Abstract
TIGAR (TP53-induced glycolysis and apoptosis regulator) is the downstream target gene of p53, contains a functional sequence similar to 6-phosphofructose kinase/fructose-2, 6-bisphosphatase (PFKFB) bisphosphatase domain. TIGAR is mainly located in the cytoplasm; in response to stress, TIGAR is translocated to nucleus and organelles, including mitochondria and endoplasmic reticulum to regulate cell function. P53 family members (p53, p63, and p73), some transcription factors (SP1 and CREB), and noncoding miRNAs (miR-144, miR-885-5p, and miR-101) regulate the transcription of TIGAR. TIGAR mainly functions as fructose-2,6-bisphosphatase to hydrolyze fructose-1,6-diphosphate and fructose-2,6-diphosphate to inhibit glycolysis. TIGAR in turn facilitates pentose phosphate pathway flux to produce nicotinamide adenine dinucleotide phosphate (NADPH) and ribose, thereby promoting DNA repair, and reducing intracellular reactive oxygen species. TIGAR thus maintains energy metabolism balance, regulates autophagy and stem cell differentiation, and promotes cell survival. Meanwhile, TIGAR also has a nonenzymatic function and can interact with retinoblastoma protein, protein kinase B, nuclear factor-kappa B, hexokinase 2, and ATP5A1 to mediate cell cycle arrest, inflammatory response, and mitochondrial protection. TIGAR might be a potential target for the prevention and treatment of cardiovascular and neurological diseases, as well as cancers.
Keywords: TIGAR, pentose phosphate pathway, NADPH, cerebral ischemia–reperfusion, neurological disorders, myocardial infarction, cancer
Glucose metabolism is one of the major energy sources and is essential for maintaining the normal physiological function of mammals. Glucose metabolism consists of a variety of biological reactions catalyzed by many enzymes. Among them, fructose-2,6-bisphosphatase (Fru-2,6-BPase) is one of the key enzymes in regulating glycolysis, and a protein named TIGAR has been reported to perform similar activity as the enzyme. In 2005, Jen and Cheung first discovered the novel TP53 regulatory gene c12orf5 in response to ionizing radiation [1]. In 2006, Bensad cloned and identified the c12orf5 gene and named it TP53-induced glycolysis and apoptosis regulator (TIGAR), reflecting its important role in maintaining the steady state of cellular energy metabolism [2]. Wong [3] and Fang [4] both regarded TIGAR as the protein encoded by the c12orf5 gene, and studied its function in a pathological state.
TIGAR gene and protein structure
The TIGAR gene is located on the short arm of chromosome 12p13-3 [3], and the sequence of the TIGAR protein is highly conserved throughout vertebrates from fish to humans. The human TIGAR gene consists of six exons, ~38,835 bp in length, and encodes a unique mRNA transcript of 813 bp [5].
The human TIGAR protein consists of 270 amino acids with a molecular weight of 30 kDa. The TIGAR protein sequence in vertebrates contains the phosphoglycerate mutase (PGM) family, which is characteristic of the RHG motif (R31, H32, and G33) [6] (Fig. 1), i.e., it is phosphorylated in the catalytic reaction and participates in the hydrolysis and phosphorylation of substrates [7]. TIGAR also contains a catalytic domain similar to that of the histidine phosphatase superfamily with hydrolysis, mutase, and phosphatase activities, which has a strictly conserved catalytic core. Taking Escherichia coli SixA, a member of the family, as an example, the center is the histidine residue H8 that mediates the catalytic activity of phosphorylation and dephosphorylation; the phosphate pocket formed by two arginine residues R7 and R55, and another histidine H108 is responsible for transferring the phospho group from the substrate to the enzyme in the reaction [6]. The functional domain is well conserved in TIGAR and centers on H11, and the phosphate pocket is formed by R10, R131, and H198 [2]. TIGAR’s domain catalyzing substrate hydrolysis (Fig. 1) is composed of two histidine residues, H11 and H198, and one glutamic acid, E102 [2, 8]. This domain is similar to the bisphosphatase domain of the 6-phosphofructokinase/fructose-2,6-bisphosphatase (PFKFB) [6]. PFKFB is a bifunctional enzyme with kinase and bisphosphatase activities. The kinase domain 6-phosphofructokinase 2 (PFK-2) is located at the NH2-terminus of the enzyme, while the bisphosphatase domain Fru-2,6-BPase-2 is located within the COOH-terminal region. TIGAR lacks the kinase domain of PFKFB, but the similarity of TIGAR to PFKFB lies in the highly conserved catalytic domain of bisphosphatase [2]. The TIGAR protein has two conserved pockets (Fig. 1), which can bind to phosphate molecules on the substrate to mediate dephosphorylation. According to the crystal structure analysis of TIGAR in zebrafish, one pocket is near the histidine residue in the core and consists of His11, His183, Arg10, Arg61, and Glu89, and the other is near Arg10 and Asn217 in the C-terminus. The two pockets are open and positively charged to bind with fructose-2,6-diphosphate (Fru-2,6-P2) or fructose-1,6-diphosphate (Fru-1,6-P2) [8], hydrolyzing these small molecules to regulate the flux of glucose metabolism.
Fig. 1. TIGAR gene structure diagram, with different colors to mark different amino acid residue positions and unique structures, including the dPGM family characteristic motif RGH, catalytic domain, histidine phosphatase folding domain, and phosphate pocket.
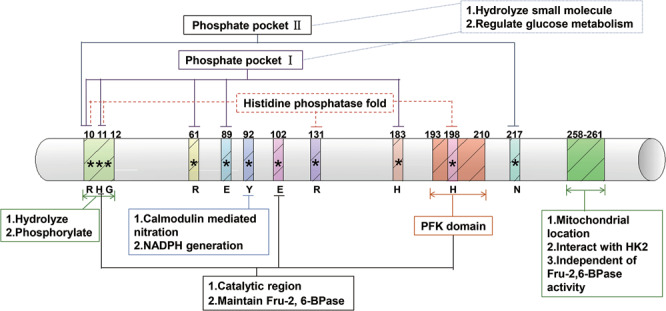
The domain and unique biological functions are annotated.
The deletion of 10RHG12 (TIGAR-∆RHG) or 193PFK210 (TIGAR-∆PFK) or the triple mutation of H11A, E102A, and H198A in TIGAR protein produced mutants that were unable to reduce U2OS cell apoptosis, and TIGAR triple mutants lost the ability to hydrolyze Fru-2,6-P2 [2]. TIGAR∆258–261 with deletion of the 258–261 region can still degrade Fru-2,6-P2, but it cannot be localized to mitochondria and cannot interact with hexokinase (HK) to protect mitochondrial function [9]. In addition, TIGAR Y92A mutants formed by the nitration of tyrosine at site 92 of TIGAR interfere with TIGAR to increase pentose phosphate pathway (PPP)-dependent nicotinamide adenine dinucleotide phosphate (NADPH) production [10].
TIGAR distribution in tissues and cells
TIGAR is expressed in almost all tissues, with high abundance in muscle [11], brain [12], and heart [11]. TIGAR is also highly expressed in some tumors, such as leukemia [13], colon cancer [14], lung cancer [15], breast cancer [16], and malignant gliomas [17]. In addition, TIGAR is highly expressed at the embryonic stage, and development and age gradually reduce TIGAR expression [18].
TIGAR is abundantly expressed in mammalian cells and located in the cytoplasm and in organelles, such as the mitochondria [9], endoplasmic reticulum (ER), and nucleus [19] (Fig. 2). TIGAR present in the cytoplasm mainly inhibits glycolysis, and promotes PPP flux to increase the production of ribose-5-phosphate and NADPH, which helps DNA repair and reduces reactive oxygen species (ROS) to maintain redox balance (see “Enzymatic activity and antioxidant effect of TIGAR” for details). When HepG2 cells are treated with epirubicin or hypoxia, TIGAR translocates to the nucleus, where it regulates cyclin-dependent kinase (CDK)-5-mediated ATM phosphorylation to induce cell cycle arrest and to promote DNA damage repair in a TP53-independent manner. The translocation of TIGAR in the nucleus thus promotes cell survival under chemotherapy and hypoxia [19]. Under hypoxic conditions, the accumulation of hypoxia-inducible factor 1 (HIF1α) promotes TIGAR to translocate to the mitochondria and binds to hexokinase 2 (HK2) in the outer mitochondrial membrane, resulting in an increase in HK2 activity. TIGAR thus reduces mitochondrial ROS production, maintains mitochondrial function, and reduces cell death via interaction with HK2. The mitochondrial localization of TIGAR depends on HK2 and HIF1α. Importantly, TIGAR’s mitochondrial localization and binding to HK2 is independent of its enzymatic activity, but depends on the HIF1α and TIGAR 258–261 domains [9]. Under ischemia–reperfusion injury in mice, TIGAR relocates to mitochondria to reduce mitochondrial ROS and maintain mitochondrial membrane potential [12]. In addition, TIGAR is highly expressed in skeletal muscles containing type II fibers. TIGAR can translocate to mitochondria to interact with ATP5A1 during exhaustive exercise and to promote mitochondrial production through the SIRT1/PGC1α axis to improve exercise tolerance, fatigue, and aging in mice [11]. Our unpublished results demonstrated that cerebral ischemia–reperfusion injury triggers ER stress, oxidative stress, and neuronal apoptosis, while TIGAR can relocate to the ER and the nucleus during ischemia–reperfusion, inhibiting ER stress-dependent oxidative stress and apoptosis, thereby mediating the protective effect against neuronal ischemic injury.
Fig. 2. TIGAR is located in the cytoplasm and enters organelles, including the mitochondria, endoplasmic reticulum, and nucleus under different stress stimuli to regulate cell function.
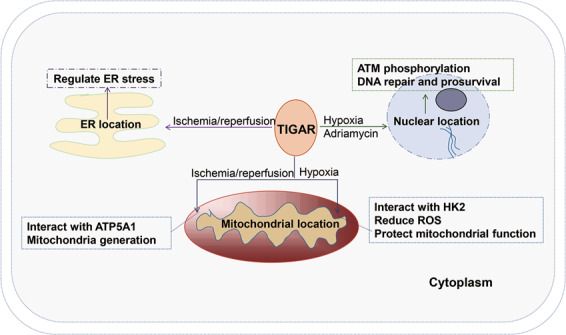
TIGAR translocates to the mitochondria during hypoxia or ischemia–reperfusion, regulates downstream HK2 to reduce ROS production, and interacts with ATP5A1 to promote mitochondrial energy production. TIGAR translocates to the nucleus to mediate ATM phosphorylation and promotes DNA repair in response to hypoxia or antitumor drug treatment. TIGAR translocates to the ER to regulate ER stress during ischemia–reperfusion.
Signals regulating TIGAR
Transcription regulation of TIGAR
There are two p53 binding sequences in the TIGAR promoter region [2]. One is located upstream of the first exon, and the other, more effective one is located inside the first intron. As a target of p53, TIGAR can be regulated to suppress tumor cell growth mediated by p53. P53 can transcriptionally regulate TIGAR and inhibit cell death during hypoxia, while TIGAR transcription and expression are inhibited in the absence of acetylation of the DNA-binding domain in the p53 mutant, resulting in failure to suppress tumor growth [20]. P73 and p63, members of the p53 family with similar structures and functions to those of p53, can activate the promoters of p53 target genes, such as p21; therefore, p63 and p73 may also regulate TIGAR expression. TIGAR expression in humans can be regulated by p53, transactivator p51A (TAp63) and p73 protein (TAp73). However, TIGAR has different regulatory signals between mice and humans. The principal p53-responsive element in the transcriptional regulatory region of TIGAR is not consistently conserved in humans and mice. The human TIGAR promoter hBS2 binds to p53 more efficiently than the mouse protein, while the responsiveness of mBS1 to p53 is more sensitive in mice. This difference leads to a lower response to TIGAR expression by p53 in mice than in humans. Moreover, the upregulation of TIGAR following cerebral ischemia–reperfusion, cerebral preconditioning, and gut regeneration is independent of p53 and p73 protein (TAp73) [21]. A SP1 binding site exists between the TIGAR promoter −54 to −4 region. The transcription factor binds to the SP1 binding site. Four GCCC nucleotides in the cis-acting element of the TIGAR promoter and SP1 then participate in the formation of a DNA–protein complex to activate the TIGAR promoter, and to increase TIGAR expression in a p53-independent manner [22]. There is a cAMP response element (CRE) site in the −4 to +13 region of the TIGAR promoter, which can be recognized and bound by CREB. Therefore, CREB functions as a transcription factor, recognized and bound by CREB-binding protein (CPB) to regulate TIGAR expression [23]. In addition, under hypoxia, HIF1α promotes TIGAR expression by binding to the HIF response element (HRE) in the TIGAR promoter [24]. However, in the absence of oxygen-sensing prolyl hydroxylase-domain protein 1 (PHD1), nuclear factor-kappa B (NF-κB)-dependent but HIF1α-independent signals increase TIGAR transcription [25]. Mechanistically, PHD1 deficiency may reduce the degradation of IκB kinase 2 (IKK2) via reduced hydroxylation [26]. IKK2 activates NF-κB signaling through the phosphorylation of IκBα (NF-κB inhibitor), and NF-κB then increases TIGAR transcription (Fig. 3). These results suggest that PHD1 and NF-κB can also regulate the transcription of TIGAR.
Fig. 3. P53 family members, such as p53, p63, and p73, some transcription factors, such as SP1 and CREB, and noncoding miRNAs, such as miR-144, miR-885-5p, and miR-101 regulate the transcription of TIGAR.
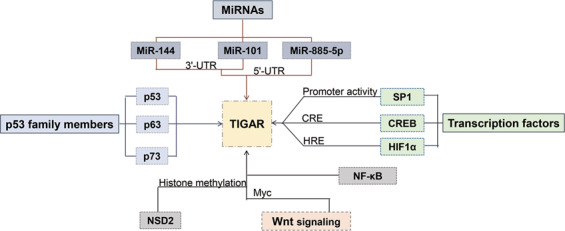
P53, p63, and p73 regulate the transcription of TIGAR by acting on the binding sites of the TIGAR promoter. SP1 can increase TIGAR transcription by binding to the TIGAR promoter. CREB increases TIGAR transcription by binding to CPB. HIF1α promotes TIGAR expression by binding to HRE. MiRNAs such as miR-144, miR-101, and miR-885-5p act on the 3′-UTR or 5′-UTR of the TIGAR promoter to inhibit TIGAR expression. In addition, NSD2 regulates TIGAR expression through histone methylation. The Wnt signaling pathway regulates TIGAR expression via Myc.
In addition to transcription factors, noncoding miRNAs, composed of 20–24 nucleotides, may mediate either a positive or negative regulatory impact on gene expression by base pairing with the mRNA ribonucleoprotein with different structures and compositions [27]. MiR-144 can bind the TIGAR 3′-UTR to reduce TIGAR expression. MiR-144 therefore inhibits cell proliferation, and promotes apoptosis and autophagy in cancer cells, suggesting that miR-144 might be used as a therapeutic target for lung cancer [28]. The TIGAR promoter region also contains two binding sites that are highly complementary to miR-885-5p, located at 1370 and 1310 bp of the transcription start site, respectively. MiR-885-5p can directly act on the miR-885-5p binding site in the TIGAR 5′-UTR region, increasing the affinity of the TIGAR chromatin conformation, and ultimately enriching RNA polymerase II in the transcriptionally active region of the promoter to regulate TIGAR expression. Pre-miR-885, the precursor of miR-885, has the same effect on TIGAR expression. In addition, the RNase III family of ribonuclease (Dicer) elicits indirect regulation of TIGAR expression by specifically recognizing double-stranded RNA to splice pre-miR-885 into mature miR-885 [29]. TIGAR is also the target gene of miR-101. MiR-101 functionally interacts with the TIGAR 3′-UTR, thus downregulating the mRNA and protein expression of TIGAR [30] (Fig. 3).
In addition, some members of the histone methyltransferase family, such as nuclear receptor-binding SET domain protein 2 (NSD2), can also regulate the TIGAR gene through the modification of histone methylation. NSD2 thus synergistically increases the catalytic activities of HK2, TIGAR, and G6PD to elevate NADPH production via PPP flux and to reduce intracellular reactive oxygen, leading to cancer cell resistance to tamoxifen therapy [31]. In contrast, DZNep (S-adenosyl homocysteine (AdoHcy) hydrolase inhibitor) induces the degradation of NSD2 protein, and reduces TIGAR expression and NADPH production, thereby inhibiting tamoxifen-resistant cancer cell growth [32]. The classic Wnt signaling pathway can be activated by the anticancer drug cisplatin. The target gene Myc in the pathway is upregulated to increase TIGAR protein expression (Fig. 3). TIGAR thus protects against cisplatin-induced cochlear ganglion damage [33].
Posttranslational modification of TIGAR
Studies have shown that the MUC1 heterodimer protein is abnormally highly expressed in colorectal cancer, and the MUC1-C subunit inhibitor (GO-203) can inhibit the phosphorylation of protein kinase B (AKT) and S6K1 to promote the expression of PDCD4, a downstream target of S6K1. GO-203 then inhibits eukaryotic initiation factor 4A (eIF4A) helicase activity, resulting in the blockade of TIGAR’s eIF4A cap-dependent translation. EIF4A can unravel the 5′-UTR end cap structure of TIGAR, allowing the preinitiation complex to bind to mRNA to initiate the translation process. The inhibition of eIF4A’s enzymatic activity reduces the translation efficiency to decrease the protein expression of TIGAR. The reduction of TIGAR then reduces GSH, increases ROS, and decreases mitochondrial transmembrane potential, which eventually leads to the inhibition of colon cancer cell growth in vivo and in vitro [34–36]. In human brain vascular endothelial cells, TIGAR can interact with calmodulin to promote TIGAR tyrosine nitration. The tyrosine 92 mutation interferes with TIGAR-dependent NADPH production, and thus abolishes the protective effect of TIGAR on the tight junction of vascular endothelial cells [10].
The biological function of TIGAR
Enzymatic activity and antioxidant effect of TIGAR
There are three known pathways of glucose metabolism in mammalian cells: aerobic metabolism, anaerobic digestion, and the PPP. TIGAR is similar to Fru-2,6-BPase of PFKFB2 isozymes in structure and function, so TIGAR can hydrolyze Fru-2,6-P2 to Fru-6-P. 6-Phosphofructokinase-1 (PFK-1) positively regulates glycolysis, while Fru-2,6-P2 is an important allosteric activator of PFK-1 to promote glycolysis. TIGAR reduces Fru-2,6-P2 levels to inhibit PFK-1 activity, thereby inhibiting glycolysis [2]. In addition to TIGAR, the cellular level of Fru-2,6-P2 is also regulated by PFKFB1–PFKFB4. Therefore, the extent to which TIGAR affects the cellular Fru-2,6-P2 level depends on its own activity, as well as on the specific PFKFB subtype that predominates in different cell types or tissues. For example, assuming that the PFKFB subtype is dominant and the kinase/bisphosphatase ratio of PFKFB is low, TIGAR will have little effect on Fru-2,6-P2 levels. PFKFB3 is an isoform with the highest kinase/bisphosphatase ratio (∼700: 1) [37]. However, the expression of PFKFB3 is extremely low in neurons [38], so neurons cannot upregulate glycolysis under stress and tend to use glucose through PPP flux to maintain their antioxidant status (see next paragraph for details). Based on this characteristic of neurons, when TIGAR is upregulated in neurons in response to ischemia/reperfusion injury, TIGAR exerts its inhibitory effect on glycolysis and increases endogenous antioxidant capacity through PPP flux, thereby protecting neurons from ischemic injury [12]. In addition, some studies have shown that the catalytic activity of TIGAR is significantly lower than that of the Fru-2,6-BPase component of the PFKFB2 isozyme in liver and muscle. These results suggest that Fru-2,6-P2 may not be the main physiological substrate of TIGAR [39]. On the other hand, TIGAR can also catalyze 2,3-bisphosphoglycerate (2,3-BPG) to 3-phosphoglycerol, while the catalytic efficiency of TIGAR for Fru-2,6-P2 is ~400-fold lower than that for 2,3-BPG. Therefore, 2,3-BPG might be more suitable as a substrate for TIGAR than Fru-2,6-P2 (Fig. 4) [39].
Fig. 4. The enzymatic activity of TIGAR.
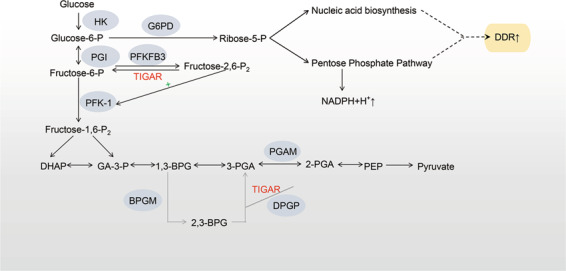
TIGAR functions as a Fru-2,6-BPase to reduce the level of Fru-2,6-P2 and inhibit the activity of PFK-1. Therefore, TIGAR can inhibit glycolysis and promote the pentose phosphate pathway and nucleotide synthesis. However, TIGAR has a stronger catalytic activity for 2,3-BPG. 1,3-BPG 1,3-diphosphoglycerate, 2,3-BPG 2,3-diphosphoglycerate, BPGM diphosphoglycerate mutase, DHAP dihydroxyacetone phosphate, DPGP 2-phosphoglycerate phosphatase, fructose-6-P fructose-6-phosphate, fructose-1,6-P2 fructose-1,6-diphosphate, fructose-2,6-P2 fructose-2,6-diphosphate, GA-3-P glyceraldehyde 3-phosphate, glucose-6-P glucose-6-phosphoric acid, HK hexokinase; PEP phosphoenolpyruvate, PFK-1 phosphofructokinase 1, PFKFB phosphofructokinase/fructose bisphosphatase, PGI glucose glucosyl isomerase, PGAM phosphoglycerate mutase, 2-PGA 2-phosphoglycerate, 3-PGA 3-phosphoglycerate.
By inhibiting glycolysis, TIGAR promotes glucose metabolism to the PPP flux by inducing Fru-6-P to produce glucose-6-phosphate (G-6-P), which will enter the PPP under catalysis by glucose-6-phosphate dehydrogenase (G6PD). In addition, research in our laboratory has found that TIGAR can increase the expression of G6PD, the PPP rate-limiting enzyme [12]. G-6-P then increases the production of NADPH and 5-ribose phosphate (R-5-P). TIGAR therefore increases the levels of NADPH and R-5-P, which is beneficial for maintaining cellular reductive redox power and reductive synthesis capability (Fig. 4) [40].
NADPH acts as a hydrogen donor to power the cell redox reaction. NADPH produces reduced GSH or Trx(SH)2 through glutathione (GSH) reductase or thioredoxin (Trx) reductase, both of which are effective in ROS elimination [41, 42]. Therefore, TIGAR shows a strong antioxidant effect, significantly reducing the level of ROS in cells [2]. The study carried out in our laboratory showed that ROS increase during cerebral ischemia–reperfusion, and ROS are believed to be an important regulator for upregulating TIGAR during ischemia–reperfusion injury [43]. In addition, NADPH and R-5-P are two key precursors for DNA synthesis and repair, so TIGAR also facilitates nucleotide synthesis and DNA repair. The CDK-5-ATM signaling pathway may be involved in TIGAR’s regulation of DNA repair [19]. TIGAR can also accelerate the reduction of the reduced protein (TRX1), thereby promoting its nuclear translocation and TRX1-dependent DNA damage repair in irradiated cells [44]. In addition, TIGAR depends on its enzymatic activity to increase acetyl-CoA levels and specifically regulate H3K9 acetylation (Fig. 5), thereby enhancing mitochondrial metabolism and the differentiation of neural stem cells (NSCs) [45].
Fig. 5. TIGAR maintains cell homeostasis.
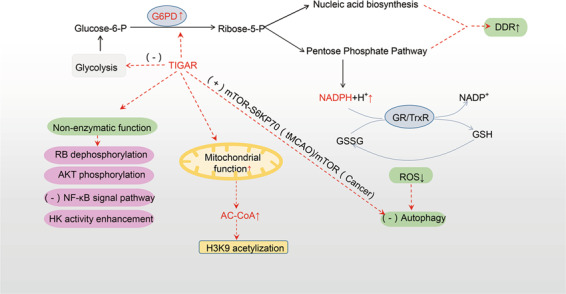
TIGAR inhibits glycolysis, directing glucose metabolism to PPP. TIGAR thus significantly reduces intracellular ROS, and promotes nucleotide synthesis and DNA damage repair. In addition to its enzymatic functions, TIGAR also has nonenzymatic functions, which affect the cell cycle, cell survival, inflammation, and mitochondrial function. TIGAR can also inhibit autophagy. These effects help maintain the energy metabolism and the stability of the internal environment. AKT protein kinase B, DDR DNA damage repair, glucose-6-P glucose-6-phosphate, G6PD glucose-6-phosphate dehydrogenase, GSSG oxidized glutathione, GSH reduced glutathione, HK hexokinase, H3K9 acetylation acetylation of histone H3 lysine residue 9, MTOR-S6KP70 rapamycin target protein-p70 ribosomal protein S6 kinase, tMCAO transient middle cerebral artery occlusion, NADPH nicotinamide adenine dinucleotide phosphate, NADP+ oxidized nicotinamide adenine dinucleotide phosphate, PPP pentose phosphate pathway, RB retinoblastoma tumor suppressor protein, ribose-5-P 5-ribose phosphate, ROS reactive oxygen species.
Interestingly, the responses of TIGAR to nitrogen species are slightly different from the responses to ROS generated from perturbations of cellular glucose metabolism. Hyperglycemia increases neuronal nitric oxide synthase (NOS) expression and NO production, while the expression of TIGAR and G6PD is downregulated. The overexpression of TIGAR effectively increases G6PD expression, but reduces the expression of NOS1 and the synthesis of NO, suggesting that TIGAR might inhibit nitrosative stress induced by hyperglycemia [46]. Significantly, in human brain microvascular endothelial cells, nitrosative stress induced by severe hypoglycemia promotes TIGAR tyrosine 92 nitration, which interferes with TIGAR-dependent NADPH production. Overexpression of TIGAR can generate NADPH by maintaining PPP and then protect the tight junctions of endothelial cells from hypoglycemic stress-induced damage [10]. Therefore, TIGAR can alleviate cellular nitrosative stress via its enzymatic activity when glucose metabolism is perturbed, but the nitration of TIGAR tyrosine by nitrogen species compromises the protective effect of TIGAR and aggravates glucose metabolic stress.
In summary, TIGAR can inhibit glycolysis and promote the PPP through its enzymatic activity. TIGAR thus shows antioxidative and DNA repair functions, helping to maintain homeostasis of the intracellular environment and metabolism.
Nonenzymatic activities of TIGAR
TIGAR’s phosphatase activity enhances antioxidative and DNA repair functions, but TIGAR also has nonenzymatic functions. TIGAR can interact with retinoblastoma protein (RB), leading to cell cycle arrest and DNA repair in cancer cells. This effect does not seem to be completely related to its phosphatase activity. Under low-level stress stimulation, TIGAR inhibits the synthesis of cyclin-dependent kinase (CDK) complex members (CDK-2, CDK-4, CDK-6, Cyclin D, and Cyclin E), resulting in RB dephosphorylation. TIGAR thus stabilizes the RB–E2F1 complex to induce p53-mediated cell cycle arrest, ultimately delaying the entry of cells into the S phase. Therefore, TIGAR-mediated cell cycle arrest can be used in combination with therapeutic drugs to accelerate the apoptosis of aggressive tumor cells [47].
The study on the progression of malignant gliomas shows that TIGAR’s role in promoting cancer metastasis is not induced by inhibiting glycolysis and NADPH production, but by interacting with AKT to phosphorylate and activate AKT. AKT then mediates the PI3K-AKT-mTOR signaling pathway, thereby promoting tumor growth and metastasis and inhibiting autophagy and apoptosis [48].
Recent studies have found that TIGAR can inhibit NF-κB signaling and inflammatory cytokines. Interestingly, TIGAR’s ability to prevent NF-κB activation is not related to TIGAR’s phosphatase activity; instead, TIGAR competes with the binding of NF-κB essential modulator (NEMO) to the HOIP subunit of the linear ubiquitin assembly complex (LUBAC). TIGAR then directly suppresses the E3-conjugating ligase activity of the LUBAC complex and prevents the linear ubiquitination of NEMO, which is required for the activation of IKKβ and other downstream targets. Thus, TIGAR plays an important physiological role in regulating adipose tissue inflammation [49].
TIGAR is transported to mitochondria under hypoxia and interacts with HK2 through a mechanism independent of its FBPase activity. TIGAR then enhances HK2 activity, reduces mitochondrial ROS, and maintains cell survival under hypoxia. This effect may promote cell survival during malignant progression [9]. In addition, research in our laboratory also found that TIGAR can translocate to mitochondria to interact with ATP5A1, thereby promoting mitochondrial function to increase ATP and reduce oxidative stress. TIGAR also promotes mitochondrial production through the SIRT1-PGC1α pathway [11]. These studies indicate that TIGAR may promote mitochondrial production and mitochondrial function through nonenzymatic effects.
Therefore, in addition to maintaining intracellular homeostasis through enzymatic activity, TIGAR can also regulate certain signal proteins through nonenzymatic actions to affect the cell cycle, cell survival, inflammation, and mitochondrial function (Fig. 5). The role of TIGAR in maintaining homeostasis and organelle functions, and promoting cell survival might be a synergy of enzymatic and nonenzymatic functions.
TIGAR’s regulation of autophagy
Since ROS is an important factor that activates autophagy, TIGAR can inhibit autophagy by promoting the PPP to reduce intracellular ROS [50]. The research in our laboratory showed that the downregulation of TIGAR can induce protective autophagy by inhibiting the target of rapamycin (mTOR) in cancer cells [51]. TIGAR inhibits autophagy by activating the mTOR-S6KP70 signaling pathway, thereby protecting neuronal damage induced by cerebral ischemia–reperfusion [52] (Fig. 5). However, studies have also shown that a reduction in TIGAR expression may facilitate ROS production, and mediate autophagy activation independent of mTOR and TP53 [50]. Hypoxia in cultured cardiomyocytes increases TIGAR expression. TIGAR then inhibits mitophagy, prevents the removal of damaged mitochondria and increases ischemic myocardial damage [53]. Contrary to the above findings, recent evidence has shown that the overexpression of TIGAR in neurons rescues autophagic impairments and increases autophagy flux under high glucose exposure. TIGAR may prevent neuronal apoptosis by promoting autophagy during hyperglycemia [46].
Therefore, under different pathological conditions, TIGAR may have different effects on autophagy or mitophagy, and TIGAR’s regulation of autophagy or mitophagy is worthy of further investigation.
TIGAR regulates stem cell differentiation
TIGAR expression increases during neural differentiation. TIGAR can promote the differentiation of NSCs by regulating the levels of associated transcriptional activators and repressors. TIGAR increases the level of lactate dehydrogenase B, enhances mitochondrial metabolism, and promotes oxidative phosphorylation. Interestingly, TIGAR increases the expression of acetyl-CoA during NSC differentiation to increase H3K9 acetylation at the promoters of glial fibrillary acidic protein (Gfap), Neurod1, and Ngn1 [54].
TIGAR also plays an essential role in regulating intestinal architecture and survival. This is because TIGAR corrects the “deficiency” of reserve intestinal stem cells (ISCs) inhibited by β-catenin/c-MYC in an activated protein 1-dependent manner. Through this effect, TIGAR facilitates the division of reserve ISCs and efficiently promotes crypt regeneration, resulting in improved mouse survival after lethal ionizing radiation injury [55]. In addition, in Fanconi anemia (FA) hematopoietic stem cells (HSCs), the p53-TIGAR metabolic axis inhibits glycolysis, reduces ROS production and DNA damage, and ameliorates chromosomal instability. TIGAR can thus prevent the premature exhaustion of FA HSCs [56].
The role of TIGAR in diseases
Cancer
TIGAR has been shown to be highly expressed in many cancer cells to promote cell survival [2, 19, 50, 51, 57], which may be related to the role of TIGAR in decreasing ROS, reducing DNA damage, and improving DNA repair in these cells. TIGAR silencing elevates epirubicin-induced ROS levels, enhances cancer cell apoptosis, reduces tumorigenicity, and increases chemosensitivity. TIGAR also suppresses autophagy. Therefore, TIGAR might increase cancer cell survival in response to chemotherapy via its dual inhibition of apoptosis and autophagy [51]. However, TIGAR’s inhibition of oxidative stress also plays an important role in the development of invasive primary cancers. TIGAR expression is dynamically regulated during the development of pancreatic ductal adenocarcinoma, resulting in lower levels of ROS to promote tumor initiation in the premalignant condition, while higher levels of ROS enable metastatic progression. TIGAR increases in the early stage of cancer, consistent with its role in limiting ROS and promoting the survival of preinvasive cells. However, progression to invasive primary tumors is accompanied by a clear decrease in TIGAR expression, and then ROS levels are increased, which enhances the signaling of mitogen-activated protein kinase by decreasing dual-specific phosphatase 6. These effects drive the dynamic transformation of cancer from a proliferative type to an invasive type, thereby enhancing the ability of migration, invasion, and metastasis. Therefore, TIGAR-mediated changes in ROS might be a vital mechanism for regulating the occurrence and progression of many malignant cancers [58].
Stroke
In 2015, our laboratory first reported the protective effect of TIGAR in cerebral ischemia–reperfusion injury and brain preconditioning [12, 59]. TIGAR expression increased rapidly during cerebral ischemia–reperfusion or brain preconditioning [12]. Downregulation of TIGAR expression in neurons and the brain increases the neuronal damage induced by ischemia–reperfusion, and abolishes the neuroprotection of brain preconditioning, while TIGAR overexpression shows neuroprotective effects in both acute and long-term cerebral ischemia–reperfusion models. TIGAR increases the survival rate, improves motor function, and promotes cognitive function recovery in stroke animals [12]. The expression level of TIGAR gradually decreases during aging, which may be related to the increased susceptibility of neurons to ischemic injury with aging [18]. The neuroprotection of TIGAR is related to the enhancement of the antioxidant capacity of neurons, mainly because TIGAR promotes the PPP flux to generate NADPH and protects mitochondrial function [12]. TIGAR also protects the tight junctions of brain microvascular endothelial cells by maintaining NADPH production [10]. In addition, TIGAR inhibits NF-κB signaling in astrocytes by reducing ROS, thereby inhibiting NF-κB-mediated astrocyte inflammation [60]. Interestingly, deficiency in oxygen-sensitive PHD1 can enhance NF-κB activity, which increases the TIGAR level and PPP flux, ameliorating neuronal death from oxygen, and glucose deprivation [25]. These results imply that the TIGAR and NF-κB signaling pathways might have complex mutual regulation.
Based on the fact that TIGAR can increase endogenous NADPH to scavenge ROS and inhibit neuronal apoptosis, we further found that the exogenous administration of NADPH shows strong neuroprotection in rodent and monkey cerebral ischemia models through antioxidant and mitochondrial protection [61].
Heart failure and myocardial ischemia
Surprisingly, TIGAR was found to aggravate myocardial ischemic injury and heart failure in both animal and cell models. Masaki Kimata et al. found that p53 and TIGAR can regulate the energy homeostasis of cardiomyocytes in response to hypoxic stress [62]. Hypoxia increased the expression of TP53 and TIGAR in cultured cardiomyocytes. Inhibition of TIGAR expression reduces apoptosis, while overexpression of TIGAR increases cardiomyocyte apoptosis and aggravates cardiac injury after ischemia [53, 62]. The mechanism is that p53/TIGAR reduces the formation of the Bnip3 homodimer. TIGAR thus inhibits mitophagy in cardiomyocytes to prevent the removal of damaged mitochondria, resulting in increased ischemic myocardial damage [53]. In addition, TIGAR reduces glucose utilization and glycolysis-dependent ATP production, which also leads to cardiomyocyte apoptosis under ischemia [19]. Consistently, in the progressive stage of heart failure, TIGAR knockdown promotes glucose oxidation and glycolysis, and maintains glucose metabolism and myocardial energy, leading to a reduction in myocardial fibrosis [63].
TIGAR plays contradictory roles in promoting both death and survival during myocardial ischemia and cerebral ischemia, which may be related to the difference in energy metabolism between cardiomyocytes and neurons. Neurons are the cells with the highest energy demands, so they are the most sensitive to energy and oxidative damage. When the oxygen supply is sufficient, neurons mainly use glucose to produce ATP through aerobic metabolism. However, due to the low activity of PFK-2 in neurons, neurons cannot effectively perform glycolysis under hypoxia, which is one of the reasons neurons are sensitive to hypoxia [64, 65]. However, neurons can utilize lactic acid released by glia using glycolysis (lactate shuttle). When the oxygen supply is restored, this lactic acid can enter the tricarboxylic acid cycle to provide energy for neurons [66]. In addition, excess ROS produced after cerebral ischemia and reperfusion [67, 68] can aggravate calcium overload [69], impair mitochondrial function [70], and promote neuronal inflammation [71]. If these excess ROS cannot be eliminated in time, they will lead to neuronal death during cerebral ischemia and reperfusion [72]. For the above reasons, neurons are highly vulnerable to ROS, and the glucose in neurons may preferentially rely on PPP to combat ROS during hypoxia [73]. Therefore, TIGAR produces antioxidants through the PPP pathway, thus exhibiting neuroprotection against cerebral ischemia–reperfusion injury [12]. On the other hand, the substrates of normal myocardial energy metabolism include the aerobic metabolism of free fatty acids (FFAs, 60%–90%) and glucose (10%–40%). During myocardial ischemia, the aerobic metabolism of FFAs and pyruvate is significantly inhibited, and cardiomyocytes tend to rely on anaerobic glycolysis to produce ATP [74–77], so the inhibitory effect of TIGAR on glycolysis may block the energy supply to cardiomyocytes. In addition, the heart has abundant mitochondria, whose structural and functional stability are essential for maintaining energy metabolism and survival of cardiomyocytes [78]. Therefore, the inhibition of mitophagy and reduction in the effective removal of damaged mitochondria by TIGAR may aggravate myocardial ischemic injury and heart failure.
Vascular disease
TIGAR exerts different effects on peripheral capillaries and central vascular endothelial cells, which may be related to the dual roles of TIGAR in regulating autophagy under different conditions. In pulmonary epithelial cells, PM2.5 exposure induces LKB1/p53/TIGAR-dependent autophagy. TIGAR triggers autophagy to upregulate VEGF transcription and protein synthesis, thereby exacerbating chronic pulmonary inflammation [79]. However, in brain microvascular endothelial cells, TIGAR may be involved in the maintenance of endothelial energy homeostasis downstream of ErbB4 (erb-b2 receptor tyrosine kinase 2), independent of the inflammatory response [80]. TIGAR, as a regulator of glucose metabolism, can maintain the balance of glycolysis and PPP. TIGAR thus inhibits excessive autophagy and the degradation of tight junction proteins by promoting PPP flux and NADPH production, and TIGAR then protects the structural integrity of the tight junctions of brain microvascular endothelial cells [10]. Therefore, due to differences in the regulation of autophagy, inflammation, and energy homeostasis, TIGAR may have opposite effects on peripheral and central capillaries.
Glycolipid metabolism
Fru-2,6-P2 can promote glycolysis and inhibit gluconeogenesis, thereby normalizing the blood glucose levels of diabetic animals [81, 82]. The increase in liver Fru-2,6-P2 levels reduces hepatic glucose production and lowers blood glucose in diabetic patients [83]. By contrast, glucagon reduces Fru-2,6-P2 levels, and promotes gluconeogenesis and glycogenolysis, resulting in increased blood glucose [10, 82]. Similarly, TIGAR can reduce Fru-2,6-P2 levels and inhibit glycolysis, which may lead to increased blood glucose and insulin resistance [84]. TIGAR-mediated decline in Fru-2,6-P2 is related to metabolic disorders in alcoholic fatty liver. In a rat model of type 2 diabetes, exercise reduces TIGAR and improves insulin resistance [85]. The above research demonstrates that targeting TIGAR to increase Fru-2,6-P2 is a promising strategy for the treatment of diabetes.
In addition, in a study regarding maternal obesity-induced meiotic defects in oocytes, TIGAR protein expression was reduced in oocytes of mice fed a high-fat diet. TIGAR-specific depletion led to a significant increase in ROS and activation of autophagy, leading to spindle defects or misaligned chromosomes during meiosis. The study revealed that TIGAR can reduce the abnormality of meiosis in oocytes induced by maternal obesity [86].
Alzheimer’s disease
TP53 can inhibit glycolysis by activating TIGAR, directing glucose metabolism to the PPP to maintain a sufficient level of reducing molecules and to protect against DNA damage-induced apoptosis. At different stages of severity in Alzheimer’s disease (AD) dementia, TIGAR protein levels were found to be decreased, but the ATM signaling and DNA damage response network was activated. This finding suggests a loss of protection initiated by ATM-p53 signaling against intensifying oxidative stress in AD. The progressive decrease in TIGAR expression is in agreement with the findings of altered posttranslational modification of TP53, which result in increased formation of functionally inactive TP53 monomers and dimers, but not functionally active TP53 tetramers in AD brains. Similarly, TIGAR protein expression is significantly reduced in patients with dementia. The strong downregulation of TIGAR expression is related to the severity of dementia, but not to histopathological changes in AD, such as neuritic plaques and nerve fiber twining [87].
Parkinson’s disease
Since oxidative damage to dopaminergic nigrostriatal neurons plays an important role in the pathogenesis of Parkinson’s disease (PD), TIGAR, which is clearly associated with neuronal glucose metabolism, PPP activity and oxidative stress, is likely to be involved in the pathogenesis of PD [88]. Exogenous NADPH administration prevents 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) toxicity to dopaminergic neurons by increasing GSH, reducing ROS and inhibiting neuroinflammation in the substantia nigra pars compacta of mice. NADPH therefore improves the motor function of PD animals [54, 89]. Moreover, overexpression of G6PD in mouse dopaminergic neurons shows a protective effect against MPTP toxicity [90]. The above findings suggest that TIGAR may become a potential target for PD treatment by promoting PPP, increasing NADPH production, and reducing oxidative stress.
However, contrary evidence suggests that TIGAR may have a negative effect on the pathogenesis of PD. Loss of function mutations in PTEN-induced kinase 1 (PINK1) typically lead to early onset PD. In pink1(−/−) zebrafish, TigarB is upregulated. TigarB inactivation rescued dopaminergic neuron damage. Moreover, TIGAR has also been identified as a negative regulator of mitophagy, whereas impaired mitophagy is currently considered to be a crucial mechanism in the pathogenesis of early onset of PD [91]. In addition, TIGAR is present in Lewy bodies in the substantia nigra of the alpha synucleinopathies PD and Lewy body dementia. These studies suggest a potential role of TIGAR in the pathogenesis of PD and Lewy body disorders [92]. In summary, the paradoxical role of TIGAR in PD remains to be further investigated.
Conclusions
TIGAR is one of the target genes of p53, and the expression of its encoded protein is related to tissue types, developmental stages, and physiological and pathological status. TIGAR expression is regulated in a p53-dependent or p53-independent manner. Transcription factors such as SP1, CREB, and HIF1α can interact with the corresponding sites of the TIGAR promoter to increase TIGAR expression. TIGAR is distributed in the cytoplasm, mitochondria, ER, and nucleus. TIGAR exerts its functions via both enzymatic and nonenzymatic activity. TIGAR mainly exerts enzymatic activity to hydrolyze Fru-2,6-P2 to promote PPP. TIGAR interacts with several signaling proteins to promote mitochondrial function or cell survival. TIGAR has multiple important biological functions, and might become a potential therapeutic target for cardiovascular and neurological diseases and cancers through different interventions (Table 1).
Table 1.
Role and mechanism of TIGAR in different disease models.
| Disease | Model | Modulation on TIGAR | Mechanism of action | Effects | Reference |
|---|---|---|---|---|---|
| Cancer | Leukemia | ↑ | ROS ↓, DNA damage ↓, DNA repair ↑, autophagy ↓ | Promote cancer cell survival | [13] |
| Lung cancer | [15, 28] | ||||
| Breast carcinoma | [16] | ||||
| Glioma | [17] | ||||
| Hepatoma | [19, 50] | ||||
| Colon cancer | [34–36] | ||||
| Nasopharyngeal cancer | [56] | ||||
| Stroke | Neuronal oxygen glucose deprivation–reperfusion (OGD/R) | ↑ | Pentose phosphate pathway flux ↑, NADPH ↑ | Promote neuronal survival | [12, 18, 60] |
| Transient middle cerebral artery occlusion (tMCAO) | [12, 18, 60] | ||||
| Heart failure and myocardial ischemia | Coronary artery ligation | ↑ | Mitophagy in cardiomyocytes ↓, apoptosis ↑ | Aggravate cardiomyocytes death | [53] |
| Transverse aortic constriction (TAC) | [63] | ||||
| Vascular disease | Human brain microvascular endothelial cells | ↑ | Excessive autophagy ↓ | Neurovascular protection | [10] |
| Bronchial epithelial cells | Autophagy ↑ | Induction of airway inflammatory responses | [79] | ||
| Endothelial ErbB4 conditional knockout mice | ↓ | Pentose phosphate pathway flux ↑ | Improve energy metabolism | [80] | |
| Glycolipid metabolism | Type 2 diabetes (T2DM) rat | ↑ | Fru-2,6-P2 levels↓, (−) glycolysis | Increase blood glucose and insulin resistance | [84] |
| Alzheimer’s disease | Human AD brain | ↑ | Antioxidant function | Potential protection for AD | [87] |
| Parkinson’s disease | Pink1−/− zebrafish | ↑ | Negatively regulate mitochondrial function | Promote dopaminergic neurons death | [91] |
| PD patients′ brains | [92] |
Although research has revealed the structure and multiple functions of TIGAR, many unknown issues remain to be elucidated. For example, the regulatory mechanisms of TIGAR localization in nonmitochondrial subcellular organelles, such as the ER and nucleus, remain unclear. These mechanisms may depend on the recruitment of TIGAR by certain signaling molecules or on specific sequences in the TIGAR structure. The mechanism involved in the regulation of TIGAR relocation in the ER and nucleus in response to stress is an important issue worthy of further study. In addition, since TIGAR intervention may be a potential target for cardiovascular diseases, neurological disorders, and cancer, there is an urgent need to find pharmacological agents that directly target TIGAR for the prevention and treatment of these diseases. For example, TIGAR inducers may be used in the treatment of cerebrovascular diseases and neurological diseases, while TIGAR inhibitors may have important therapeutic effects in cancer. These studies may provide innovative insights into the role of TIGAR in human health and diseases, and provide drug candidates for the treatment of certain diseases.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81673421, 81973315 and 81730092), Natural Science Foundation of Jiangsu Higher Education (20KJA310008), Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), and the Priority Academic Program Development of the Jiangsu Higher Education Institutes (PAPD).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jie Tang, Lei Chen
References
- 1.Jen KY, Cheung VG. Identification of novel p53 target genes in ionizing radiation response. Cancer Res. 2005;65:7666–73.. doi: 10.1158/0008-5472.CAN-05-1039. [DOI] [PubMed] [Google Scholar]
- 2.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20.. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 3.Wong EY, Wong SC, Chan CM, Lam EK, Ho LY, Lau CP, et al. TP53-induced glycolysis and apoptosis regulator promotes proliferation and invasiveness of nasopharyngeal carcinoma cells. Oncol Lett. 2015;9:569–74.. doi: 10.3892/ol.2014.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang P, De Souza C, Minn K, Chien J. Genome-scale CRISPR knockout screen identifies TIGAR as a modifier of PARP inhibitor sensitivity. Commun Biol. 2019;2:335. doi: 10.1038/s42003-019-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee P, Vousden KH, Cheung EC. TIGAR, TIGAR, burning bright. Cancer Metab. 2014;2:1. doi: 10.1186/2049-3002-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigden DJ. The histidine phosphatase superfamily: structure and function. Biochem J. 2008;409:333–48.. doi: 10.1042/BJ20071097. [DOI] [PubMed] [Google Scholar]
- 7.Bazan JF, Fletterick RJ, Pilkis SJ. Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Proc Natl Acad Sci USA. 1989;86:9642–6. doi: 10.1073/pnas.86.24.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Jogl G. Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator) J Biol Chem. 2009;284:1748–54.. doi: 10.1074/jbc.M807821200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung EC, Ludwig RL, Vousden KH. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc Natl Acad Sci USA. 2012;109:20491–6. doi: 10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CK, Ahmed MM, Jiang Q, Lu NN, Tan C, Gao YP, et al. Melatonin ameliorates hypoglycemic stress-induced brain endothelial tight junction injury by inhibiting protein nitration of TP53-induced glycolysis and apoptosis regulator. J. Pineal Res. 2017;63:e12440. [DOI] [PMC free article] [PubMed]
- 11.Geng J, Wei M, Yuan X, Liu Z, Wang X, Zhang D, et al. TIGAR regulates mitochondrial functions through SIRT1-PGC1alpha pathway and translocation of TIGAR into mitochondria in skeletal muscle. FASEB J. 2019;33:6082–98.. doi: 10.1096/fj.201802209R. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Sun M, Cao L, Gu JH, Ge J, Chen J, et al. A TIGAR-regulated metabolic pathway is critical for protection of brain ischemia. J Neurosci. 2014;34:7458–71.. doi: 10.1523/JNEUROSCI.4655-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian S, Li J, Hong M, Zhu Y, Zhao H, Xie Y, et al. TIGAR cooperated with glycolysis to inhibit the apoptosis of leukemia cells and associated with poor prognosis in patients with cytogenetically normal acute myeloid leukemia. J Hematol Oncol. 2016;9:128. doi: 10.1186/s13045-016-0360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung EC, Athineos D, Lee P, Ridgway RA, Lambie W, Nixon C, et al. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev Cell. 2013;25:463–77.. doi: 10.1016/j.devcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Lu F, Gong Y, Zhao C, Pan Q, Ballantyne S, et al. High expression of synthesis of cytochrome c oxidase 2 and TP53-induced glycolysis and apoptosis regulator can predict poor prognosis in human lung adenocarcinoma. Hum Pathol. 2018;77:54–62. doi: 10.1016/j.humpath.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Won KY, Lim SJ, Kim GY, Kim YW, Han SA, Song JY, et al. Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Hum Pathol. 2012;43:221–8. doi: 10.1016/j.humpath.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Wanka C, Steinbach JP, Rieger J. Tp53-induced glycolysis and apoptosis regulator (TIGAR) protects glioma cells from starvation-induced cell death by up-regulating respiration and improving cellular redox homeostasis. J Biol Chem. 2012;287:33436–46.. doi: 10.1074/jbc.M112.384578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao L, Chen J, Li M, Qin YY, Sun M, Sheng R, et al. Endogenous level of TIGAR in brain is associated with vulnerability of neurons to ischemic injury. Neurosci Bull. 2015;31:527–40.. doi: 10.1007/s12264-015-1538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu HP, Xie JM, Li B, Sun YH, Gao QG, Ding ZH, et al. TIGAR regulates DNA damage and repair through pentosephosphate pathway and Cdk5-ATM pathway. Sci Rep. 2015;5:9853. doi: 10.1038/srep09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, et al. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–73.. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee P, Hock AK, Vousden KH, Cheung EC. p53- and p73-independent activation of TIGAR expression in vivo. Cell Death Dis. 2015;6:e1842. doi: 10.1038/cddis.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou S, Gu Z, Ni P, Liu X, Wang J, Fan Q. SP1 plays a pivotal role for basal activity of TIGAR promoter in liver cancer cell lines. Mol Cell Biochem. 2012;359:17–23. doi: 10.1007/s11010-011-0993-0. [DOI] [PubMed] [Google Scholar]
- 23.Zou S, Wang X, Deng L, Wang Y, Huang B, Zhang N, et al. CREB, another culprit for TIGAR promoter activity and expression. Biochem Biophys Res Commun. 2013;439:481–6. doi: 10.1016/j.bbrc.2013.08.098. [DOI] [PubMed] [Google Scholar]
- 24.Rajendran R, Garva R, Ashour H, Leung T, Stratford I, Krstic-Demonacos M, et al. Acetylation mediated by the p300/CBP-associated factor determines cellular energy metabolic pathways in cancer. Int J Oncol. 2013;42:1961–72.. doi: 10.3892/ijo.2013.1907. [DOI] [PubMed] [Google Scholar]
- 25.Quaegebeur A, Segura I, Schmieder R, Verdegem D, Decimo I, Bifari F, et al. Deletion or Inhibition of the oxygen sensor PHD1 protects against ischemic stroke via reprogramming of neuronal metabolism. Cell Metab. 2016;23:280–91.. doi: 10.1016/j.cmet.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholz CC, Taylor CT. Hydroxylase-dependent regulation of the NF-kappaB pathway. Biol Chem. 2013;394:479–93.. doi: 10.1515/hsz-2012-0338. [DOI] [PubMed] [Google Scholar]
- 27.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Li P, Li J, Wang Y, Du Y, Chen X, et al. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem. 2015;35:997–1007. doi: 10.1159/000369755. [DOI] [PubMed] [Google Scholar]
- 29.Zou S, Rao Y, Chen W. miR-885-5p plays an accomplice role in liver cancer by instigating TIGAR expression via targeting its promoter. Biotechnol Appl Biochem. 2019;66:763–71.. doi: 10.1002/bab.1767. [DOI] [PubMed] [Google Scholar]
- 30.Huang S, Yang Z, Ma Y, Yang Y, Wang S. miR-101 enhances cisplatin-induced dna damage through decreasing nicotinamide adenine dinucleotide phosphate levels by directly repressing Tp53-induced glycolysis and apoptosis regulator expression in prostate cancer cells. DNA Cell Biol. 2017;36:303–10.. doi: 10.1089/dna.2016.3612. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Duan Z, Nugent Z, Zou JX, Borowsky AD, Zhang Y, et al. Reprogramming metabolism by histone methyltransferase NSD2 drives endocrine resistance via coordinated activation of pentose phosphate pathway enzymes. Cancer Lett. 2016;378:69–79. doi: 10.1016/j.canlet.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Zheng J, Zou JX, Xu J, Han F, Xiang S, et al. S-adenosylhomocysteine (AdoHcy)-dependent methyltransferase inhibitor DZNep overcomes breast cancer tamoxifen resistance via induction of NSD2 degradation and suppression of NSD2-driven redox homeostasis. Chem-Biol Interact. 2020;317:108965. doi: 10.1016/j.cbi.2020.108965. [DOI] [PubMed] [Google Scholar]
- 33.Liu W, Xu X, Fan Z, Sun G, Han Y, Zhang D, et al. Wnt signaling activates TP53-induced glycolysis and apoptosis regulator and protects against cisplatin-induced spiral ganglion neuron damage in the mouse cochlea. Antioxid redox Signal. 2019;30:1389–410.. doi: 10.1089/ars.2017.7288. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad R, Alam M, Hasegawa M, Uchida Y, Al-Obaid O, Kharbanda S, et al. Targeting MUC1-C inhibits the AKT-S6K1-elF4A pathway regulating TIGAR translation in colorectal cancer. Mol Cancer. 2017;16:33. doi: 10.1186/s12943-017-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GongSun X, Zhao Y, Jiang B, Xin Z, Shi M, Song L, et al. Inhibition of MUC1-C regulates metabolism by AKT pathway in esophageal squamous cell carcinoma. J Cell Physiol. 2019;234:12019–28. doi: 10.1002/jcp.27863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45.. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakakibara R, Uemura M, Hirata T, Okamura N, Kato M. Human placental fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase: its isozymic form, expression and characterization. Biosci Biotechnol Biochem. 1997;61:1949–52.. doi: 10.1271/bbb.61.1949. [DOI] [PubMed] [Google Scholar]
- 38.Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–52.. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- 39.Gerin I, Noel G, Bolsee J, Haumont O, Van Schaftingen E, Bommer GT. Identification of TP53-induced glycolysis and apoptosis regulator (TIGAR) as the phosphoglycolate-independent 2,3-bisphosphoglycerate phosphatase. Biochem J. 2014;458:439–48.. doi: 10.1042/BJ20130841. [DOI] [PubMed] [Google Scholar]
- 40.Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer Biol Ther. 2006;5:1610–3. doi: 10.4161/cbt.5.12.3617. [DOI] [PubMed] [Google Scholar]
- 41.Blacker TS, Duchen MR. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic Biol Med. 2016;100:53–65. doi: 10.1016/j.freeradbiomed.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green DR, Chipuk JE. p53 and metabolism: inside the TIGAR. Cell. 2006;126:30–2. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 43.Sun M, Li M, Huang Q, Han F, Gu JH, Xie J, et al. Ischemia/reperfusion-induced upregulation of TIGAR in brain is mediated by SP1 and modulated by ROS and hormones involved in glucose metabolism. Neurochem Int. 2015;80:99–109. doi: 10.1016/j.neuint.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Gu C, Yu J, Wang Z, Yuan X, Yang L, et al. Radiosensitization of glioma cells by TP53-induced glycolysis and apoptosis regulator knockdown is dependent on thioredoxin-1 nuclear translocation. Free Radic Biol Med. 2014;69:239–48. doi: 10.1016/j.freeradbiomed.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 45.Zhou W, Zhao T, Du J, Ji G, Li X, Ji S, et al. TIGAR promotes neural stem cell differentiation through acetyl-CoA-mediated histone acetylation. Cell Death Dis. 2019;10:198. doi: 10.1038/s41419-019-1434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Yao Y, Li J, Wu D, Zhao M, Yan Z, et al. TIGAR attenuates high glucose-induced neuronal apoptosis via an autophagy pathway. Front Mol Neurosci. 2019;12:193. doi: 10.3389/fnmol.2019.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madan E, Gogna R, Kuppusamy P, Bhatt M, Pati U, Mahdi AA. TIGAR induces p53-mediated cell-cycle arrest by regulation of RB-E2F1 complex. Br J Cancer. 2012;107:516–26.. doi: 10.1038/bjc.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Z, He Z. TIGAR promotes growth, survival and metastasis through oxidation resistance and AKT activation in glioblastoma. Oncol Lett. 2019;18:2509–17.. doi: 10.3892/ol.2019.10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y, Kwon H, Neel BA, Kasher-Meron M, Pessin JB, Yamada E, et al. The fructose-2,6-bisphosphatase TIGAR suppresses NF-kappaB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. J Biol Chem. 2018;293:7578–91.. doi: 10.1074/jbc.RA118.002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–26.. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie JM, Li B, Yu HP, Gao QG, Li W, Wu HR, et al. TIGAR has a dual role in cancer cell survival through regulating apoptosis and autophagy. Cancer Res. 2014;74:5127–38.. doi: 10.1158/0008-5472.CAN-13-3517. [DOI] [PubMed] [Google Scholar]
- 52.Zhang DM, Zhang T, Wang MM, Wang XX, Qin YY, Wu J, et al. TIGAR alleviates ischemia/reperfusion-induced autophagy and ischemic brain injury. Free Radic Biol Med. 2019;137:13–23. doi: 10.1016/j.freeradbiomed.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, et al. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–84.. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Zhou JS, Zhu Z, Wu F, Zhou Y, Sheng R, Wu JC, et al. NADPH ameliorates MPTP-induced dopaminergic neurodegeneration through inhibiting p38MAPK activation. Acta Pharmacol Sin. 2019;40:180–91.. doi: 10.1038/s41401-018-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F, Zhang Y, Hu S, Shi X, Wang Z, Deng Z, et al. TIGAR/AP-1 axis accelerates the division of Lgr5(-) reserve intestinal stem cells to reestablish intestinal architecture after lethal radiation. Cell Death Dis. 2020;11:501. doi: 10.1038/s41419-020-2715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, Wu L, Zopp M, Kopelov S, Du W. p53-TP53-induced glycolysis regulator mediated glycolytic suppression attenuates DNA damage and genomic instability in fanconi anemia hematopoietic stem cells. Stem Cells. 2019;37:937–47.. doi: 10.1002/stem.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lui VW, Wong EY, Ho K, Ng PK, Lau CP, Tsui SK, et al. Inhibition of c-Met downregulates TIGAR expression and reduces NADPH production leading to cell death. Oncogene. 2011;30:1127–34.. doi: 10.1038/onc.2010.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, et al. Dynamic ROS control by TIGAR regulates the initiation and progression of pancreatic cancer. Cancer Cell. 2020;37:168–82.e4. doi: 10.1016/j.ccell.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou JH, Zhang TT, Song DD, Xia YF, Qin ZH, Sheng R. TIGAR contributes to ischemic tolerance induced by cerebral preconditioning through scavenging of reactive oxygen species and inhibition of apoptosis. Sci Rep. 2016;6:27096. doi: 10.1038/srep27096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Zhang DM, Feng X, Wang J, Qin YY, Zhang T, et al. TIGAR inhibits ischemia/reperfusion-induced inflammatory response of astrocytes. Neuropharmacology. 2018;131:377–88.. doi: 10.1016/j.neuropharm.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Li M, Zhou ZP, Sun M, Cao L, Chen J, Qin YY, et al. Reduced nicotinamide adenine dinucleotide phosphate, a pentose phosphate pathway product, might be a novel drug candidate for ischemic stroke. Stroke. 2016;47:187–95.. doi: 10.1161/STROKEAHA.115.009687. [DOI] [PubMed] [Google Scholar]
- 62.Kimata M, Matoba S, Iwai-Kanai E, Nakamura H, Hoshino A, Nakaoka M, et al. p53 and TIGAR regulate cardiac myocyte energy homeostasis under hypoxic stress. Am J Physiol Heart Circ Physiol. 2010;299:H1908–16. doi: 10.1152/ajpheart.00250.2010. [DOI] [PubMed] [Google Scholar]
- 63.Okawa Y, Hoshino A, Ariyoshi M, Kaimoto S, Tateishi S, Ono K, et al. Ablation of cardiac TIGAR preserves myocardial energetics and cardiac function in the pressure overload heart failure model. Am J Physiol Heart Circ Physiol. 2019;316:H1366–H77.. doi: 10.1152/ajpheart.00395.2018. [DOI] [PubMed] [Google Scholar]
- 64.Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb Symp Quant Biol. 2011;76:211–6. doi: 10.1101/sqb.2011.76.010868. [DOI] [PubMed] [Google Scholar]
- 65.Chesney J. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and tumor cell glycolysis. Curr Opin Clin Nutr Metab Care. 2006;9:535–9. doi: 10.1097/01.mco.0000241661.15514.fb. [DOI] [PubMed] [Google Scholar]
- 66.Genc S, Kurnaz IA, Ozilgen M. Astrocyte-neuron lactate shuttle may boost more ATP supply to the neuron under hypoxic conditions-in silico study supported by in vitro expression data. BMC Syst Biol. 2011;5:162. doi: 10.1186/1752-0509-5-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Awooda HA, Lutfi MF, Sharara GG, Saeed AM. Oxidative/nitrosative stress in rats subjected to focal cerebral ischemia/reperfusion. Int J health Sci. 2015;9:17–24. doi: 10.12816/0024679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmed E, Donovan T, Yujiao L, Zhang Q. Mitochondrial targeted antioxidant in cerebral ischemia. J Neurol Neurosci. 2015;6:17. [DOI] [PMC free article] [PubMed]
- 69.Baines CP. The mitochondrial permeability transition pore and ischemia-reperfusion injury. Basic Res Cardiol. 2009;104:181–8. doi: 10.1007/s00395-009-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tajeddine N. How do reactive oxygen species and calcium trigger mitochondrial membrane permeabilisation? Biochim Biophys Acta. 2016;1860:1079–88.. doi: 10.1016/j.bbagen.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Kvietys PR, Granger DN. Role of reactive oxygen and nitrogen species in the vascular responses to inflammation. Free Radic Biol Med. 2012;52:556–92.. doi: 10.1016/j.freeradbiomed.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugawara T, Chan PH. Reactive oxygen radicals and pathogenesis of neuronal death after cerebral ischemia. Antioxid Redox Signal. 2003;5:597–607. doi: 10.1089/152308603770310266. [DOI] [PubMed] [Google Scholar]
- 73.Fernandez-Fernandez S, Almeida A, Bolanos JP. Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem J. 2012;443:3–11. doi: 10.1042/BJ20111943. [DOI] [PubMed] [Google Scholar]
- 74.Xu KY, Zweier JL, Becker LC. Functional coupling between glycolysis and sarcoplasmic reticulum Ca2+ transport. Circ Res. 1995;77:88–97. doi: 10.1161/01.RES.77.1.88. [DOI] [PubMed] [Google Scholar]
- 75.Bertero E, Maack C. Metabolic remodelling in heart failure. Nat Rev Cardiol. 2018;15:457–70.. doi: 10.1038/s41569-018-0044-6. [DOI] [PubMed] [Google Scholar]
- 76.Lopaschuk GD, Ussher JR. Evolving concepts of myocardial energy metabolism: more than just fats and carbohydrates. Circ Res. 2016;119:1173–6. doi: 10.1161/CIRCRESAHA.116.310078. [DOI] [PubMed] [Google Scholar]
- 77.Boyman L, Karbowski M, Lederer WJ. Regulation of mitochondrial ATP production: Ca2+ signaling and quality control. Trends Mol Med. 2020;26:21–39. doi: 10.1016/j.molmed.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertero E, Maack C. Calcium signaling and reactive oxygen species in mitochondria. Circ Res. 2018;122:1460–78.. doi: 10.1161/CIRCRESAHA.118.310082. [DOI] [PubMed] [Google Scholar]
- 79.Xu H, Xu X, Wang H, Qimuge A, Liu S, Chen Y, et al. LKB1/p53/TIGAR/autophagy-dependent VEGF expression contributes to PM2.5-induced pulmonary inflammatory responses. Sci Rep. 2019;9:16600. doi: 10.1038/s41598-019-53247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu G, Liu XX, Lu NN, Liu QB, Tian Y, Ye WF, et al. Endothelial ErbB4 deficit induces alterations in exploratory behavior and brain energy metabolism in mice. CNS Neurosci Ther. 2017;23:510–7. doi: 10.1111/cns.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monge L, Mojena M, Ortega JL, Samper B, Cabello MA, Feliu JE. Chlorpropamide raises fructose-2,6-bisphosphate concentration and inhibits gluconeogenesis in isolated rat hepatocytes. Diabetes. 1986;35:89–96. doi: 10.2337/diab.35.1.89. [DOI] [PubMed] [Google Scholar]
- 82.Rodriguez-Gil JE, Gomez-Foix AM, Fillat C, Bosch F, Guinovart JJ. Activation by vanadate of glycolysis in hepatocytes from diabetic rats. Diabetes. 1991;40:1355–9. doi: 10.2337/diab.40.10.1355. [DOI] [PubMed] [Google Scholar]
- 83.Choi IY, Wu C, Okar DA, Lange AJ, Gruetter R. Elucidation of the role of fructose 2,6-bisphosphate in the regulation of glucose fluxes in mice using in vivo [13C] NMR measurements of hepatic carbohydrate metabolism. Eur J Biochem. 2002;269:4418–26.. doi: 10.1046/j.1432-1033.2002.t01-1-03125.x. [DOI] [PubMed] [Google Scholar]
- 84.Derdak Z, Lang CH, Villegas KA, Tong M, Mark NM, de la Monte SM, et al. Activation of p53 enhances apoptosis and insulin resistance in a rat model of alcoholic liver disease. J Hepatol. 2011;54:164–72.. doi: 10.1016/j.jhep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qi Z, He J, Zhang Y, Shao Y, Ding S. Exercise training attenuates oxidative stress and decreases p53 protein content in skeletal muscle of type 2 diabetic Goto-Kakizaki rats. Free Radic Biol Med. 2011;50:794–800. doi: 10.1016/j.freeradbiomed.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 86.Wang H, Cheng Q, Li X, Hu F, Han L, Zhang H, et al. Loss of TIGAR induces oxidative stress and meiotic defects in oocytes from obese mice. Mol Cell Proteomics. 2018;17:1354–64.. doi: 10.1074/mcp.RA118.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katsel P, Tan W, Fam P, Purohit DP, Haroutunian V. Cell cycle checkpoint abnormalities during dementia: a plausible association with the loss of protection against oxidative stress in Alzheimer’s disease [corrected]. PLoS One. 2013;8:e68361. doi: 10.1371/journal.pone.0068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dunn L, eld VF, Daham S, Bolaños JP, Heales SJ. Pentose-phosphate pathway disruption in the pathogenesis of Parkinson’s disease. Transl Neurosci. 2014;5:179–84. doi: 10.2478/s13380-014-0221-y. [DOI] [Google Scholar]
- 89.Zhou Y, Wu J, Sheng R, Li M, Wang Y, Han R, et al. Reduced nicotinamide adenine dinucleotide phosphate inhibits MPTP-induced neuroinflammation and neurotoxicity. Neuroscience. 2018;391:140–53.. doi: 10.1016/j.neuroscience.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 90.Mejias R, Villadiego J, Pintado CO, Vime PJ, Gao L, Toledo-Aral JJ, et al. Neuroprotection by transgenic expression of glucose-6-phosphate dehydrogenase in dopaminergic nigrostriatal neurons of mice. J Neurosci. 2006;26:4500–8. doi: 10.1523/JNEUROSCI.0122-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flinn LJ, Keatinge M, Bretaud S, Mortiboys H, Matsui H, De Felice E, et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann Neurol. 2013;74:837–47.. doi: 10.1002/ana.23999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lopez KLR, Simpson JE, Watson LC, Mortiboys H, Hautbergue GM, Bandmann O, et al. TIGAR inclusion pathology is specific for Lewy body diseases. Brain Res. 2019;1706:218–23.. doi: 10.1016/j.brainres.2018.09.032. [DOI] [PubMed] [Google Scholar]


