Abstract
In most organisms, transition metal ions are necessary cofactors of ribonucleotide reductase (RNR), the enzyme responsible for biosynthesis of the 2′-deoxynucleotide building blocks of DNA. The metal ion generates an oxidant for an active site cysteine (Cys), yielding a thiyl radical that is necessary for initiation of catalysis in all RNRs. Class I enzymes, widespread in eukaryotes and aerobic microbes, share a common requirement for dioxygen in assembly of the active Cys oxidant and a unique quaternary structure, in which the metallo- or radical-cofactor is found in a separate subunit, β, from the catalytic α subunit. The first class I RNRs, the class Ia enzymes, discovered and characterized more than 30 years ago, were found to use a diiron(III)-tyrosyl-radical Cys oxidant. Although class Ia RNRs have historically served as the model for understanding enzyme mechanism and function, more recently, remarkably diverse bioinorganic and radical cofactors have been discovered in class I RNRs from pathogenic microbes. These enzymes use alternative transition metal ions, such as manganese, or posttranslationally installed tyrosyl radicals for initiation of ribonucleotide reduction. Here we summarize the recent progress in discovery and characterization of novel class I RNR radical-initiating cofactors, their mechanisms of assembly, and how they might function in the context of the active class I holoenzyme complex.
Keywords: electron transfer, enzyme structure, iron, manganese, radical
Abbreviations: Cys, cysteine; DOPA, dihydroxyphenylalanine; EPR, electron paramagnetic resonance; HU, hydroxyurea; Lys, lysine; PCET, proton-coupled electron transfer; RNR, ribonucleotide reductase; RT, radical translocation; Trp, tryptophan; Tyr, tyrosine; Tyr•, tyrosyl radical; XAS, X-ray absorption spectroscopy
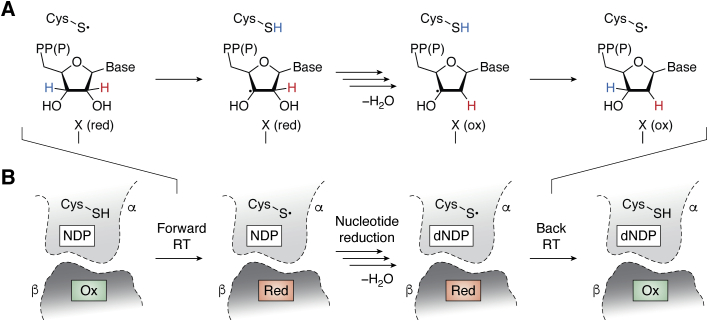
In nearly all organisms, replication and repair of genetic material require transition metal ions, specifically via the action of ribonucleotide reductases (RNRs) (1, 2, 3). RNRs provide the only de novo source of 2′-deoxyribonucleotide building blocks for DNA biosynthesis by performing a chemically challenging dehydroxylation reaction upon a ribonucleotide di- or triphosphate substrate (4). The role of the transition metal cofactor in this process is to transiently oxidize an active site cysteine (Cys) side chain to a thiyl radical (Cys•) (2), which then initiates reaction with substrate by radical activation of the inert 3′-C-H bond in the ribose ring (Fig. 1A) (5). In this approach, an activating oxidation step at an adjacent site induces the desired cleavage of the neighboring 2′-C-O bond. At the end of the reaction, the thiyl radical is regenerated and the net reduction reaction is balanced by provision of reducing equivalents from two additional Cys residues in the active site or the external reductant, formate (6).
Figure 1.
Schematic of ribonucleotide reduction and radical translocation (RT) in class I RNR.A, universal mechanism for nucleotide reduction in RNRs. B, diagram of the steps involved in radical translocation in class I RNRs.
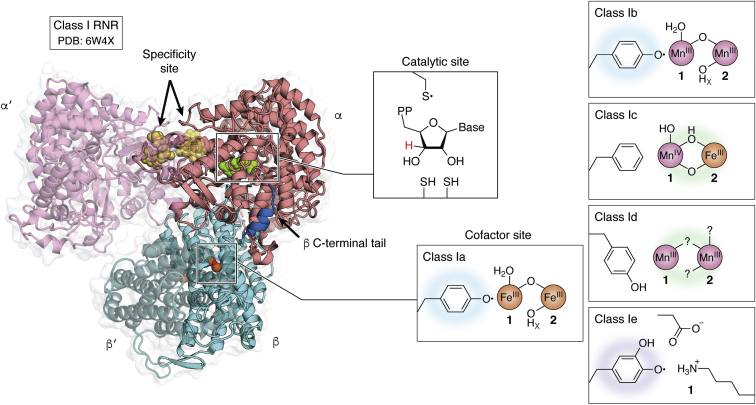
While all RNRs share this universal mechanism of ribonucleotide reduction, the metal-ion-driven method of thiyl radical generation varies substantially (2). The identity of the Cys oxidant provides the basis for classification of RNRs into three classes, I to III. Class II enzymes use a Co-dependent adenosylcobalamin cofactor to oxidize the active site Cys directly (7). Class III enzymes use an iron-dependent [4Fe-4S]-activase to generate a stable glycyl radical in the RNR active site (8), which then forms the thiyl radical locally. These enzymes are found exclusively in microbes and their metallocofactors are either agnostic to oxygen (class II) or potently inactivated by it (class III). By contrast, class I RNRs operate in all three domains of life, including humans and other eukaryotes (1, 2). Class I enzymes share a reliance on dioxygen for assembly of the Cys oxidant cofactor and a complex quaternary structure, in which the metallo- or radical cofactor is located in a separate enzyme subunit (9). This enzyme architecture necessitates long-range radical translocation (RT) for thiyl radical generation (Fig. 1B) (3, 10). The first class I RNRs to be characterized biochemically, class Ia enzymes, contain a diiron(III)-tyrosyl-radical (Fe2III/III-Y•) metallocofactor (Fig. 2) (11). More recent characterization of sequence-divergent class I RNRs has revealed remarkably varied inorganic chemistry associated with this enzyme class (2). Five class I RNR subclasses (class Ia–e) have now been reported (2, 12, 13, 14, 15, 16), four of which (Ib–e) deviate from the Fe2III/III-Y• cofactor of well-characterized Ia enzymes. The features that differentiate these five known class I RNRs include 1) the number and type of metal ions present in the active β subunit for cofactor maturation, 2) the oxidant used to generate the active cofactor (dioxygen or superoxide), 3) the involvement of additional activase proteins, and 4) the identity of the stable oxidant in β (metal- or Tyr•-based) that forms the Cys• in α (17). Here, in the context of the canonical class Ia RNR, we discuss progress in understanding cofactor assembly in Mn-dependent class Ib and Ic RNRs. We also summarize the discovery and characterization of the two most newly reported subclasses, Mn-dependent Id, and the apparently metal-free Ie enzymes. Finally, we present a bioinformatic analysis of the class I RNR β subunits, highlighting areas of sequence space that remain uncharacterized and might serve as fertile ground for discovery of other novel cofactors and their mechanisms of O2-mediated assembly.
Figure 2.
Quaternary structure of the active holoenzyme complex in class I RNR (PDB accession code6W4X).Insets show the location of the active site in the catalytic α subunit (middle top) and the metallo- or radical cofactor (middle bottom and far right) in the β subunit.
Fe-dependent class Ia RNR
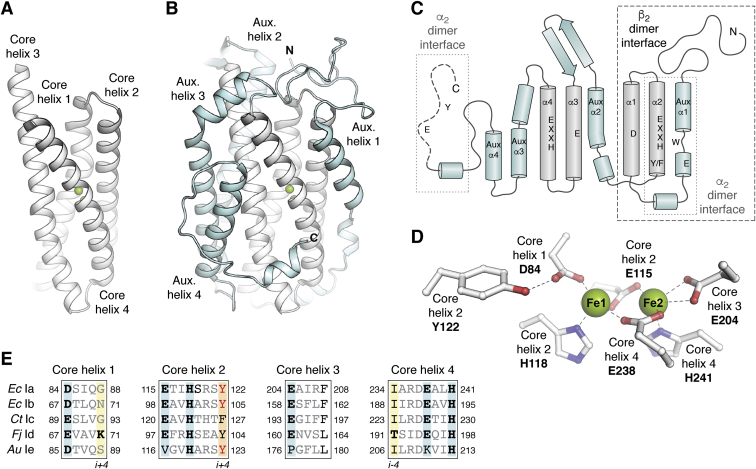
Class I RNRs assemble a Cys oxidant cofactor in a β subunit. All β subunits characterized to date belong to the ferritin structural superfamily (9, 18), using a core four-helix-bundle motif to bind and assemble the active oxidant (Fig. 3, A–C). In class Ia RNRs, this core unit is symmetrical, with two pairs of α-helices arranged head-to-tail to fully bury the Fe2III/III-Y• oxidant and protect it from solvent. Each helix in the pair contributes a D/E or EXXH metal-binding motif (Fig. 3D). In addition to protection from solvent, the regular helical core structure also enables provision of second-sphere side chains that can contribute significantly to cofactor assembly and reactivity, often located one helical turn away from metal-binding side chains in i ± 3 or i ± 4 positions (Fig. 3E). For example, the tyrosine (Tyr) side chain that harbors the Y• in the activated form of class Ia RNR is located in the i + 4 position of core helix 2 relative to the EXXH iron-binding motif. Patterns of conservation in amino acids important for cofactor function can vary among class I RNR enzyme subclasses.
Figure 3.
Structural features of the class I RNR β subunits.A, core four-helix bundle implicated in metal binding and/or radical cofactor assembly (white) and B, auxiliary helices (cyan). C, topology diagram showing the locations of amino acids in class Ia RNR important for metal-binding, Tyr• formation, or radical translocation. D, metal-binding site in FeII2-β of class Ia RNR (PDB accession code 1PIY). Selected amino acids shown in stick format and FeII ions shown as green spheres. Coordination interactions or hydrogen bonds shown as dashed lines. E, a sequence alignment showing conservation of important metal-binding (cyan) or radical formation sites (orange) in all known class I RNR subclasses. Amino acids important for oxidant access and suppression of Tyr• formation in class Id RNR are shown in boldface and highlighted in yellow.
The core helical bundle of class I RNRs is surrounded by additional secondary structures, typically four auxiliary α-helices and flexible N- and C-termini (17, 18) (Fig. 3, B and C). These auxiliary structures are important for mediating interactions with other polypeptides. All class I RNRs rely on such interactions for catalysis (1, 2). The β subunit itself operates as a homodimer (Fig. 2), with an interface that involves the N-terminal half of the β-subunit-fold (9). In the most well-characterized class Ia RNR from Escherichia coli, the active holoenzyme adopts an α2β2 quaternary structure (Fig. 2) (19, 20) in which the α subunit contains the active site for nucleotide reduction and one or more allosteric regulatory nucleotide-binding sites (21). Formation of this complex involves N-terminal auxiliary secondary structures in β as well as the C-terminus of the protein (20). Interestingly, the final ∼30 amino acids of the β subunit are flexible and remain persistently disordered in X-ray crystal structures of β alone (9). Despite this disorder, the C-terminus of β forms a key part of the interface with the catalytic subunit and contains two amino acids essential for turnover, E350 and Y356 (E. coli Ia numbering) (20).
The primary function of the α-β subunit interface in class I RNRs is reversible translocation of an oxidizing equivalent, or radical, from the cofactor in β to the active site in the α subunit (3, 10) (Fig. 1B). This process occurs with remarkable speed, up to 10 s−1 in the fastest class I enzymes (22), and over a great distance, estimated to span a ∼35 to 40 Å distance in docking models of the two subunits (21, 23). The distance between oxidant source and destination requires involvement of aromatic amino acids in RT (10). Conserved Tyr and tryptophan (Trp) amino acids, spanning both subunits, were initially implicated as these intermediates based on conservation and requirement for activity (21, 24). Initiation of RT and formation of these pathway intermediates must be tightly controlled. For example, the oxidizing equivalent in β is only deployed once substrate is bound to the α subunit (25). This step and subsequent Tyr oxidation steps are proposed to involve proton-coupled electron transfer (PCET) events (10). In E. coli class Ia RNR, the entire radical translocation process is gated by a slow conformational change within the holoenzyme complex (22, 25), and this feature prevents direct detection of pathway radical intermediates in the wild-type enzyme. To overcome this issue, key Tyr side chains in E. coli class Ia RNR have been successfully substituted with noncanonical analogs of altered reduction potential (26, 27). These substitutions slow down the radical transfer steps such that they are no longer masked by conformational change, allowing for observation of pathway Tyr• at sites such as Y356 in the C-terminus of β (28). Recent advances in cryo-electron microscopy technology, together with insight from experiments to perturb radical translocation, enabled capture of the structure of the active form of the E. coli class Ia RNR holoenzyme complex just last year (20). Here, we also summarize the key findings of this tour-de-force study and its implications for understanding radical translocation in other class I RNR subclasses.
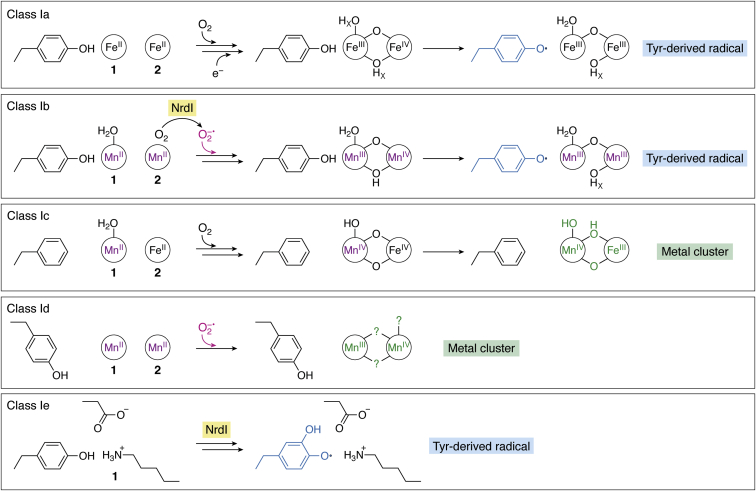
Assembly of the active cofactor in the β subunit is an important maturation step in class I RNR catalysis, one that likely provides additional opportunities for regulation of enzyme activity (2). Cofactor maturation is best understood in the Fe2III/III-Y•-utilizing E. coli class Ia RNR (Fig. 4). In this enzyme, the β subunit can be activated in vitro in a self-assembly reaction that only requires purified apo β protein, FeII, oxygen, and a reductant. Inside the cell, this process probably involves other components, including factors that control metallation and cofactor oxidation state (29). The former step is important because mismetallation with other metal ions, such as MnII, occurs readily and potently inhibits the enzyme (30). During the in vitro self-assembly reaction, apo β loads with two equivalents of FeII, and this form of the protein has been structurally characterized (31). The dinuclear cofactor is coordinated by the four carboxylate and two histidine ligands contributed by the four-helix core of the ferritin fold (Fig. 3D) (31). The Tyr122 side chain that ultimately becomes oxidized to a Y• in the activated cofactor resides near one of the metal ions, Fe1. The metal-binding site is fully buried from solvent but an internal hydrophobic cavity and adjacent open coordination positions suggest an access route for oxygen (Fig. 3D) (31, 32). The next step in the cofactor maturation reaction is rapid addition of O2 to the Fe2II/II cluster, yielding a short-lived peroxo-Fe2III/III intermediate (33, 34). A conserved second-sphere aromatic, Trp48, donates an electron to reductively cleave the peroxide unit forming a Fe2III/IV intermediate, X (35, 36). The resulting Trp48+• is reduced by the necessary external reducing agent (36). Intermediate X oxidizes the adjacent Tyr122 to form the Fe2III/III-Y• cofactor (Fig. 4) (35, 36), capable of initiating RT to the α subunit to oxidize the active site Cys. Forward RT results in a Fe2III/III state that can reversibly reform the active Fe2III/III-Y• after nucleotide reduction is completed (Fig. 1B) (2). The RT product complex is likely distinct from a separate Fe2III/III (met) form of the active cofactor that forms via unproductive 1-electron reduction of the Y•. The difference between these two cluster forms could involve subtle changes in cofactor structure and/or protonation state. The ferritin fold and its buried cofactor site insulate against proton transfer in and out of the interior of the protein, allowing for control of RT initiation via carefully controlled PCET steps. The met Ia cofactor is stable and conversion back to its active form requires reduction of the FeIII ions (37).
Figure 4.
Cofactor assembly mechanisms for class I RNRs. Manganese-dependent enzymes are highlighted in purple. Superoxide-dependent RNRs are highlighted in hot pink. Subclasses that require an NrdI activase are indicated with a yellow box. Metal-centered Cys oxidants shown in green and Tyr-derived radical Cys oxidants shown in blue.
Recent advances in class Ia RNR holoenzyme characterization
The active metallo- or radical cofactor in class I RNR β subunits is ultimately used to generate a transient thiyl radical in the α subunit to initiate the ribonucleotide reduction reaction (1). This intersubunit radical translocation (RT) is triggered in the β subunit upon substrate binding to the active site in α. RT occurs on every turnover of the enzyme, and it is reversible, with the oxidizing equivalent returning to the β subunit after the reaction is finished. Docking models and pulse electron paramagnetic resonance (EPR) measurements estimated an RT distance of 35 to 40 Å for this process (21, 23, 38). To account for the rate of class I RNR turnover (∼1–10 s−1), which is too fast for single-step electron transfer, RT is proposed to involve smaller proton-coupled electron transfer steps mediated by conserved aromatic amino acid side chains arranged in a pathway between the cofactors in each subunit (2). In the past decade, a series of elegant studies by Stubbe and coworkers (10, 27, 28) have established functional roles for conserved aromatics along the RT pathway in E. coli class Ia RNR by showing that modulation of reduction potentials of pathway Tyr via incorporation of noncanonical amino acid analogs can slow RT sufficiently to detect transient radical intermediates localized to these sites (10).
The capacity for rapid and reversible electron transfer across the subunit interface has inspired a great deal of interest in the structure of the active holoenzyme complex in any class Ia RNR. Efforts to obtain this information in the E. coli Ia system have been hindered by the dynamic nature of subunit interaction (39) and a propensity to form allosterically inhibited higher-order structures at high protein concentration (19). For example, the koff for α-β dissociation is estimated to be >60 s−1 (39). However, some of the modifications used to characterize pathway Tyr radicals also stabilize the holoenzyme complex for several minutes (40). In combination with technical advances in cryo-electron microscopy approaches, which allow for sample analysis on the seconds-to-minutes timescale at low protein concentration, Stubbe, Drennan, and coworkers recently reported a 3.6-Å-resolution structure of the E. coli class Ia α2β2 holoenzyme complex by using a E52Q/(2,3,5)-F3-Y122-substituted version of β (20) (Fig. 5A). This structure significantly advances our understanding of class I RNR radical translocation by showing how the active complex assembles to arrange PCET components upon substrate binding and disassembles to permit product release. Here we review aspects that are likely to be conserved among all class I RNRs and contrast those with the details that might differ in class Ib–e enzymes, thereby warranting further study.
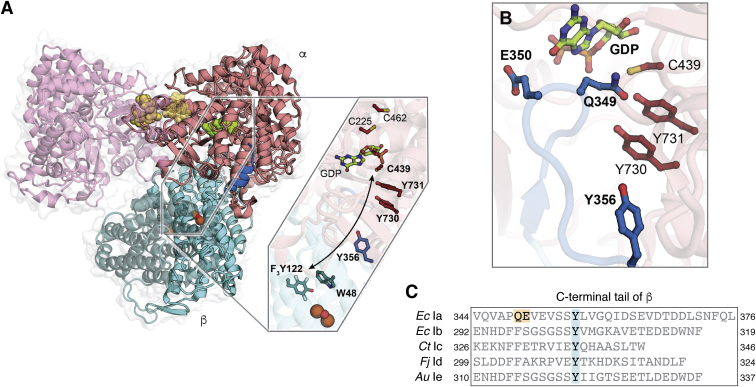
Figure 5.
Structure of the active holoenzyme complex in class Ia RNR (PDB accession code6W4X).A, the engaged half of the holoenzyme complex reveals the arrangement of aromatic amino acid side chains along the RT pathway (indicated by arrow and boldface labels). B, a zoomed-in view of the ordered C-terminus of the β subunit shows important interactions with the GDP substrate. Panels A and B adapted from refs. (3, 20). C, a sequence alignment of the β subunit C-terminal tail in representative members of class Ia–e subclasses. RT pathway intermediate Y356 is conserved among all class I RNRs but the polar side chains implicated in substrate binding in class Ia RNR differ in Ib–e RNRs.
One of the most striking features of the E. coli class Ia RNR active holoenzyme complex is its asymmetry (20) (Fig. 5A). Only one half of the subunit interface engages at a time. Therefore, the RT pathway cannot simultaneously assemble in both halves of the α2β2 complex. This result is consistent with experimental observations of half-of-sites reactivity in class Ia RNRs (41). Other subclasses also exhibit evidence of this phenomenon. In class Ib RNR, a low-resolution X-ray crystal structure of an α2β2 complex shows a similar asymmetric intersubunit interaction (42). And in class Ic RNR, detection of two kinetic phases of hydroxyurea-(HU)-mediated cofactor reduction during enzyme turnover supports half-of-sites reactivity and complex asymmetry in this subclass (43). The fast phase is interpreted as HU-mediated interception of a pathway Tyr• in the engaged half of the holoenzyme while a slow phase represents direct reaction of HU with the MnIVFeIII cofactor in the other subunit. While half-of-sites reactivity has not been investigated to date in class Id or class Ie RNR, class Ie enzymes exhibit ∼50% sequence identity to class Ib enzymes, suggesting they likely engage the α subunit similarly.
All class I RNRs characterized to date conserve a Tyr residue (Y356, E. coli Ia numbering) in a C-terminal tail of the β subunit that is typically disordered in X-ray structures of the β2 complex alone (2). This residue is essential for RNR activity because it mediates RT between the two subunits of the holoenzyme complex, but its functional location had never been visualized previously. On one side of the E. coli class Ia holoenzyme complex, the entire C-terminus orders to reveal the location of Y356 and all other residues in this previously disordered tail (20). The side chain of Y356 is found adjacent to a pair of conserved Tyr side chains in the α subunit, also implicated in PCET, consistent with an RT mediator role for Y356 (Fig. 5A). Interestingly, Y356 does not appear to have a dedicated amino acid proton donor/acceptor. Instead, water molecules at the subunit interface are implicated in this process, although these waters cannot be detected directly in the cryo-EM structure. While all class I RNRs conserve a Y356 equivalent, we cannot yet infer its exact structural context, including proximity to interfacial water molecules, in other subclasses.
The ordering of the β subunit C-terminus revealed by the cryo-EM structure of the E. coli class Ia holoenzyme complex also shows conformational changes that could communicate substrate binding in the α subunit to the Cys oxidant in the β subunit (20). The C-terminus of the β subunit folds into the active site of the catalytic subunit to make both direct and indirect contacts to substrate (Fig. 5B). This observation shows that the β subunit detects the status of the active site (substrate, product, nothing bound) directly via ordering of the β subunit C-terminal tail. This mechanism of intersubunit communication also explains the dynamic nature of subunit interaction. Because the tail blocks the active site, the complex must fully dissociate to release product and rebind substrate. Interestingly, the two key amino acids in the β subunit C-terminus that contact substrate in E. coli class Ia RNR, Q349 and E350, are not universally conserved in other subclasses (Fig. 5C), suggesting differences in the exact structure of the C-terminus of the β subunit may exist in other RNRs that use alternative cofactors.
These conformational changes are likely an important part of the event (or events) that initiate radical translocation from the β subunit cofactor upon substrate binding in α. The mechanism of RT initiation still remains elusive due to the low local resolution of the cryo-EM structure in the vicinity of the metallocofactor (∼5.5 Å resolution) and the nature of the mutations needed to trap a stable complex (20). Analysis of the α subunit active site structure suggests that the disengaged half of the holoenzyme complex has undergone a single turnover, due to observation of a disulfide bond in that subunit. The other half of the complex appears to be organized for radical translocation but the oxidizing equivalent in β has not yet deployed to the α subunit. The link between the substitutions made in this system and the structural observations remains unclear—but the effects on Y122 reduction potential and pKa conferred by the fluoro groups of the (2,3,5)-F3 modification are likely the key perturbations dictating outcome (27, 44). Two other recent studies of class Ia and Ic RNR provide some clues about the molecular details of RT initiation in class I enzymes. Mössbauer analysis of the RT product in E. coli class Ia RNR suggests that Fe1-mediated proton transfer to the Y• at position 122 involving a terminal water ligand is the first step in oxidant deployment (45). And the aforementioned study of HU-induced cofactor reduction in class Ic RNR concludes that the key chemical event responsible for Cys oxidant excursion occurs in the β subunit and local to the cofactor (43). Interestingly, these proposals would indicate that the chemical details of the first step of radical translocation in other subclasses could differ substantially. The metal-centered oxidants of class Ic and Id enzymes lack the Tyr• proton destination (12, 14) and class Ie enzymes lack a metal-coordinated water molecule in their active forms (15, 16).
Iron-dependent class Ia RNRs are dominant in eukaryotes, including humans (46), and widespread in bacteria (47). Among prokaryotes, acquisition of iron to drive DNA replication can present a challenge in nutrient limited environments (48). To adapt, other RNR subclasses have evolved in bacterial and archaeal organisms employing different metallocofactors and/or radical components (2, 3). These distinctions necessitate novel mechanisms of cofactor maturation (Fig. 4), including requirement for additional proteins or alternative O2-derived metal/Tyr oxidants. The next sections of this review focus on the alternative metallo- or radical cofactors found among class I RNRs, with an emphasis on subclasses that use transition metal ions other than iron and their cofactor assembly mechanisms. Efforts to understand these aspects of RNR catalysis are significant because Ib–e enzymes dominate in aerobic bacterial pathogens. Evolution of new Cys oxidants likely provides a competitive advantage against human and animal hosts that rely on iron-dependent RNRs.
Mn-dependent class Ib RNR
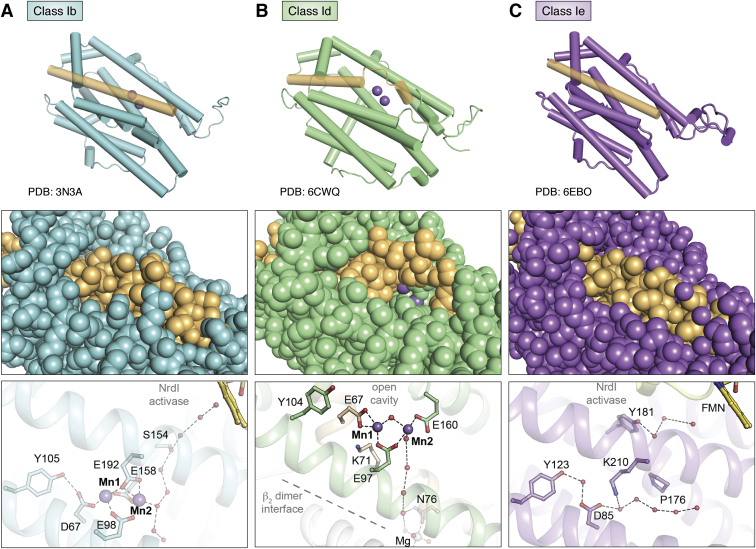
Class Ib RNRs were first differentiated from class Ia enzymes on the basis of sequence identity (49). Despite early indications of a reliance upon manganese for activity (50), particularly in enzymes isolated from their native host organisms (51, 52), this subclass was initially proposed to assemble an Fe2III/III-Y• (53). This form is active in vitro (13, 54), albeit to a lesser extent than typical class Ia RNRs (22). The importance of manganese in class Ib RNR function was definitively established in 2010 by Cotruvo and Stubbe (13). A highly active Mn2III/III-Y• can be formed by reconstitution of Ib β with MnII, oxygen, and the reduced form of a flavoprotein, NrdI (13) (Fig. 4). Consistent with an essential role in active cofactor assembly, NrdI is universally encoded in the genomes of class-Ib-utilizing organisms, typically as part of the same operon as the Ib α and β subunits. The activase binds to the Ib β subunit, an interaction that has been characterized crystallographically in two different class Ib homologs (32, 55). The activation complex contains a hydrophilic channel that links the flavin cofactor in NrdI to the Mn2II/II site in the β subunit (Fig. 6A) (32). This structural feature shows a feasible route to Mn-dependent class Ib RNR cofactor assembly involving flavin-mediated remote generation of a reduced form of O2 (superoxide or peroxide) that is channeled within the activation complex to oxidize a reduced dimanganese cluster. In this model, the function of the universally encoded NrdI activase is to overcome the inherent inertness of O2 to reaction with MnII by providing a more reactive form of the cofactor oxidant. This model also represents a rare example of use of normally toxic reactive oxygen species to perform a beneficial function inside the cell.
Figure 6.
Comparison of overall structure (top), cofactor solvent exposure (middle), and location of solvent channels for superoxide access (bottom) in class Ib (A) (PDB accession code3N3A), class Id (B) (PDB accession code6CWQ), and class Ie RNR (C) (PDB accession code6EBO). Selected amino acids shown in stick format and MnII ions shown as purple spheres. Coordination interactions or hydrogen bonds shown as dashed lines. Water molecules shown as red spheres.
More recently, several different research groups have pursued open questions associated with the biological significance of class Ib Mn-dependent RNR activity, the detailed mechanism of Mn2III/III-Y• cofactor assembly and its optimization, and the structural and mechanistic diversity of class Ib RNRs. The observation of significant in vitro activity with either iron or manganese in these systems led to a series of investigations into which form is predominantly used in native organisms. In 2011, two different groups investigated class Ib RNR usage in E. coli, a facultative anaerobe encoding two aerobic RNRs, class Ia and Ib (56, 57). Martin and Imlay demonstrated that Ib RNR is only expressed upon derepression of its operon by the FeII-responsive transcription factor, Fur (56). This observation is consistent with a biological role for class Ib RNR in enabling cell replication when iron levels are low. Cotruvo and Stubbe reached a similar conclusion, showing that class Ib RNR isolated from E. coli strains deficient in five different iron uptake systems contains only manganese (57). Importantly, this work represented the first isolation and characterization of the active cofactor in an Ib RNR obtained without overexpression. Class Ib RNRs have also been isolated with active Mn2III/III-Y• cofactors from homologous overexpression systems in two different native organisms (Cornyebacterium ammoniagenes and Bacillus subtilis) that rely on the Ib enzyme as their sole aerobic source of deoxyribonucleotides (51, 52, 58). The active enzyme from C. ammoniagenes was characterized comprehensively by EPR spectroscopy and X-ray crystallography (52), providing the most complete description to date of an activated Ib RNR. Metal ion usage by class Ib RNR has also been investigated in the opportunistic pathogen, Streptococcus sanguinis, which encodes an Ib enzyme as its sole aerobic RNR (59, 60). In a rabbit model of S. sanguinis-mediated infective endocarditis, the gene encoding NrdI, required for Mn-dependent Ib activity but not iron-dependent activity, was shown to be essential for growth and survival of the pathogen (60). A recent transcriptomics study of S. sanguinis response to Mn-depletion under aerobic conditions shows downregulation of the Ib operon accompanied by diminished growth (61). These results are consistent with inability of the S. sanguinis Ib RNR to substitute iron for manganese in vivo.
In 2013, a kinetic dissection of the metallocofactor assembly in B. subtilis class Ib RNR provided important information about the identity of the MnII oxidant generated by NrdI and a key intermediate responsible for Tyr oxidation (Fig. 4) (62). Stopped-flow absorption spectroscopy analysis of NrdI flavin oxidation showed that only the two-electron-reduced, NrdIhq, form of the activase reacts with O2 quickly enough to account for the overall rate of cofactor assembly. This observation implicates a single equivalent of superoxide as the MnII oxidant. Rapid freeze-quench EPR (RFQ-EPR) spectroscopy analysis of the first observable oxidized metal-centered intermediate in this system reveals a spectrum assigned as a MnIIIMnIV cluster. This species decays at a rate that matches that of Tyr• appearance, as measured in the stopped-flow absorption analysis. Provision of a single equivalent of superoxide for cofactor assembly is both a creative and economical solution to class Ib cofactor assembly (62). Not only does it generate a form of O2 that reacts readily with MnII, but it also provides the exact number of oxidizing equivalents required to generate two MnIII ions and a Tyr•.
Mechanistic studies of class Ib RNRs have been hindered by diversity in yields of active cofactor and differences in structure, β subunit affinity, and redox properties of the NrdI activases. The divergence of biochemical properties is perhaps not surprising because phylogenetic analysis of NrdI sequences reveals three distinct groups of Ib enzymes (63). The structurally well-characterized E. coli and C. ammoniagenes Ib systems belong to a group that consistently yields low levels (0.2–0.3 Tyr•/β) of active cofactor upon isolation or in vitro reconstitution (13, 52). NrdIs from a second phylogenetic group, including proteins from B. subtilis and related organisms, exhibit variable redox potentials for different forms of the flavin cofactor. While E. coli and B. subtilis NrdIs cannot stabilize significant quantities of one-electron-reduced NrdIsq (54, 62), Bacillus anthracis NrdI can stabilize quantitative amounts of the semiquinone, similar to traditional flavodoxins (64). Based on this observation, maturation of B. anthracis Mn2II/II-β has been proposed to initiate from a NrdIsq state. However, detailed kinetic analysis of the assembly mechanism, as performed for B. subtilis class Ib RNR, would be required to definitively establish this distinction. In 2014, Lofstad et al. (65) reported an important advance in enhancing levels of Tyr• in Bacillus cereus NrdI-dependent cofactor assembly by screening candidate flavodoxin reductases for those that enhance yield of active cofactor. The approach produced 0.6 Tyr• per β, a threefold improvement in yield over initial reconstitutions in the absence of a reductase. Application of this tactic to other class Ib enzymes could facilitate mechanistic study and biophysical characterization of active Mn cofactors or oxidized intermediates.
Mn- and Fe-dependent class Ic RNR
The class Ic enzyme from Chlamydia trachomatis was the first class I RNR shown to use a metal ion other than iron in its active cofactor (12). It was also the first class I RNR demonstrated to use a metal-based Cys oxidant rather than a Tyr•-derived oxidant. In 2007, Jiang et al. showed that maximal enzyme activity could be achieved with in vitro cofactor assembly in the presence of MnII and FeII (Fig. 4). The active cofactor is a MnIVFeIII complex, formed by reaction of a MnIIFeII precursor with O2. By an alternative mechanism, the C. trachomatis RNR can also be activated by reaction of MnIIFeII with two equivalents of H2O2 (66). Both the MnIVFeIII state and a preceding MnIVFeIV intermediate have been extensively characterized by Mössbauer, EPR, and X-ray absorption spectroscopy (XAS) (67). These efforts were instrumental in establishing the identities and oxidation/spin states of these complexes. Recently, researchers have focused on understanding additional structural features of the class Ic active cofactor and its assembly pathway.
Crystallographic characterization and application of advanced spectroscopic techniques to the Mn-bound forms of class Ic RNR have enabled detailed assignment of metal ion and proton location in the active cofactor and its precursors (68, 69, 70). Two different crystallographic analyses of activated Ic enzymes revealed that the MnIV ion resides in site 1 (68, 70), the metal-binding site closest to the Tyr• in class Ia/b enzymes (Fig. 4). This assignment is consistent with functional substitution of the redox-active Tyr by an oxidized Mn ion in the class Ic enzymes. The Mn site 1 assignment was also corroborated by spectroscopic studies on activated enzyme samples in solution (69). The crystallographic studies further revealed partial occupancy of site 1 by iron and site 2 by manganese, particularly when the enzyme was reconstituted with excess of either metal ion (68, 70). This finding is consistent with a known propensity for mismetallation of this scaffold.
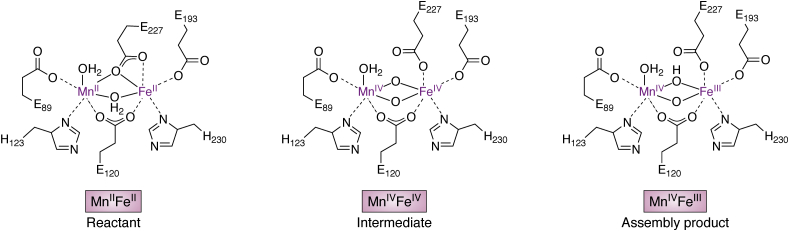
In a subsequent crystallographic study of the reactant MnIIFeII complex, issues with site-selective metal ion binding were overcome via extended anaerobic incubation of apo class Ic RNR β with MnII and FeII (71). In this experiment, each metal ion was present in 1:1 ratio per β monomer, resulting in strict partitioning of Mn to site 1 and Fe to site 2. Interestingly, structures of apo β co-crystallized with MnII show occupancy of both sites, even at substoichiometric metallation. Selective assembly of the relevant precursor to the active cofactor might be driven, therefore, by a preference of iron for site 2 in class Ic RNR. Interestingly, analyses of metal ion binding in E. coli class Ia RNR are also consistent with higher affinity of iron for site 2 (72, 73). The structure of MnIIFeII-β additionally provides insight into the details of the first coordination sphere prior to O2 addition (71). The reactant state contains two water ligands, one bound terminally to Mn1 and another that bridges the two metal ions (Fig. 7). The structure also reveals a μ-η1,η2 binding mode for one of two bridging Glu ligands, an interaction seen in the fully reduced forms of other dimetal-binding class I RNR β subunits. Interestingly, this ligand undergoes significant conformational change upon cofactor oxidation, the sole protein ligand to do so in class Ic RNR.
Figure 7.
Structures of reactant (left), intermediate (middle), and assembly product (right) in MnIVFeIIIcofactor activation in class Ic RNR.
The relatively small number of conformational and coordination changes upon conversion to the active form of class Ic RNR have made it possible to fully map the structures of intermediate and product states via spectroscopic and computational approaches (Fig. 7). XAS and pulse EPR spectroscopic studies of the MnIVFeIV intermediate in class Ic RNR are consistent with a di-μ-oxo bridging structure and a terminal MnIV hydroxide ligand (74), suggesting that O2 inserts between the metal ions to displace the μ-η1,η2-bridging Glu ligand, accompanied by deprotonation of the solvent ligand to Mn1. A recent valence-to-core X-ray emission study of this intermediate and the active MnIVFeIII cofactor confirms this assignment for the MnIVFeIV complex (75). Spectral comparisons show elongation of one Mn-O bond concomitant with contraction of the other upon 1-electron reduction, consistent with protonation of one of the two oxo bridges. The structure of the further reduced MnIIIFeIII product of the RT reaction remains to be defined in detail, but it may be accompanied by similar proton transfers to the active cofactor, a phenomenon that is probably universal to reaction initiation in all class I RNRs.
Mn-dependent class Id RNR
Class Id ribonucleotide reductases were initially distinguished from previously characterized class I RNRs by a unique Tyr/Glu pairing in the first and second coordination sphere, as opposed to a Tyr/Asp pairing in Ia/Ib and Phe/Glu pairing in Ic β (Fig. 3E) (14). In the biochemical characterization of Id β from Flavobacterium johnsoniae, Rose et al. demonstrated that efficient activation of metal-depleted Id β occurs in the presence of MnII, O2 and superoxide-generating compounds, such as naphthoquinol or hydroquinol. Cofactor assembly is inhibited by the presence of the O2− scavenger, superoxide dismutase, confirming the ability of Id β to utilize environmental sources of O2−. Using a combination of XAS, EPR, and UV–visible spectroscopy experiments, the active cofactor was confirmed as a stable Mn2III/IV cluster. Even though the class Id RNRs universally conserve a second-sphere Tyr in the location that harbors a radical in class Ia and Ib enzymes, F. johnsoniae Id RNR does not oxidize this side chain to form a stable Tyr• during cofactor assembly. The oxidized Mn2III/IV cluster serves as the radical initiator in this system, making Id enzymes the second example of a metal ion-based Cys oxidant after the heterodimeric MnIVFeIII cofactor of Ic RNR (14). Two other class Id RNR β subunits from Facklamia ignava and Leeuwenhoekiella blandensis have been characterized in their activated forms by EPR, yielding similar spectral features to those of Mn2III/IV F. johnsoniae β (76, 77)
Class Id RNR cofactor assembly bears some resemblance to class Ib cofactor maturation, the only other solely Mn-dependent class I RNR identified to date (62). The class Id cofactor assembly mechanism is arguably simpler because it requires only the addition of MnII and O2− to metal-depleted preparations of β. Class Ib RNR cofactor assembly also involves superoxide-dependent oxidation of a di-manganese cluster, but the metal ion oxidant is generated by the flavoprotein activase, NrdI (13), generally encoded adjacent to the Ib α and β subunits. Of the >260 Id RNR β sequences identified to date, none are found encoded near an NrdI (14). Additionally, Id β can be activated in vitro independent of an accessory activase. Class Ib enzymes also share the ability to achieve a Mn2III/IV oxidation state, but only as a transient intermediate, which then decays to a Mn2III/III state concomitant with Tyr• formation (62). Interestingly, the active Id cofactor easily converts back to a Mn2II/II state poised for reactivation, via cooperative three-electron reduction of the Mn2III/IV complex (14). This behavior contrasts with the reductive decay pathways of class Ia–c cofactors (2). In these systems, the one-electron reduced metal ion clusters (Fe2III/III, Mn2III/III, or MnIIIFeIII) are inert to further reduction and likely require a dedicated reductase inside the cell to reactivate the enzyme.
Structural characterization of class Id Mn2II/II-β subunits has provided insight into the structural basis for direct superoxide scavenging and use of a metal ion-based Cys oxidant. Id β from F. johnsoniae was the first enzyme in this subclass to be structurally characterized in a defined metal ion-bound form, revealing a solvent-exposed metal center resulting from an unwinding of core helix 1 and a distinctive orientation for the second-sphere Tyr (Fig. 6B and 8, A–C) (14). The helical distortion occurs immediately after Mn1 ligand Glu67 (F. johnsoniae numbering), exposing a ∼300 Å3 cavity and the metal cluster to solvent (14). This distortion and cavity have also been observed in metal ion-bound structures of L. blandensis and F. ignava β (76, 77). In the former system, the structural deviation seems to be the result of metal ion loading because the corresponding helix is uninterrupted in apo structures of L. blandensis β (77). A conserved positively charged side chain, lysine (Lys71) (F. johnsoniae numbering), within the unwound portion of helix 1 resides within the vicinity of the metal center—the first incidence of a positive charge adjacent to a dimetal center in any class I β (Fig. 8) (14). Rose et al., proposed that Lys71 and the open cavity function in electrostatic attraction of negatively charged superoxide.
Figure 8.
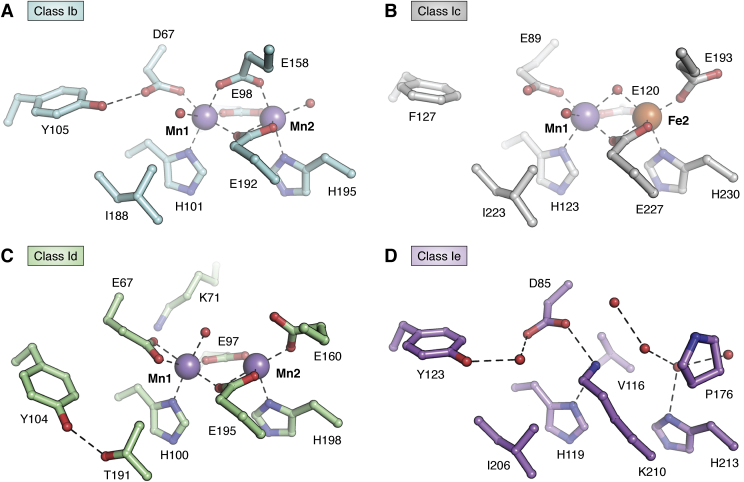
Comparison of metal-binding sites in class Ib (A) (PDB accession code3N3A), class Ic (B) (PDB accession code4M1I), class Id (C) (PDB accession code6CWP), and class Ie RNR (D) (PDB accession code6EBO). Selected amino acids shown in stick format. MnII ions shown as purple spheres, FeII ions shown as orange spheres. Coordination interactions or hydrogen bonds shown as dashed lines. Water molecules shown as red spheres.
Tyr104 (F. johnsoniae numbering), the Tyr•-harboring residue in Ia and Ib RNR, possesses a novel hydrogen bonding partner, Thr191, in class Id enzymes (Fig. 8C). This interaction sequesters Tyr104 ∼7 Å away from Mn1. By contrast, the corresponding Tyr in class Ia and Ib RNR is positioned much closer (∼4–5 Å) to the metal cluster and in H-bonding contact with a unique Asp ligand (Figs. 3D and 8, A and C) (31, 32). These interactions could facilitate Tyr oxidation, whereas the alternative orientation and interactions in Id RNR are likely linked to the inability to form and stabilize a Tyr• in activated Id β. Surprisingly, the Tyr104 side chain remains essential for RNR activity, playing an as-yet unknown role in the radical translocation process (14). When compared with class Ia–c RNRs, the significant structural differences associated with the reactant Mn2II/II form of the β subunit in class Id enzymes underscore the distinctive cofactor assembly and maintenance pathway utilized by this subclass.
Certain class Id β subunits have additional functional domains appended to their N-terminus. F. ignava and L. blandensis Id β subunits contain ATP cones (77), an overall activity regulatory domain typically found at the N-terminus of the α subunit in class Ia and Ic RNRs (1). In class Ia RNR, ATP-binding to the ATP cone promotes formation of active α2 dimers with exposed β2 binding sites while dATP association stabilizes inhibited higher-order oligomers incapable of functional interaction with β2 (19). All class Id RNR α subunits identified to date lack an ATP cone domain (78), but this regulatory unit is found instead in selected β subunits (77). In X-ray crystal structures of ATP-cone bearing L. blandensis β, the subunits form tetramers when bound to dATP. While this quaternary structure superficially resembles the inactivated α4 structures observed in dATP-bound E. coli class Ia RNR (19), the mechanism of ATP-cone-mediated β subunit inhibition remains unclear. The class Id β4 tetramer forms exclusively via ATP cone interactions, leaving the predicted α binding site unobstructed (77). Nucleotide-independent regulation of the α subunit activity may also be possible in Id systems lacking ATP cones entirely (78). F. johnsoniae α can form tetramers in solution, but the physiological relevance of these structures and implications for activity are open questions.
Metal-free class Ie RNR
Class Ie RNRs were codiscovered in 2018 by two independent research teams (15, 16). The Ie β subunit sequences are similar in overall sequence identity to class Ib enzymes, but with a striking substitution of three metal ion coordinating Glu side chains (Figs. 3E and 8D) (15, 16, 79). Interestingly, RNR β subunits with this property were first reported by Roca et al. (79), 10 years earlier. In this study, two Ib-like RNR operons were identified within the Streptococcus pyogenes genome, each encoding a catalytic α subunit, a β subunit, and an NrdI homolog. One contained a true Ib β with the anticipated D…EXXH…E…EXXH metal-binding motif while the other encoded a β sequence with the aforementioned substitutions at three of six metal-coordinating Glu residues (Fig. 3E). The components of the latter operon showed no activity in vitro but could complement an E. coli strain possessing a temperature-sensitive class Ia RNR mutation (79). This early study did not, however, identify the Cys oxidant assembled in the unusual β subunit. More recently, these experiments were replicated with Aerococcus urinae and Mesoplasma florum Ie homologs, showing that each is capable of nucleotide reduction in E. coli, provided that all three components (α, β, NrdI) were coexpressed (15, 16). Bioinformatic analyses uncovered more than 400 bacterial RNR β sequences with the characteristic substitutions of Ie enzymes, some of which represent the sole aerobic RNR encoded in the organism’s genome (15). Together, these observations suggest that class Ie RNRs are capable of ribonucleotide reduction activity despite the significant changes to the predicted metal-binding motif.
Although involvement of an NrdI activase in cofactor assembly is a shared feature with class Ib β subunits, the identity of the class Ie radical-initiating cofactor distinguishes these enzymes from other class I RNR subclasses (15, 16). Activated β subunits can be isolated from NrdI coexpression cultures for the S. pyogenes, A. urinae, and M. florum Ie homologs. These preparations yield active enzyme in in vitro activity assays (0.18–0.35 s−1 per β subunit), but the β subunit contains <0.2 equivalents of any first-row transition metal. X-ray crystallography, EPR, and mass spectrometry analysis of activated Ie β subunits revealed that the active Cys oxidant is a 3,4-dihydroxyphenylalanine radical (DOPA•), generated via posttranslational modification of Tyr123, the site of Tyr• formation in class Ia/b RNRs. X-ray structures of activated Ie β subunits show no evidence of a transition metal component, leading to the conclusion that the active form of β is metal ion-free. Interestingly, a conserved Lys residue (Figs. 3E, 6C and 8D) appears to substitute metal site 1, suggesting that Ie enzymes may use this alternative cation as a surrogate for the oxidized metals of other class I RNR active cofactors.
The identity of the Ie radical initiator is surprising because the predicted redox potential of an anionic DOPA• (∼0.6 V) would be inadequate to oxidize a Cys side chain (∼0.9 V) (28). To illustrate this point, semisynthetic incorporation of DOPA on the protein surface at the position of Tyr356 in E. coli Ia β results in a trapped DOPA• radical and an inactive enzyme (28). However, pulse EPR analysis of activated class Ie β established that the buried DOPA• in this system is instead found in a neutral form (15). We propose that the DOPA• redox potential is tuned by its protonation state and surrounding protein environment to render it a sufficiently potent oxidant for the Cys.
Class Ie RNRs have not yet been successfully reconstituted in vitro from purified components. All active preparations of Ie β studied to date were generated by coexpression with NrdI inside the cell (15, 16). A full understanding of the components and conditions necessary for DOPA• maturation remains to be elucidated. Once these factors are defined, we can begin to understand the order of events in cofactor maturation—radical generation versus Tyr hydroxylation—and maintenance. Srinivas et al. (16), showed that after reductive inactivation of DOPA• by HU, ∼10% of the inactivated enzyme could be reconstituted via redox cycling in the presence of NrdI and O2, suggesting that it is possible to form active radical from DOPA via the action of NrdI alone. However, the exact mechanism of the reactivation and its implications for DOPA• maturation from Tyr are not known. We also do not understand the conditions in which class Ie RNR is used inside the cell, although a metal-free radical initiating cofactor would seem advantageous for pathogens susceptible to iron or manganese limitation induced by the host immune system. In support of this hypothesis, Do et al. (80) demonstrated that increased intracellular Fe and Mn represses Ie RNR expression in S. pyogenes, an organism that encodes both a Mn-dependent class Ib RNR and a Ie system. This observation would be consistent with a role for the Ie enzyme as a backup or failsafe for DNA replication when trace metal ions are scare. Work to more fully decipher the conditions in which Ie RNRs are expressed in native organisms could reveal additional cofactor maturation requirements and information about the biological significance of this unusual RNR subclass.
Bioinformatic analysis of class I RNR β subunits
Sequence comparisons have often provided crucial initial clues about use of novel cofactors and assembly pathways in class I RNR β subunits. A map of sequence space for annotated class I RNR β proteins highlights the areas in which cofactor usage and biochemical properties remain to be explored. Figure 9 shows the top nine clusters (ranked by number of nodes, in which each node represents a set of sequences with >50% identity) in a sequence similarity network (SSN) (81) generated by analysis of >31,000 annotated class I RNR β sequences (82) extracted from UniProtKB (September 2019 release). We annotated the SSN by genome neighbor patterns (node size) and conservation of predicted metal-binding residues (node color). Enlarged nodes indicate a neighboring class I RNR α catalytic subunit verified in the genome. Node color indicates conservation of an Ia/b-like (dark gray), a Ic-like (purple), a Id-like (yellow), or a Ie-like metal-binding motif (light green). Clusters are shaded according to the dominant subclass present in that cluster. Note that at the current alignment score of 80, this SSN groups sequences with Ic-like metal binding together with Ia-like homologs, and Ib-like sequences are found together with Ie-like counterparts (indicated as purple/gray shading in cluster 2, dark green/light green shading in cluster 3). Biochemically characterized class I RNR β subunits discussed in this review are indicated as boxes.
Figure 9.
A sequence similarity network (SSN) (alignment score = 80, nodes represent sequences that share >50% identity) showing the top nine clusters, ranked by number of nodes, for annotated class I RNR β subunits. All clusters have a significant number of β subunits with an adjacent catalytic subunit encoded. Nodes are colored by conservation of subclass-specific metal-binding motif or conservation of the radical-harboring Tyr. Outlined nodes denote sequences characterized structurally. Labeled nodes indicate sequences with experimentally validated cofactor assignments discussed in this review. Shading indicates subclass assignment of a sequence cluster based on assessments of sequence and genomic context.
Class Ia-like sequences can be found in clusters 1, 2, 4, 5 to 6, and 8 to 9. Of these sequence clusters, only cluster 4 contains a comprehensively characterized enzyme (E. coli Ia RNR) in terms of cofactor assembly, stability, and holoenzyme complex structure. This cluster predominately contains other closely related proteobacterial sequences. The other sequence groups are largely unexplored even though they contain eukaryotic and viral RNRs (cluster 1) and enzymes from important bacterial pathogens such as Pseudomonas aeruginosa (cluster 2), Campylobacter jejuni (cluster 6), and Anaplasma phagocytophilum (cluster 5). Clusters 8 and 9 are dominated by environmental actinobacterial and bacteriophage sequences, respectively. These untapped areas of class I β subunit sequence space might harbor unexpected bioinorganic and/or radical chemistry that could be exploited in development of novel drugs that target class I RNRs.
Future directions and outlook
The reliance, for the most part, of ribonucleotide reduction on transition metal ions provides a window into how organisms adapt to changing availability of trace elements in the environment or in complex consortia of microbes and their hosts. Among class I RNRs, the discovery of alternative metal-ion- or radical-based cofactors for DNA biosynthesis has led to elucidation of novel biochemistry associated with cofactor assembly and radical translocation. Important research objectives for the future include (1) continued mining of class I RNR sequence space for novel cofactors; (2) investigation of holoenzyme complex structures beyond the E. coli class Ia RNR model system; (3) understanding pathways of cofactor assembly, regulation, and maintenance inside the cell; and (4) exploration of the evolutionary relationships among class I RNR subclasses.
Conflict of interest
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
We thank Kai Hu for assistance with preparation of Figure 9. Our work on this topic has been supported by grants from the NIH (GM100011 and GM119707).
Author contributions
T. B. R. and A. K. B., writing–original draft; T. B. R. and A. K. B., writing–review and editing.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ruma Banerjee
References
- 1.Nordlund P., Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Cotruvo J.A., Jr., Stubbe J. Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 2011;80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greene B.L., Kang G., Cui C., Bennati M., Nocera D.G., Drennan C.L., Stubbe J. Ribonucleotide reductases: Structure, chemistry, and metabolism suggest new therapeutic targets. Annu. Rev. Biochem. 2020;89:45–75. doi: 10.1146/annurev-biochem-013118-111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofer A., Crona M., Logan D.T., Sjöberg B.-M. DNA building blocks: Keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stubbe J., Ackles D. On the mechanism of ribonucleoside diphosphate reductase from Escherichia coli. Evidence for 3'-CH bond cleavage. J. Biol. Chem. 1980;255:8027–8030. [PubMed] [Google Scholar]
- 6.Mao S.S., Holler T.P., Yu G.X., Bollinger J.M., Jr., Booker S., Johnston M.I., Stubbe J. A model for the role of multiple cysteine residues involved in ribonucleotide reduction: Amazing and still confusing. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 7.Licht S., Gerfen G.J., Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- 8.Sun X., Harder J., Krook M., Jornvall H., Sjöberg B.-M., Reichard P. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: Nucleotide sequence of the cloned nrdD gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:577–581. doi: 10.1073/pnas.90.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordlund P., Sjöberg B.-M., Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 10.Minnihan E.C., Nocera D.G., Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc. Chem. Res. 2013;46:2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkin C.L., Thelander L., Reichard P., Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein β2 subunit of the Escherichia coli enzyme. J. Biol. Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 12.Jiang W., Yun D., Saleh L., Barr E.W., Xing G., Hoffart L.M., Maslak M.A., Krebs C., Bollinger J.M., Jr. A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 13.Cotruvo J.A., Jr., Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry. 2010;49:1297–1309. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose H.R., Ghosh M.K., Maggiolo A.O., Pollock C.J., Blaesi E.J., Hajj V., Wei Y., Rajakovich L.J., Chang W.-c., Han Y., Hajj M., Krebs C., Silakov A., Pandelia M.-E., Bollinger J.M., Jr. Structural basis for superoxide activation of Flavobacterium johnsoniae class I ribonucleotide reductase and for radical initiation by its dimanganese cofactor. Biochemistry. 2018;57:2679–2693. doi: 10.1021/acs.biochem.8b00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaesi E.J., Palowitch G.M., Hu K., Kim A.J., Rose H.R., Alapati R., Lougee M.G., Kim H.J., Taguchi A.T., Tan K.O., Laremore T.N., Griffin R.G., Krebs C., Matthews M.L., Silakov A. Metal-free class Ie ribonucleotide reductase from pathogens initiates catalysis with a tyrosine-derived dihydroxyphenylalanine radical. Proc. Natl. Acad. Sci. U. S. A. 2018;115:10022–10027. doi: 10.1073/pnas.1811993115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivas V., Lebrette H., Lundin D., Kutin Y., Sahlin M., Lerche M., Eirich J., Branca R.M.M., Cox N., Sjöberg B.-M., Högbom M. Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens. Nature. 2018;563:416–420. doi: 10.1038/s41586-018-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose H.R., Palowitch G.M., Hu K., Gandhi A.V., Boal A.K. In: Comprehensive Natural Products III. Liu H.-W., Begley T.P., editors. Elsevier; Amsterdam, the Netherlands: 2020. Structure-function relationships in assembly of the radical-initiating cofactors of class Ia-e ribonucleotide reductase; pp. 415–441. [Google Scholar]
- 18.Cooley R.B., Arp D.J., Karplus P.A. Evolutionary origin of a secondary structure: Pi-helices as cryptic but widespread insertional variations of alpha-helices that enhance protein functionality. J. Mol. Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ando N., Brignole E.J., Zimanyi C.M., Funk M.A., Yokoyama K., Asturias F.J., Stubbe J., Drennan C.L. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21046–21051. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang G., Taguchi A.T., Stubbe J., Drennan C.L. Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex. Science. 2020;368:424–427. doi: 10.1126/science.aba6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlin U., Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 22.Ge J., Yu G., Ator M.A., Stubbe J. Pre-steady-state and steady-state kinetic analysis of E. coli class I ribonucleotide reductase. Biochemistry. 2003;42:10071–10083. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 23.Bennati M., Weber A., Antonic J., Perlstein D.L., Robblee J., Stubbe J. Pulsed ELDOR spectroscopy measures the distance between the two tyrosyl radicals in the R2 subunit of the E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2003;125:14988–14989. doi: 10.1021/ja0362095. [DOI] [PubMed] [Google Scholar]
- 24.Climent I., Sjöberg B.-M., Huang C.Y. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry. 1992;31:4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 25.Minnihan E.C., Seyedsayamdost M.R., Uhlin U., Stubbe J. Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases. J. Am. Chem. Soc. 2011;133:9430–9440. doi: 10.1021/ja201640n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seyedsayamdost M.R., Yee C.S., Reece S.Y., Nocera D.G., Stubbe J. pH-rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: Evidence that Y356 is a redox-active amino acid along the radical propagation pathway. J. Am. Chem. Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama K., Uhlin U., Stubbe J. A hot oxidant, 3-NO2Y122 radical, unmasks conformational gating in ribonucleotide reductase. J. Am. Chem. Soc. 2010;132:15368–15379. doi: 10.1021/ja1069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seyedsayamdost M.R., Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the beta2 subunit of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 29.Wu C.H., Jiang W., Krebs C., Stubbe J. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry. 2007;46:11577–11588. doi: 10.1021/bi7012454. [DOI] [PubMed] [Google Scholar]
- 30.Atta M., Nordlund P., Aberg A., Eklund H., Fontecave M. Substitution of manganese for iron in ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 1992;267:20682–20688. [PubMed] [Google Scholar]
- 31.Voegtli W.C., Sommerhalter M., Saleh L., Baldwin J., Bollinger J.M., Jr., Rosenzweig A.C. Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2. J. Am. Chem. Soc. 2003;125:15822–15830. doi: 10.1021/ja0370387. [DOI] [PubMed] [Google Scholar]
- 32.Boal A.K., Cotruvo J.A., Jr., Stubbe J., Rosenzweig A.C. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong W.H., Chen S., Lloyd S.G., Edmondson D.E., Huynh B.H., Stubbe J. Mechanism of assembly of the diferric cluster-tyrosyl radical cofactor of Escherichia coli ribonucleotide reductase from the diferrous form of the R2 subunit. J. Am. Chem. Soc. 1996;118:2107–2108. [Google Scholar]
- 34.Moënne-Loccoz P., Baldwin J., Ley B.A., Loehr T.M., Bollinger J.M., Jr. O2 activation by non-heme diiron proteins: Identification of a symmetric m-1,2-peroxide in a mutant of ribonucleotide reductase. Biochemistry. 1998;37:14659–14663. doi: 10.1021/bi981838q. [DOI] [PubMed] [Google Scholar]
- 35.Bollinger J.M., Jr., Edmondson D.E., Huynh B.H., Filley J., Norton J.R., Stubbe J. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science. 1991;253:292–298. doi: 10.1126/science.1650033. [DOI] [PubMed] [Google Scholar]
- 36.Bollinger J.M., Tong W.H., Ravi N., Huynh B.H., Edmondson D.E., Stubbe J. Mechanism of assembly of the tyrosyl radical-diiron(III) cofactor of Escherichia coli ribonucleotide reductase. 3. Kinetics of the limiting Fe2+ reaction by optical, EPR, and Mössbauer spectroscopies. J. Am. Chem. Soc. 1994;116:8024–8032. [Google Scholar]
- 37.Hristova D., Wu C.H., Jiang W., Krebs C., Stubbe J. Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry. 2008;47:3989–3999. doi: 10.1021/bi702408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livada J., Martinie R.J., Dassama L.M., Krebs C., Bollinger J.M., Jr., Silakov A. Direct measurement of the radical translocation distance in the class I ribonucleotide reductase from Chlamydia trachomatis. J. Phys. Chem. B. 2015;119:13777–13784. doi: 10.1021/acs.jpcb.5b04067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minnihan E.C., Ando N., Brignole E.J., Olshansky L., Chittuluru J., Asturias F.J., Drennan C.L., Nocera D.G., Stubbe J. Generation of a stable, aminotyrosyl radical-induced a2b2 complex of Escherichia coli class Ia ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3835–3840. doi: 10.1073/pnas.1220691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Q., Parker M.J., Taguchi A.T., Ravichandran K., Kim A., Kang G., Shao J., Drennan C.L., Stubbe J. Glutamate 52-beta at the alpha/beta subunit interface of Escherichia coli class Ia ribonucleotide reductase is essential for conformational gating of radical transfer. J. Biol. Chem. 2017;292:9229–9239. doi: 10.1074/jbc.M117.783092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artin E., Wang J., Lohman G.J., Yokoyama K., Yu G., Griffin R.G., Bar G., Stubbe J. Insight into the mechanism of inactivation of ribonucleotide reductase by gemcitabine 5'-diphosphate in the presence or absence of reductant. Biochemistry. 2009;48:11622–11629. doi: 10.1021/bi901590q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uppsten M., Davis J., Rubin H., Uhlin U. Crystal structure of the biologically active form of class 1b ribonucleotide reductase small subunit from Mycobacterium tuberculosis. FEBS Lett. 2004;569:117–122. doi: 10.1016/j.febslet.2004.05.059. [DOI] [PubMed] [Google Scholar]
- 43.Jiang W., Xie J., Varano P.T., Krebs C., Bollinger J.M., Jr. Two distinct mechanisms of inactivation of the class Ic ribonucleotide reductase from Chlamydia trachomatis by hydroxyurea: Implications for the protein gating of intersubunit electron transfer. Biochemistry. 2010;49:5340–5349. doi: 10.1021/bi100037b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seyedsayamdost M.R., Yee C.S., Stubbe J. Use of 2,3,5-F3Y-b2 and 3-NH2Y-a2 to study proton-coupled electron transfer in Escherichia coli ribonucleotide reductase. Biochemistry. 2011;50:1403–1411. doi: 10.1021/bi101319v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wörsdörfer B., Conner D.A., Yokoyama K., Livada J., Seyedsayamdost M., Jiang W., Silakov A., Stubbe J., Bollinger J.M., Jr., Krebs C. Function of the Diiron cluster of Escherichia coli class Ia ribonucleotide reductase in proton-coupled electron transfer. J. Am. Chem. Soc. 2013;135:8585–8593. doi: 10.1021/ja401342s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou B., Shao J., Su L., Yuan Y.C., Qi C., Shih J., Xi B., Chu B., Yen Y. A Dityrosyl-Diiron radical cofactor center is essential for human ribonucleotide reductases. Mol. Cancer Ther. 2005;4:1830–1836. doi: 10.1158/1535-7163.MCT-05-0273. [DOI] [PubMed] [Google Scholar]
- 47.Lundin D., Torrents E., Poole A.M., Sjöberg B.-M. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics. 2009;10:589. doi: 10.1186/1471-2164-10-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zygiel E.M., Nolan E.M. Transition metal sequestration by the host-defense protein calprotectin. Annu. Rev. Biochem. 2018;87:621–643. doi: 10.1146/annurev-biochem-062917-012312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eriksson M., Jordan A., Eklund H. Structure of Salmonella typhimurium NrdF ribonucleotide reductase in its oxidized and reduced forms. Biochemistry. 1998;37:13359–13369. doi: 10.1021/bi981380s. [DOI] [PubMed] [Google Scholar]
- 50.Schimpff-Weiland G., Follmann H., Auling G. A new manganese-activated ribonucleotide reductase found in gram-positive bacteria. Biochem. Biophys. Res. Commun. 1981;102:1276–1282. doi: 10.1016/s0006-291x(81)80149-0. [DOI] [PubMed] [Google Scholar]
- 51.Stølle P., Barckhausen O., Oehlmann W., Knobbe N., Vogt C., Pierik A.J., Cox N., Schmidt P.P., Reijerse E.J., Lubitz W., Auling G. Homologous expression of the nrdF gene of Corynebacterium ammoniagenes strain ATCC 6872 generates a manganese-metallocofactor (R2F) and a stable tyrosyl radical (Y ) involved in ribonucleotide reduction. FEBS J. 2010;277:4849–4862. doi: 10.1111/j.1742-4658.2010.07885.x. [DOI] [PubMed] [Google Scholar]
- 52.Cox N., Ogata H., Stolle P., Reijerse E., Auling G., Lubitz W. A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 2010;132:11197–11213. doi: 10.1021/ja1036995. [DOI] [PubMed] [Google Scholar]
- 53.Högbom M., Huque Y., Sjöberg B.-M., Nordlund P. Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes. Biochemistry. 2002;41:1381–1389. doi: 10.1021/bi011429l. [DOI] [PubMed] [Google Scholar]
- 54.Cotruvo J.A., Jr., Stubbe J. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14383–14388. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammerstad M., Hersleth H.P., Tomter A.B., Røhr A.K., Andersson K.K. Crystal structure of Bacillus cereus class Ib ribonucleotide reductase di-iron NrdF in complex with NrdI. ACS Chem. Biol. 2014;9:526–537. doi: 10.1021/cb400757h. [DOI] [PubMed] [Google Scholar]
- 56.Martin J.E., Imlay J.A. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cotruvo J.A., Jr., Stubbe J. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry. 2011;50:1672–1681. doi: 10.1021/bi101881d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Stubbe J. Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry. 2011;50:5615–5623. doi: 10.1021/bi200348q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makhlynets O., Boal A.K., Rhodes D.V., Kitten T., Rosenzweig A.C., Stubbe J. Streptococcus sanguinis class Ib ribonucleotide reductase: High activity with both iron and manganese cofactors and structural insights. J. Biol. Chem. 2014;289:6259–6272. doi: 10.1074/jbc.M113.533554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhodes D.V., Crump K.E., Makhlynets O., Snyder M., Ge X., Xu P., Stubbe J., Kitten T. Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis. J. Biol. Chem. 2014;289:6273–6287. doi: 10.1074/jbc.M113.533620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puccio T., Kunka K.S., Zhu B., Xu P., Kitten T. Manganese depletion leads to multisystem changes in the transcriptome of the opportunistic pathogen Streptococcus sanguinis. Front. Microbiol. 2020;11:592615. doi: 10.3389/fmicb.2020.592615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cotruvo J.A., Jr., Stich T.A., Britt R.D., Stubbe J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: Enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J. Am. Chem. Soc. 2013;135:4027–4039. doi: 10.1021/ja312457t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson R., Torrents E., Lundin D., Sprenger J., Sahlin M., Sjoberg B.M., Logan D.T. High-resolution crystal structures of the flavoprotein NrdI in oxidized and reduced states--an unusual flavodoxin. FEBS J. 2010;277:4265–4277. doi: 10.1111/j.1742-4658.2010.07815.x. [DOI] [PubMed] [Google Scholar]
- 64.Berggren G., Duraffourg N., Sahlin M., Sjoberg B.M. Semiquinone-induced maturation of Bacillus anthracis ribonucleotide reductase by a superoxide intermediate. J. Biol. Chem. 2014;289:31940–31949. doi: 10.1074/jbc.M114.592535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lofstad M., Gudim I., Hammerstad M., Røhr A.K., Hersleth H.P. Activation of the class Ib ribonucleotide reductase by a flavodoxin reductase in Bacillus cereus. Biochemistry. 2016;55:4998–5001. doi: 10.1021/acs.biochem.6b00699. [DOI] [PubMed] [Google Scholar]
- 66.Jiang W., Xie J., Nørgaard H., Bollinger J.M., Jr., Krebs C. Rapid and quantitative activation of Chlamydia trachomatis ribonucleotide reductase by hydrogen peroxide. Biochemistry. 2008;47:4477–4483. doi: 10.1021/bi702085z. [DOI] [PubMed] [Google Scholar]
- 67.Jiang W., Yun D., Saleh L., Bollinger J.M., Jr., Krebs C. Formation and function of the manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Biochemistry. 2008;47:13736–13744. doi: 10.1021/bi8017625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dassama L.M., Boal A.K., Krebs C., Rosenzweig A.C., Bollinger J.M., Jr. Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1. J. Am. Chem. Soc. 2012;134:2520–2523. doi: 10.1021/ja211314p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwak Y., Jiang W., Dassama L.M., Park K., Bell C.B., 3rd, Liu L.V., Wong S.D., Saito M., Kobayashi Y., Kitao S., Seto M., Yoda Y., Alp E.E., Zhao J., Bollinger J.M., Jr. Geometric and electronic structure of the Mn(IV)Fe(III) cofactor in class Ic ribonucleotide reductase: Correlation to the class Ia binuclear non-heme iron enzyme. J. Am. Chem. Soc. 2013;135:17573–17584. doi: 10.1021/ja409510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersson C.S., Öhrström M., Popovic-Bijelic A., Graslund A., Stenmark P., Högbom M. The manganese ion of the heterodinuclear Mn/Fe cofactor in Chlamydia trachomatis ribonucleotide reductase R2c is located at metal position 1. J. Am. Chem. Soc. 2012;134:123–125. doi: 10.1021/ja209678x. [DOI] [PubMed] [Google Scholar]
- 71.Dassama L.M., Krebs C., Bollinger J.M., Jr., Rosenzweig A.C., Boal A.K. Structural basis for assembly of the Mn(IV)/Fe(III) cofactor in the class Ic ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2013;52:6424–6436. doi: 10.1021/bi400819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pierce B.S., Elgren T.E., Hendrich M.P. Mechanistic implications for the formation of the diiron cluster in ribonucleotide reductase provided by quantitative EPR spectroscopy. J. Am. Chem. Soc. 2003;125:8748–8759. doi: 10.1021/ja021290h. [DOI] [PubMed] [Google Scholar]
- 73.Bollinger J.M., Jr., Chen D., Parkin S.E., Mangravite L.M., Ley B.A., Edmondson D.E., Huynh B.H. Differential iron(II) affinity of the sites of the diiron cluster in protein R2 of Escherichia coli ribonucleotide reductase: Tracking the individual sites through the O2 activation sequence. J. Am. Chem. Soc. 1997;119:5976–5977. [Google Scholar]
- 74.Martinie R.J., Blaesi E.J., Krebs C., Bollinger J.M., Jr., Silakov A., Pollock C.J. Evidence for a di-mu-oxo diamond core in the Mn(IV)/Fe(IV) activation intermediate of ribonucleotide reductase from Chlamydia trachomatis. J. Am. Chem. Soc. 2017;139:1950–1957. doi: 10.1021/jacs.6b11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinie R.J., Blaesi E.J., Bollinger J.M., Jr., Krebs C., Finkelstein K.D., Pollock C.J. Two-color valence-to-core x-ray emission spectroscopy tracks cofactor protonation state in a class I ribonucleotide reductase. Angew. Chem. Int. Ed. Engl. 2018;57:12754–12758. doi: 10.1002/anie.201807366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rozman Grinberg I., Berglund S., Hasan M., Lundin D., Ho F.M., Magnuson A., Logan D.T., Sjöberg B.-M., Berggren G. Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading. J. Biol. Inorg. Chem. 2019;24:863–877. doi: 10.1007/s00775-019-01697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rozman Grinberg I., Lundin D., Hasan M., Crona M., Jonna V.R., Loderer C., Sahlin M., Markova N., Borovok I., Berggren G., Hofer A., Logan D.T., Sjöberg B.-M. Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit. Elife. 2018;7 doi: 10.7554/eLife.31529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rose H.R., Maggiolo A.O., McBride M.J., Palowitch G.M., Pandelia M.-E., Davis K.M., Yennawar N.H., Boal A.K. Structures of class Id ribonucleotide reductase catalytic subunits reveal a minimal architecture for deoxynucleotide biosynthesis. Biochemistry. 2019;58:1845–1860. doi: 10.1021/acs.biochem.8b01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roca I., Torrents E., Sahlin M., Gibert I., Sjöberg B.-M. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J. Bacteriol. 2008;190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Do H., Makthal N., Chandrangsu P., Randall J., Kumaraswami M. Metal sensing and regulation of adaptive responses to manganese limitation by MtsR is critical for group A streptococcus virulence. Nucleic Acids Res. 2019;47:7476–7493. doi: 10.1093/nar/gkz524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zallot R., Oberg N., Gerlt J.A. The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry. 2019;58:4169–4182. doi: 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rose H.R., Palowitch G.M., Hu K., Gandhi A.V., Boal A.K. In: Comprehensive Natural Products III: Chemistry and Biology. Liu H.-W., Begley T.P., editors. Elsevier; Amsterdam: 2020. Structure-function relationships in assembly of the radical-initiating cofactors of class Ia-e ribonucleotide reductases; pp. 415–441. [Google Scholar]