Abstract
Background & Aims
The interorgan crosstalk between the liver and the intestine has been the focus of intense research. Key in this crosstalk are bile acids, which are secreted from the liver into the intestine, interact with the microbiome, and upon absorption reach back to the liver. The bile acid-activated farnesoid X receptor (Fxr) is involved in the gut-to-liver axis. However, liver-to-gut communication and the roles of bile acids and Fxr remain elusive. Herein, we aim to get a better understanding of Fxr-mediated liver-to-gut communication, particularly in colon functioning.
Methods
Fxr floxed/floxed mice were crossed with cre-expressing mice to yield Fxr ablation in the intestine (Fxr-intKO), liver (Fxr-livKO), or total body (Fxr-totKO). The effects on colonic gene expression (RNA sequencing), the microbiome (16S sequencing), and mucus barrier function by ex vivo imaging were analysed.
Results
Despite relatively small changes in biliary bile acid concentration and composition, more genes were differentially expressed in the colons of Fxr-livKO mice than in those of Fxr-intKO and Fxr-totKO mice (3272, 731, and 1824, respectively). The colons of Fxr-livKO showed increased expression of antimicrobial genes, Toll-like receptors, inflammasome-related genes and genes belonging to the ‘Mucin-type O-glycan biosynthesis’ pathway. Fxr-livKO mice have a microbiome profile favourable for the protective capacity of the mucus barrier. The thickness of the inner sterile mucus layer was increased and colitis symptoms reduced in Fxr-livKO mice.
Conclusions
Targeting of FXR is at the forefront in the battle against metabolic diseases. We show that ablation of Fxr in the liver greatly impacts colonic gene expression and increased the colonic mucus barrier. Increasing the mucus barrier is of utmost importance to battle intestinal diseases such as inflammatory bowel disease, and we show that this might be done by antagonising FXR in the liver.
Lay summary
This study shows that the communication of the liver to the intestine is crucial for intestinal health. Bile acids are key players in this liver-to-gut communication, and when Fxr, the master regulator of bile acid homoeostasis, is ablated in the liver, colonic gene expression is largely affected, and the protective capacity of the mucus barrier is increased.
Keywords: Farnesoid X receptor, Liver-specific Fxr-KO mouse, Intestine-specific Fxr-KO mouse, Colon, Gut microbiome, Mucus layer, Liver–gut axis
Abbreviations: BAs, bile acids; DSS, dextran sodium sulfate; Fgfr4, fibroblast growth factor receptor 4; FITC, fluorescein isothiocyanate; fpkm, fragments per kilobase of transcript per million mapped reads; Fxr, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout; Fxr-totKO, whole body Fxr knockout; GO, Gene Ontology; HID, high-iron diamine; IBD, inflammatory bowel disease; KEGG, Kyoto Encyclopedia of Genes and Genomes; RT qPCR, real-time quantitative PCR
Graphical abstract
Highlights
-
•
Fxr ablation in the mouse liver has a major impact on colonic gene expression.
-
•
Fxr signalling is induced in the colons of liver Fxr knockout (Fxr-livKO) mice.
-
•
In Fxr-livKO colons, expression of antimicrobial and mucus genes is increased.
-
•
Microbiome of Fxr-livKO mice is indicative of enhanced mucus barrier function.
-
•
Fxr-livKO mice have an increased mucus barrier.
Introduction
The gut–liver axis refers to the crosstalk between the intestine and the liver, particularly mediated by microbiome- and diet-derived metabolites. The (patho)physiological relevance of this crosstalk is increasingly recognised. Key in this crosstalk are bile acids (BAs), which circulate between these organs within the enterohepatic circulation through the portal venous (gut-to-liver) and biliary (liver-to-gut) systems. In the liver, BAs are synthesised and conjugated with glycine or taurine, which increase their solubility. BAs are subsequently stored in the gallbladder. After food intake, bile is released into the intestine to facilitate absorption of dietary lipids. The majority (∼95%) of BAs is reabsorbed in the ileum and reaches the liver via the portal vein. Unabsorbed BAs enter the colon and are subsequently deconjugated and converted by bacteria into secondary BAs.1
Farnesoid X receptor (Fxr), which is the key regulator of BA transport, signalling, and metabolism2 is highly expressed in the gut–liver axis. In the liver, Fxr determines the synthesis of the primary BAs and their transport from liver cells into the biliary system by regulating transcription of several key genes, including Cyp7a1, Cyp8b1, Shp, and Bsep.1 In the ileum, where Fxr is activated by absorbed BAs, Fxr regulates transcription of Ibabp, Ostα, and β, which are involved in BA uptake,1 and of FGF15 (FGF19 in humans), which is a key enterokine involved in gut–liver communication.3
Fgf15/19 is secreted at the basolateral membrane of enterocytes and reaches the liver via the portal vein. Here it binds to the Fibroblast growth factor receptor 4 (Fgfr4)/β-klotho complex on hepatocytes, which subsequently represses BA synthesis via Cyp7a1.3 Next to regulation of BA synthesis, FXR-mediated expression of FGF15/19 has other functions. Mice lacking Fgf15 fail to maintain glucose homoeostasis.4 Pharmacological administration or overexpression of FGF19 restores glucose homoeostasis and glycogen metabolism.5,6 In conclusion, FXR-mediated FGF15/19 expression plays a key role in gut-to-liver communication.
By contrast to gut-to-liver communication, much less is known about liver-to-gut communication. It is known that decreased bile flow from the liver to the intestine in humans and mice results in bacterial overgrowth, which is reversed by oral BA treatment.7,8 Additionally, BAs contribute to the exclusion of certain bacteria, favouring others.7,8 Some pathobionts, such as Bilophila wadsworthia, are not only BA resistant, but their growth is favoured by taurine-conjugated, but not glycine-conjugated, BA,9 indicating that hepatic BA output influences the gut microbiota composition, potentially impacting intestinal functions. However, the role of liver Fxr in this respect, for example, by modulating biliary BA concentration and composition, as well as the potential consequences on intestinal integrity and function remain elusive.
To gain a better understanding of liver-to-gut communication by Fxr-regulated BA synthesis and secretion, we generated mice in which we ablated Fxr tissue specifically in either the liver or the intestine and studied colonic gene expression, the microbiome and mucus barrier characteristics.
Materials and methods
Mice
The experiment was approved by the ethics committee of the University Medical Center Utrecht and was in accordance with European law. To generate whole-body (Fxr-totKO), liver-specific (Fxr-livKO), and intestine-specific (Fxr-intKO) Fxr-null mice, homozygous Fxr-floxed mice (C57Bl6J Fxr fl/fl, kindly provided by K. Schoonjans, Ecole Polytechnique Federale de Lausanne, Switzerland10) were crossed with C57Bl6J mice harbouring a cre-recombinase allele under the control of the meox2 promoter (Meox2-cre mice, stock 003755), albumin promoter (Alb-Cre mice, stock 003574), and villin promoter (Villin-Cre mice, stock 004586) from The Jackson Laboratory, respectively. Fxr-floxed mice were used as control. For genotyping, DNA was isolated from the mouse ear. Genotyping was performed as described previously10 and by protocols supplied by Jackson Laboratory. Ablation of Fxr mRNA in the colonic tissue was validated by real-time quantitative PCR (RT-qPCR) using primers Fw: 5′-tgagaacccacagcatttcg-3′, Rv: 5′-gcgtggtgatggttgaatgtc-3′. qPCR was performed on a CFX384 Real-Time system by using FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland).
Male mice of 9–12 weeks old were included. The number of mice used per experiment is indicated in the figure legends. Mice were fed a purified diet (AIN-93M, Research Diets, Wijk bij Duurstede, The Netherlands) and drinking water ad libitum. Mice were individually housed in a room with controlled temperature (20–24°C), relative humidity (55% ± 15%), and a 12-h light–dark cycle. Mice were acclimatised for 1 week; after that, the intervention period of 1 week started in which mice remained on the same diet and body weight was recorded. Mice were fasted 4 h before sacrifice, and 0.6 mg/g bodyweight of fluorescein isothiocyanate (FITC)-conjugated dextran (Sigma, St. Louis, MO, USA; molecular mass 3–5 kDa) was given to measure intestinal permeability, which is a measurement to determine epithelial leakage by dysfunctional tight junctions. After 4 h, blood was collected to measure FITC plasma concentrations. The colon was excised, mesenteric fat was removed, and the colon was opened longitudinally, washed in PBS, and cut into 3 parts. The middle 1.5-cm colon tissue was formalin fixed and paraffin embedded for histology. The remaining proximal and distal parts were scraped. Scrapings include the epithelial lining and lamina propria, but not the muscle layer. These scrapings were pooled per mouse, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis. Colonic contents were sampled and snap-frozen for microbiota analysis. Colitis was induced by administration of 2.5% (w/v) dextran sodium sulfate (DSS; molecular mass 36–50 kDa; MP Biomedicals, Solon, Ohio, USA) in drinking water for 10 days. Rectal bleeding was scored on a scale from 0 to 5, indicating no (0) to very severe (5) rectal bleeding.
Western blot analysis
Western blot on the liver, colon, and kidney tissue was performed as described in the Supplementary information.
RNA isolation, sequencing, and qPCR
Total RNA from the colon was isolated for sequencing and qPCR; see Supplementary information and File S1. Sequencing data can be found in Gene Expression Omnibus under accession number GSE163157.
Histology and immunohistochemistry
H&E, Muc2, and high-iron diamine (HID) stainings and quantification are described in the Supplementary information.
Plasma and gall bladder BAs
Plasma and gall bladder BAs were determined as described in the Supplementary information.
Bacterial DNA isolation and 16S rRNA gene sequencing and analyses
Bacterial DNA was isolated for 16S rRNA gene sequencing, as described in the Supplementary information.
Mucus layer measurements
Mucus barrier function was determined in n = 5 Fxr-intKO and Fxr-livKO and n = 7 control mice. See the Supplementary information.
Statistical analysis
Differences between the groups were tested by Student’s t test using GraphPad Prism 8 (GraphPad Software, San Diego, California, USA). RNA and bacterial sequencing data were analysed statistically as indicated in the respective methods descriptions.
Results
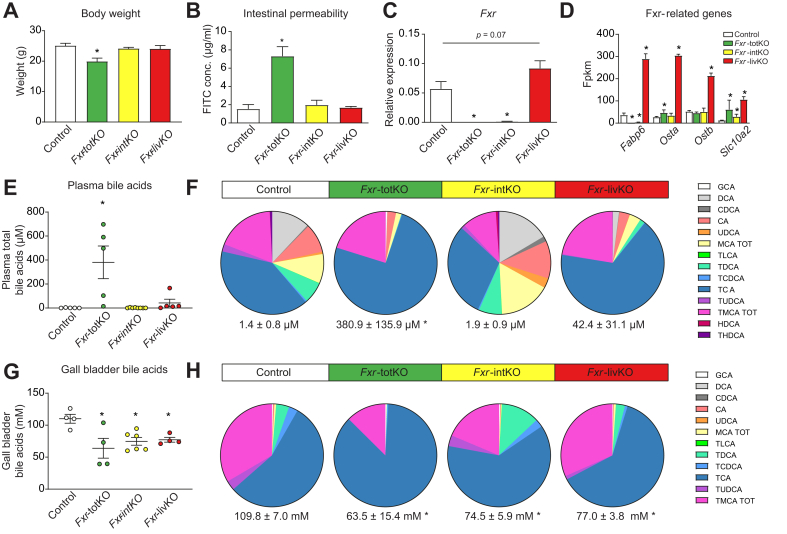
To study the Fxr-dependent communication between the liver and the intestine, we compared Fxr floxed (control) with whole-body (Fxr-totKO), intestine-specific (Fxr-intKO), and liver-specific (Fxr-livKO) Fxr knockout mice. Absence of Fxr protein was validated by Western blot (Fig. S1). Ablation of Fxr in either the liver or the intestine had no effect on body weight (Fig. 1A) or intestinal permeability (Fig. 1B). However, the Fxr-totKO mice had a significantly decreased body weight and an increased intestinal permeability. Ablation of Fxr in the colons of the Fxr-intKO and Fxr-totKO was checked by qPCR (Fig. 1C). Remarkably, Fxr expression in colonocytes of the Fxr-livKO tended to increase. Colon-specific Fxr-target genes are currently unidentified, but we see a significant increase of known ileal Fxr-target genes in the colons of the Fxr-livKO (Fig. 1D), implying that Fxr signalling is increased in colonocytes of the Fxr-livKO mice.
Fig. 1.
Characteristics of the animal models.
(A) Body weight of the different models. (B) Intestinal permeability as determined by FITC dextran plasma concentrations. (C) Relative FXR expression in colon determined by qPCR. (D) Expression of Fxr-related genes in fpkm as determined by RNA sequencing. (E) Plasma bile concentrations and (F) relative plasma bile composition. (G) Gall bladder bile concentration and (H) relative gall bladder bile composition. All data are shown as mean ± SEM (n = 4–8 mice per group, except for (D) in which n = 3). Values for Fxr-intKO, Fxr-livKO, and Fxr-totKO vs. controls were tested using Student’s t test, and ∗ indicates p <0.05. FITC, fluorescein isothiocyanate; fpkm, fragments per kilobase of transcript per million mapped reads; FXR, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout; Fxr-totKO, whole-body Fxr knockout; qPCR, quantitative PCR.
Because Fxr is the main regulator of BA synthesis, we investigated plasma and gall bladder BA levels and composition. In plasma, the serum BA concentration was not different between the Fxr-livKO, Fxr-intKO, and control mice (Fig. 1E). This is in line with previous data,11 showing that BA levels can be maintained when Fxr is ablated in either the liver or the intestine. The total BA concentration was strongly increased in the Fxr-totKO, mainly caused by elevated levels of conjugated BA (Fig. 1F), which indicates that this model has severe liver problems leading to spillover of BA to the systemic circulation. A previous study also found increased plasma bile concentrations in the Fxr-totKO, but not in the Fxr-livKO and Fxr-intKO,12 although the total plasma concentration was lower, which might be explained by a different diet or the difference in the Fxr KO model (exon 11 vs. exon 6 ablation).
The total BA concentration in the gall bladder bile, which enters the intestine after gall bladder contraction, was decreased in the Fxr-totKO, Fxr-intKO, and Fxr-livKO compared with that in the controls (Fig. 1G). However, the relative composition of the gall bladder bile was not different between the Fxr-livKO and the controls (Fig. 1H). Overall, this showed that BA concentration and composition can be maintained in the absence of Fxr in either the liver or the intestine. The ablation of FXR in both tissues, or the accompanying ablation of FXR in other tissues in the Fxr-totKO, causes the deregulation of BA homoeostasis.
Ablation of liver Fxr has a major impact on colonic gene expression
Next, we used RNA sequencing to investigate the effect of Fxr ablation on colonic gene expression. A distribution of the significant changes in expression for the Fxr-totKO, Fxr-intKO, and Fxr-livKO compared with the controls is visualised in volcano plots (Fig. 2A–C). The Fxr-livKO group shows the largest number of differentially expressed genes as well as the highest log2 fold changes (Fig. 2C). Remarkably, the effect of ablating Fxr in the liver on colonic gene expression is much larger than the effect of intestine-specific ablation or whole-body Fxr ablation. The Venn diagram (Fig. 2D) illustrates that the colons of the Fxr-livKO show in total 3,272 differentially expressed genes, of which 47% is specific for this model and not shared by the Fxr-totKO and Fxr-intKO. Ablation of liver Fxr is thus a major determinant of colonic gene expression, underlying the importance of liver-to-gut communication.
Fig. 2.
Ablation of Fxr in the liver has a major impact on colonic gene expression.
Volcano plots of RNA sequencing data of colons of (A) Fxr-totKO, (B) Fxr-intKO, and (C) Fxr-livKO mice compared with controls. X-axes: genes located on the right-hand side of zero have a higher expression in the KO model compared with control, whereas points on the left of zero are more highly expressed in the control. Grey: |fold change| <2, q-value >0.05. Green: |fold change| >2, q-value >0.05. Blue: |fold change| <2, q-value <0.05. Red: |fold change| >2, q-value <0.05. The q-value is calculated by cufflink test statistics. (D) Proportional Venn diagram showing numbers of significantly changed genes in the colon of the different KO models compared with controls. Fxr, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout; Fxr-totKO, whole-body Fxr knockout; KO, knockout.
Ablation of liver Fxr changes colonic defence response to bacteria
To get more insight into the affected pathways in the colons of the Fxr-livKO and Fxr-intKO compared with the controls, we performed Gene Ontology (GO)-term enrichment analysis. For reasons indicated above, the Fxr-totKO group was excluded from further analysis. The main GO categories that were enriched in the Fxr-livKO compared with the controls involved defence response to bacteria, metabolism, and inflammation (Fig. 3A). All these pathways were regulated to a lower extent or not significant in the Fxr-intKO compared with the controls. With regard to the GO term ‘response to bacterium’, 165 out of 367 genes were significantly different in the Fxr-livKO vs. controls, compared with only 31 in the Fxr-intKO. We performed hierarchical clustering on all genes included in the ‘response to bacterium’ GO term (GO: 0009617) (Fig. 3B and qPCR validation in Fig. S2). The controls and Fxr-intKO cluster separately from the Fxr-livKO. Remarkably, the antimicrobial molecules regenerating islet-derived 3 beta and gamma (Reg3β and Reg3γ, respectively) were increased in the Fxr-livKO but decreased in the Fxr-intKO compared with the controls. The expression of Reg3β and Reg3γ is stimulated by activation of Toll-like receptors (Tlrs). Notably, Tlr2, Tlr3, and Tlr5 are upregulated specifically in the Fxr-livKO. Nlrp6, Casp1, and Pycard are inflammasome-related genes activated in the Fxr-livKO, and this Nlrp6 inflammasome is involved in goblet cell mucus secretion.13 Another mucus-related gene present in this GO term and decreased in the Fxr-livKO was B3galt5. Furthermore, several cell-type markers and immune-related genes were regulated.
Fig. 3.
GO-term analysis shows enrichment for ‘response to bacterium’ in the Fxr-livKO.
(A) GO terms of interest that were enriched in Fxr-livKO. Bars show the −log10 of the Fisher’s exact test p value from the GO enrichment in Fxr-livKO (red) and Fxr-intKO (yellow). Vertical dotted line represents the significant p value cut-off of 0.05 (1.3 on the −log10 scale). Analysis included RNA sequencing data of n = 3 mice per group. (B) Heatmap of the normalised expression per sample of a selection of genes belonging to the GO term ‘response to bacterium’ (GO:0009617, unsupervised clustering of the groups). N = 3 mice per group. (C) Heatmap of genes belonging to the KEGG pathway ‘Mucin type O-glycan biosynthesis’ (mmu00512) (unsupervised clustering). In brackets behind the gene name, significance is indicated with ‘a’ meaning different in Fxr-livKO compared with controls and ‘b’ indicating a difference in Fxr-intKO compared with controls (q <0.05). The average expression for the controls is set to 1. N = 3 mice per group. Fxr, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Overall, genes involved in the defence against and (immunological) response to bacteria were altered in the Fxr-livKO, hinting towards possible differences in mucus characteristics. Genes belonging to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway Mucin-type O-glycan biosynthesis (mmu00512) were visualised in a heatmap (Fig. 3C and qPCR validation in Fig. S2), and 12 out of 21 identified genes belonging to this pathway were significantly different in the Fxr-livKO. Therefore, mucus glycosylation is likely altered in the Fxr-livKO, with potential effects on the gut microbiome composition and mucus barrier characteristics (as reviewed by Corfield14).
Fxr-livKO have a microbiome composition with possible beneficial effects on the mucus barrier
The gut microbiome of the Fxr-intKO and Fxr-livKO was investigated by 16s sequencing and compared with that of the controls. The relative bacterial abundance per mouse for the 10 most abundant genera (Fig. 4A) showed that at a global level, microbial community structures were similar in all groups of mice. This is also apparent when examining the Shannon diversity index (Fig. 4B), which showed no significant differences between the Fxr-livKO and the Fxr-intKO compared with the controls. When differences in relative abundance for all genera present were analysed, a number of taxa were found to be either enriched or depleted in 1 or more of the groups (Fig. 4C). In the Fxr-livKO, the butyrate producer Roseburia is increased in abundance as well as Bifidobacterium and Clostridium sensu stricto 1, which are all described to be beneficial for the mucus layer.[15], [16], [17] In addition, in the Fxr-livKO, there was a decreased abundance of the genus Turicibacter and a member of the Ruminococcaceae family. Both taxa are predicted mucin degraders. A study on the degradation of human-synthetised mucin glycans and utilisation of the derived monosaccharides showed that 2 analysed Turicibacter genomes contained several glycosyl hydrolases, as well as catabolic pathways, for mucus-derived monosaccharides.18 Most of the same functions were also found in the known mucin degrader Akkermansia muciniphila (Table S2). Based on this information, we infer that Turicibacter is a mucus-degrading bacterium. Likewise, various members of the Ruminococcaceae family have been described to live in and degrade host mucin glycans.19 This information suggests that the microbiota of the Fxr-livKO mice contain a microbiota signature that may increase the protective capacity of the mucus barrier.
Fig. 4.
Fxr-livKO have a microbial profile indicative of increased mucus barrier integrity.
(A) Relative abundance of microbiota at the genus level in individual samples (mice were individually housed). Depicted are the 10 most abundant genera. (B) Shannon diversity index (n = 5 for controls; n = 6 for Fxr-intKO and Fxr-livKO). (C) Bacteria genera that show statistically significant differences in relative abundance, as analysed with the statistical framework ANCOM-BC (Analysis of Compositions of Mircobiomes with Bias Correction) (n = 5 for controls; n=6 for Fxr-intKO and Fxr-livKO). Y-axis show the log10 relative abundance. ∗, ∗∗, and ∗∗∗ indicate p <0.05. p <0.01, and p <0.001, respectively. Fxr, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout.
Fxr-livKO have an increased mucus barrier
The mucus barrier quality is determined by mucus production in the goblet cells of the intestinal epithelium as well as by mucus breakdown at the luminal side by bacteria and proteases. Both the RNA sequencing data and the microbiome analysis point towards increased protective capacity of the mucus barrier of the Fxr-livKO. As a measure for mucus production, we first investigated the amount of goblet cells, by staining colon sections for Muc2 (Fig. 5A) and quantifying the number of goblet cells per crypt (Fig. 5B). There was no difference in the amount of goblet cells per crypt in the Fxr-intKO and Fxr-livKO compared with the controls. We performed HID staining (Fig. 5A) to investigate sulfation of mucins (brown). There was no difference in the percentage of sulfated mucins present in goblet cells (Fig. 5C). There was also no apparent change in filling of the goblet cells. Gene expression of Muc1 was decreased and that of Muc2 increased in the Fxr-livKO compared with the controls (Fig. 5D). To investigate differences in mucus barrier characteristics, we determined the mucus barrier permeability ex vivo by using upright microscopy and red fluorescent beads with the size of bacteria (1 μm).20 The distance between the beads and the colonic epithelium (in green) represents the thickness of the colonic inner impermeable mucus layer, which was increased in the Fxr-livKO compared with the controls and Fxr-intKO (Fig. 5E–G). This implies that the colonic epithelial cells of the Fxr-livKO are better protected against threats (e.g. bacteria and toxins) from the luminal side. This implication is in contrast to that for the Fxr-intKO, which have a more permeable and less protective mucus barrier. These data are in line with gene expression of the defence- and mucus-related genes (Fig. 3) and with the microbiota signature (Fig. 4) in the Fxr-livKO. We investigated if the Fxr-livKO are better protected against DSS-induced colitis. Rectal bleeding, a hallmark of inflammatory bowel disease (IBD), was lower in the Fxr-livKO than in the controls at Day 11 (Fig. 5H). Lymphoid follicle hyperplasia scores suggest that the Fxr-livKO have, next to a thicker mucus layer, a more active adaptive immune system. Expression levels of the protective Reg3β and Reg3γ were significantly increased at baseline and tended to increase even further upon DSS treatment in the Fxr-livKO compared with wild-type mice. Together, these data suggest the ablation of liver Fxr decreased the effects of DSS-induced colitis.
Fig. 5.
Fxr-livKO show an increased mucus barrier function.
(A) Representative pictures of Muc2-staining and HID staining (sulfated mucins in brown and carboxylated mucins in blue). (B) Quantification of the number of Muc2-positive cells per crypt. Data are represented as mean ± SEM (n = 6 for controls, n = 7 for Fxr-intKO, and n = 5 for Fxr-livKO). Differences between Fxr-intKO and Fxr-livKO vs. controls were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). (C) Quantification of the percentage of goblet cells filled with sulfated mucins per crypt. Data are represented as mean ± SEM (n = 5 for controls, n = 7 for Fxr-intKO, and n = 5 for Fxr-livKO). Differences between Fxr-intKO and Fxr-livKO vs. controls were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). (D) Expression of the transmembrane mucins Muc1, Muc4, and Muc13 and the secreted mucin Muc2 in fpkm from RNA sequencing data (n = 3 per group). Differences between Fxr-intKO and Fxr-livKO vs. controls were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). Expression levels of Muc5ac, Muc5b, Muc6, Muc15, Muc19, and Muc20 were also included in the RNA sequencing dataset; however, these were expressed at very low levels or completely absent and therefore not considered. (E) Permeability of the distal colonic mucus layer was visualised by adding 1-μm fluorescent beads (red) to the epithelial tissue (green). Shown are representative maximum projections of a resliced z-stack, showing a summarised view of the sample from the side. (F) The distance between beads and the epithelium was measured, and the 10% quantile, the closest to the epithelium, was depicted (average of n = 7 for controls, n = 5 for Fxr-intKO, and n = 5 for Fxr-livKO). Differences between Fxr-intKO and Fxr-livKO vs. controls were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). (G) Violin plot of the distance of a random subset of 130 identified beads to the epithelial cells plotted for all the individual mice (n = 7 for controls, n = 5 for Fxr-intKO, and n = 5 for Fxr-livKO). (H) Rectal bleeding scores as marker for inflammation in DSS-treated mice (n = 8 controls and n = 6 for Fxr-livKO). Differences between Fxr-livKO vs. controls were tested per timepoint using Student’s t test, and ∗ indicates significant differences (p <0.05). (I) Lymphoid follicle hyperplasia scores in controls and Fxr-livKO with and without DSS treatment. Differences between Fxr-livKO and controls, and between with or without DSS within the animal model were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). (J) Expression of Reg3β and Reg3γ in fpkm from RNA sequencing data (n = 3 per group). Differences between Fxr-livKO and controls and between with or without DSS within the animal model were tested using Student’s t test, and ∗ indicates significant differences (p <0.05). DSS, dextran sodium sulfate; fpkm, fragments per kilobase of transcript per million mapped reads; Fxr, farnesoid X receptor; Fxr-intKO, intestine-specific Fxr knockout; Fxr-livKO, liver-specific Fxr knockout; HID, high-iron diamine.
Discussion
In this study, we aimed to get a better understanding of liver-to-gut communication mediated via Fxr. We show that ablation of Fxr in the liver is a major determinant of colonic gene expression and leads to an improved colonic mucus barrier function. This is in sharp contrast to Fxr ablation in the intestine, which resulted in worsening in mucus barrier function compared with the controls.
There are several reasons that might explain the major effect of liver Fxr ablation on the colon. Firstly, Fxr-target genes are induced in the colons of the Fxr-livKO, This upregulation is indicative of increased Fxr signalling in colonocytes of the Fxr-livKO, which might contribute to the changes in metabolic, inflammatory, and bacterial response pathways. Currently, we have no explanation for the upregulation of colonic Fxr-target genes caused by liver Fxr ablation, as we found no large differences in biliary BA concentration and composition in the Fxr-livKO compared with the controls. At this point, we cannot exclude that the total amount of bile secreted into the intestine and/or the spillover from the ileum to the colon in the Fxr-livKO is increased and may contribute to the upregulation of Fxr-target genes in the colons of these mice. Previously, increased plasma cholesterol levels were found in the Fxr-livKO,21 which might stimulate BA production.
Secondly, with regard to mucus synthesis, BAs have been shown to increase Cdx2 and Muc2 expression via the FXR/NF-κB signalling pathway in gastric epithelial cells.22 Cdx2 expression was increased 4-fold in the Fxr-livKO, as was the expression of the major secreted mucin Muc2 (3-fold). The absence of expression of the transmembrane mucin Muc1 is in line with the increased mucus barrier, as it was shown that Muc1 is upregulated under inflammatory conditions,23 and in diverse cancers, including breast, ovarian, lung, and colon cancer. Therefore, decreased Muc1 expression is suggestive of increased colon health. It is thought that Muc1, via its intracellular domain, stimulates various signalling pathways involved in cell survival through alterations of cell growth, proliferation, and cell death.24 Furthermore, Muc1 is a negative regulator of TLR signalling (reviewed in Dhar and McAuley23) and lower expression in the Fxr-LivKO coincides with increased expression of Tlr2, Tlr3, and Tlr5 in this model. We speculate that intestinal Fxr and Fxr-target genes play a role in the increased mucus barrier. Healthy mucus is glycosylated and often heavily sulfated to maintain barrier function by conferring resistance to bacterial enzymatic degradation. Decreased sulfation25 and decreased glycosylation26 is found in patients with IBD. The Fxr-livKO showed increased expression of several key glycosylating genes such as Gcnt3 and Gcnt4.
Importantly, Reg3γ and Reg3β were upregulated in the Fxr-livKO but downregulated in the Fxr-intKO. Reg3β and Reg3γ are antimicrobial molecules involved in maintaining distance between bacteria and the epithelial lining.[27], [28], [29] Reg3γ knockout mice display an altered mucus distribution, resulting in increased bacterial–epithelial contact, and increases in expression of innate immune response genes in the ileum,30 but not in the colon. Our data do show an association of the impermeable mucus thickness with the expression of Reg3γ in the colon of the mouse models. Furthermore, the Fxr-LivKO showed reduced rectal bleeding upon DSS administration, which was in line with increases in expression of the protective Reg3γ and Reg3β.
It has been shown that mucus secretion is regulated by the inflammasome.13 Deletion of NLRP6, and key components caspase 1 and ASC (Pycard) involved in inflammasome signalling, leads to the protrusion of mucus granules, which do not fuse with the apical basement membrane and release their mucins, but the entire cells are sloughed off into the lumen. We showed an increased expression of inflammasome-related genes in the Fxr-livKO, potentially stimulating mucus secretion. Lymphoid follicle hyperplasia scores suggest that the Fxr-livKO have, next to a thicker mucus layer, a more active adaptive immune system. Interestingly, it has recently been shown that the microbiome modulates adaptive immunity in mice by formation of secondary BA species that act on RORγ+ regulatory T cells via the vitamin D receptor, thereby lowering the vulnerability for chemically induced colitis.31
The microbiota signature that we found in the Fxr-livKO is supportive of an increased mucus barrier function, with several bacterial taxa present that might enforce the mucus barrier (Roseburia, Bifidobacterium, and Clostridium sensu stricto 1) and several taxa absent that might degrade mucins (Turicibacter and a member of the Ruminococcaceae family). Remarkably, in patients with IBD, the mucus barrier is often compromised,32 and decreased abundance of Roseburia, Bifidobacterium, and Clostridium are reported in (early-onset) IBD.33,34
Overall, we can conclude that ablation of Fxr in the liver has a major effect on colonic gene expression and improved mucus barrier characteristics. An impermeable inner mucus layer is important for the protection against diseases such as IBD.32 Therapeutic strategies to fortify the mucus barrier are therefore of utmost importance to battle IBD, and we show here the first indications that this may be done by antagonising FXR in the liver. Most likely, the increased Fxr signalling in the colonocytes of the Fxr-livKO contributes to this, as Fxr activation can inhibit inflammation and contributes to intestinal barrier preservation.35 As there are currently no liver-specific FXR antagonists available, giving an FXR antagonist to inhibit FXR function in the liver will also inhibit FXR function in the intestine, with possible consequences for the mucus barrier, and therefore, there is yet no therapeutic application for these new findings.
Targeting of FXR is currently at the forefront in the battle against metabolic diseases.36,37 In that respect, our study may imply that activation of FXR in the liver may have repercussions for mucus barrier integrity in the colon. Gastrointestinal problems have not been reported in clinical studies with FXR agonists, which may be because agonists simultaneously activate FXR in the intestine, inducing protection against intestinal inflammation.35 Together, we show that liver Fxr mediates colonic health, which, together with the already known communication from the intestine to the liver, points towards an FXR-dependent reciprocal communication between the liver and the intestine.
Financial support
NI is supported by the MLDS Career Development grant (CDG16-04) and by the Wilhelmina Children’s Hospital Research Fund. NI and YV are supported by the Unusual Collaborations Grant from CUCo (Center for Unusual Collaborations; Strategic Alliance between Eindhoven University of Technology, Wageningen University & Research, Utrecht University, and the University Medical Center Utrecht). MRdZ is supported by a VIDI grant from the Netherlands Organization for Scientific Research (ZonMW, grant 917.15.377) and the Utrecht Exposome Hub of Utrecht Life Sciences. MB and YV are supported by NWO Gravitation 024.001.028. SvM is supported by the Netherlands Organization for Scientific Research (NWO) project ZonMW VICI (917.11.365) and project ZonMW Aspasia program (015.015.013).
Authors’ contributions
Designed the study: NI, SvM. Interpreted the data: NI, KR, JP, SM, SvM. Conducted the animal experiment: NI, KR, EW. Conducted, analysed, and interpreted RNA sequencing data: JP, SM. Analysed and interpreted mucus barrier measurements: NI, MB, YV. Conducted bile acid measurements: CC, RP, FK. Performed, analysed, and interpreted bacterial sequencing data: MZ, SM, PS. Performed and analysed DSS colitis scoring: SY, AdB. Wrote the manuscript: NI. Critically revised the manuscript for important intellectual content: SvM, YV, FK.
Data availability statement
Sequencing is submitted to Gene Expression Omnibus under accession number GSE163157. The 16S rRNA gene raw reads were deposited at the European Nucleotide Archive under the following project accession number: PRJEB47071.
Conflicts of interest
Authors declare no conflict of interest, except for RP, CEO at TES Pharma; CC, employed by TESS Pharma; and SY, currently working at Janssen Pharmaceuticals.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
The authors would like to thank Alexandra Milona, Ingrid Bijsmans, Vittoria Massafra, Suzanne van der Veen, Monique Appelman, Anja van der Sar, and Trudy Oosterveld-Romijn for (technical) assistance; Martijn Koehorst for bile acid analysis; Livio Kleij for microscopy assistance; Bas Oldenburg and Karin Strijbis for discussions; the lab of Malin Johansson; and Gunnar Hansson for hosting us and teaching us the ex vivo mucus measurement techniques. We thank Nico Lansu and Noortje van den Dungen (Epigenomics Facility) and the Utrecht Sequencing Facility for the preparation of sequencing libraries and Illumina sequencing.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100344.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Hegyi P., Maléth J., Walters J.R., Hofmann A.F., Keely S.J. Guts and Gall: bile acids in regulation of intestinal epithelial function in health and disease. Physiol Rev. 2018;98:1983–2023. doi: 10.1152/physrev.00054.2017. [DOI] [PubMed] [Google Scholar]
- 2.Massafra V., van Mil S.W.C. Farnesoid X receptor: a “homeostat” for hepatic nutrient metabolism. Biochim Biophys Acta Mol Basis Dis. 2018;1864:45–59. doi: 10.1016/j.bbadis.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Potthoff M.J., Boney-Montoya J., Choi M., He T., Sunny N.E., Satapati S. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson E., Fu L., John L., Hultgren B., Huang X., Renz M. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–1747. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- 6.Kir S., Beddow S.A., Samuel V.T., Miller P., Previs S.F., Suino-Powell K. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam K.B., Fukiya S., Hagio M., Fujii N., Ishizuka S., Ooka T. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 8.Inagaki T., Moschetta A., Lee Y.K., Peng L., Zhao G., Downes M. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milona A., Owen B.M., van Mil S., Dormann D., Mataki C., Boudjelal M. The normal mechanisms of pregnancy-induced liver growth are not maintained in mice lacking the bile acid sensor Fxr. Am J Physiol Gastrointest Liver Physiol. 2010;298:G151–G158. doi: 10.1152/ajpgi.00336.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim I., Ahn S.-H., Inagaki T., Choi M., Ito S., Guo G.L. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S., Tanaka N., Fukami T., Xie C., Yagai T., Kim D. Role of farnesoid X receptor and bile acids in hepatic tumor development. Hepatol Commun. 2018;2:1567–1582. doi: 10.1002/hep4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wlodarska M., Thaiss C.A., Nowarski R., Henao-Mejia J., Zhang J.-P., Brown E.M. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corfield A.P. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms. 2018;6:78. doi: 10.3390/microorganisms6030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder B.O., Birchenough G.M.H., Ståhlman M., Arike L., Johansson M.E.V., Hansson G.C. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 2018;23 doi: 10.1016/j.chom.2017.11.004. 27–40.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wlodarska M., Willing B.P., Bravo D.M., Finlay B.B. Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci Rep. 2015;5:9253. doi: 10.1038/srep09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Rosa S.L., Leth M.L., Michalak L., Hansen M.E., Pudlo N.A., Glowacki R. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat Commun. 2019;10:905. doi: 10.1038/s41467-019-08812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravcheev D.A., Thiele I. Comparative genomic analysis of the human gut microbiome reveals a broad distribution of metabolic pathways for the degradation of host-synthetized mucin glycans and utilization of mucin-derived monosaccharides. Front Genet. 2017;8:111. doi: 10.3389/fgene.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tailford L.E., Crost E.H., Kavanaugh D., Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustafsson J.K., Ermund A., Johansson M.E., Schütte A., Hansson G.C., Sjövall H. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol. 2012;302:G430–G438. doi: 10.1152/ajpgi.00405.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt J., Kong B., Stieger B., Tschopp O., Schultze S.M., Rau M. Protective effects of farnesoid X receptor (FXR) on hepatic lipid accumulation are mediated by hepatic FXR and independent of intestinal FGF15 signal. Liver Int. 2015;35:1133–1144. doi: 10.1111/liv.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J.-H., Zheng J.-B., Qi J., Yang K., Wu Y.-H., Wang K. Bile acids promote gastric intestinal metaplasia by upregulating CDX2 and MUC2 expression via the FXR/NF-κB signalling pathway. Int J Oncol. 2019;54:879–892. doi: 10.3892/ijo.2019.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhar P., McAuley J. The role of the cell surface mucin MUC1 as a barrier to infection and regulator of inflammation. Front Cell Infect Microbiol. 2019;9:117. doi: 10.3389/fcimb.2019.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bafna S., Kaur S., Batra S.K. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raouf A.H., Tsai H.H., Parker N., Hoffman J., Walker R.J., Rhodes J.M. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83:623–626. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- 26.Theodoratou E., Campbell H., Ventham N.T., Kolarich D., Pučić-Baković M., Zoldoš V. The role of glycosylation in IBD. Nat Rev Gastroenterol Hepatol. 2014;11:588–600. doi: 10.1038/nrgastro.2014.78. [DOI] [PubMed] [Google Scholar]
- 27.Cash H.L., Whitham C.V., Behrendt C.L., Hooper L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaishnava S., Behrendt C.L., Ismail A.S., Eckmann L., Hooper L.V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stelter C., Käppeli R., König C., Krah A., Hardt W.-D., Stecher B. Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PloS One. 2011;6 doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loonen L.M., Stolte E.H., Jaklofsky M.T., Meijerink M., Dekker J., van Baarlen P. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 31.Song X., Sun X., Oh S.F., Wu M., Zhang Y., Zheng W. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ijssennagger N., van der Meer R., van Mil S.W.C. Sulfide as a mucus barrier-breaker in inflammatory bowel disease? Trends Mol Med. 2016;22:190–199. doi: 10.1016/j.molmed.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K., Nishida A., Fujimoto T., Fujii M., Shioya M., Imaeda H. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn's disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 34.Kowalska-Duplaga K., Gosiewski T., Kapusta P., Sroka-Oleksiak A., Wedrychowicz A., Pieczarkowski S. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn's disease. Sci Rep. 2019;9:18880. doi: 10.1038/s41598-019-55290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadaleta R.M., van Erpecum K.J., Oldenburg B., Willemsen E.C., Renooij W., Murzilli S. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 36.Stofan M., Guo G.L. Bile acids and FXR: novel targets for liver diseases. Front Med (Lausanne) 2020;7:544. doi: 10.3389/fmed.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuschwander-Tetri B.A. Therapeutic landscape for NAFLD in 2020. Gastroenterology. 2020;158 doi: 10.1053/j.gastro.2020.01.051. 1984–1998e1983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing is submitted to Gene Expression Omnibus under accession number GSE163157. The 16S rRNA gene raw reads were deposited at the European Nucleotide Archive under the following project accession number: PRJEB47071.