It was not until the ingenious works of Anton van Leeuwenhoek in the 17th century that bacteria were first discovered and understood to be a “new” form of life on Earth (1). For the next 200 y, bacteria were largely ignored and thought to simply be “bags of proteins,” even though it was recognized that they could have enormous impacts on human health. It is now widely appreciated that bacteria are incredibly complex organisms that can function in a manner that mimics a multicellular organism (2, 3). The ability of bacterial cells to constantly produce and remodel their cell wall is no exception to this, and myriad crucial findings about this process have been made over the last century. One of the most seminal of these discoveries came in the 1940s when β-lactam−containing molecules were found to combat bacterial infections. More recently, the targets of these drugs, the penicillin-binding proteins, as well as other protein machinery have been identified as essential elements for construction of the complex mesh that keeps these cells intact, the peptidoglycan (PG). Still, many mysteries remain about exactly how bacterial cells grow and divide. In particular, while enzymes that cleave and tailor portions of the PG are known to play important roles in microbial survival, they are dramatically understudied. Taguchi et al. (4) shed light on two enzymes in Streptococcus pneumoniae, MpgA and MpgB, shown to cleave a nascent carbohydrate strand to liberate newly synthesized PG. While previous studies postulated that this occurred in a fashion similar to that utilized by a close relative, the lytic glycosylase MltG in Escherichia coli [1,6-anhydroMurNac product (5)], Taguchi et al. instead discover that MpgA and MpgB act as muramidases [saccharide product with reducing end (6)]. Amazingly, they determined that a single amino acid is largely responsible for the difference in catalytic mechanism between MltG and MpgA/B (Fig. 1).
Fig. 1.
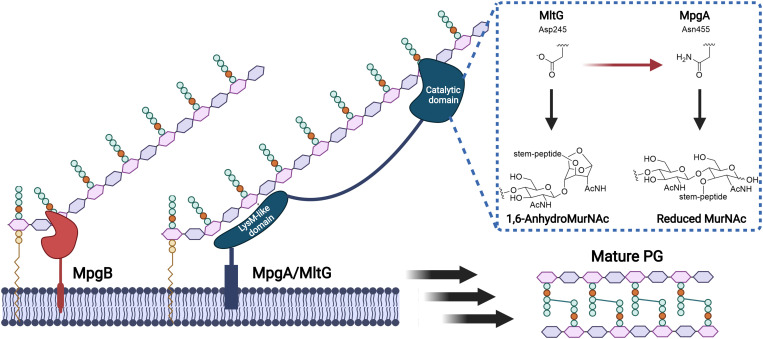
Release of newly synthesized PG from the cell membrane by muramidases MpgA and MpgB and lytic transglycosylase MltG using different enzymatic activities. MpgB cleaves nascent PG near the lipid anchor, while MpgA and MltG utilize a LysM-like domain to recognize oligosaccharide units farther from the lipid anchor. The ability of MpgA to cleave PG as a muramidase is due to the switch from an aspartate to an asparagine in the catalytic active site, while MltG possessing an aspartate works as a lytic transglycosylase. Despite differences in enzymatic activity, both enzymes function to release newly synthesized PG to be incorporated into mature PG, ensuring cell wall integrity throughout the bacterial cell cycle. Figure created with BioRender.
The PG is a polymeric, mesh-like structure that plays an essential role in maintaining cellular integrity and providing cell shape and structure in bacteria. It comprises repeating units of N-acetylglucosamine (GlcNac) and N-acetylmuramic acid (MurNac) connected via β-1,4 linkages, and decorated with stem peptides (7–11). Lipid II is the substrate for PG biosynthesis, which is a unit of GlcNac−MurNac containing the stem peptide and an undecaprenyl diphosphate group that anchors the entire structure into the cytoplasmic membrane. Lipid II is utilized by SEDS (shape, elongation, division, sporulation) proteins and penicillin-binding proteins (PBPs) to carry out the highly orchestrated process of producing nascent PG, which can then be incorporated into mature PG within the periphery of the cell (side wall) and the cell division site (septum) (12–16). While the SEDS proteins and PBPs synthesize new PG from Lipid II, there are additional proteins, called the hydrolases, which are required to break down mature PG and/or liberate newly synthesized PG strands from the cytoplasmic membrane.
To elucidate how MpgA and MpgB cleave nascent PG to different lengths, Taguchi et al. examined the LysM-like domain found in MpgA that has significant homology to the LysM-like domain in MltG.
Taguchi et al. (4) detail their findings about two such hydrolases in S. pneumoniae, an opportunistic, Gram-positive bacterium that is the leading cause of community-acquired pneumoniae worldwide and is a primary contributor to many other diseases, including otitis media and meningitis (17). The biosynthesis of S. pneumoniae PG has been extensively studied (8), yet there remain key gaps in knowledge about how this process is regulated and the roles of individual enzymes within the protein machinery complexes (i.e., divisome and elongasome). Previous work had identified two proteins, here named MpgA and MpgB, as important factors in maintaining the integrity of the pneumococcal cell wall (18–20). It was proposed that MpgA was a lytic transglycosylase with importance for peripheral cell wall synthesis, and MpgB was a bacterial lytic transglycosylase or muramidase that was required for cell shape and division. Taguchi et al. expand upon these findings and show that MpgA and MpgB are, in fact, both muramidases capable of cleaving nascent but not mature PG, releasing the polysaccharide strand from its membrane anchor. These studies were only possible because of earlier work by Walker and coworkers (21, 22) to devise an elegant strategy for the isolation of significant quantities of Lipid II, which can be chemically modified with a fluorophore for easy visualization. Polymerized Lipid II was incubated with either MpgA or MpgB, and the cleavage products were analyzed using liquid chromatography−mass spectrometry. In both cases, MpgA- or MpgB-treated substrate was broken down into oligosaccharides of varying lengths with a reducing end, indicative of muramidase activity and not lytic glycosylase activity (anhydroMurNac product). Indeed, anhydroMurNac species have not been detected in S. pneumoniae cells (18, 20), providing further support that MpgA and MpgB do not act as lytic transglycosylases. This evidence was strengthened by the construction of engineered S. pneumoniae ΔmpgA strains that produced either E. coli MltG or wild-type MpgA under ectopic expression. Following isolation of the PG from these strains, 1,6-anhydroMurNac species were detected only in cells expressing MltG. Taken together, these data demonstrate that MpgA and MpgB are muramidases and not lytic transglycosylases, as was previously predicted.
Given the high structural homology between MpgA and the YceG family of proteins, including MltG, it is perplexing that the activity of these enzymes is different. The authors (4) analyzed over 10,000 sequences of YceG family proteins to identify conserved residues and compared these to the sequence of MpgA. Within the YceG domain, there is a conserved glutamate thought to be required for catalysis, which is also observed in MpgA. Near this conserved residue is an aspartate that is found in 95% of YceG-containing proteins. However, in MpgA, an asparagine resides at this position. The authors speculated that this change from aspartate to asparagine may be sufficient to explain the differences in enzymatic activity between MltG and MpgA. To test this, they generated MltGD245N and found that, instead of the native anhydroMurNAc products, this mutant enzyme generated only muramidase products from polymerized Lipid II. Thus, a single amino acid change dramatically altered the mechanism of this enzyme (Fig. 1). They also constructed MpgAN455D but found that it still did not produce anhydroMurNAc species. They postulated the evolutionary divergence of MpgA-like muramidases from lytic transglycosylases, although it remains to be determined why this would be advantageous.
While these studies show that MpgA and MpgB act as muramidases on the nascent PG substrate, the resulting products differ dramatically in size. MpgB cleavage results in loss of the lipid tail and a disaccharide product, while MpgA generates a product corresponding to seven disaccharide units. To elucidate how MpgA and MpgB cleave nascent PG to different lengths, Taguchi et al. (4) examined the LysM-like domain found in MpgA that has significant homology to the LysM-like domain in MltG. A recent report by Sassine et al. (23) demonstrated that MltG from Bacillus subtilis and E. coli cleaved nascent PG to yield products that were seven disaccharide units in length. Given this similarity and the absence of this domain in MpgB, Taguchi et al. propose that it plays a vital role in the recognition of substrate, enabling cleavage at a distance farther from the lipid anchor (∼70 Å). When the LysM-like domain was removed from MpgA, the enzyme kept near-native levels of activity, but the resulting PG cleavage products were half the length of those from native MpgA. These data imply that the LysM-like domain is important for recognition of the extended substrate and/or simply required as a spacer to place the catalytic domain farther away from the membrane (Fig. 1). The role of this domain was further tested in a strain of S. pneumoniae that expressed inducible wild-type MpgA and MpgA lacking the LysM-like domain (mpgAΔLysM). These cells were found to be shorter and rounder than wild type, similar to those expressing the catalytically compromised mutant, MpgAN455D, highlighting the importance of the LysM-like domain and MpgA in determining cell shape and size.
The authors (4) propose that MpgA and MpgB perform important roles in PG maturation, enabling the release of newly incorporated PG from the cytoplasmic membrane. This serves as critical evidence that, unlike most cell wall hydrolases that are not membrane bound and function as PG degraders for cell division, MpgA and MpgB may represent a vastly understudied group of membrane-bound glycosidases that are present in most bacterial cells and serve an essential role in cell development. The authors also demonstrate that a single amino acid change results in the difference between lytic glycosidase and muramidase activity, providing a potent reminder than even a few atoms can alter the function and activity of closely related enzymes.
Acknowledgments
This work was supported by NIH Grant R01 GM140486-01 and the University of Minnesota, Department of Chemistry.
Footnotes
The authors declare no competing interest.
See companion article, “Biochemical reconstitution defines new functions for membrane-bound glycosidases in assembly of the bacterial cell wall,” 10.1073/pnas.2103740118.
References
- 1.Gest H., The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes Rec. R. Soc. Lond. 58, 187–201 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Hunter P., Not so simple after all. A renaissance of research into prokaryotic evolution and cell structure. EMBO Rep. 9, 224–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons N. A., Kolter R., On the evolution of bacterial multicellularity. Curr. Opin. Microbiol. 24, 21–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taguchi A., Page J. E., Tsui H.-C. T., Winkler M. E., Walker S., Biochemical reconstitution defines new functions for membrane-bound glycosidases in assembly of the bacterial cell wall. Proc. Natl. Acad. Sci. U.S.A. 118, e2103740118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dik D. A., Marous D. R., Fisher J. F., Mobashery S., Lytic transglycosylases: Concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol. 52, 503–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermassen A., et al., Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 10, 331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher J. F., Mobashery S., Constructing and deconstructing the bacterial cell wall. Protein Sci. 29, 629–646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vollmer W., Massidda O., Tomasz A., The cell wall of Streptococcus pneumoniae. Microbiol. Spectr. 7, 10.1128/microbiolspec.GPP3-001802018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan A. J., Cleverley R. M., Peters K., Lewis R. J., Vollmer W., Regulation of bacterial cell wall growth. FEBS J. 284, 851–867 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopal M., Walker S., Envelope structures of Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyachiro M. M., Contreras-Martel C., Dessen A., Penicillin-binding proteins (PBPs) and bacterial cell wall elongation complexes. Subcell. Biochem. 93, 273–289 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P., The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Meeske A. J., et al., SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclercq S., et al., Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 7, 43306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taguchi A., et al., FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiser J. N., Ferreira D. M., Paton J. C., Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsui H. C. T., et al., Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol. Microbiol. 100, 1039–1065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagliero E., et al., The inactivation of a new peptidoglycan hydrolase Pmp23 leads to abnormal septum formation in Streptococcus pneumoniae. Open Microbiol. J. 2, 107–114 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacq M., et al., The cell wall hydrolase Pmp23 is important for assembly and stability of the division ring in Streptococcus pneumoniae. Sci. Rep. 8, 7591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao Y., et al., Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nat. Chem. Biol. 13, 793–798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taguchi A., Kahne D., Walker S., Chemical tools to characterize peptidoglycan synthases. Curr. Opin. Chem. Biol. 53, 44–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassine J., Pazos M., Breukink E., Vollmer W., Lytic transglycosylase MltG cleaves in nascent peptidoglycan and produces short glycan strands. Cell Surf. 7, 100053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]