Significance
Global change threatens the genetic diversity of economically important and foundational ecosystem-building species such as corals. We tested whether cryopreserved coral sperm could be used to transfer genetic diversity among genetically isolated populations of the critically endangered Caribbean elkhorn coral, Acropora palmata. Here we report successful assisted gene flow (AGF) in corals using cryopreserved sperm, yielding the largest living wildlife population ever created from cryopreserved cells. Furthermore, we produced direct evidence that genetically distinct populations of Caribbean coral can interbreed. Thus, we demonstrated that sperm cryopreservation can enable efficient, large-scale AGF in corals. This form of assisted genetic migration can enhance genetic diversity and help critically endangered species adapt to local environments in the face of rapid global change.
Keywords: assisted gene flow, coral reproduction, cryopreservation, endangered species, Acropora palmata
Abstract
Assisted gene flow (AGF) is a conservation intervention to accelerate species adaptation to climate change by importing genetic diversity into at-risk populations. Corals exemplify both the need for AGF and its technical challenges; corals have declined in abundance, suffered pervasive reproductive failures, and struggled to adapt to climate change, yet mature corals cannot be easily moved for breeding, and coral gametes lose viability within hours. Here, we report the successful demonstration of AGF in corals using cryopreserved sperm that was frozen for 2 to 10 y. We fertilized Acropora palmata eggs from the western Caribbean (Curaçao) with cryopreserved sperm from genetically distinct populations in the eastern and central Caribbean (Florida and Puerto Rico, respectively). We then confirmed interpopulation parentage in the Curaçao–Florida offspring using 19,696 single-nucleotide polymorphism markers. Thus, we provide evidence of reproductive compatibility of a Caribbean coral across a recognized barrier to gene flow. The 6-mo survival of AGF offspring was 42%, the highest ever achieved in this species, yielding the largest wildlife population ever raised from cryopreserved material. By breeding a critically endangered coral across its range without moving adults, we show that AGF using cryopreservation is a viable conservation tool to increase genetic diversity in threatened marine populations.
Assisted gene flow (AGF) is a conservation genetic intervention to accelerate the adaptation of plant and animal populations to environmental change (1–7). As a form of assisted migration, AGF involves the translocation of individual organisms or their germplasm across a species’ current range to transfer naturally occurring allelic diversity into a local population and thus support its adaptation to changing conditions (1, 4, 5). Climate change disproportionately threatens fragile, sessile, and slow-growing species such as reef-building corals, which have suffered widespread losses in past decades (8, 9). Unable to migrate to safer habitats, corals must adapt to global change by relying primarily on standing genetic variation (10) and the import of new genetic variation via larval recruitment, yet pervasive reproductive failure, recruitment failure, and population declines impede this (11).
AGF is a promising intervention to enhance standing genetic diversity in threatened species because it moves alleles among historically or recently isolated populations in different environmental conditions while adding new, sexually produced genotypes. This facilitated crossbreeding approach relies on the presumption that populations are locally adapted to historical environmental conditions and that donor populations can thus contribute valuable alleles to help recipient populations adapt to changing environments. For example, by moving alleles from warmer coral reefs to rapidly warming reefs, the pace of thermal adaptation could be accelerated. Importantly, this is possible even when the genetic architecture of adaptive traits is not known (10, 12).
As proof-of-concept that AGF can accelerate thermal adaptation, adults from one Pacific coral species were moved from a warmer to cooler habitat (13). Crossbreeding with the local population produced offspring with heritable increases in thermal tolerance. However, moving reproductive adult colonies proved costly (13) and imposed damage on the source reef. Furthermore, translocating adult corals can facilitate the spread of pathogens and invasive species because corals harbor diverse microbes, fungi, parasites, and endolithic fauna (14, 15). In contrast, translocating gametes would represent a less-destructive, less-costly, and lower-risk approach to large-scale AGF, but the nature of gamete release in wild corals has prevented this until now.
Most coral species release gametes during spawning events on a few days per year (16, 17). Unlike plant seeds, freshly released coral eggs and sperm are only viable for minutes to hours (18–20); hence, they cannot be transported to achieve AGF. However, recent cryopreservation progress has enabled the freezing, storage, transport, and thawing of live coral sperm (21, 22). Using two Pacific coral species, we previously demonstrated that freshly collected eggs could be fertilized using frozen-thawed (FT) sperm and the resulting larvae had equal settlement success compared to larvae produced from conspecifics using fresh sperm (23). These advances built upon decades of cryopreservation work in other endangered species, including the black-footed ferret (24) and the cheetah (25).
The Caribbean elkhorn coral, Acropora palmata (Fig. 1A), is a formerly dominant, shallow-water species that has declined by over 95% since the 1980s (8). Its populations have been compromised by habitat loss, poor water quality, physical damage, predation, algal overgrowth, and temperature-induced bleaching (26, 27), leading to its designations as “threatened” on the US Endangered Species List (28) and “critically endangered” on the International Union for Conservation of Nature (IUCN) Red List (29). The US government recovery plan for A. palmata mandates conservation of genetic diversity and active restoration of populations, but both have been hampered by practical and technical barriers including asynchronous spawning times, low gamete quality, high juvenile mortality after settlement, and the population genetic structure of the species itself, which is highly clonal at many sites (30–33).
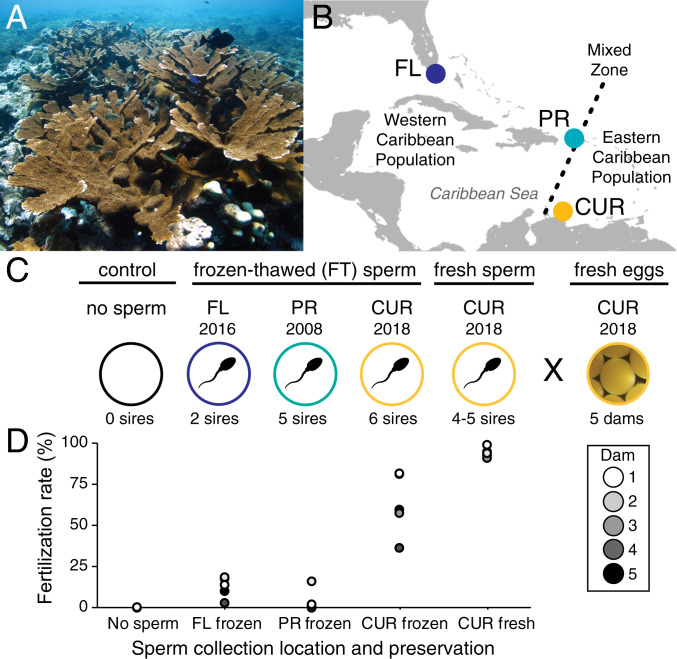
Fig. 1.
Study species, experimental crosses, and fertilization rates. (A) Study species: elkhorn coral A. palmata. (B) Map of the Caribbean Sea and genetically distinct A. palmata populations. Circles indicate the locations of gamete collection and cryopreservation of sperm samples from Florida (FL, n = 2 sires), Puerto Rico (PR, n = 5 sires), and Curaçao (CUR, n = 6 sires). FT sperm was cryopreserved in the year listed and thawed immediately before use in in vitro fertilization experiments with freshly collected eggs from Curaçao (n = 5 dams). Freshly collected sperm from Curaçao (n = 4 or 5 sires, depending on spawning night) was used for comparison. (C) Summary of in vitro crosses conducted to test the feasibility of AGF in A. palmata. (D) Mean fertilization success for A. palmata eggs from five donor colonies (dams) when mixed with each of the four sperm pools or a no-sperm control (n = 1 to 9 containers per dam × sperm pool cross; replicate egg containers were preferentially assigned to the three FT crosses and the two interpopulation (AGF) crosses based on the lower fertilization rates expected from FT sperm and the higher conservation value of the AGF crosses). Sperm concentration and handling details are summarized in SI Appendix, Table S2. For all FT treatments, fertilization was attempted at two different sperm concentrations (n = 1 to 6 containers per concentration per cross); this bracketing approach during gamete handling was employed to maximize the overall chance of achieving fertilization using cryopreserved material (SI Appendix, Tables S2 and S3 for details).
Region-wide surveys show there are two genetically isolated populations of A. palmata in the Caribbean and northwestern Atlantic with a central, mixed genetic zone in Puerto Rico (Fig. 1B) (30, 31). Over evolutionary time, thermal and oceanographic gradients across the Caribbean region (34) likely imposed different local selective pressures, yielding regionally distinct allele pools; these alleles could be harnessed to accelerate region-wide adaptation to climate change (10) and other stressors, if only they could be transferred between populations. However, if insurmountable reproductive barriers exist, each population must be managed as a separate biological species, making their conservation status all the more severe and eliminating AGF as a viable tool to fortify either population.
To test the feasibility of AGF in Caribbean coral and to test the reproductive compatibility of A. palmata across its genetic barrier, we conducted in vitro fertilization experiments crossing fresh A. palmata eggs from Curaçao with cryopreserved sperm from Florida, Puerto Rico, and Curaçao. This demonstrates the successful use of cryopreserved coral sperm to achieve AGF across genetically isolated populations of coral.
Results
In 2018, in vitro fertilization crosses were performed using freshly collected A. palmata eggs from five donor colonies (i.e., dams) in Curaçao (Fig. 1C and SI Appendix, Tables S1 and S2). Sperm samples were obtained from Florida (FL), Puerto Rico (PR), and Curaçao (CUR). Sperm were either used fresh within hours of collection or cryopreserved on the night of collection, stored for up to 10 y, and thawed immediately before use [i.e., Frozen-Thawed (FT)]. Across two nights and five coral dams (Fig. 1D and SI Appendix, Table S3), mean fertilization success per dam ranged from 91 to 99% for CUR×CUR (Fresh sperm), 37 to 82% for CUR×CUR (FT sperm), 3 to 18.5% for CUR×FL (FT sperm), and 0 to 16% for CUR×PR (FT sperm).
Embryos and larvae from the interpopulation (AGF) crosses developed normally (Days 1 to 3 postfertilization). At Day 6 and 7 postfertilization, swimming larvae were transported by air to grow out facilities in Florida with no observable mortality seen during transport. After 1 wk in settlement containers, settlement rates were 46% for CUR×CUR (fresh sperm; 3,105 settlers), 45% for CUR×CUR (FT sperm; 3,577 settlers), 57% for CUR×FL (FT; 1,247 settlers), and 42% for CUR×PR (FT; 233 settlers), yielding 5,057 settlers from the three FT sperm crosses and a total of 1,480 settlers from the two interpopulation (AGF) crosses.
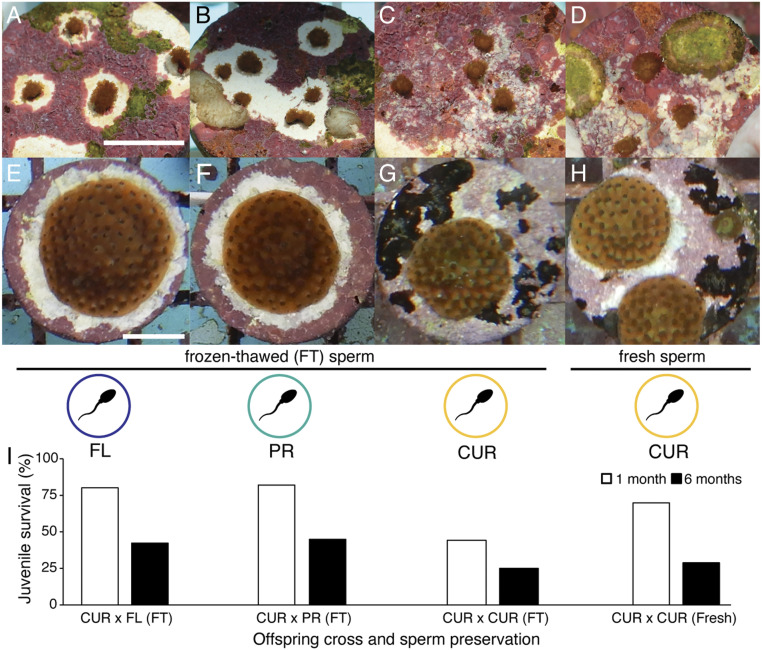
Development was consistent across all four cohorts; settlers began uptake of symbiotic dinoflagellates (Symbiodiniaceae spp.) within 5 to 10 d. Within 1 mo, over 75% of settlers had well-established symbiont populations, indicated by dark brown tissue coloration (Fig. 2 A–D). At 6 mo, there were no observable developmental deficits in any of the cohorts. Total survival was 29% for the CUR×CUR cohort (fresh sperm; 888 juveniles), 25% for CUR×CUR (FT sperm; 903 juveniles), 42% for CUR×FL (FT; 522 juveniles), and 45% for CUR×PR (FT; 104 juveniles) (Fig. 2 E–I and SI Appendix, Table S4). To our knowledge, this is the highest 6-mo survival ever achieved in A. palmata breeding. Survival of the AGF cohorts was higher overall, reflecting the greater care directed to these cohorts given their conservation value.
Fig. 2.
Survival and growth of A. palmata juveniles reared from cryopreserved sperm. Photographs of (A–D) 1- and (E–H) 6-mo-old juvenile colonies of the coral A. palmata, reared from cryopreserved (FT) or fresh sperm crossed with freshly collected eggs from Curaçao (n = 5 dams). FT sperm was collected in Florida (FL, n = 2 sires), Puerto Rico (PR, n = 5 sires), and Curaçao (CUR, n = 6 sires). Fresh sperm from Curaçao was used for comparison (n = 4 or 5 sires, depending on spawning night) (Scale bars, 1 cm). (A and E) CUR×FL (FT sperm), (B and F) CUR×PR (FT sperm), (C and G) CUR×CUR (FT sperm), and (D and H) CUR×CUR (Fresh sperm). Due to their conservation value, the interpopulation (AGF) juveniles (A, B, E, and F) were given increased care and more space per juvenile beginning at the time of settlement. White areas around the juveniles are spaces where the coral inhibited encroaching coralline algae or coralline algae was removed to facilitate coral growth. No differences were apparent between the four cohorts in juvenile polyp morphology or colony growth pattern. (I) Survival of juveniles by cohort at 1 and 6 mo after settlement (n = 233 to 3,577 initial settlers per cohort at n = 2 separate rearing facilities; data were pooled across facilities to determine overall survival at each time point). The overall higher survival of the interpopulation (AGF) juveniles reflects the greater care directed to these cohorts due to their conservation value (SI Appendix, Table S4).
During spawning and handling, errant sperm can be introduced through open mesh collection nets or through sample cross-contamination. For all crosses, no-sperm controls showed that high levels of self-fertilization or cross-contamination were unlikely; no-sperm control fertilization was 0.0% for three dams and 0.1 to 0.7% for two dams (SI Appendix, Table S3). To further confirm that offspring were indeed fertilized by the intended sperm pool only, we conducted genetic analyses of two juvenile cohorts [(CUR×CUR (FT) and CUR×FL (FT), n = 15 per cohort; Fig. 3 and SI Appendix, Table S5)].
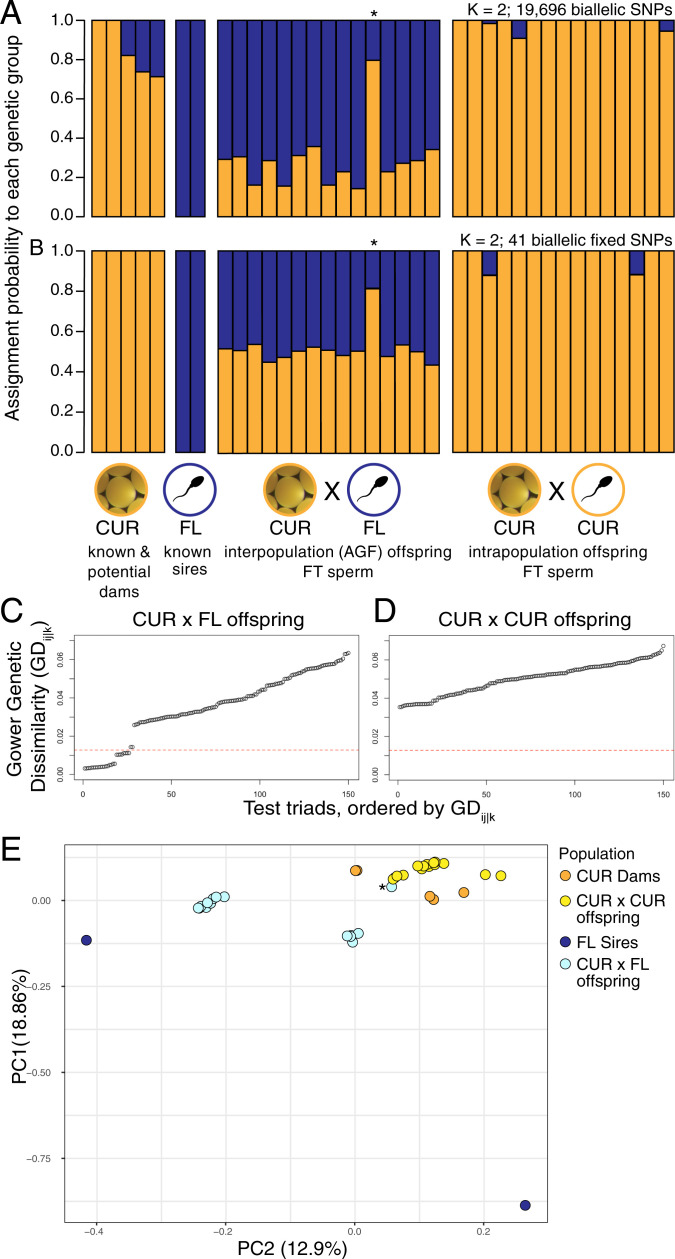
Fig. 3.
Parentage analysis of A. palmata juveniles reared from cryopreserved sperm demonstrating interpopulation parentage and successful AGF. (A and B) Genetic admixture plots for sires, dams, and offspring. The offspring genotyped for this analysis were drawn from two cohorts: CUR×FL (FT sperm, n = 15 offspring bred from n = 5 dams and n = 2 sires) and CUR×CUR (FT sperm, n = 15 offspring bred from n = 5 dams and n = 6 sires). The dataset also includes both of the known sires from Florida and five of the known and putative dams from Curaçao. The bars represent the probability of assignment (x-axis) for each individual (y-axis) to each of K = 2 genetic groups (A and B). (A) Probability of assignment using the full set of 19,696 genotyping SNPs for K = 2 populations. (B) Probability of assignment using only SNPs fixed between the Florida versus Curaçao parents (n = 41) for K = 2 populations. In (A and B), one of the juveniles from the CUR×FL cohort had a higher than expected probability of assignment to CUR (orange cluster) and is thought to be the result of self-fertilization or a labeling error (denoted with a *). (C and D) Triad assignment plots for the juveniles genotyped and analyzed Above. The data points represent the genetic dissimilarity among hypothetical triads of one sire, one dam, and one offspring. (C) Assignment plot of triads with FL sires, CUR dams, and only interpopulation (CUR×FL) offspring. A total of 13 of 15 juveniles from the interpopulation cross between FL sires (FT sperm) and CUR dams (fresh eggs) were successfully assigned to two specific parents in the sample set, shown by data points below the 0.0118 Gower genetic dissimilarity threshold. (D) Assignment plot of triads using FL sires, CUR dams, and only intrapopulation (CUR×CUR) offspring. As expected, no juveniles in the CUR×CUR cohort were assigned CUR×FL parentage (SI Appendix, Table S5 for details). (E) PCA of genetic structure among all coral dams, sires, and offspring genotyped in the analyses Above. PCA analysis was conducted using all 19,696 genotyping SNPs (R Package SNPrelate). All CUR×CUR offspring clustered with the CUR dams, while CUR×FL offspring formed a cluster in between their known FL sires and the known and putative CUR dams. As in (A and B), one CUR×FL juvenile clustered with the CUR×CUR offspring instead of with the other CUR×FL offspring, likely the result of selfing or mislabeling (denoted with a *).
Juveniles were sampled for genetic analysis at 9 and 14 mo (i.e., once they were large enough to survive fragmentation). To assign parentage to the offspring, tissue was also collected from the two Florida sires and from five of the known and putative Curaçao dams (SI Appendix). The Curaçao sires, Puerto Rico sires, and other putative Curaçao dams were not mapped during gamete collection and therefore not sampled. Samples were genotyped [Axiom Coral Genotyping Array 550962 (35), Applied Biosystems] and 19,696 single-nucleotide polymorphism (SNP) probes were used to calculate the probability of assignment for each of the sampled sires, dams, and juveniles to either the Curaçao or Florida population.
For the CUR×CUR offspring, the probability of assignment to the Curaçao population was 90 to 100%, as expected for these intrapopulation juveniles. For the CUR×FL offspring, the probability of assignment was mixed between both Curaçao and Florida populations, as expected for offspring with one parent from each population [Fig. 3A, Admixture version 1.3 (36), SI Appendix]. The probability of assignment in this cohort was skewed toward the Florida population, which reflects the bias of the genotyping array toward polymorphisms common in Florida, where the most deeply sequenced genome of A. palmata was obtained (10). Alternatively, transmission ratio distortion may have occurred with a bias toward converting Curaçao to Florida alleles (37, 38). Among the two Florida sires and five known and putative Curaçao dams, there were 41 SNP loci with fixed allelic differences between the populations. When assignment probabilities were calculated based on this probe set, ancestry was inferred as near 100% Curaçao in the CUR×CUR (FT) cohort and 45 to 55% Curaçao in the CUR×FL (FT) cohort (Fig. 3B), with the exception of one juvenile in the CUR×FL (FT) cohort, likely representing a selfing event or mislabeled juvenile (Fig. 3 A and B, denoted by *).
To further confirm interpopulation parentage, we calculated the genetic dissimilarity among all possible dam–sire–offspring triads for the CUR×CUR (FT) and CUR×FL (FT) juveniles. In total, 13 of the 15 CUR×FL offspring were assigned with high certainty to a specific CUR dam and one of the two FL sires (<0.0118 Gower genetic dissimilarity; Fig. 3C). All 13 of these offspring were half-siblings from the same CUR dam; 9 were sired by one FL sire and 4 by the other sire. The remaining 2 CUR×FL offspring could not be assigned to both a Florida and Curaçao parent via the parentage analysis. As in the previous analysis, one of these CUR×FL offspring appeared to have full Curaçao ancestry, likely the result of self-fertilization or mislabeling. The other unassigned offspring appeared to be of mixed Curaçao and Florida ancestry, indicating that this individual was indeed a result of an interpopulation cross, but it could not be assigned to a specific Curaçao dam. It is possible that this individual was the offspring of a Curaçao dam not genotyped here. Triad assignment for the CUR×CUR offspring was not possible because the dataset contained too few of the putative parents (Fig. 3D). A principal components analysis (PCA) of genetic similarity confirmed interpopulation parentage of the AGF juveniles (Fig. 3E, R package SNPrelate).
Overall, three lines of genetic evidence demonstrated that the CUR×FL (FT) cohort was indeed sired by the cryopreserved sperm from Florida, thus confirming that AGF was achieved using cryopreserved sperm and further confirming that the two genetically distinct populations are reproductively compatible.
Discussion
As a form of managed breeding, AGF is a viable conservation intervention to genetically enrich threatened coral populations and thus facilitate their adaptation to changing environments. Previously, AGF has been tested in corals by moving whole adult colonies (13), but it has never been attempted in Caribbean coral nor has cryopreserved material been used to crossbreed genetically isolated coral populations. Given the historic >95% decline in A. palmata populations, it is encouraging to observe that crossbreeding is feasible in this critically endangered species across its geographic range. By conducting in vitro fertilization using frozen sperm, we produced the largest living wildlife population ever created from cryopreserved material, thus demonstrating the potential to use cryopreservation to increase the scale of endangered species conservation.
Cryopreservation is now a field-ready tool to safeguard much of the extant genetic diversity of corals. For threatened species, genome resource banks containing cryopreserved material can serve to 1) preserve existing large gene pools for current and future rescue of genetically depleted populations; 2) transfer genetic material between widely dispersed living populations; and 3) improve the efficiency and effectiveness of germplasm research and endangered species breeding. Although most coral populations still retain high allelic diversity (10), ongoing population decline and the increasing application of captive breeding can both contribute to inbreeding and future genetic bottlenecks; these consequences can be prevented through cryopreservation and AGF (12).
Cryopreservation does impose a substantial stress on the cells involved. In corals, this can result in a reduced concentration of motile sperm after thawing, a longer time until cell cleavage begins, and higher variability in fertilization success from trial to trial (21, 23). Encouragingly, the stressful effects of cryopreservation are only evident during the early hours of fertilization and embryogenesis (39). Once embryos develop into motile larvae, development appears unaffected, and standard propagation methods can be applied. To date, we have not observed any abnormalities or deficits in the swimming behavior, settlement rate, or postsettlement growth of juvenile corals produced with FT sperm (23). Nevertheless, because of the wide variability in fertilization success with cryopreserved material, the fertilization data presented here (Fig. 1D) should be interpreted with caution. Most importantly, the observed differences in fertilization do not necessarily indicate the existence of gametic or genetic barriers between A. palmata populations (31) as this could be caused by other factors.
Superimposed on the stress that cryopreservation places on cells are natural reproductive barriers. Even when using fresh gametes collected from a single population, the fertilization compatibility of paired A. palmata genets ranges from 0 to >90%; these incompatibilities have been observed in neighboring genets from individual reefs in Florida, Puerto Rico, and Curaçao (10, 40, 41). It was therefore encouraging to observe that the occasional incompatibility observed between genets did not translate into complete interpopulation incompatibility, thus allowing AGF juveniles to be bred and raised. Despite the success of transferring alleles between populations in this study, the genetic data do hint at a possible allelic transmission ratio distortion wherein Curaçao alleles were apparently converted to Florida alleles in the surviving CUR×FL offspring. This process has been observed in plants and animals (42, 43) and can lead to asymmetric introgression.
When introducing alleles into a population, there is a risk of outbreeding depression (i.e., the introduction of alleles unsuitable for a local environment and a consequent reduction in local population adaptation). In corals, the risk of outbreeding depression is considered low given their high extant genetic diversity and generally outcrossed population genetic structure (10, 44). Future experimental tests for outbreeding depression in corals would be informative, albeit time consuming, as this can be only detected in F2 or F3 generations. In the case of A. palmata, which faces widespread reproductive and recruitment failure, the risk of a decade of conservation inaction might considerably outweigh the risk of outbreeding depression. Importantly, AGF does not involve any manipulation of genes other than genetic assortment during reproduction, nor does it require knowledge of the genetics underlying environmental adaptation. This makes AGF a more practical and accessible tool for managing wildlife genetics compared to other interventions such as gene editing. In addition, AGF can be conducted without selecting for individual phenotypes at the expense of others. In sum, AGF represents a holistic approach to managed breeding that can bolster allelic diversity in populations, preserve the overall genetic diversity of species, and accelerate adaptation to local environments.
We produced direct evidence that gene flow is possible between genetically isolated populations of the threatened Caribbean coral A. palmata, and in doing so, we demonstrated that cryopreserved sperm can be used to preserve and move genetic diversity in corals. This represents a powerful option for coral breeding and restoration, both regionally and worldwide. Furthermore, this creates the foundation for future experiments in which sperm from thermally tolerant corals could be used to transfer heritable thermal tolerance to other populations (10). The corals reared in this study remain under human care as they undergo screening for increased thermal tolerance and as they await outplanting permissions from management authorities.
Priority next steps for the Caribbean include 1) identifying coral populations that are flourishing despite severe local stress, cryopreserving sperm from these populations, and transferring alleles via crossbreeding to populations currently collapsing due to these stressors (or predicted to experience them in the future); 2) testing AGF offspring for thermal tolerance and other stress-resistance traits in the laboratory and in in situ nurseries; 3) identifying the genetic basis underlying stress-resistance traits in corals in order to target future gene banking efforts and assess the potential conservation value of samples currently held in repositories; 4) expanding dialogue and building consensus with ecosystem managers and stakeholders to allow controlled in situ testing of AGF corals; and 5) eventual outplanting and long-term monitoring of large populations of AGF coral on natural reefs.
Thus, the progress reported here represents both an important scientific proof of concept as well as the impetus for a wider conversation about genetic risk tolerance in coral restoration, including how to safely augment current coral reef management practices and build community consensus around new conservation technologies.
Materials and Methods
Collection Locations, Sperm Cryopreservation, and Gamete Crosses.
The coral reproduction and sperm cryopreservation methods applied in this study are described in previous work (21, 23, 45, 46) and in the SI Appendix, Extended Materials and Methods. Briefly, coral egg–sperm bundles were collected in the field using tents at two locations in Curaçao: Spanish Water (12°4'13.11”N, 68°52’18.22”W) and the Curaçao Sea Aquarium (12°4'59.94”N, 68°53’42.47”W). Divers surveyed between 25 and 100 A. palmata colonies per night for a total of 30 nights (Month 1: 2 d before the full moon (BFM) to 11 d after the full moon (AFM) in late July 2018, and Month 2: 2 d BFM to 13 d AFM in late August 2018, SI Appendix, Table S1). In the laboratory, eggs and sperm were separated, then eggs were rinsed multiple times and allocated into fertilization containers. Sperm was assessed for motility and concentration, pooled, cryopreserved and thawed (if applicable), and then allocated into fertilization containers. The 2018 lunar cycle produced a split spawn in Curaçao, dividing the spawning period over two consecutive months. Thus, FT donor sperm from Curaçao (6 sires) was frozen in Month 1 for use in Month 2. Fresh donor sperm was collected and used in Month 2 (4 or 5 sires, depending on the night). In Month 2, 12 colonies produced enough eggs for fertilization; however, 7 of these colonies had high levels of self-fertilization; these samples were omitted from analysis, leaving 5 total dams. For cryopreservation in Month 1, A. palmata sperm from Curaçao was pooled, mixed 1:1 with 20% dimethyl sulfoxide in filtered seawater, frozen at 20 °C/min, and held in liquid nitrogen until it was thawed for in vitro fertilization in Month 2. For the AGF crosses, A. palmata sperm from Florida (n = 2 sires, stored for 2 y) and from Puerto Rico (n = 5 sires, stored for 10 y) was frozen in an identical manner, stored at the United States Department of Agriculture National Animal Germplasm Program, sent via dry shipper to Curaçao, thawed, and used for in vitro fertilization (see details in SI Appendix, Table S1).
Statistical Analysis: Propagation Data.
Fertilization rates from replicate fertilization containers were used to calculate summary statistics per cross (Fig. 1D) and for low and high sperm concentration treatments within each cross (SI Appendix, Table S3). Gamete density varied due to differences in colony fecundity and containers were not interspersed (to prevent cross-contamination); therefore, inferential statistics and significance tests were not appropriate for these data. Larvae were settled and propagated at two facilities in Florida, where each team applied different methods to maximize settlement and survival. Settlement and propagation methods from each facility are described in detail in SI Appendix, Extended Materials and Methods. Small-scale tank replication and interspersion was not conducted, as this would have compromised survival of the endangered coral juveniles; therefore, inferential statistics and significance tests were not applied (Fig. 2 and SI Appendix, Table S4).
DNA Sampling and Genotyping.
Tissue samples for genetic analysis were collected from adult coral colonies using a hammer and chisel (fragments ∼1 cm3; n = 5 putative and known Curaçao dams; n = 2 known Florida sires) and from coral offspring using a diamond saw (fragments ∼0.5 cm3; n = 25 juveniles per cohort, CUR×CUR (FT) and CUR×FL (FT) cohorts). Tissue was preserved in 96% nondenatured ethanol in 1.8-mL cryovials. DNA was extracted from the tissue samples using the DNeasy Blood and Tissue kit (Qiagen) following published protocol (8). DNA quantities were determined via PicoGreen (ThermoFisher) and ranged from 1.27 to 15.25 ng/µL (SI Appendix, Table S5). All 50 juvenile tissue samples yielded DNA of adequate quality and quantity. From this set, 15 samples per cohort were randomly selected for genotyping. DNA was genotyped by ThermoFisher using the Applied Biosystems Axiom Coral Genotyping Array 550962 (35). Quality control (QC) analysis was performed by ThermoFisher, where all arrays with a dish QC value of 0.82 or greater and a QC call rate of 97% or greater were considered to pass. All samples analyzed passed QC, with an average cluster call rate of 99.85% and sample reproducibility of 99.88%. Of all 32,123 probes on the array, 25,257 (78.63%) passed QC. Of the 32,123 probes on the array, 19,696 probes were designed to resolve population genomic structure of A. palmata and A. cervicornis. This genotyping probe set was used in subsequent analysis. Raw SNP data has been submitted to the European Variation Archive (Project: PRJEB41095, Analyses: ERZ1669087; SI Appendix, Table S5).
Statistical Analysis: Genetic Data.
Admixture.
To estimate ancestry proportions, two sets of loci were used. First, ADMIXTURE version 1.3 (47) was run with the full set of 19,696 probes designed for resolution of population genomic structure as previously described (35). Population SNPs were previously identified based on pairwise comparisons of samples subjected to shallow genome sequencing from four collection sites (Florida, Curaçao, US Virgin Islands, and Belize) (35). These SNPs were filtered such that all samples from one site shared an allele with a frequency of 0.8 or greater and differed from the samples of the other site with the alternative allele at a frequency of 0.8 or greater (35). After identifying loci that were alternatively fixed in the Florida or Curaçao parents, this subset of 41 “diagnostic” probes was also used in the admixture analysis to estimate ancestry proportions present in the offspring uniquely contributed by the parents. Ancestry proportions were estimated using ADMIXTURE version 1.3 (47). ADMIXTURE was run assuming the number of groups to be K = 2 to 7, with a cross-validation of five folds run for every estimate of K. Ancestry proportions for K = 2 using the “diagnostic” loci are reported in SI Appendix, Table S5.
Parentage Analysis.
Parentage assignment was performed using the R package apparent. Parentage assignment was analyzed for two sets of samples. Parentage analysis was run at an alpha level of 0.01, with a minimum acceptable number of loci used to compute pairwise genetic distance of 500. First, offspring from the CUR×CUR (FT) and CUR×FL (FT) crosses as well as both sets of putative parents were included in the parentage analysis. In this analysis, 13 of the CUR×FL (FT) offspring were assigned with high confidence to a set of two parents, one from FL and one from CUR. However, parentage analysis considering only Curaçao samples failed to assign offspring to a unique set of parents for the CUR×CUR (FT) offspring because the sample set contained too few of the putative donor colonies. Putative Assigned Parents as well as genetic dissimilarity metrics are included in SI Appendix, Table S5.
PCA.
To further estimate genetic structure between putative parents and their offspring, a PCA was performed (48). Eigenvalues were calculated using the R package SNPRelate from the set of 19,696 SNP loci. The snpgdsPCA function was run using standard settings. The first two principal components were extracted and plotted. Principal component 1 represented 18.86% of variation among individuals, and principal component 2 represented 12.9% of variation among individuals.
Supplementary Material
Acknowledgments
This work was supported by the Paul G. Allen Family Foundation. Additional support was provided by the Smithsonian Conservation Biology Institute, the Hawaii Institute of Marine Biology (HIMB), and the Volgenau Fitzgerald Family Fund (to M.H.), National Geographic (to K.L.M.), NSF (OCE-1537959 to I.B.B. and IOS-1848671 to K.L.M.), and the Government of Curaçao [to Caribbean Research and Management of Biodiversity (CARMABI) Foundation]. Funders did not influence the interpretation of the project data or the decision to publish the findings. We are grateful for the support of colleagues at each of our institutions, especially the logistical support and advice from R. C. Barnes, J. Blokzeijl, G. Boecker, J. Bouwmeester, Coral Restoration Foundation, F. Dilrosun, C. Engelsma, K. Fitzgerald, R.-J. Geertsma, S. Gordon, S. Graves, M. Hooftijzer, E. Houtepen, J. Huckeba, K. Latijnhouwers, K. Leeper, M. Miller, A. Moura, A. Muskat, R. Robbins, Sexual Coral Reproduction (SECORE) International, A. Shantz, K. Stankiewicz, The Florida Aquarium Center for Conservation, United States Department of Agriculture National Animal Germplasm Program (USDA NAGP), C. van Bijnen, K. Vasquez Kuntz, D. Vaughan, C. Wilson, C. Winterdaal, and S. Winters. M. Matz and anonymous reviewers provided valuable insights on a draft manuscript. Special thanks to V. L. Carter for cryopreserving and banking the A. palmata sperm over time. K.L.M. also thanks C.E. Marhaver for help with thermodynamic testing of the larval transport containers and for providing wisdom and encouragement from Day 1. Coral sperm were transported under Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) Certificate #18US59512C/9. Coral larvae were transported under CITES permits #18CW007 and #18CW012 issued by the Curaçao Ministerie van Gezondheid, Milieu, en Natuur. This manuscript is HIMB contribution # 1863.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110559118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Aitken S., Whitlock M., Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367–388 (2013). [Google Scholar]
- 2.Palumbi S. R., Barshis D. J., Traylor-Knowles N., Bay R. A., Mechanisms of reef coral resistance to future climate change. Science 344, 895–898 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Anthony K., et al., New interventions are needed to save coral reefs. Nat. Ecol. Evol. 1, 1420–1422 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Saremi N. F., et al., Puma genomes from North and South America provide insights into the genomic consequences of inbreeding. Nat. Commun. 10, 4769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weeks A. R., et al., Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population. Nat. Commun. 8, 1071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoegh-Guldberg O., et al., Assisted colonization and rapid climate change. Science 321, 345–346 (2008). [DOI] [PubMed] [Google Scholar]
- 7.van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D., Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer K. L., et al., Widespread loss of Caribbean acroporid corals was underway before coral bleaching and disease outbreaks. Sci. Adv. 6, eaax9395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schutte V. G. W., Selig E., Bruno J., Regional spatio-temporal trends in Caribbean coral reef benthic communities. Mar. Ecol. Prog. Ser. 402, 115–122 (2010). [Google Scholar]
- 10.Baums I., et al., Considerations for maximizing the adaptive potential of restored coral populations in the western Atlantic. Ecol. Appl. 29, e01978 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes T., Tanner J., Recruitment failure, life histories, and long-term decline of Caribbean corals. Ecology 81, 2250–2263 (2000). [Google Scholar]
- 12.National Academies of Sciences, Engineering, and Medicine , A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs (The National Academies Press, Washington, DC, 2019). [Google Scholar]
- 13.Dixon G. B., et al., Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Knowlton N., Rohwer F., Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am. Nat. 162 (suppl. 4), S51–S62 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Marhaver K. L., Edwards R. A., Rohwer F., Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szmant A. M., Reproductive ecology of Caribbean reef corals. Coral Reefs 5, 43–53 (1986). [Google Scholar]
- 17.Gleason D., Hofmann D., Coral larvae: From gametes to recruits. J. Exp. Mar. Biol. Ecol. 408, 42–57 (2011). [Google Scholar]
- 18.Levitan D. R., et al., Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution 58, 308–323 (2004). [PubMed] [Google Scholar]
- 19.Fogarty N. D., Lowenberg M., Ojima M. N., Knowlton N., Levitan D. R., Asymmetric conspecific sperm precedence in relation to spawning times in the Montastraea annularis species complex (Cnidaria: Scleractinia). J. Evol. Biol. 25, 2481–2488 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Harrison P. L., “Sexual reproduction of scleractian corals” in Coral Reefs: An Ecosystem in Transition, Dubinsky Z., Stambler N., Eds. (Springer, 2011), pp. 59–85. [Google Scholar]
- 21.Hagedorn M., et al., Preserving and using germplasm and dissociated embryonic cells for conserving Caribbean and Pacific coral. PLoS One 7, e33354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagedorn M., et al., First frozen repository for the Great Barrier Reef coral created. Cryobiology 65, 157–158 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Hagedorn M., et al., Producing coral offspring with cryopreserved sperm: A tool for coral reef restoration. Sci. Rep. 7, 14432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard J. G., et al., Recovery of gene diversity using long-term cryopreserved spermatozoa and artificial insemination in the endangered black-footed ferret. Anim. Conserv. 19, 102–111 (2016). [Google Scholar]
- 25.Crosier A. E., et al., Improved quality of cryopreserved cheetah (Acinonyx jubatus) spermatozoa after centrifugation through Accudenz. J. Androl. 30, 298–308 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Williams D. E., Miller M. W., Attributing mortality among drivers of population decline in Acropora palmata in the Florida Keys (USA). Coral Reefs 31, 369–382 (2012). [Google Scholar]
- 27.Williams D. E., Miller M. W., Bright A. J., Pausch R. E., Valdivia A., Thermal stress exposure, bleaching response, and mortality in the threatened coral Acropora palmata. Mar. Pollut. Bull. 124, 189–197 (2017). [DOI] [PubMed] [Google Scholar]
- 28.National Marine Fisheries Service , Endangered and threatened species: Final listing determinations for Elkhorn coral and Staghorn coral. Fed. Regist. 71, 26852–26872 (2006). [Google Scholar]
- 29.Aronson R., et al. , Acropora palmata. The IUCN Red List of Threatened Species 2008 (2008).
- 30.Devlin-Durante M. K., Baums I. B., Genome-wide survey of single-nucleotide polymorphisms reveals fine-scale population structure and signs of selection in the threatened Caribbean elkhorn coral, Acropora palmata. PeerJ 5, e4077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baums I., Hughes C., Hellberg M. E., Mendelian microsatellite loci for the Caribbean coral, Acropora palmata. Mar. Ecol. Prog. Ser. 288, 115–127 (2005). [Google Scholar]
- 32.Boström-Einarsson L., et al., Coral restoration – A systematic review of current methods, successes, failures and future directions. PLoS One 15, e0226631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Marine Fisheries Service , Recovery plan for Elkhorn (Acropora palmata) and Staghorn (A. cervicornis) corals (Prepared by the Acropora Recovery Team for the National Marine Fisheries Service, Silver Spring, MD, 2015). [Google Scholar]
- 34.Muñiz-Castillo A. I., et al., Three decades of heat stress exposure in Caribbean coral reefs: A new regional delineation to enhance conservation. Sci. Rep. 9, 11013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitchen S. A., et al., STAGdb: A 30K SNP genotyping array and Science Gateway for Acropora corals and their dinoflagellate symbionts. Sci. Rep. 10, 12488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duret L., Galtier N., Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10, 285–311 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Halldorsson B. V., et al., The rate of meiotic gene conversion varies by sex and age. Nat. Genet. 48, 1377–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagedorn M., et al., Potential bleaching effects on coral reproduction. Reprod. Fertil. Dev. 28, 1061–1071 (2016). [Google Scholar]
- 40.Miller M. W., et al., Clonal structure and variable fertilization success in Florida Keys broadcast-spawning corals. Coral Reefs 37, 239–249 (2018). [Google Scholar]
- 41.Baums I. B., et al., Genotypic variation influences reproductive success and thermal stress tolerance in the reef building coral, Acropora palmata. Coral Reefs 32, 703–717 (2013). [Google Scholar]
- 42.Seymour D. K., Chae E., Arioz B. I., Koenig D., Weigel D., Transmission ratio distortion is frequent in Arabidopsis thaliana controlled crosses. Heredity 122, 294–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gòdia M., et al., Whole genome sequencing identifies allelic ratio distortion in sperm involving genes related to spermatogenesis in a swine model. DNA Res. 27, dsaa019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matz M. V., Treml E. A., Aglyamova G. V., Bay L. K., Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLoS Genet. 14, e1007220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagedorn M., et al., Successful demonstration of assisted gene flow in the threatened coral Acropora palmata across genetically-isolated Caribbean populations using cryopreserved sperm. bioRxiv [Preprint] (2018) 10.1101/492447 (Accessed 6 June 2021). [DOI]
- 46.Chamberland V. F., et al., The reproductive biology and early life ecology of a common Caribbean brain coral, Diploria labyrinthiformis (Scleractinia: Faviinae). Coral Reefs 36, 83–94 (2017). [Google Scholar]
- 47.Alexander D. H., Lange K., Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12, 246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X., et al., A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.