulais É., Lachaine R., Lebrun J.-J., Meunier M. Off-resonance plasmonic enhanced femtosecond laser optoporation and transfection of cancer cells. Biomaterials. 2012;33:2345–2350. doi: 10.1016/j.biomaterials.2011.11.062. [DOI] [PubMed] [Google Scholar]
- 16.Xiong R., Drullion C., Verstraelen P., Demeester J., Skirtach A.G., Abbadie C., De Vos W.H., De Smedt S.C., Braeckmans K. Fast spatial-selective delivery into live cells. J. Control. Release. 2017;266:198–204. doi: 10.1016/j.jconrel.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 17.Van Hoecke L., Raes L., Stremersch S., Brans T., Fraire J.C., Roelandt R., Declercq W., Vandenabeele P., Raemdonck K., Braeckmans K., Saelens X. Delivery of Mixed-Lineage Kinase Domain-Like Protein by Vapor Nanobubble Photoporation Induces Necroptotic-Like Cell Death in Tumor Cells. Int. J. Mol. Sci. 2019;20:4254. doi: 10.3390/ijms20174254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraire J.C., Houthaeve G., Liu J., Raes L., Vermeulen L., Stremersch S., Brans T., García-Díaz Barriga G., De Keulenaer S., Van Nieuwerburgh F. Vapor nanobubble is the more reliable photothermal mechanism for inducing endosomal escape of siRNA without disturbing cell homeostasis. J. Control. Release. 2020;319:262–275. doi: 10.1016/j.jconrel.2019.12.050. [DOI] [PubMed] [Google Scholar]
- 19.Bošnjak B., Permanyer M., Sethi M.K., Galla M., Maetzig T., Heinemann D., Willenzon S., Förster R., Heisterkamp A., Kalies S. CRISPR/Cas9 Genome Editing Using Gold-Nanoparticle-Mediated Laserporation. Adv. Biosyst. 2018;2:1700184. [Google Scholar]
- 20.Bak R.O., Dever D.P., Porteus M.H. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat. Protoc. 2018;13:358–376. doi: 10.1038/nprot.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X., Liu Z., Jo M.C., Zhang K., Li Y., Zeng Z., Li N., Zu Y., Qin L. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 2015;1:e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki A., Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Z., Shi L., Zhang W., Han J., Zhang S., Fu Z., Cai J. CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes. Oncotarget. 2017;9:5208–5215. doi: 10.18632/oncotarget.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raes L., De Smedt S.C., Raemdonck K., Braeckmans K. Non-viral transfection technologies for next-generation therapeutic T cell engineering. Biotechnol. Adv. 2021;49:107760. doi: 10.1016/j.biotechadv.2021.107760. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M., Ma Z., Selliah N., Weiss G., Genin A., Finkel T.H., Cron R.Q. The impact of Nucleofection® on the activation state of primary human CD4 T cells. J. Immunol. Methods. 2014;408:123–131. doi: 10.1016/j.jim.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey S.R., Maus M.V. Gene editing for immune cell therapies. Nat. Biotechnol. 2019;37:1425–1434. doi: 10.1038/s41587-019-0137-8. [DOI] [PubMed] [Google Scholar]
- 27.Levine B.L., Miskin J., Wonnacott K., Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. Methods Clin. Dev. 2016;4:92–101. doi: 10.1016/j.omtm.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol. Ther. Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hultquist J.F., Hiatt J., Schumann K., McGregor M.J., Roth T.L., Haas P., Doudna J.A., Marson A., Krogan N.J. CRISPR-Cas9 genome engineering of primary CD4+ T cells for the interrogation of HIV-host factor interactions. Nat. Protoc. 2019;14:1–27. doi: 10.1038/s41596-018-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiTommaso T., Cole J.M., Cassereau L., Buggé J.A., Hanson J.L.S., Bridgen D.T., Stokes B.D., Loughhead S.M., Beutel B.A., Gilbert J.B. Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. Proc. Natl. Acad. Sci. USA. 2018;115:E10907–E10914. doi: 10.1073/pnas.1809671115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart M.P., Sharei A., Ding X., Sahay G., Langer R., Jensen K.F. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 35.Stewart M.P., Langer R., Jensen K.F. Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev. 2018;118:7409–7531. doi: 10.1021/acs.chemrev.7b00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimbrel E.A., Lanza R. Next-generation stem cells - ushering in a new era of cell-based therapies. Nat. Rev. Drug Discov. 2020;19:463–479. doi: 10.1038/s41573-020-0064-x. [DOI] [PubMed] [Google Scholar]
- 37.Steinbeck J.A., Studer L. Moving stem cells to the clinic: potential and limitations for brain repair. Neuron. 2015;86:187–206. doi: 10.1016/j.neuron.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barry F.P., Murphy J.M. Mesenchymal stem cells: clinical applications and biological characterization. Int. J. Biochem. Cell Biol. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Savukinas U.B., Enes S.R., Sjöland A.A., Westergren-Thorsson G. Concise Review: The Bystander Effect: Mesenchymal Stem Cell-Mediated Lung Repair. Stem Cells. 2016;34:1437–1444. doi: 10.1002/stem.2357. [DOI] [PubMed] [Google Scholar]
- 40.Klapper S.D., Sauter E.J., Swiersy A., Hyman M.A.E., Bamann C., Bamberg E., Busskamp V. On-demand optogenetic activation of human stem-cell-derived neurons. Sci. Rep. 2017;7:14450. doi: 10.1038/s41598-017-14827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motta-Mena L.B., Reade A., Mallory M.J., Glantz S., Weiner O.D., Lynch K.W., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao J., Wang M., Yu G., Zhu S., Yu Y., Heng B.C., Wu J., Ye H. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2018;115:E6722–E6730. doi: 10.1073/pnas.1802448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golchin A., Shams F., Karami F. In: Turksen K., editor. Springer; 2019. Advancing mesenchymal stem cell therapy with CRISPR/Cas9 for clinical trial studies; pp. 89–100. (Cell Biology and Translational Medicine, vol. 8). [DOI] [PubMed] [Google Scholar]
- 44.Hamann A., Nguyen A., Pannier A.K. Nucleic acid delivery to mesenchymal stem cells: a review of nonviral methods and applications. J. Biol. Eng. 2019;13:7. doi: 10.1186/s13036-019-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harizaj A., Wels M., Raes L., Stremersch S., Goetgeluk G., Brans T., Vandekerckhove B., Sauvage F., De Smedt S.C., Lentacker I. Photoporation with Biodegradable Polydopamine Nanosensitizers Enables Safe and Efficient Delivery of mRNA in Human T Cells. Adv. Funct. Mater. 2021;31:2102472. [Google Scholar]
- 46.Singh N., Shi J., June C.H., Ruella M. Genome-Editing Technologies in Adoptive T Cell Immunotherapy for Cancer. Curr. Hematol. Malig. Rep. 2017;12:522–529. doi: 10.1007/s11899-017-0417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukianova-Hleb E.Y., Samaniego A.P., Wen J., Metelitsa L.S., Chang C.-C., Lapotko D.O. Selective gene transfection of individual cells in vitro with plasmonic nanobubbles. J. Control. Release. 2011;152:286–293. doi: 10.1016/j.jconrel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong R., Raemdonck K., Peynshaert K., Lentacker I., De Cock I., Demeester J., De Smedt S.C., Skirtach A.G., Braeckmans K. Comparison of gold nanoparticle mediated photoporation: vapor nanobubbles outperform direct heating for delivering macromolecules in live cells. ACS Nano. 2014;8:6288–6296. doi: 10.1021/nn5017742. [DOI] [PubMed] [Google Scholar]
- 49.Humblet-Baron S., Sather B., Anover S., Becker-Herman S., Kasprowicz D.J., Khim S., Nguyen T., Hudkins-Loya K., Alpers C.E., Ziegler S.F. Wiskott-Aldrich syndrome protein is required for regulatory T cell homeostasis. J. Clin. Invest. 2007;117:407–418. doi: 10.1172/JCI29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turtle C.J., Hanafi L.-A., Berger C., Gooley T.A., Cherian S., Hudecek M., Sommermeyer D., Melville K., Pender B., Budiarto T.M. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 2016;126:2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi B.D., Yu X., Castano A.P., Darr H., Henderson D.B., Bouffard A.A., Larson R.C., Scarfò I., Bailey S.R., Gerhard G.M. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunother. Cancer. 2019;7:304. doi: 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017;7:737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rejman J., Tavernier G., Bavarsad N., Demeester J., De Smedt S.C. mRNA transfection of cervical carcinoma and mesenchymal stem cells mediated by cationic carriers. J. Control. Release. 2010;147:385–391. doi: 10.1016/j.jconrel.2010.07.124. [DOI] [PubMed] [Google Scholar]
- 55.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.