Abstract
RNA interference-based gene silencing drugs are attracting attention for treating various diseases. Lipid nanoparticles (LNPs) are carriers that efficiently deliver small interfering RNA (siRNA) to the cytoplasm of target cells. Recently, we developed potent and well-tolerated biodegradable LNPs with asymmetric ionizable lipids. Here, we evaluated the effect of LNPs on immune cells in mice. After intravenous administration, LNPs were efficiently incorporated into several tissue-resident macrophages, including liver macrophages, through an apolipoprotein E (ApoE)-independent mechanism. Administration of LNP-encapsulated siRNA against Irf5, encoding the transcription factor critical for inflammatory responses, sharply reduced its expression in macrophages in vivo, and persisted for as long as 7 days. The therapeutic potential of Irf5 siRNA-loaded LNPs in inflammatory diseases was tested in a concanavalin A (Con A)-induced hepatitis model, whose pathogenic mechanisms are dependent on cytokine secretion from macrophages. We found that Con A-induced liver injury was significantly attenuated after LNP injection. Serum aspartate transaminase, alanine aminotransferase, and inflammatory cytokine levels were significantly reduced in mice injected with Irf5 siRNA-loaded LNPs compared to those injected with control siRNA-loaded LNPs. Our results suggest that administering biodegradable LNPs to deliver siRNA is a promising strategy for treating inflammatory disorders.
Keywords: drug delivery system, siRNA, lipid nanoparticles, macrophages, IRF5, hepatitis
Graphical abstract
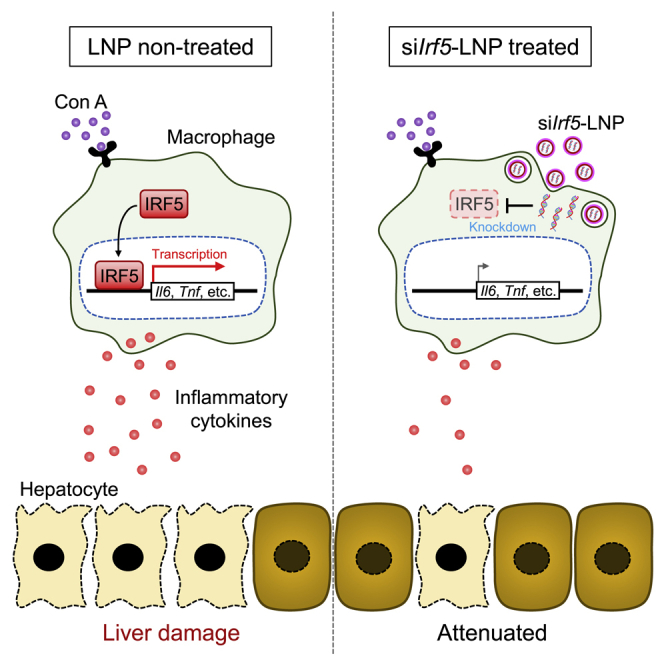
Kawase et al. report that ionizable lipid-based biodegradable LNPs are effectively incorporated into macrophages in vivo. Administration of LNP-encapsulated siRNA against Irf5, encoding a transcription factor mediating inflammatory responses, resulted in long-term IRF5 downregulation in liver macrophages and prevented cytokine production and liver damage in a hepatitis model.
Introduction
RNA interference (RNAi) is a fundamental eukaryotic mechanism that induces targeted mRNA degradation, which is mediated by small interfering RNA (siRNA).1 RNAi-based therapeutic agents can potentially inhibit “undruggable” molecules that are difficult to target with small molecule compounds and antibodies.2 Developing siRNA delivery systems that overcome properties of siRNA that make them easily degraded by nucleases in vivo is indispensable for using RNAi-based therapeutic drugs.3,4
In 2018, the first siRNA-based medicine was approved by the United States Food and Drug Administration.5,6 This drug utilizes lipid nanoparticles (LNPs) as the siRNA carrier. LNPs efficiently deliver siRNA to the cytoplasm and are mainly composed of ionizable lipids, phospholipids, cholesterol, and polyethylene glycol (PEG) lipids that protect siRNA from nucleases.7,8 The characteristics of LNPs are largely dependent on the types of ionizable lipids.7,9 Ionizable lipids are critical for endosomal disruption and siRNA release from LNPs in target cells,9 but they can cause cytotoxicity due to non-specific binding to proteins.7 There are various types of ionizable lipids, such as symmetric ionizable lipids,10 asymmetric ionizable lipids,11 and lipidoids.12 Among them, asymmetric ionizable lipids effectively lead to target gene silencing and are less cytotoxic than other ionizable lipids.11,13,14 Recently, we developed biodegradable LNPs comprised of novel ionizable lipids, L120, with an asymmetric lipid tail.15 The ester bond in the ionizable lipid tail is hydrolyzed in vivo, leading to rapid lipid degradation and reduced cytotoxicity. We have shown that our biodegradable LNPs were successfully incorporated into liver hepatocytes and led to effective knockdown of target mRNA expression.15
Apolipoprotein E (ApoE) and low-density lipoprotein receptor (LDLR) are involved in the incorporation of LNPs into hepatocytes in vivo.16 Plasma ApoE preferentially binds to PEG lipids of the LNP surface. LDLR proteins on hepatocytes recognize ApoE-LNP complexes, resulting in LNP incorporation via endocytosis. LNPs become cationic due to reduced pH during endosome maturation, leading to LNP fusion to the endosomal membrane.17 siRNA is finally released into the cytoplasm, thereby inducing RNAi-mediated knockdown of target molecules in hepatocytes.
Mononuclear phagocytes, including macrophages and monocytes, can also take up LNPs.12,18, 19, 20, 21 These cells are critical for host defense against pathogens, but they are also involved in pathogenic mechanisms of various diseases, such as inflammatory diseases, cardiac diseases, and cancers.22, 23, 24, 25, 26 Therefore, gene suppression in mononuclear phagocytes by LNPs may be an effective treatment strategy for such disorders.
In this study, we comprehensively analyzed the effects of our asymmetric ionizable lipid-based biodegradable LNPs on immune cells. LNPs were strongly incorporated into tissue-resident macrophages, including liver macrophages. Analysis of ApoE-deficient mice revealed that LNP uptake by liver macrophages is not dependent on ApoE expression. Administering LNPs encapsulating siRNA targeting interferon regulatory factor-5 (IRF5) (siIrf5-LNPs), an essential transcription factor in triggering inflammatory responses, reduced IRF5 expression in macrophages in vivo. Furthermore, prophylactic administration of siIrf5-LNPs was effective in preventing liver damage in concanavalin A (Con A)-induced hepatitis. These results indicate that our biodegradable LNPs are a useful tool for analyzing macrophage function in vivo, and they can be used in novel therapies for treating inflammatory diseases.
Results
Biodegradable LNPs are mainly incorporated by macrophages in immune cell populations
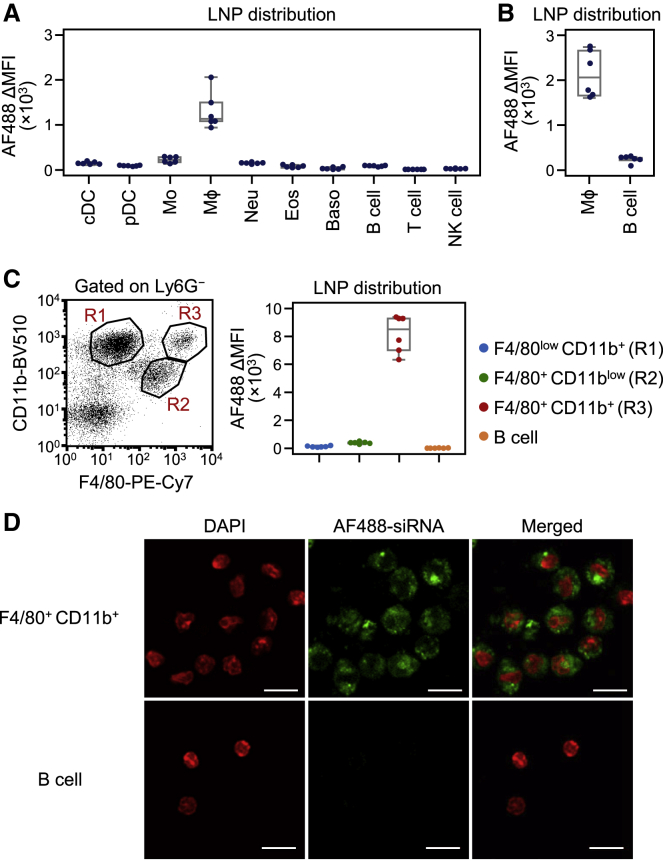
To examine the in vivo distribution of biodegradable LNPs in immune cells, we administered biodegradable LNPs containing Alexa Fluor 488 (AF488)-labeled siRNA into mice through intravenous injections and analyzed bone marrow cells and splenocytes (Figure S1). Flow cytometry analysis revealed that the fluorescence signal of AF488-labeled siRNA was most abundant in splenic macrophages, and to a lesser degree in monocytes 3 h after LNP injection. Bone marrow hematopoietic stem and progenitor cells, splenic lymphocytes, dendritic cells, neutrophils, eosinophils, and basophils did not effectively take up the LNPs (Figures 1A, S2, and S3A).
Figure 1.
Incorporation of biodegradable LNPs into immune cells
(A–C) Flow cytometry analysis of AF488-labeled siRNA distribution following intravenous injection of LNPs in mice. Immune cells in the spleen (A), peritoneal exudate cells (B), and liver (C) were analyzed 3 h after intravenous injection of LNPs encapsulating AF488-labeled siRNA at a siRNA dose of 0.8 mg/kg. The MFI of AF488 in each immune cell population is shown in the boxplots. Values from two independent experiments are shown. A representative dot plot of liver macrophage populations is shown (C, left). R1, R2, and R3 indicate F4/80lowCD11b+, F4/80+CD11blow, and F4/80+CD11b+ cells, respectively (C, right). (D) Liver F4/80+CD11b+ macrophages and B cells were sorted by FACSAria II 3 h after intravenous injection of LNPs containing AF488-labeled siRNA and analyzed by confocal microscopy. Nuclei were stained with DAPI. Data are representative of two independent experiments with similar results. Scale bars represent 10 μm. cDC, classical dendritic cell; pDC, plasmacytoid dendritic cell; Mo, monocyte; Mϕ, macrophage; Neu, neutrophil; Eos, eosinophil; Baso, basophil; MFI, mean fluorescent intensity. See also Figures S1, S2, and S3.
Next, we investigated the distribution of biodegradable LNPs in peritoneal and liver immune cells. Three hours after intravenous administration of LNPs, peritoneal exudate cells were collected and analyzed by flow cytometry. We found that the biodegradable LNPs were taken up by peritoneal macrophages, but not by B lymphocytes (Figures 1B and S3B). There are several macrophage and monocyte subpopulations in the liver.27, 28, 29 We confirmed that cells expressing F4/80, a typical marker for macrophages, were mainly composed of F4/80lowCD11b+ cells (gated in R1), F4/80+CD11blow cells (R2), and F4/80+CD11b+ cells (R3) (Figure 1C). Previous studies suggested that the F4/80lowCD11b+ population corresponds to monocytes and monocyte-derived macrophages, while F4/80+CD11blow and F4/80+CD11b+ populations mainly correspond to liver-resident Kupffer cells.28,30 Among the three F4/80-expressing populations, biodegradable LNPs were mainly detected in F4/80+CD11b+ cells. The uptake of LNPs in F4/80lowCD11b+ cells, F4/80+CD11blow cells, and B cells was less prominent (Figures 1C and S3C). To assess whether siRNAs in LNPs were incorporated into the cytoplasm of target cells, we isolated liver F4/80+CD11b+ macrophages and B cells after injection of LNPs and then observed localization of AF488-labeled siRNA using confocal microscopy. As expected, AF488-labeled siRNA was present in the cytoplasm of liver F4/80+CD11b+ macrophages, but not in B cells (Figure 1D). These results suggest that the biodegradable LNPs are efficiently incorporated into resident macrophage populations, such as liver F4/80+ CD11b+ macrophages. Notably, we previously compared the uptake of our LNPs between liver macrophages and hepatocytes. The rates of LNPs incorporated by macrophages and hepatocytes were approximately 40% and 60%, respectively,31 indicating that most LNPs are taken up by hepatocytes in the liver.
Biodegradable LNP administration induces knockdown of a target molecule in macrophages in vivo
To assess the gene silencing effect of biodegradable LNPs in macrophages, we selected interferon regulatory factor-5 (IRF5) as a target molecule. The transcription factor IRF5 is strongly expressed in macrophages, monocytes, dendritic cells, and B cells.32,33 Pathogen-associated molecular patterns such as single-stranded RNA and CpG DNA activate IRF5 via Toll-like receptor (TLR)-mediated signaling.34,35 Activated IRF5 translocates from the cytoplasm to the nucleus, leading to expression of type I interferons and proinflammatory cytokines such as tumor necrosis factor (TNF)-α and interleukin (IL)-6.33,35 To investigate the therapeutic effect of the biodegradable LNPs in inflammatory disorders, we generated LNPs containing siRNA against Irf5 mRNA (siIrf5-LNPs) (see Figure S1).
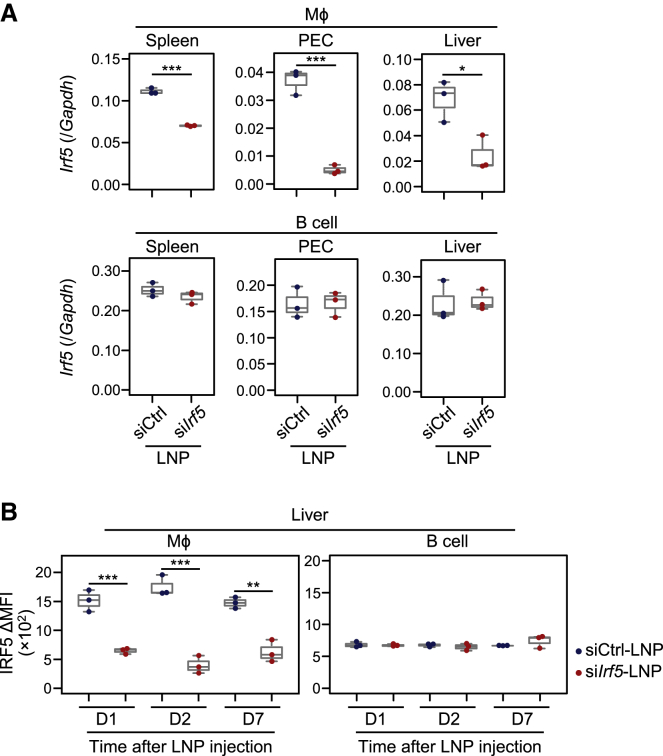
We first determined the optimal dosage of siIrf5-LNPs in vivo. The data indicated that the effect of suppressing Irf5 expression in peritoneal macrophages reached a plateau at a dose of 5.0 mg/kg (Figures 2A and S4). Irf5 mRNA expression was also downregulated significantly in splenic and liver macrophages (Figure 2A). In contrast, Irf5 mRNA expression in B cells was not affected by siIrf5-LNP administration. No Irf genes other than Irf5 were suppressed in liver macrophages by the siIrf5-LNPs, indicating the specific knockdown of Irf5 (Figure S5). Consistent with quantitative reverse transcriptase PCR (qRT-PCR) data, IRF5 protein expression in liver F4/80+CD11b+ macrophages was significantly reduced 1, 2, and 7 days after administration (Figure 2B). These results suggest that administration of biodegradable LNPs can induce long-term gene silencing in tissue-resident macrophages in vivo. Notably, IRF5 expression in non-immune liver CD45– cells, possibly including hepatocytes, sinusoidal endothelial cells, and stellate cells, was low and not significantly downregulated by siIrf5-LNP administration (Figure S6).
Figure 2.
IRF5 silencing effect of biodegradable LNPs in macrophages
IRF5 expression analysis following intravenous injection of siIrf5-LNPs in mice. Mice were injected with siIrf5-LNPs or siCtrl-LNPs at a siRNA dose of 5.0 mg/kg. (A) Twenty-four hours after LNP administration, Irf5 mRNA expression in Mϕs and B cells in the spleen, PECs, and liver were analyzed by qRT-PCR. Values were normalized to Gapdh levels using the ΔΔCt method. (B) After LNP administration, IRF5 protein expression in liver Mϕs and B cells was analyzed by immunofluorescent staining at indicated time points. Boxplots indicate MFI of IRF5 expression. Data from three independent experiments are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test). PEC, peritoneal exudate cell. See also Figures S4, S5, and S6.
LNPs are incorporated into macrophages via an ApoE-independent mechanism
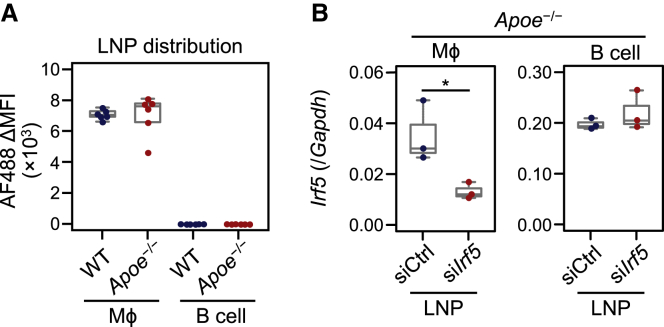
Previous reports have suggested that biodegradable LNPs are incorporated into hepatocytes via the ApoE-LDLR pathway.16,36 To investigate whether the ApoE-LDLR pathway is involved in incorporating biodegradable LNPs used in this study into macrophages, we injected them into ApoE-deficient mice. ApoE-deficient F4/80+CD11b+ macrophages incorporated the LNPs encapsulating AF488-labeled siRNA as much as did wild-type macrophages (Figure 3A). In addition, we found that Irf5 mRNA expression in ApoE-deficient macrophages, but not B cells, was significantly reduced after administration of siIrf5-LNPs (Figure 3B). These results indicate that ApoE is not involved in the uptake of biodegradable LNPs in liver F4/80+CD11b+ macrophages.
Figure 3.
Role of ApoE in biodegradable LNP uptake into macrophages
(A) Flow cytometry analysis of AF488-labeled siRNA distribution following intravenous injection of LNPs in Apoe−/− mice. F4/80+CD11b+ macrophages and B cells in the liver were analyzed 3 h after intravenous injection of LNP-encapsulated AF488-labeled siRNA at a siRNA dose of 0.8 mg/kg. MFI of AF488 in each cell population is shown in the boxplots. Values from two independent experiments are shown. (B) Irf5 mRNA expression analysis following intravenous injection of siIrf5-LNPs in Apoe−/− mice. Mice were injected with siIrf5-LNPs or siCtrl-LNPs at a siRNA dose of 5.0 mg/kg. Twenty-four hours after LNP administration, Irf5 mRNA expression in liver F4/80+CD11b+ Mϕs and B cells were analyzed by qRT-PCR. Data from three independent experiments are shown. ∗p < 0.05 (Student’s t test). WT, wild-type.
Con A-induced hepatitis is attenuated in IRF5-deficient mice
The above results revealed that IRF5 expression in liver macrophages can be efficiently downregulated upon injection of biodegradable LNPs. Therefore, we decided to use a Con A-induced hepatitis model, in which liver macrophages play a critical role in pathogenesis. Con A administrated through intravenous injection binds to and activates liver macrophages.37, 38, 39 Subsequently, these cells produce inflammatory cytokines and chemokines, such as TNF-α and CXCL2, leading to the activation of T cells and natural killer (NK) T cells, and recruitment of neutrophils to the liver.40, 41, 42, 43 Liver necrosis is eventually caused by these inflammatory cytokines and by direct cytotoxic effects such as Fas-mediated cell death.44, 45, 46
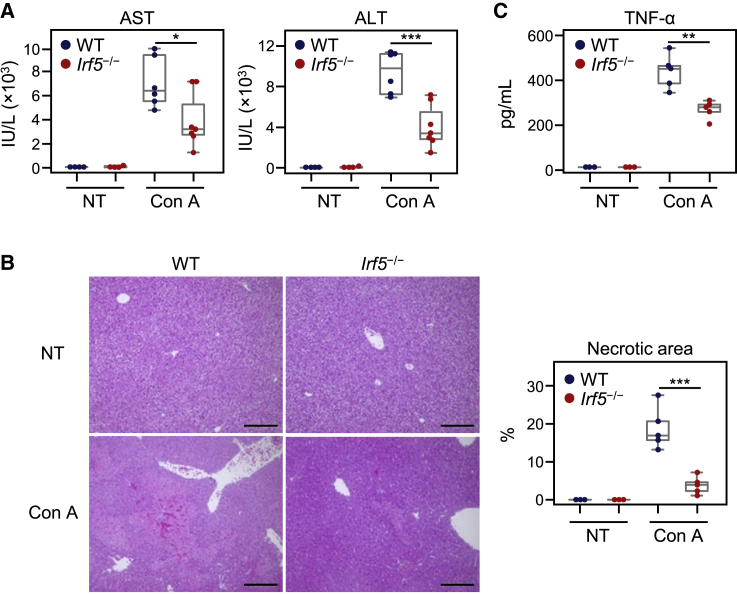
To evaluate the validity of targeting IRF5 in this model, we injected Con A into IRF5-deficient mice. As expected, Con A administration induced a significant increase in blood aspartate transaminase (AST) and alanine aminotransferase (ALT) levels, as well as hepatocyte necrosis, in wild-type mice (Figures 4A and 4B). We found that levels of AST and ALT, as well as liver damage, were significantly ameliorated in IRF5-deficient mice. Furthermore, the production of TNF-α, an essential cytokine involved in the pathogenesis of Con A-induced hepatitis, was significantly reduced in IRF5-deficient mice compared to wild-type mice (Figure 4C). To our knowledge, this is the first demonstration that IRF5 is a critical regulator of Con A-induced hepatitis progression. These results suggest that IRF5 is a candidate therapeutic target in this model.
Figure 4.
Con A-induced hepatic injury in Irf5−/− mice
(A) Twelve hours after Con A administration, plasma AST and ALT levels were measured by a 7180 clinical analyzer. WT or Irf5−/− mice were injected with 20 mg/kg Con A. (B) Twelve hours after Con A administration, livers were collected. H&E staining of liver sections from mice injected with Con A is shown (left). Scale bars represent 200 μm. Percentages of necrotic area were calculated using ImageJ software (right). (C) Two hours after Con A administration, serum TNF-α levels were determined by ELISA. Data are shown using boxplots. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test). NT, no treatment.
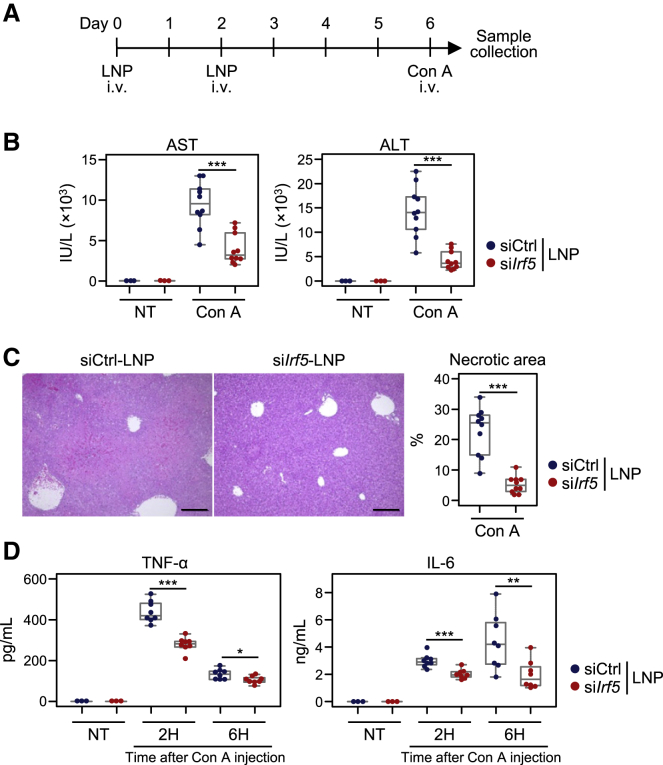
Con A-induced hepatitis is ameliorated by siIrf5-LNP administration
To investigate the prophylactic effect of siIrf5-LNPs, we administered LNPs twice and then challenged the mice with Con A 4 days after LNP injection (Figure 5A). We found that AST and ALT levels were significantly reduced in these mice compared to control LNP-treated mice (Figure 5B). Consistent with these results, liver necrosis and bleeding were suppressed by siIrf5-LNP treatment (Figure 5C). Furthermore, production of TNF-α and IL-6 was significantly downregulated in mice injected with siIrf5-LNPs (Figure 5D). Taken together, our results indicate that siIrf5-LNP administration reduces IRF5 expression in macrophages, thereby suppressing production of pro-inflammatory cytokines and liver damage after Con A injection.
Figure 5.
Inhibition of IRF5 on Con A-induced hepatic injury
(A) Mice were injected with siIrf5-LNPs or siCtrl-LNPs at a siRNA dose of 5.0 mg/kg twice. Four days after LNP injection, mice were either untreated or administered 20 mg/kg Con A through intravenous injection. (B) Twelve hours after Con A injection, plasma AST and ALT levels were measured. (C, left) H&E staining of the liver sections 12 h after Con A injection was performed. Scale bars represent 200 μm. (C, right) Percentages of necrotic area were calculated using ImageJ software. (D) Two and 6 h after Con A administration, serum levels of TNF-α and IL-6 were determined by ELISA. Data are shown using boxplots. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test).
Discussion
LNPs have been primarily used to target hepatocytes in mouse disease models and clinical trials. In this study, we investigated the effects of biodegradable LNPs on immune cells, as well as their potential application in preventing inflammatory diseases. Biodegradable LNPs were strongly incorporated by tissue-resident macrophages, especially liver macrophages. LNP uptake was also observed in ApoE-deficient macrophages, suggesting that LNPs are incorporated through a mechanism independent of the ApoE-LDLR pathway. We also showed that IRF5 is essential for pathogenesis in Con A-induced hepatitis. Furthermore, siIrf5-LNP injection resulted in long-term downregulation of IRF5 in liver macrophages and prevented inflammatory cytokine production and liver damage in the Con A-induced hepatitis model.
The molecular mechanisms underlying uptake of biodegradable LNPs into macrophages remain unclear. Previous studies have shown that incorporation of LNPs with symmetric ionizable lipids and lipidoids into macrophages is mediated by phagocytosis, but not micropinocytosis or endocytosis.19 Binding of plasma proteins, such as complements and immunoglobulins, to LNPs can also assist phagocytosis by macrophages in vivo.17,47,48 These findings imply that our biodegradable LNPs are incorporated via phagocytosis in tissue-resident macrophages.
A time-course analysis revealed that knockdown effects of the biodegradable LNPs in liver macrophages persisted for as long as 7 days. Previous reports suggested that most tissue-resident macrophages, including liver Kupffer cells, are generated from fetal macrophage progenitors during embryogenesis.49 Fetal progenitor-derived liver macrophages are not replenished by bone marrow cells, such as monocytes, and they maintain their numbers by slow proliferation in adult mice.49 Therefore, siRNA incorporated in macrophages may be sustained due to slow cell division. Likewise, the knockdown effects of LNPs persist longer in hepatocytes, which barely proliferate in steady-state conditions, compared to fast-growing cancer cells.15,50 Long-term knockdown by biodegradable LNPs will enable us to analyze the function of target molecules in tissue-resident macrophages in vivo.
The molecular basis for IRF5 activation in Con A-induced hepatitis remains unclear. IRF5 is activated by TLR-MyD88 signaling pathways.35,51 Con A administration induces expression of TLR2, TLR3, TLR4, and TLR9 in macrophages in vivo.52, 53, 54, 55 In addition, Con A-induced TNF-α expression in macrophages is downregulated in MyD88-deficient mice.53 It is well known that damage-associated molecular pattern molecules (DAMPs) are released from cells undergoing necrotic cell death, and then recognized by TLRs and other pattern recognition receptors.56 Liver macrophages can sense DAMPs, thereby producing inflammatory cytokines.57 From these reports and our results, we speculate that Con A administration induces the release of DAMPs from hepatocytes, activating the TLR-MyD88-IRF5 signaling cascade in liver macrophages.
Further modifications to the biodegradable LNPs may allow them to be more efficiently incorporated into macrophages and other immune cell populations. LNPs larger than 200 nm in diameter are more readily taken up by peritoneal macrophages compared to smaller LNPs.58 As the size of our LNPs is approximately 70–80 nm (see Figure S1), increasing their size might further improve the efficacy. In addition, mannosylated LNPs are effectively incorporated into mannose receptor-expressing macrophages such as tumor-associated macrophages.59 LNPs coated with antibodies targeting DEC205 and CD4 were capable of efficiently delivering siRNA to a dendritic cell subpopulation and CD4+ T cells in mice, respectively.60,61 Such modifications could improve the efficiency and versatility of our LNPs and extend their potential applications for clinical use in the future.
Materials and methods
Mice
Male and female wild-type, Irf5−/−,35 and Apoe−/− mice 8–12 weeks old in a C57BL/6 background were used. All animal experiments were performed in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan), and all protocols were approved by the Institutional Review Boards of Yokohama City University (protocols F-A-17-018 and F-A-20-043).
siRNA synthesis
siIrf5 and siRNA targeting luciferase (siCtrl) have been described in previous reports.16,18 AF488-labeled siRNA (sense, 5′-AcAuGAAGcAGcACGACuUdTsdT-3′; antisense, 5′-AAGUCGUGCUGCUUcAUGUdTsdT-aminoC6 linker-AF488-3′), siIrf5 (sense, 5′-cuGcAGSGSSuSScccuGAdTsdT-3′; antisense, 5′-UcAGGGUuAUUCUCUGcAGdTsdT-3′), and siCtrl (sense, 5′-cuuAcGcuGAGuAcuucGAdTsdT-3′; antisense, 5′-UCGAAGuACUcAGCGuAAGdTsdT-3′) were synthesized by GeneDesign (Japan). Lowercase letters denote 2′-O-methyl (2′-OMe)-modified residues.
Formulation of biodegradable LNPs
Biodegradable LNPs were prepared as previously described.15 Briefly, siRNA was dissolved in 10 mM sodium citrate (0.45 mg/mL). Biodegradable ionized lipid L120,31 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol (PEG-DMG) at a molar ratio of 60:8.5:30:1.5 were then dissolved in ethanol. The total lipid concentration was 40 mM. Using two syringe pumps, siRNA solutions and lipid solutions were mixed at a flow rate of 3 and 1 mL/min, respectively. The mixing ratio of siRNA and lipids was 0.06 w/w. After overnight dialysis with PBS (pH 7.4) using a 100-kDa dialysis tube, the solution was sterilized using a 0.22-μm membrane filter. The physical properties of our LNPs are shown in Figure S1.
Cell isolation and flow cytometry
Hepatic immune cells were prepared as previously described.62 Briefly, after perfusion with 0.5 mg/mL collagenase type 4 (Worthington Biochemical) via the portal vein, liver tissue was minced and homogenized using a 70-μm cell strainer (Corning Life Sciences). Liver cells were resuspended in 45% Percoll solution and centrifuged at 50 × g for 10 min to enrich liver-infiltrating immune cells. Spleen cells were obtained with Liberase and DNase I (Roche) treatment.63 Bone marrow cells were harvested by flushing femurs and tibias.63 Splenocytes and bone marrow cells were treated with red blood cell lysis solution. Peritoneal cells were obtained by washing the peritoneal cavity with RPMI 1640 media containing 10% fetal bovine serum (FBS). These cells were stained with fluorochrome-labeled antibodies. Flow cytometry was performed using FACSCanto II and FACSAria II (BD Biosciences), and data were analyzed using FlowJo software (FlowJo).
Markers and antibodies used for flow cytometric analysis
Markers used for flow cytometry in this study were as follows: hematopoietic stem cells, lineage markers (Lin; CD5, B220, CD11b, Ly6G/C, 7-4, Ter-119)–Sca-1+CD117+IL-7R–CD150+CD34low; lymphoid-primed multipotent progenitors, Lin–Sca-1+CD117+IL-7R–CD150–CD34+Flt3+; common myeloid progenitors, Lin–CD117hiIL-7R–CD115–CD34+CD16/32low; granulocyte monocyte progenitors, Lin–CD117hiIL-7R–CD115–CD34+CD16/32hi; megakaryocyte erythrocyte progenitors, Lin–CD117hiIL-7R–CD115–CD34–CD16/32–; monocyte dendritic cell progenitors, Lin–CD117hiIL-7R–CD115+Flt3+; common dendritic cell progenitors, Lin–CD117int/lowIL-7R–CD115+Flt3+; committed classical dendritic cell precursors, Lin–major histocompatibility complex (MHC) class II–CD11c+Flt3+; common monocyte progenitors, CD3–CD19–Ly6G–NK1.1–CD115+CD117hiFlt3–Ly6ChiCD11blow; classical dendritic cells, MHC class IIhiCD11chi; plasmacytoid dendritic cells, B220+PDCA-1+CD11blowCD11chi; monocytes, CD11bhiCD115+Ly6G–; neutrophils, CD11bhiLy6Ghi; eosinophils, Ly6G–F4/80–CD11b+Siglec F+; basophils, CD117–CD49b+CD200R3+; B cells, CD3ε–B220+CD19+CD11b–CD11c–; T cells, CD3ε+TCR-β+CD11c–; and NK cells, NK1.1+CD49b+. Macrophages are identified by F4/80 and CD11b staining. Antibodies used in this study and their clone names are as follows: anti-B220 (RA3-6B2), CD115 (AFS98), CD117 (2B8), CD11b (M1/70), CD11c (N418), CD150 (TC15-12F12.2), CD16/32 (93), CD200R3 (Ba160), CD3ε (145-2C11), CD4 (GK1.5), CD49b (DX5), CD8 (53-6.7), CD19 (6D5), F4/80 (BM8), Flt3 (A2F10), IL-7R (A7R34), Ly6C (HK1.4), Ly6G (1A8), MHC class II (AF6-120.1), NK1.1 (PK136), PDCA-1 (927), Sca-1 (E13-161.7), TCR-β (H57-597) (BioLegend), CD34 (RAM34) (eBioscience), and Siglec F (E50-2440) (BD Biosciences).
Intracellular IRF5 staining
Liver cells were stained with antibodies for surface markers as described above. The cells were then fixed in 4% paraformaldehyde and permeabilized with 90% methanol. Rabbit anti-IRF5 (ab21689; Abcam) and R-phycoerythrin-conjugated donkey anti-rabbit antibodies (Jackson ImmunoResearch) were used as primary and secondary antibodies, respectively.
qRT-PCR
Total mRNA was extracted using an RNeasy micro kit (QIAGEN) and reverse transcribed using the PrimeScript RT reagent kit (Takara) according to the manufacturer’s instructions. The following primers were used in qRT-PCR analyses: Irf1 (sense, 5′-CCC AGG GCT GAT CTG GAT CAA TAA A-3′; antisense, 5′-ACA GAC AGG CAT CCT TGT TGA TGT G-3′), Irf2 (sense, 5′-TTC AAC TGA CGG GCT TTC ATT TCC A-3′; antisense, 5′-CAC CGG CAT GGT ACC CTC TCA A-3′), Irf3 (sense, 5′-TGC GAG TCT CAG AAC TAC TGT TTG G-3′; antisense, 5′-GTT TCC ATG CTC TAG CCA GGG G-3′), Irf4 (sense, 5′-AAG GCC CAT CTT GTG AAA ATG GTT G-3′; antisense, 5′-CTT ATG CTT GGC TCA ATG GGG ATT C-3′), Irf5 (sense, 5′-CAG TGG GTC AAC GGG GAA AAG AAA C-3′; antisense, 5′-CTT TAG CCC AGG CCT TGA AGA TGG-3′), Irf6 (sense, 5′-CAC GGA CTG AGG CCA GAT CAT-3′; antisense, 5′-TGG AAG CGT TTG GAA TCT CTG TGT A-3′), Irf7 (sense, 5′-ACA GCA CAG GGC GTT TTA TC-3′; antisense, 5′-GAG CCC AGC ATT TTC TCT TG-3′), Irf8 (sense, 5′-GCA GGA TGT GTG ACC GAA AC-3′; antisense, 5′-GGA TAC GGA ACA TGG TCT TCT CAT C-3′), Irf9 (sense, 5′-AGG GGT ATG GTA AGG AGA AGG ATG G-3′; antisense, 5′-CAG GGA ATC CGG AAC ATG GTC TT-3′), Gapdh (sense, 5′-GTG TTC CTA CCC CCA ATG T-3′; antisense, 5′-TGT CAT CAT ACT TGG CAG GTT TC-3′). Data were analyzed using the ΔΔCt method and normalized against Gapdh expression levels.
Induction of Con A-induced liver injury
Con A (Vector Laboratories) was dissolved in PBS and administered in mice through intravenous injection (20 mg/kg body weight).
Quantification of AST and ALT levels
Serum AST and ALT levels were quantified by a 7180 clinical analyzer (Hitachi) at Oriental Yeast.
Histology
Livers were harvested 12 h after Con A injection. Tissues were fixed in 10% buffered formalin. Formalin-fixed and paraffin-embedded liver sections were cut and stained with hematoxylin and eosin at the Sapporo General Pathology Laboratory. Necrotic areas were measured in each section using ImageJ software (https://imagej.nih.gov/ij/).
Enzyme-linked immunosorbent assay (ELISA)
After administration of Con A, blood was collected through tail veins at specified time points. Serum was analyzed using the mouse TNF-α ELISA MAX standard kit and mouse IL-6 ELISA MAX standard kit (BioLegend).
Acknowledgments
The authors thank Dr. Masako Kikuchi, Masahiro Yoshinari, and Ayano Shinozuka for their help with experiments. This work was supported by Grants-in-Aid (KAKENHI) from the Japan Society for the Promotion of Science (JSPS)/Ministry of Education, Culture, Sports, Science and Technology (MEXT) (nos. JP21H02954 [to T. Tamura] and JP19H03691 [to D.K.]); the Fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from the MEXT/Japan Science and Technology Agency (to T. Tamura), which accompanies a matching fund from Eisai Co., Ltd.; and a joint research fund from Eisai Co., Ltd. (to T. Tamura). W.K. was supported by the PhD Scholarship (Kibou Project) from the Japanese Society for Immunology.
Author contributions
W.K., D.K., and T. Tamura designed the study. W.K., D.K., Y.S., and J.I. performed the experiments. W.K., D.K., and Y.S. analyzed the data. W.K., D.K., and T. Tamura wrote the manuscript. Y.S., H.I., T.B., G.R.S., H.Y., T. Taniguchi, and K.T. provided key resources. T. Tamura supervised the project. All authors have approved the final version of the manuscript.
Declaration of interests
Y.S., H.I., and K.T. are employees of Eisai Co., Ltd. T. Tamura received a joint research fund from Eisai Co., Ltd. The remaining authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.023.
Contributor Information
Daisuke Kurotaki, Email: kurotaki@yokohama-cu.ac.jp.
Tomohiko Tamura, Email: tamurat@yokohama-cu.ac.jp.
Supplemental information
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatakeyama H., Akita H., Kogure K., Harashima H. A novel nonviral gene delivery system: Multifunctional envelope-type nano device. Adv. Biochem. Eng. Biotechnol. 2010;119:197–230. doi: 10.1007/10_2008_40. [DOI] [PubMed] [Google Scholar]
- 5.Hoy S.M. Patisiran: First global approval. Drugs. 2018;78:1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 6.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 7.Barros S.A., Gollob J.A. Safety profile of RNAi nanomedicines. Adv. Drug Deliv. Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez-Trabado J., Diebold Y., Sanchez A. Designing lipid nanoparticles for topical ocular drug delivery. Int. J. Pharm. 2017;532:204–217. doi: 10.1016/j.ijpharm.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Allen T.M., Cullis P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Fan H., Levorse D.A., Crocker L.S. Interaction of cholesterol-conjugated ionizable amino lipids with biomembranes: Lipid polymorphism, structure-activity relationship, and implications for siRNA delivery. Langmuir. 2011;27:9473–9483. doi: 10.1021/la201464k. [DOI] [PubMed] [Google Scholar]
- 12.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J. Control. Release. 2015;220(Pt A):44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Gindy M.E., Feuston B., Glass A., Arrington L., Haas R.M., Schariter J., Stirdivant S.M. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol. Pharm. 2014;11:4143–4153. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki Y., Hyodo K., Suzuki T., Tanaka Y., Kikuchi H., Ishihara H. Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates. Int. J. Pharm. 2017;519:34–43. doi: 10.1016/j.ijpharm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki Y., Ishihara H. Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid. Int. J. Pharm. 2016;510:350–358. doi: 10.1016/j.ijpharm.2016.06.124. [DOI] [PubMed] [Google Scholar]
- 17.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courties G., Heidt T., Sebas M., Iwamoto Y., Jeon D., Truelove J., Tricot B., Wojtkiewicz G., Dutta P., Sager H.B. In vivo silencing of the transcription factor IRF5 reprograms the macrophage phenotype and improves infarct healing. J. Am. Coll. Cardiol. 2014;63:1556–1566. doi: 10.1016/j.jacc.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novobrantseva T.I., Borodovsky A., Wong J., Klebanov B., Zafari M., Yucius K., Querbes W., Ge P., Ruda V.M., Milstein S. Systemic RNAi-mediated gene silencing in nonhuman primate and rodent myeloid cells. Mol. Ther. Nucleic Acids. 2012;1:e4. doi: 10.1038/mtna.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leuschner F., Dutta P., Gorbatov R., Novobrantseva T.I., Donahoe J.S., Courties G., Lee K.M., Kim J.I., Markmann J.F., Marinelli B. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat. Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uemura Y., Naoi T., Kanai Y., Kobayashi K. The efficiency of lipid nanoparticles with an original cationic lipid as a siRNA delivery system for macrophages and dendritic cells. Pharm. Dev. Technol. 2019;24:263–268. doi: 10.1080/10837450.2018.1469149. [DOI] [PubMed] [Google Scholar]
- 22.Morawski P.A., Bolland S. Expanding the B cell-centric view of systemic lupus erythematosus. Trends Immunol. 2017;38:373–382. doi: 10.1016/j.it.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Visser K.E., Eichten A., Coussens L.M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 24.Guilliams M., Bruhns P., Saeys Y., Hammad H., Lambrecht B.N. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014;14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 25.Yáñez A., Coetzee S.G., Olsson A., Muench D.E., Berman B.P., Hazelett D.J., Salomonis N., Grimes H.L., Goodridge H.S. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity. 2017;47:890–902.e4. doi: 10.1016/j.immuni.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruhns P., Samuelsson A., Pollard J.W., Ravetch J.V. Colony-stimulating factor-1-dependent macrophages are responsible for IVIG protection in antibody-induced autoimmune disease. Immunity. 2003;18:573–581. doi: 10.1016/s1074-7613(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 27.Sheng J., Ruedl C., Karjalainen K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity. 2015;43:382–393. doi: 10.1016/j.immuni.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Lai S.M., Sheng J., Gupta P., Renia L., Duan K., Zolezzi F., Karjalainen K., Newell E.W., Ruedl C. Organ-specific fate, recruitment, and refilling dynamics of tissue-resident macrophages during blood-stage malaria. Cell Rep. 2018;25:3099–3109.e3. doi: 10.1016/j.celrep.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 29.Kim S.Y., Jeong J.M., Kim S.J., Seo W., Kim M.H., Choi W.M., Yoo W., Lee J.H., Shim Y.R., Yi H.S. Pro-inflammatory hepatic macrophages generate ROS through NADPH oxidase 2 via endocytosis of monomeric TLR4-MD2 complex. Nat. Commun. 2017;8:2247. doi: 10.1038/s41467-017-02325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagemeyer N., Kierdorf K., Frenzel K., Xue J., Ringelhan M., Abdullah Z., Godin I., Wieghofer P., Costa Jordão M.J., Ulas T. Transcriptome-based profiling of yolk sac-derived macrophages reveals a role for Irf8 in macrophage maturation. EMBO J. 2016;35:1730–1744. doi: 10.15252/embj.201693801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., Yamazaki K., Ishida T., Ishihara H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: Faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020;588:119792. doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
- 32.Tamura T., Yanai H., Savitsky D., Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 33.Ban T., Sato G.R., Tamura T. Regulation and role of the transcription factor IRF5 in innate immune responses and systemic lupus erythematosus. Int. Immunol. 2018;30:529–536. doi: 10.1093/intimm/dxy032. [DOI] [PubMed] [Google Scholar]
- 34.Schoenemeyer A., Barnes B.J., Mancl M.E., Latz E., Goutagny N., Pitha P.M., Fitzgerald K.A., Golenbock D.T. The interferon regulatory factor, IRF5, is a central mediator of Toll-like receptor 7 signaling. J. Biol. Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 35.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T.W., Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 36.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heymann F., Hamesch K., Weiskirchen R., Tacke F. The concanavalin A model of acute hepatitis in mice. Lab. Anim. 2015;49(1, Suppl):12–20. doi: 10.1177/0023677215572841. [DOI] [PubMed] [Google Scholar]
- 38.Schümann J., Wolf D., Pahl A., Brune K., Papadopoulos T., van Rooijen N., Tiegs G. Importance of Kupffer cells for T-cell-dependent liver injury in mice. Am. J. Pathol. 2000;157:1671–1683. doi: 10.1016/S0002-9440(10)64804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto T., Maeda O., Tsuzuike N., Hara K. Effect of gadolinium chloride treatment on concanavalin A-induced cytokine mRNA expression in mouse liver. Jpn. J. Pharmacol. 1998;78:101–103. doi: 10.1254/jjp.78.101. [DOI] [PubMed] [Google Scholar]
- 40.Takeda K., Hayakawa Y., Van Kaer L., Matsuda H., Yagita H., Okumura K. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc. Natl. Acad. Sci. USA. 2000;97:5498–5503. doi: 10.1073/pnas.040566697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko Y., Harada M., Kawano T., Yamashita M., Shibata Y., Gejyo F., Nakayama T., Taniguchi M. Augmentation of Vα14 NKT cell-mediated cytotoxicity by interleukin 4 in an autocrine mechanism resulting in the development of concanavalin A-induced hepatitis. J. Exp. Med. 2000;191:105–114. doi: 10.1084/jem.191.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyabe S., Seki S., Iiai T., Takeda K., Shirai K., Watanabe H., Hiraide H., Uchiyama M., Abo T. Requirement of IL-4 and liver NK1+ T cells for concanavalin A-induced hepatic injury in mice. J. Immunol. 1997;159:1537–1542. [PubMed] [Google Scholar]
- 43.Tiegs G., Hentschel J., Wendel A. A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J. Clin. Invest. 1992;90:196–203. doi: 10.1172/JCI115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan H.A., Ahmad M.Z., Khan J.A., Arshad M.I. Crosstalk of liver immune cells and cell death mechanisms in different murine models of liver injury and its clinical relevance. Hepatobiliary Pancreat. Dis. Int. 2017;16:245–256. doi: 10.1016/S1499-3872(17)60014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fullerton A.M., Roth R.A., Ganey P.E. Pretreatment with TCDD exacerbates liver injury from concanavalin A: Critical role for NK cells. Toxicol. Sci. 2013;136:72–85. doi: 10.1093/toxsci/kft174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima H., Kinoshita M., Nakashima M., Habu Y., Shono S., Uchida T., Shinomiya N., Seki S. Superoxide produced by Kupffer cells is an essential effector in concanavalin A-induced hepatitis in mice. Hepatology. 2008;48:1979–1988. doi: 10.1002/hep.22561. [DOI] [PubMed] [Google Scholar]
- 47.Nagayama S., Ogawara K., Fukuoka Y., Higaki K., Kimura T. Time-dependent changes in opsonin amount associated on nanoparticles alter their hepatic uptake characteristics. Int. J. Pharm. 2007;342:215–221. doi: 10.1016/j.ijpharm.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 48.Dobrovolskaia M.A., Aggarwal P., Hall J.B., McNeil S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perdiguero E.G., Geissmann F. The development and maintenance of resident macrophages. Nat. Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Judge A.D., Robbins M., Tavakoli I., Levi J., Hu L., Fronda A., Ambegia E., McClintock K., MacLachlan I. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J. Clin. Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balkhi M.Y., Fitzgerald K.A., Pitha P.M. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol. Cell. Biol. 2008;28:7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao X., Zhao P., Rodriguez-Pinto D., Qi D., Henegariu O., Alexopoulou L., Flavell R.A., Wong F.S., Wen L. Inflammatory regulation by TLR3 in acute hepatitis. J. Immunol. 2009;183:3712–3719. doi: 10.4049/jimmunol.0901221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ojiro K., Ebinuma H., Nakamoto N., Wakabayashi K., Mikami Y., Ono Y., Po-Sung C., Usui S., Umeda R., Takaishi H. MyD88-dependent pathway accelerates the liver damage of concanavalin A-induced hepatitis. Biochem. Biophys. Res. Commun. 2010;399:744–749. doi: 10.1016/j.bbrc.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 54.Jiang W., Sun R., Zhou R., Wei H., Tian Z. TLR-9 activation aggravates concanavalin A-induced hepatitis via promoting accumulation and activation of liver CD4+ NKT cells. J. Immunol. 2009;182:3768–3774. doi: 10.4049/jimmunol.0800973. [DOI] [PubMed] [Google Scholar]
- 55.Tu C.T., Han B., Yao Q.Y., Zhang Y.A., Liu H.C., Zhang S.C. Curcumin attenuates concanavalin A-induced liver injury in mice by inhibition of Toll-like receptor (TLR) 2, TLR4 and TLR9 expression. Int. Immunopharmacol. 2012;12:151–157. doi: 10.1016/j.intimp.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 57.Su L., Li N., Tang H., Lou Z., Chong X., Zhang C., Su J., Dong X. Kupffer cell-derived TNF-α promotes hepatocytes to produce CXCL1 and mobilize neutrophils in response to necrotic cells. Cell Death Dis. 2018;9:323. doi: 10.1038/s41419-018-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsui H., Sato Y., Hatakeyama H., Akita H., Harashima H. Size-dependent specific targeting and efficient gene silencing in peritoneal macrophages using a pH-sensitive cationic liposomal siRNA carrier. Int. J. Pharm. 2015;495:171–178. doi: 10.1016/j.ijpharm.2015.08.044. [DOI] [PubMed] [Google Scholar]
- 59.Nimje N., Agarwal A., Saraogi G.K., Lariya N., Rai G., Agrawal H., Agrawal G.P. Mannosylated nanoparticulate carriers of rifabutin for alveolar targeting. J. Drug Target. 2009;17:777–787. doi: 10.3109/10611860903115308. [DOI] [PubMed] [Google Scholar]
- 60.Katakowski J.A., Mukherjee G., Wilner S.E., Maier K.E., Harrison M.T., DiLorenzo T.P., Levy M., Palliser D. Delivery of siRNAs to dendritic cells using DEC205-targeted lipid nanoparticles to inhibit immune responses. Mol. Ther. 2016;24:146–155. doi: 10.1038/mt.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramishetti S., Kedmi R., Goldsmith M., Leonard F., Sprague A.G., Godin B., Gozin M., Cullis P.R., Dykxhoorn D.M., Peer D. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano. 2015;9:6706–6716. doi: 10.1021/acsnano.5b02796. [DOI] [PubMed] [Google Scholar]
- 62.Katayama S., Tateno C., Asahara T., Yoshizato K. Size-dependent in vivo growth potential of adult rat hepatocytes. Am. J. Pathol. 2001;158:97–105. doi: 10.1016/S0002-9440(10)63948-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurotaki D., Kawase W., Sasaki H., Nakabayashi J., Nishiyama A., Morse H.C., 3rd, Ozato K., Suzuki Y., Tamura T. Epigenetic control of early dendritic cell lineage specification by the transcription factor IRF8 in mice. Blood. 2019;133:1803–1813. doi: 10.1182/blood-2018-06-857789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.