Significance
There is abundant evidence that environmental insults, such as exposure to anesthetics, at early-life stages impair brain functions in later life, but the underlying mechanisms are poorly understood. Neural stem cells (NSCs) express inhibitory neurotransmitter γ-aminobutyric acid type A receptors that are the main target of most anesthetics, including midazolam (MDZ). We found that early-life exposure to MDZ persistently alters chromatin accessibility and global transcription to reinforce long-lasting NSC dormancy in the mouse hippocampus until adulthood, resulting in reduced neurogenesis and cognitive decline. Furthermore, a simple physical activity, running, could overcome these adverse outcomes by normalizing MDZ-induced transcriptomic alterations. Our study provides insights for further understanding and developing therapeutic strategies for neurological disorders induced by early-life exposure to anesthetics.
Keywords: neurogenesis, neural stem cell, midazolam, cognitive function, chromatin accessibility
Abstract
Linkage between early-life exposure to anesthesia and subsequent learning disabilities is of great concern to children and their families. Here we show that early-life exposure to midazolam (MDZ), a widely used drug in pediatric anesthesia, persistently alters chromatin accessibility and the expression of quiescence-associated genes in neural stem cells (NSCs) in the mouse hippocampus. The alterations led to a sustained restriction of NSC proliferation toward adulthood, resulting in a reduction of neurogenesis that was associated with the impairment of hippocampal-dependent memory functions. Moreover, we found that voluntary exercise restored hippocampal neurogenesis, normalized the MDZ-perturbed transcriptome, and ameliorated cognitive ability in MDZ-exposed mice. Our findings thus explain how pediatric anesthesia provokes long-term adverse effects on brain function and provide a possible therapeutic strategy for countering them.
Use of general anesthetics during early childhood is usually unavoidable to ensure safety during operations. Several human epidemiological studies have demonstrated that repeated (but possibly not single) early-life exposure to anesthesia is associated with adverse neurobehavioral outcomes in later life, such as a higher risk of learning disability (1) and attention deficit hyperactivity disorder (2). Based on evidence using experimental animals, causes of such neurocognitive impairment in later life are considered to be dysfunction or death of cells in the central nervous system (CNS) that occur shortly after anesthesia (3–5). However, it remains unclear whether transient anesthetic exposure establishes cellular memory alterations, such as epigenetic changes, that could play a role in long-term CNS cell dysfunction.
Slowly dividing quiescent neural stem cells (NSCs) in the subgranular zone of the hippocampal dentate gyrus (DG) are the source of granule neurons, which contribute to hippocampus-dependent memory processing (6, 7). The inhibitory neurotransmitter γ-aminobutyric acid (GABA), released from local and long-range GABAergic projection neurons, participates in the homeostatic control of quiescence of NSCs expressing GABAA receptors (GABAARs) (8, 9). Since GABAARs are the main targets of most anesthetics, including midazolam (MDZ) (10), we examined whether early-life exposure to MDZ in mice disturbs the balance between quiescence and activation of NSCs; having found that it does, we went on to elucidate the mechanism underlying this MDZ-induced long-lasting imbalance, which is implicated in cognitive impairments in later life.
MDZ Anesthesia Reinforces NSC Dormancy in the Hippocampus
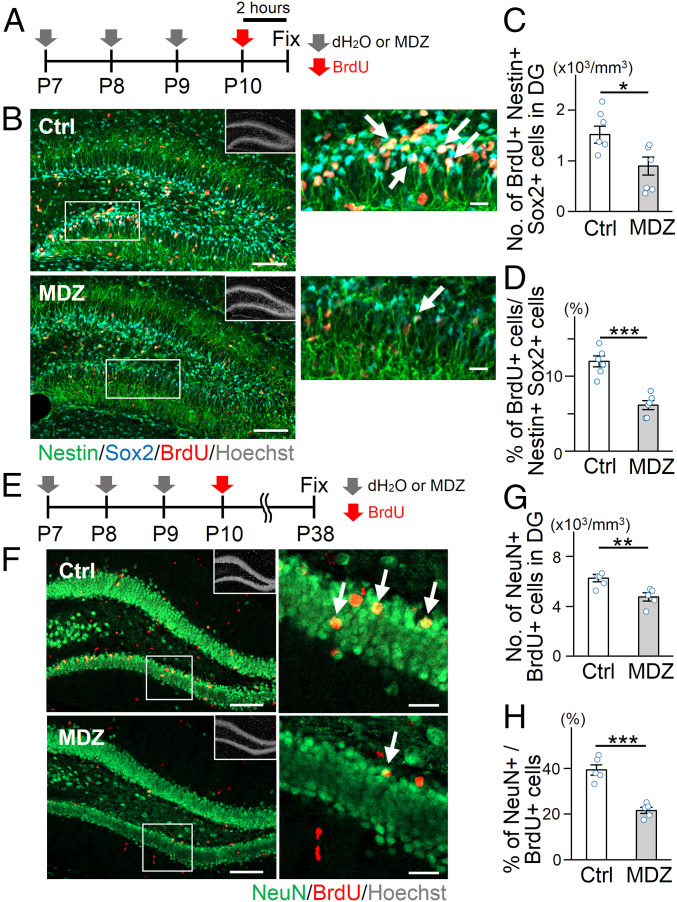
MDZ, a positive allosteric modulator of synaptic GABAARs, is commonly used for general anesthesia and sedation in pediatric patients (11). MDZ is also employed to manage status epilepticus in children and is easier to administer than other anesthetics, such as diazepam, due to its higher water solubility (12). In addition, more than three exposures to anesthetics during childhood have been shown to yield a higher incidence of neurocognitive impairment in later life compared to that inflicted by two (1, 13), although the difference in adverse effect between three and four or more exposures has not been extensively explored. Experimental studies using nonhuman primates also revealed that three applications of anesthetics at the early postnatal stage impair later cognitive function (14). We therefore decided to administer MDZ intraperitoneally to 7-d-old (P7) mice daily for 3 consecutive days (MDZ mice) (Fig. 1A) to investigate whether repeated early postnatal exposure to anesthetics affects NSC behavior in the hippocampal DG. The required dose of MDZ for anesthesia in P7 mice was determined based on the anesthetic score (15, 16), and MDZ treatment at 10 mg/kg was sufficient to exert its anesthetic effects without causing apparent signs of nutritional deficiency, such as obvious loss of body weight (SI Appendix, Fig. S1 A–C). As Nestin- and Sox2-positive (Nestin+ Sox2+) NSCs with radial glia-like morphology in the hippocampal DG comprise cells in both quiescent and active states, we injected bromodeoxyuridine (BrdU) intraperitoneally into mice 2 h before sacrifice to distinguish the two types of NSCs (Fig. 1B). We found a decrease in the number and proportion of BrdU+-activated NSCs in MDZ mice compared with control (Ctrl) (Fig. 1 C and D). The number of Ki67+ Tbr2+ intermediate neuronal progenitor cells also decreased after MDZ exposure (SI Appendix, Fig. S2 A and B). Although a previous study showed that early-life exposure to a high dose of anesthetics triggers apoptosis of neuronal cells (5), we rarely observed active Caspase 3+ NSCs in the two groups (SI Appendix, Fig. S2 C and D). To determine whether MDZ directly affects NSC behavior, MDZ was added to cultures of NSCs obtained from three different species: mouse, rat, and human. We found that MDZ suppressed the proliferation of all three species’ NSCs without inducing cell death (SI Appendix, Fig. S2 E–M), indicating that the effect of MDZ on NSCs is direct and evolutionarily conserved. Moreover, diazepam (another GABAAR modulator) also inhibited NSC proliferation (SI Appendix, Fig. S2 K–M). These data suggest that MDZ directly inhibits NSC activation and augments their dormancy, possibly through GABA signaling. Since inhibiting NSC activation generally leads to a reduction in the number of newly generated neurons in the DG (17) and previous studies demonstrated that early-life exposure to anesthetics including MDZ reduces neurogenesis (4, 15, 18), we examined neurogenesis in our experimental setting (Fig. 1E). In agreement with previous reports (15, 18), we observed a reduction of BrdU+ NeuN+ newly generated mature neurons in MDZ mice compared to Ctrl (Fig. 1 F–H), associated with a decrease in the volume of the granule cell layer (GCL) in MDZ mice (SI Appendix, Fig. S2N). Thus, these results indicate that MDZ exposure reinforces NSC dormancy during the early-life period, resulting in reduced neurogenesis in the hippocampus.
Fig. 1.
Early postnatal exposure to MDZ suppresses NSC proliferation and neurogenesis in the DG. (A) Experimental scheme for investigating the effect of MDZ on NSC proliferation shortly after exposure from P7 to P9. (B) Representative tile scan confocal images of staining for Nestin (green), Sox2 (cyan), BrdU (red), and Hoechst (gray; insets). The areas outlined by a white rectangle are enlarged to the right. Arrows indicate Nestin+ Sox2+ BrdU+ activated NSCs. (Scale bars, 100 μm [Left] and 20 μm [Right].) (C and D) Quantification of the number of BrdU+ Nestin+ Sox2+ activated NSCs (C) and the proportion of BrdU+ Sox2+ Nestin+ cells among Sox2+ Nestin+ cells (D) in the early postnatal DG (n = 6 animals per group). (E) Experimental scheme for investigating the effect on neurogenesis. (F) Representative tile scan confocal images of immunostaining of NeuN (green), BrdU (red), and Hoechst (gray; insets) in the DG 4 wk after BrdU injection at P10. The areas outlined by a white rectangle are enlarged to the right. Arrows indicate BrdU+ NeuN+ cells. (Scale bars, 100 μm [Left] and 20 μm [Right].) (G and H) Quantification of the number of BrdU+ NeuN+ mature neurons (G) and the proportion of BrdU+ NeuN+ mature neurons among BrdU+ cells (H) in the DG at P38 (n = 5 animals per group). *P < 0.05, **P < 0.01, and ***P < 0.001 by t test.
MDZ-Induced Dormancy in NSCs Is Long-Lasting
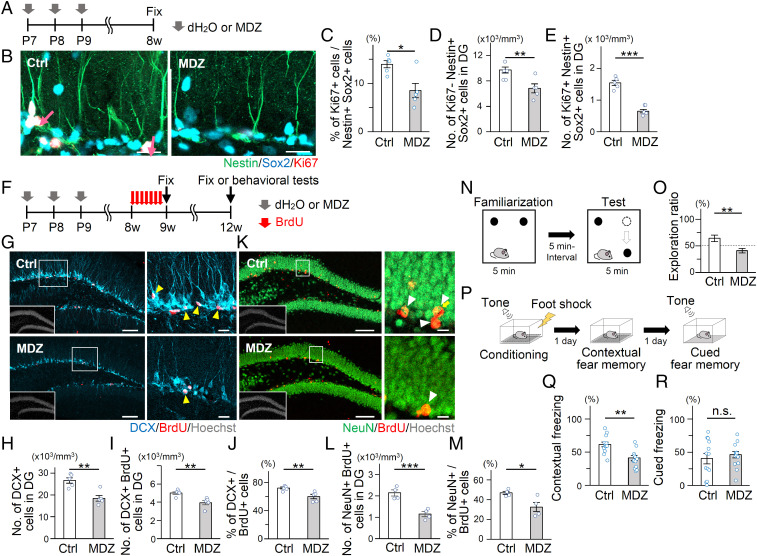
We then assessed whether MDZ-induced reinforcement of NSC dormancy during the early postnatal period is sustained until adulthood (Fig. 2A). Interestingly, the proportion of active NSCs (Ki67+ Nestin+ Sox2+) among total NSCs (Nestin+ Sox2+) was lower in the DG of 8-wk-old (8w) MDZ mice (Fig. 2 B and C), indicating that MDZ-induced NSC dormancy is long-lasting. MDZ mice also displayed a smaller quiescent NSC pool (Ki67− Nestin+ Sox2+) in the adult DG (Fig. 2D), associated with the reduced number of proliferating NSCs (Fig. 2E). Consistent with the lasting effects of MDZ on NSC dormancy, we found a decrease in the number of Ki67+ Tbr2+ intermediate progenitor cells in the adult DG of MDZ mice (SI Appendix, Fig. S3 A and B). To investigate the impact of these defects on adult neurogenesis, we labeled newborn neurons by administering BrdU to 8w Ctrl and MDZ mice for 7 consecutive days and killed the animals 1 d or 21 d post-BrdU injection (Fig. 2F). Quantification of DCX+ BrdU+ cells showed a significantly lower number of newborn immature neurons in the adult DG of 9w MDZ mice than in Ctrl (Fig. 2 G–J). Accordingly, there was a significant reduction of total NeuN+ BrdU+ mature neurons in 12w MDZ mice (Fig. 2 K and M). Moreover, GCL volume was reduced in MDZ mice (SI Appendix, Fig. S3C), which was probably due to a sustained impairment of neurogenesis until adulthood. These results indicate that early postnatal exposure to MDZ confers long-term dormancy on NSCs, resulting in a persistent decline in hippocampal neurogenesis.
Fig. 2.
Early postnatal exposure to MDZ inhibits NSC activation and neurogenesis in the adult DG. (A) Experimental scheme for investigating the proliferation of adult NSCs at 7 wk after early postnatal exposure to MDZ. (B) Representative images of Nestin (green), Sox2 (cyan), Ki67 (red), and Hoechst (gray) in the DG of 8w Ctrl and MDZ-exposed mice. Arrows indicate Nestin+ Sox2+ Ki67+ activated NSCs. (Scale bars, 20 μm.) (C) Percentage of Ki67+ Nestin+ Sox2+ activated NSCs among Sox2+ Nestin+ cells in the adult DG of Ctrl and MDZ mice (n = 5 animals per group). (D and E) Quantification of the number of Nestin+ Sox2+ Ki67− cells (D) and Nestin+ Sox2+ Ki67+ cells (E) in B (n = 5 animals per group). (F) Experimental scheme for investigating adult neurogenesis after early postnatal exposure to MDZ. (G–I) Representative tile scan images (G) and quantifications of DCX+ (H) or DCX+ BrdU+ (I) cells in the adult DG (n = 5 animals per group). The areas outlined by a white rectangle are enlarged to the right. Arrowheads indicate DCX+ BrdU+ cells. (Scale bars, 100 μm [Left] and 10 μm [Right].) (J) Percentage of DCX+ BrdU+ cells among BrdU+ cells in the adult DG of Ctrl and MDZ mice. (K and L) Representative tile scan images (K) and quantifications of NeuN+ BrdU+ cells (L) in the adult DG of Ctrl and MDZ mice (n = 4 animals per group). The areas outlined by a white rectangle in K are enlarged to the right. Arrowheads indicate NeuN+ BrdU+ cells. (Scale bars, 100 μm [Left] and 10 μm [Right].) (M) Percentage of NeuN+ BrdU+ cells among BrdU+ cells in the adult DG of Ctrl and MDZ mice. (N) Schematic diagram of the NLR task. (O) Exploration ratio for time spent in exploring the relocated object during the test phase in Ctrl and MDZ mice (n = 12 animals per group). (P) Schematic diagram of the contextual and cued FC test. (Q and R) Quantification of freezing time, showing a reduced freezing response in MDZ-exposed mice in the contextual FC test (Q) (n = 12 animals per group) but not in the cued FC test (R) (Ctrl group: n = 13; MDZ group: n = 14). *P < 0.05, **P < 0.01, and ***P < 0.001 by t test. n.s., not significant.
Adult neurogenesis in the DG is essential for hippocampus-dependent cognitive functions. Therefore, we next conducted two hippocampus-dependent memory tests (19–21), i.e., a novel location recognition (NLR) task and a contextual fear conditioning (FC) test (Fig. 2F). In the NLR task, 12w MDZ mice showed a reduced preference for an object placed at the novel location compared with Ctrl (Fig. 2 N and O and SI Appendix, Fig. S4A), although locomotor activity was not different between Ctrl and MDZ mice (SI Appendix, Fig. S4 C–E). MDZ mice also displayed significantly decreased contextual fear memory compared with Ctrl (Fig. 2 P and Q and SI Appendix, Fig. S4B). Cued fear memory, for which the amygdala is more responsible (22), was similar between Ctrl and MDZ mice (Fig. 2R and SI Appendix, Fig. S4B). These data indicate that hippocampus-dependent memory function is impaired in adult MDZ mice.
MDZ Establishes Molecular Signatures of Quiescence in NSCs
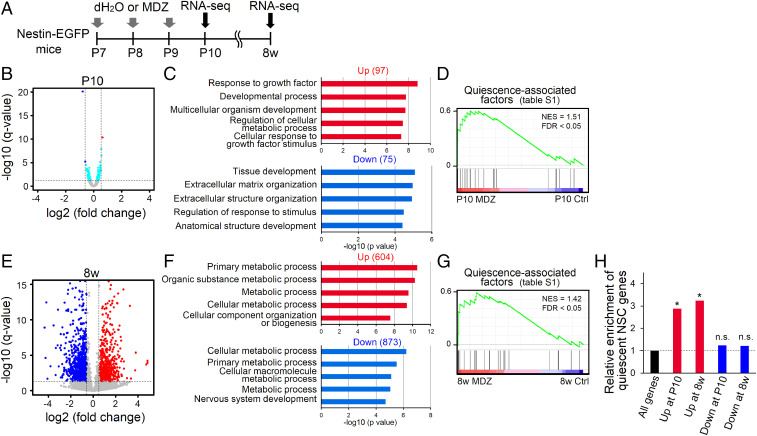
To gain insights into how early postnatal MDZ exposure induces long-lasting NSC dormancy, we performed RNA sequencing (RNA-seq) using EGFP+ NSCs and their progeny isolated from Nestin-EGFP mice at P10 and 8w with or without early-life MDZ exposure (Fig. 3A). Unexpectedly, RNA-seq analysis uncovered only a few genes whose expression changed in P10 MDZ mice with criteria fulfilling q value <0.05 and fold change ≥1.5 (Fig. 3B). Therefore, it is conceivable that behavioral alteration in NSCs in response to MDZ exposure is attributable not to prominent changes in the expression of specific genes but instead to the cumulative or cooperative effects of small expression changes in genes that critically regulate NSC activity. We then identified 172 differentially expressed genes (DEGs) without considering fold change, comprising 97 and 75 significantly up- and down-regulated genes, respectively (Fig. 3B). These DEGs were subjected to gene ontology (GO) analysis of biological processes, which showed that genes whose expression was altered by MDZ exposure are significantly associated with development-related terms (Fig. 3C). A literature survey identified a set of genes that reportedly play important roles in the induction and maintenance of NSC quiescence (hereafter referred to as quiescence-associated factors) (SI Appendix, Table S1). Gene set enrichment analysis (GSEA) showed a significant increase in the expression of these quiescence-associated factors in P10 MDZ mice compared with Ctrl (Fig. 3D). Next, we compared the gene expression profile of 8w MDZ mice with that of 8w Ctrl. We identified 604 significantly up-regulated and 873 down-regulated genes in 8w MDZ mice (q value < 0.05, fold change ≥ 1.5) (Fig. 3E), indicating that although MDZ exposure initially triggers only a small gene expression difference, the number of both up- and down-regulated DEGs expands at a later stage. GO analysis showed that down-regulated genes were associated with “nervous system development” (Fig. 3F), which could explain the decrease in adult neurogenesis in 8w MDZ mice (Fig. 2 G–M). Furthermore, the expression of quiescence-associated factors remained significantly higher in 8w MDZ mice, long after their exposure at the early postnatal stage (Fig. 3G). These data suggest that MDZ exposure evokes long-lasting dormancy in NSCs, most likely through the sustained expression of quiescence-associated factors. In support of this, we observed that genes highly expressed in quiescent NSCs of the hippocampus (hereafter referred to as quiescent NSC genes) (23) were significantly enriched in up-regulated but not down-regulated genes in MDZ mice at both P10 and 8w (Fig. 3H).
Fig. 3.
Transcriptome analysis of Nestin-EGFP+ cells in the early postnatal and adult DG. (A) Experimental scheme for transcriptomic analysis of EGFP+ cells isolated from Nestin-EGFP mice at P10 and 8w with or without early-postnatal MDZ exposure. EGFP+ cells are isolated by fluorescence-activated cell sorting. (B) Scatter plot of genes expressed in Nestin-EGFP+ cells from Ctrl and MDZ mice at P10. Cyan dots indicate DEGs (q value < 0.05; n = 172); red (n = 1) and blue (n = 2) dots indicate DEGs up- or down-regulated >1.5-fold, respectively. (C) The top 5 GO terms in significantly up-regulated (n = 97) or down-regulated (n = 75) DEGs (n = 172) at P10 are displayed. (D) GSEA plot of genes expressed in Nestin-EGFP+ cells of P10 Ctrl and MDZ mice for the list of quiescence-associated factors (SI Appendix, Table S1). FDR, false discovery rate; NES, normalized enrichment score. (E) Scatter plot of genes expressed in Nestin-EGFP+ cells from Ctrl and MDZ mice at 8w. Up-regulated (red) and down-regulated (blue) genes in MDZ mice (q value < 0.05, fold change ≥ 1.5) are highlighted. (F) The top five GO terms in up- or down-regulated genes at 8w are displayed. (G) GSEA in 8w MDZ mice showed enrichment of gene sets for quiescence-associated factors. (H) Enrichment analysis of quiescent NSC genes, identified in a previous report (23), on up- or down-regulated genes at two distinct stages (P10, 8w). *P < 0.01 by hypergeometric distribution. n.s., not significant.
MDZ Exposure Modifies the Chromatin Accessibility Landscape in NSCs
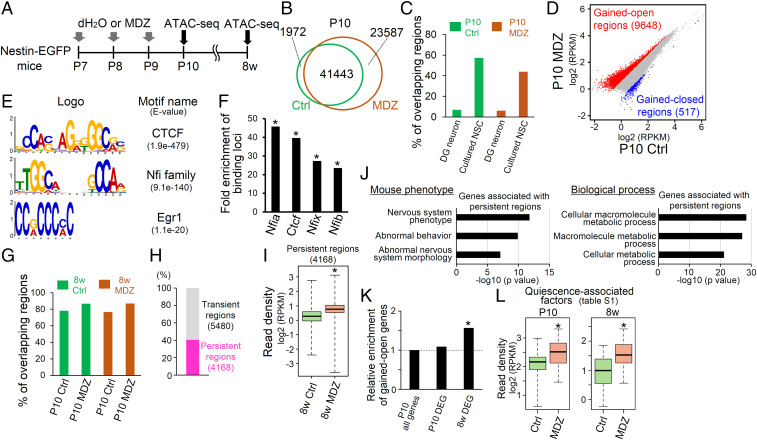
Having observed an MDZ-induced sustained molecular signature of quiescence in NSCs, we next assessed whether MDZ exposure induces a long-lasting epigenetic change. To this end, we analyzed changes in chromatin accessibility in NSCs and their progeny in Nestin-EGFP mice with or without MDZ exposure by assay for transposase-accessible chromatin (ATAC)-seq (Fig. 4A). This analysis revealed 43,415 and 65,030 open chromatin regions in Ctrl and MDZ mice at P10, respectively (Fig. 4B). We obtained public ATAC-seq data for mouse primary cultured NSCs and DG neurons (24, 25) and confirmed that the signature of open chromatin regions in EGFP+ cells of Ctrl and MDZ mice at P10 is closer to that of NSCs than of neurons more fully (Fig. 4C). We then identified 9,648 “gained-open” and 517 “gained-closed” regions in P10 MDZ mice compared with Ctrl (Fig. 4D). Motif discovery analysis of gained-open regions using MEME-ChIP showed that CTCF and cell quiescence–regulating transcription factors (NFI family members and Egr1) (26, 27) were enriched in these regions (Fig. 4E). The most enriched motif is for CTCF, a known insulator protein associated with boundaries of topologically associated domains (TADs) of chromatin (28). These results implied that an MDZ-induced increment of chromatin accessibility occurred in TADs. We also employed public chromatin immunoprecipitation sequencing (ChIP-seq) data for CTCF and NFI family members from NSCs and neural progenitor cells (26, 29, 30), allowing us to compare the enrichment of these factors in gained-open regions in P10 MDZ mice with that in randomly selected regions from the whole genome (Fig. 4F). These data suggest that MDZ increases the accessibility of transcription factors involved in cell quiescence (NFI family members and Egr1) to their target sites within TADs.
Fig. 4.
Persistent increase in chromatin accessibility of quiescence-associated genes in response to early postnatal MDZ expsure. (A) Schematic representation of sample preparation for ATAC-seq analysis at P10 and 8w using MDZ-exposed Nestin-EGFP mice. (B) Venn diagram of open chromatin regions of EGFP+ cells obtained from Ctrl and MDZ-exposed mice at P10. (C) Bar graph showing the overlap of open chromatin regions between EGFP+ cells obtained from Ctrl and MDZ-exposed mice at P10 and neural cells (DG neurons and primary cultured NSCs) obtained from publicly available data. (D) Dot plot showing a comparison of open chromatin profiles of EGFP+ cells of P10 mice between Ctrl and MDZ mice. Gained-open and gained-closed regions are shown in red and blue, respectively (fold change >1.5; n = 2 in each group). (E) Motif enrichment analysis for gained-open regions. (F) Enrichment of binding loci of the indicated factors in gained-open regions relative to randomly generated regions. *P < 0.01 by hypergeometric distribution. (G) Bar graph showing the percentage of open chromatin regions in 8w mice that overlap with open chromatin regions in P10 mice. (H) Gained-open regions at P10 include 5,480 transient (P10-specific) and 4,168 persistent (common between gained-open regions at P10 and open regions at 8w) regions. (I) Box plot showing the increased chromatin accessibility in persistent regions (n = 4,168) of 8w MDZ mice relative to Ctrl. *P < 2.23 × 10−16 by Wilcoxon rank-sum test. (J) Functional annotation of genes associated with gained-open regions (n = 719) in mouse phenotype (Upper) and biological process (Lower). (K) Enrichment analysis of gained-open genes in DEGs at P10 and 8w. *P < 0.01 by hypergeometric distribution. (L) Box plot showing the increased chromatin accessibility around quiescence-associated factors at both P10 and 8w in MDZ mice relative to Ctrl. *P < 0.001 by Wilcoxon rank-sum test.
To check whether early-life exposure to MDZ influences chromatin conformation in NSCs at the adult stage, we analyzed ATAC-seq data from Ctrl and MDZ mice at 8w and found that ∼80% of open chromatin regions overlapped with those in P10 mice (Fig. 4G), indicating that P10 and 8w NSCs have similar accessible chromatin landscapes. Among these gained-open regions in P10 MDZ mice, 5,480 (57%) are transient (P10-specific gained-open regions), whereas 4,168 regions (43%) are persistent (common between gained-open regions at P10 and open regions at 8w) (Fig. 4H). These persistently open regions were more accessible in 8w MDZ mice relative to Ctrl (Fig. 4I). These data suggest that MDZ exposure at the early postnatal stage establishes a chromatin accessibility landscape in NSCs that perpetuates throughout life. In addition, among the 4,168 persistent regions, 719 were annotated as being within 2.5 kb from the transcription start site (TSS) of individual genes belonging to the “abnormal behavior” and “nervous system phenotype” mouse phenotype categories of GO analysis (Fig. 4J), which may implicate these genes in the impairment of neurogenesis and cognition observed in MDZ mice (Fig. 2 F–R).
Since the augmentation of chromatin accessibility is associated with both increased and decreased gene expression (31), we next determined whether MDZ-induced changes in chromatin accessibility affect gene expression. Genes associated with persistent gained-open regions (Fig. 4J) showed significant enrichment in DEGs at 8w but not P10 (Fig. 4K), indicating that persistent alterations in chromatin accessibility affect gene expression in the adult stage. Given that we observed an increase in the expression of quiescence-associated factors (Fig. 3 D and G), we then examined whether MDZ increased chromatin accessibility around genes encoding these factors. We found that regions within 2.5 kb from the TSS of these genes are significantly more accessible in P10 and 8w MDZ mice than in Ctrl (Fig. 4L), in agreement with their increased expression (Fig. 3 D and G). Overall, these data indicate that although most of the changes in chromatin accessibility induced by MDZ prime the associated genes for later expression, those changes associated with genes such as quiescence-associated factors persistently affect their expression from early postnatal to adult stages, contributing to the induction of long-lasting dormancy of NSCs.
Egr1 Expression Induces NSC Dormancy
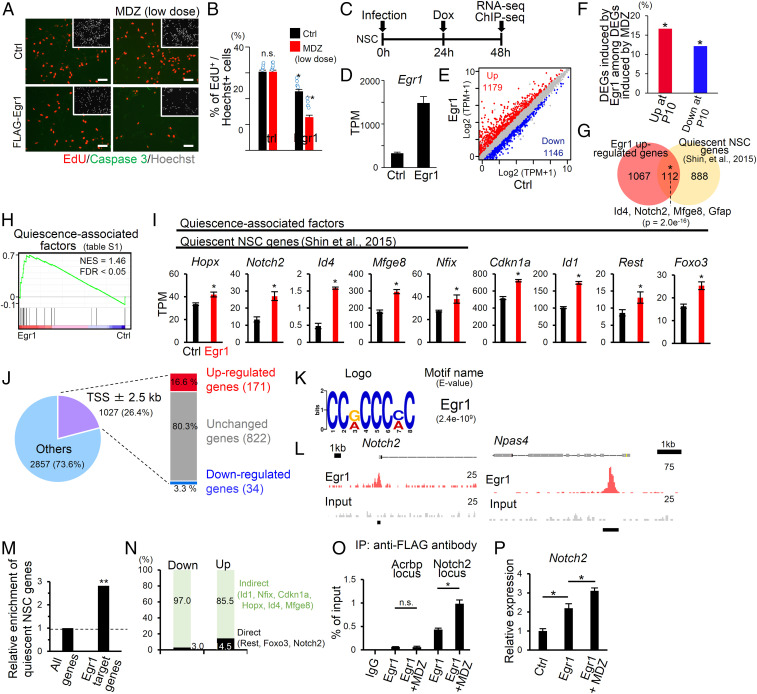
To obtain a clue to elucidating the mechanism underlying MDZ-induced NSC dormancy, we focused on Egr1, which has been reported to be associated with stem cell quiescence (27), among the top 10 up- or down-regulated genes in P10 MDZ mice (SI Appendix, Table S2). Egr1 expression was up-regulated in mouse, rat, and human NSCs after MDZ treatment, indicating that MDZ directly stimulates NSCs to express Egr1 (SI Appendix, Fig. S5 A–C). To investigate whether Egr1 induces NSC quiescence, FLAG-tagged Egr1 was expressed in cultured NSCs under the control of the doxycycline-inducible tetracycline response element promoter with a high infection rate (>96%) (SI Appendix, Fig. S5 D and E). We found that forced expression of Egr1 inhibited proliferation of mouse NSCs without inducing apoptosis (Fig. 5 A and B). Since NSCs in the hippocampus receive GABA tonically from connecting GABAergic neurons (8), Egr1-transduced NSCs were treated with MDZ at a low dose that did not inhibit proliferation of Ctrl NSCs. We observed that proliferation of Egr1-transduced cells was reduced even further with the low dose of MDZ (Fig. 5 A and B), indicating that MDZ enhances Egr1 activity in addition to inducing its expression.
Fig. 5.
Implication of Egr1 in MDZ-induced NSC dormancy. (A) Representative images of EdU (red), active Caspase 3 (green), and Hoechst (gray; insets) staining in Egr1 or Ctrl virus-infected mouse NSCs treated with or without a low dose of MDZ (5 μM). Cells were fixed 1 d after doxycycline addition. (Scale bars, 50 μm.) (B) Quantification of EdU+ cells in A. *P < 0.05 and **P < 0.01 by ANOVA with Tukey post hoc tests. n.s., not significant. (C) Experimental scheme for investigating the molecular mechanism underlying the reduction of NSC proliferation by Egr1. NSCs infected with lentivirus encoding M2rtTA were used as Ctrl. (D) Bar graph showing the expression levels of Egr1 in Egr1 or Ctrl virus-infected mouse NSCs. (E) Scatter plot of genes expressed in Egr1 or Ctrl virus-infected mouse NSCs. (F) Percentage of Egr1-induced up- or down-regulated genes among MDZ-induced up-regulated (red) or down-regulated (blue) genes, respectively, at P10. *P = 1.0e−5 (Left) and 0.006 (Right) by hypergeometric distribution. (G) Venn diagram showing significant overlap between genes that are up-regulated in Egr1-transduced cells relative to Ctrl (fold change ≥ 1.5, q < 0.05) and quiescent NSC genes identified in a previous report (the 1,000 most highly expressed genes in quiescent NSCs) (23). (H) GSEA in Egr1-transduced cells showed enrichment of gene sets for quiescence-associated factors (SI Appendix, Table S1). (I) Bar graphs showing the expression levels of quiescence-associated factors and quiescent NSC genes (23) after Egr1 transduction (n = 3). *q < 0.05. (J) Pie chart showing the percentage of Egr1-occupied loci within 2.5 kb of a TSS. These Egr1-bound genes were classified into three groups based on their gene expression pattern relative to the Ctrl: down-regulated (fold change ≤ 0.66, q < 0.05), up-regulated (fold change ≥ 1.5, q < 0.05), and unchanged genes. (K) Motif enrichment analysis against Egr1-bound sites ± 200 bp around a TSS. (L) Alignment of Egr1 ChIP-seq (red) and input (gray) data near the Notch2 and Npas4 loci. The Egr1 target site is displayed by a black bar. (M) Relative to randomly selected genes, Egr1-bound genes are significantly enriched with quiescent NSC genes (23). **P < 0.01 by hypergeometric distribution. (N) Bar graph showing the percentage of Egr1 target (Direct) and nontarget genes (Indirect) among up- or down-regulated genes in Egr1-transduced mouse NSCs. (O) ChIP-qPCR analyses of Egr1 enrichment in Egr1-binding sites around the Notch2 locus in Egr1-transduced NSCs treated with or without a low dose of MDZ (5 μM) (n = 4). The Acrbp locus served as a negative Ctrl. *P < 0.01 by ANOVA with Tukey post hoc tests. n.s., not significant. (P) Relative expression of Notch2 in the indicated cells (n = 4). *P < 0.01 by ANOVA with Tukey post hoc tests. n.s., not significant.
Since we found that MDZ inhibits neuronal differentiation of NSCs in the DG (Fig. 2 J and M), we next tested whether MDZ-downstream Egr1 affects neuronal differentiation of NSCs (SI Appendix, Fig. S6A). To do this, we induced Egr1 expression in NSCs and cultured them in the presence or absence of a low dose of MDZ. Five days after Egr1 induction, we performed immunostaining using immature and mature neuronal markers (βIII-tubulin and Map2ab, respectively) and found that Egr1 repressed neuronal differentiation of NSCs (SI Appendix, Fig. S6 B–E). In contrast to the case of NSC proliferation, low-dose MDZ treatment did not affect neuronal differentiation of Egr1-transduced NSCs (SI Appendix, Fig. S6 B–E), suggesting that MDZ specifically enhances the inhibitory effect of Egr1 on NSC proliferation. Moreover, βIII-tubulin+ Map2ab− cells were observed in control virus-infected NSCs, whereas almost all neurons that differentiated from Egr1-transduced NSCs became Map2ab+ cells (SI Appendix, Fig. S6 B–D). These data suggest that, even though Egr1 can inhibit neuronal differentiation, once Egr1-expressing cells are committed to the neuronal lineage, they mature quickly relative to control, in agreement with a previous report showing that Egr1 promotes maturation of newborn neurons (32).
To validate Egr1-induced transcriptional regulation in NSC dormancy, we analyzed the transcriptome of Egr1-transduced mouse NSCs 24 h after the induction of Egr1 expression (Fig. 5C). Egr1 transduction of NSCs increased the Egr1 expression level (Fig. 5D). We identified genes whose expression was either up-regulated (n = 1,179) or down-regulated (n = 1,146) in Egr1-transduced cells relative to Ctrl (fold change ≥ 1.5, q < 0.05) (Fig. 5E). Sixteen percent of the up-regulated genes and 12% of the down-regulated genes at P10 after MDZ treatment displayed statistically significant overlaps with Egr1-induced up-regulated or down-regulated genes, respectively (Fig. 5F), suggesting that at least a proportion of the MDZ-induced gene expression changes were mediated through Egr1 expression. We also found that up-regulated genes in Egr1-transduced cells significantly overlapped with quiescent NSC genes of the adult hippocampus identified in a previous study (23), including Id4 and Notch2 (Fig. 5G), both of which have been shown to induce NSC quiescence in the adult DG (33). GSEA also showed a significant increase in the expression of the quiescence-associated factors (SI Appendix, Table S1) in Egr1-transduced cells compared with Ctrl (Fig. 5 H and I). Taking these observations together, it is conceivable that Egr1 induces NSC dormancy through the expression of these genes.
To find target genes of Egr1, we performed ChIP-seq for Egr1 in mouse NSCs at 24 h after Egr1 transduction (Fig. 5C) and identified 3,884 Egr1-occupied loci using input DNA as Ctrl (Fig. 5J). We confirmed that the most enriched motif in these loci was Egr1 (Fig. 5K), and that Egr1 occupied the region around the TSS of a known Egr1 target gene, Npas4 (34) (Fig. 5L). Approximately 26% of Egr1-occupied loci were annotated within 2.5 kb from a TSS, and 16.6% of such annotated loci neighbored up-regulated genes (fold change ≥ 1.5, q < 0.05) in Egr1-transduced cells relative to Ctrl, whereas only 3.3% neighbored down-regulated genes (Fig. 5J). These Egr1-occupied and up-regulated genes were enriched for quiescent NSC genes (23) such as Notch2 (Fig. 5 L and M), suggesting that Egr1 directly induces the expression of these genes. We also found that most of the genes (85.5%) that were up-regulated in Egr1-transduced cells lacked Egr1 occupancy around their TSS (Fig. 5N), implying that Egr1 can also increase the expression of genes associated with NSC quiescence indirectly, possibly through the induction of Egr1 target gene expression.
We next asked why low-dose MDZ treatment can further enhance NSC quiescence in Egr1-transduced cells (Fig. 5 A and B). When we examined the binding of Egr1 to the whole genome by ChIP-qPCR analysis, Egr1 occupancy was increased around the TSS of the Egr1 target gene Notch2 in the presence of a low dose of MDZ (Fig. 5O). Moreover, Notch2 expression was more strongly up-regulated in Egr1-transduced cells treated with a low dose of MDZ than in Egr1-transduced cells without MDZ treatment (Fig. 5P). Thus, MDZ augments the expression of Egr1 and its binding to target genes associated with NSC quiescence, resulting in the reinforcement of NSC dormancy.
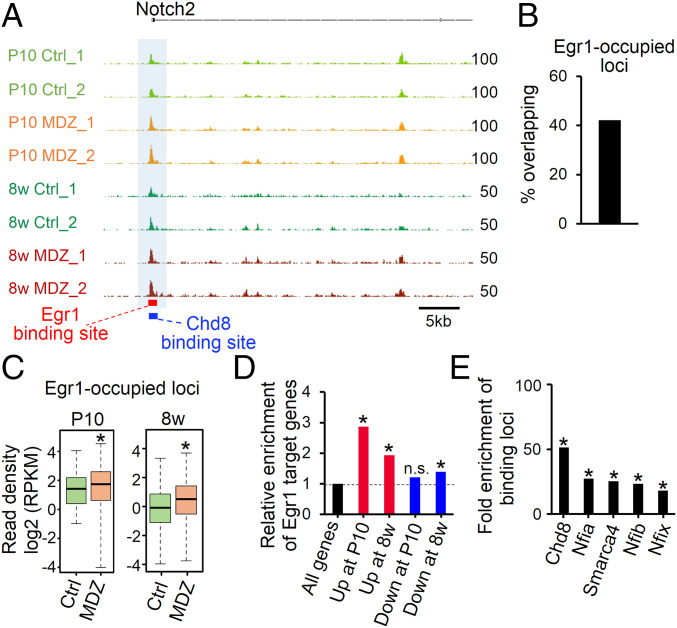
MDZ Exposure Persistently Increases Chromatin Accessibility in Egr1-Occupied Loci
Egr1 interacts with epigenetic factors such as Tet1 to establish and maintain the epigenetic signature in the brain (35). Having observed increased Egr1 expression and chromatin accessibility after MDZ exposure, we next checked chromatin accessibility at Egr1-occupied loci in P10 and 8w MDZ mice. Approximately 40% of Egr1-occupied loci were located in accessible chromatin regions (Fig. 6B), and chromatin accessibility at these loci, including the Notch2 locus, was persistently higher, from P10 to 8w, in NSCs of MDZ mice than in Ctrl (Fig. 6 A and C). Furthermore, Egr1 target genes were significantly enriched in up-regulated genes at both P10 and 8w (Fig. 6D), suggesting that Egr1 up-regulates its target genes persistently, even though the up-regulation of Egr1 expression itself was transient after MDZ exposure (SI Appendix, Fig. S7 A and B). Integrative analysis of public ChIP-seq data obtained with NSCs suggested that Egr1 co-occupied loci with Chd8 and Smarca4 (Fig. 6E), chromatin remodeling factors that augment chromatin accessibility (36, 37). Moreover, NFI family proteins appeared to be enriched in Egr1-occupied loci (Fig. 6E). These findings suggest that Egr1 acts in concert with chromatin remodeling proteins (Chd8 and Smarca4) and quiescence-associated factors (NFI) to cause the protracted increment of chromatin accessibility and long-term NSC dormancy, regardless of its transient up-regulation after MDZ exposure.
Fig. 6.
Increase in chromatin accessibility of Egr1-occupied loci in response to early postnatal MDZ exposure. (A) Alignment data for ATAC-seq at the Notch2 locus in P10 and 8w MDZ mice (n = 2). Egr1 and Chd8 target sites are displayed as red and blue bars, respectively. (B) Bar graph showing the percentage of overlapping Egr1-occupied loci in open chromatin regions in P10 Ctrl mice. (C) Box plots showing the chromatin accessibility in P10 and 8w Ctrl and MDZ mice for the overlapping Egr1-occupied loci shown in B (n = 1,635). *P < 0.001 by Wilcoxon rank-sum test. (D) Enrichment analysis of Egr1 target genes (Fig. 5M) in DEGs at P10 and 8w. *P < 0.01 by hypergeometric distribution. n.s., not significant. (E) Enrichment of binding loci for the indicated factors in the Egr1 binding loci shown in B and C (n = 1,635) relative to randomly generated loci. *P < 0.01 by hypergeometric distribution.
Voluntary Exercise Rescues Impaired Hippocampal Neurogenesis and Cognitive Functions
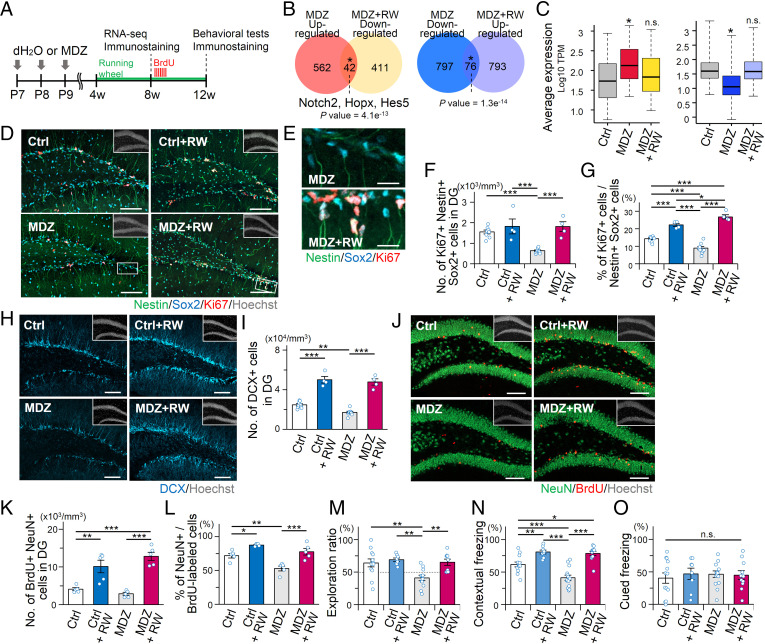
Voluntary exercise in a running wheel, a widely accepted neurogenic stimulus (38, 39), ameliorates long-lasting neurocognitive sequelae following anesthesia in rodents (40, 41), although the mechanisms underlying these effects are yet to be unveiled. To explore whether voluntary running can restore the disrupted NSC behavior and impaired memory function in MDZ mice, we evaluated the effect of voluntary exercise on the transcriptional profile of NSCs and their progeny using MDZ-exposed Nestin-EGFP mice (Fig. 7A). Notably, voluntary running significantly amended subfractions of positively (42 genes) and negatively (76 genes) disrupted gene expression, including that of the Egr1 target Notch2, to the opposite direction in the adult MDZ mice (Fig. 7B). Indeed, the expression of these genes was normalized to levels comparable to Ctrl by the physical activity (Fig. 7C).
Fig. 7.
Voluntary exercise rescues MDZ-induced adverse effects. (A) Schematic diagram of preparation for immunostaining and RNA-seq analysis in 8w MDZ mice with or without a running wheel. (B) Venn diagrams depicting significant overlapping of genes between up-regulated at 8w MDZ mice relative to 8w Ctrl mice (fold change > 1.5, q < 0.05) and down-regulated at 8w MDZ mice housed with running wheel (MDZ + RW) relative to 8w MDZ mice (fold change > 1.5, q < 0.05) and vice versa (Left and Right, respectively). *P = 4.1e−13 (Left) and 1.3e−14 (Right) by hypergeometric distribution. (C) Boxplots showing transcripts per millions (TPM) values of genes amended negatively (Left; n = 42) or positively (Right; n = 76), indicated in B. *P < 0.001 by Wilcoxon rank-sum test. (D–G) Representative tile scan images (D and E) and quantifications (F and G) of Nestin+ Sox2+ Ki67+ activated NSCs in the DG of 8w Ctrl (n = 8), Ctrl + RW (n = 4), MDZ (n = 9), and MDZ + RW (n = 4) mice. Magnified images of the white boxes in D are shown in E. (Scale bars, 100 μm [D] and 20 μm [E].) (H and I) Representative tile scan images (H) and quantification (I) of DCX+ immature neurons in the adult DG. (Scale bars, 100 μm.) (J and K) Representative images (J) and quantification of NeuN+ BrdU+ mature neurons (K) in the DG of Ctrl and MDZ-exposed mice at 3 wk after BrdU injection (n = 5 animals per group). (Scale bars, 100 μm.) (L) Percentage of NeuN+ BrdU+ cells among BrdU+ cells in the adult DG. (M) Exploration ratio for time spent in exploring the relocated object during the test phase in Ctrl (n = 13), Ctrl + RW (n = 8), MDZ (n = 14), and MDZ + RW (n = 11) mice. (N and O) Quantification of freezing time showing an increased freezing response in MDZ + RW mice (n = 11) in the contextual FC test (N) compared to Ctrl mice (n = 13) but not in the cued FC test (O). Data for Ctrl (n = 13) and MDZ (n = 14) mice in M–O were obtained from behavioral tests in Fig. 2 O, Q, and R. *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey post hoc tests (unless indicated otherwise). n.s., not significant.
In light of these findings, we next asked whether running rescues the MDZ-induced impairment of adult hippocampal neurogenesis and hippocampus-dependent memory. We found that running indeed restored both the number and proportion of Ki67+-activated NSCs in MDZ mice to the same levels as in Ctrl with running wheel (RW) (Ctrl + RW) (Fig. 7 D–G). These results indicate that running canceled the disrupted quiescent state of NSCs that was inflicted by MDZ exposure. We also observed that running increased the number of DCX+ immature neurons in MDZ mice, which is higher than in Ctrl because voluntary running promotes the survival of newly generated neurons (38, 39) (Fig. 7 H and I). Cell tracing experiments for newly generated neurons, identified by administering BrdU, further revealed a significant increase in total NeuN+ BrdU+ mature neurons of 12w MDZ mice as a result of running (Fig. 7 J–L). We next attempted to examine the effects of the running on the impaired memory function in MDZ mice by conducting an NLR task and a contextual FC test (Fig. 7 M–O). We observed that the hippocampal-dependent memory function of MDZ mice improved not only in the NLR task (Fig. 7M and SI Appendix, Fig. 8A) but also in the contextual FC test (Fig. 7 N and O and SI Appendix, Fig. S8 B–E), while locomotor activity was not different among the 4 groups of mice (SI Appendix, Fig. S8 F and G). Taken together, these results clearly indicate that voluntary exercise can rescue MDZ-impaired hippocampal neurogenesis and memory functions in adult mice, possibly by normalizing the MDZ-induced aberrant transcriptome in NSCs.
Discussion
We have demonstrated that exposure of mice to the anesthetic MDZ in the early postnatal stage established persistent modification of the chromatin accessibility landscape and of quiescence-associated gene expression to induce long-lasting hippocampal NSC dormancy and cognitive decline, at least in part through the transient up-regulation of Egr1 expression. We also found that Egr1 and other factors such as NFI family proteins were enriched in gained-open regions induced by MDZ exposure. Since NFI family proteins are known to maintain the global chromatin landscape to retain stem cell identity (42), these results suggest that, in addition to Egr1, other factors, including NFI family proteins, also contribute to the long-lasting NSC dormancy by sustaining the enhanced chromatin accessibility after MDZ exposure. Furthermore, although we focused on NSCs in this study, exposure to anesthetics is known to increase Egr1 expression in cells other than NSCs (34). Analogous phenomena such as altered chromatin accessibility induced by anesthetics may therefore occur in other CNS cell types.
To model cognitive decline in humans induced by anesthetic exposure during the early postnatal period, we exposed P7 mice repeatedly to MDZ. While it is difficult to align neurodevelopmental ages in mice and humans, the first to second postnatal week of mice is roughly equivalent to children under 2 y (43), which is consistent with human epidemiological studies showing an association between anesthetic exposure before 3 y old and neurocognitive impairment in later life (4). This age range in mice is considered a critical period for NSC expansion in the DG, when precise balancing of NSC proliferation and quiescence should be taking place (32). We revealed that exposure of mice to MDZ during this age range distorts the switching between the active and quiescent states of NSCs by inhibiting their proliferation in the hippocampus persistently until adulthood, resulting in cognitive impairment. We also showed that MDZ can inhibit the proliferation of NSCs from other species, namely rat and human, indicating that the effect of the anesthetic is evolutionarily conserved. Although adult hippocampal neurogenesis in humans remains controversial (44, 45), it is generally accepted that the human brain retains neurogenic potential during early childhood. Thus, hippocampal-dependent memory decline triggered by repeated exposure to anesthetics during early childhood may be caused by abnormal NSC behavior and a subsequent diminution of neurogenesis, although we cannot completely exclude the possibility that an anesthetic-induced property change in other CNS cells mediates the memory impairment.
In the present study, we have shown that voluntary exercise normalizes MDZ-perturbed gene expression in NSCs and overcomes decreased neurogenesis and impaired hippocampal-dependent memory function in MDZ-exposed mice, even when the exercising begins as late as 3 wk after the exposure. We also observed that voluntary running significantly amended both positively and negatively disrupted gene expression in NSCs of MDZ-exposed mice, an example being the Egr1 target gene Notch2, which can strongly induce NSC quiescence in the adult DG (33), implying that running reversed, at least partly, Egr1-mediated chromatin changes. However, the transcriptomic changes differ considerably between NSCs in MDZ mice and MDZ mice with running. These observations suggest that running may also ameliorate the deficits in neurogenesis and behavior of MDZ mice through as-yet-unknown mechanisms distinct from which remodels chromatin around Egr1 target genes, warranting future investigations.
Our findings, despite certain limitations, should pave the way toward a better understanding of adverse anesthetic effects on neurocognitive function and the development of a novel therapeutic strategy to preclude delayed onset of learning and memory decline in people who experienced anesthesia in their early life.
Materials and Methods
Mice.
All efforts were made to minimize animal suffering and to reduce the number of animals used. Mice on a C57BL/6 background were housed under a 12-h light/dark cycle and could access food and water ad libitum. Mice received a daily injection of MDZ (10 mg/kg, distilled water to a concentration of 2 mg/mL, Sandoz) or equivalent volumes of water intraperitoneally for 3 d (P7, P8, and P9) and then weaned at P28. To assess the proliferation of NSCs and neurogenesis, mice were injected intraperitoneally with BrdU (100 mg/kg). Both male and female mice were analyzed indiscriminately, except for behavioral analysis using 12w male mice. Nestin-EGFP mice on a C57BL/6 background were kindly provided by M. Yamaguchi (46). All experiments were carried out according to the animal experimentation guidelines of Kyushu University, which comply with the NIH Guide for the Care and Use of Laboratory Animals (47).
Measuring Anesthetic Score and Duration of MDZ Anesthesia.
The anesthetic effects of MDZ in P7 mice were evaluated by righting reflex score and duration time as described previously with some modifications (15, 16). For the righting reflex scale, a mouse was placed on its back and the response assessed. A score was given according to its response as follows: the mouse cannot be placed on its back (0), rights itself after multiple attempts (1), attempts to right itself but cannot (2), and does not move at all in response to being placed on its back (3). The righting reflex score was recorded every 15 min until anesthetized mice regained the righting reflex. The duration of MDZ anesthesia for each MDZ dose was defined as the time interval between an impaired righting reflex and recovery of the righting reflex. Three different doses (1 mg/kg, 10 mg/kg, 20 mg/kg of MDZ) were chosen for our pilot study to determine the required dose for MDZ anesthesia. During anesthesia, mice were kept on a warming pad maintained at ∼37 °C for the duration of the experiment.
Immunohistochemistry.
We performed immunohistochemistry as described previously (48). Briefly, mouse brains were fixed in 4% paraformaldehyde (PFA) and 40-μm sections were cut with a cryostat. The sections were incubated with blocking solution (5% fetal bovine serum [FBS] and 0.3% Triton X-100) for 1 h at room temperature and then incubated with primary antibodies at 4 °C overnight. The following primary antibodies were used: chicken anti-Nestin (1:500, NES, Aves Labs); mouse anti-Ki67 (1:500, 550609, BD Biosciences); goat anti-Sox2 (1:500, sc-17320, Santa Cruz Biotechnology); goat anti-DCX (1:500, sc-8066, Santa Cruz Biotechnology); rabbit anti-DCX (1:500, 4604, Cell Signaling Technology); mouse anti-NeuN (1:500, MAB-377, Millipore); rabbit anti-active Caspase 3 (1:500, AF-835, R&D Systems); chick anti-GFP (1:500, GFP-1010, Aves Labs); and rat anti-BrdU (1:500, ab-6326, Abcam). For staining of Ki67, antigen retrieval was performed by heating sections in target retrieval solution (Dako) at 105 °C for 15 min before blocking. For detection of BrdU-labeled cells, free-floating sections were treated with 2 N HCl at 37 °C for 15 min and washed with phosphate-buffered saline (PBS) before blocking and incubation with primary antibodies. After three washes in PBS, sections were incubated for 2 h with corresponding secondary antibodies: CF-488 donkey anti-mouse IgG (1:500, 20014, Biotium); CF-488 donkey anti-chicken IgG (1:500, 20166, Biotium); CF-555 donkey anti-rabbit IgG (1:500, 20038, Biotium); CF-555 donkey anti-mouse IgG (1:500, 20037, Biotium); CF-568 donkey anti-rat IgG (1:500, 20092, Biotium); CF-647 donkey anti-rabbit IgG (1:500, 20047, Biotium); and CF-647 donkey anti-goat IgG (1:500, 20048, Biotium). Nuclei were stained using Hoechst 33258 (Nacalai Tesque). Fluorescence images were obtained on a confocal laser microscope (LSM 700 and LSM 800, Zeiss). For immunostaining of cells, the cells were fixed with 4% PFA in PBS for 20 min and incubated for 2 h at 4 °C with primary antibodies diluted in blocking solution, followed by incubation with the corresponding secondary antibodies for 1 h at room temperature. Images of stained cells were taken using a Leica AF600 fluorescence microscope.
Cell Counting.
Cell counting was performed on every sixth coronal section (240 μm apart) containing DG at the same anatomical level. The total number of marker-positive cells per DG was estimated by multiplying the number counted in the series of sections by the inverse of the section sampling fraction. In brief, the number of counted cells was multiplied by 12 to obtain the total number of marker-positive cells in the bilateral DG per brain.
Cell Culture.
Mouse NSCs with multipotency and self-renewal capacity derived from the cortex at embryonic day 11 and passaged more than 20 times were used in this study. Mouse NSCs were plated on poly-L-ornithine/fibronectin-coated dishes in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 containing N2 supplement (7502048, Gibco) with 20 ng/mL basic fibroblast growth factor (bFGF; 100-18B, PeproTech), 20 ng/mL epidermal growth factor (EGF; AF-100-15, PeproTech), and 0.1 mg/mL penicillin/streptomycin/fungizone (HyClone) under 5% CO2 at 37 °C. Rat adult hippocampal NSCs (49) were plated on poly-L-ornithine/laminin–coated dishes in DMEM/F-12 containing N2 supplement, 20 ng/mL bFGF, and 0.1 mg/mL penicillin/streptomycin/fungizone (HyClone) under 5% CO2 at 37 °C. Human induced pluripotent stem cell–derived NSC cells (a generous gift from A. Smith, University of Exeter, Exeter, United Kingdom) were maintained and used as described previously (50). Human NSCs were plated on poly-L-ornithine/laminin–coated plates in maintenance medium, which consists of DMEM/F12 (Invitrogen), 2.5 μg/mL insulin (Sigma), 100 μg/mL transferrin (Sigma), 16 μg/mL putrescine (Sigma), 30 nM sodium selenite (Sigma), 20 nM progesterone (Sigma), 0.1 mg/mL penicillin/streptomycin/fungizone (HyClone), 1 μL/mL B27 (Gibco), 10 ng/mL bFGF, and 10 ng/mL EGF. HEK293T human embryonic kidney cells were grown in DMEM (Nacalai Tesque) supplemented with 10% FBS.
Ethynyldeoxyuridine (EdU) Labeling and Detection.
For EdU experiments, 10 mM EdU was added to the culture medium, and the cells were fixed 1 h later. EdU staining was performed using a Click-iT EdU Alexa Fluor 555 Imaging Kit (Life Technologies) according to the supplier’s protocol. Images of stained cells were taken using a Leica AF600 fluorescence microscope.
Overexpression Experiments.
For overexpression of Egr1, we used a lentiviral vector (Tet-O-FUW; #30130, Addgene) that controls gene expression under the tetracycline operator. Complementary DNA (cDNA) for Flag-tagged Egr1 was cloned into the Tet-O-FUW vector using EcoRI sites. Lentiviruses were produced by transfecting HEK293T cells with the lentivirus constructs pCMV-VSV-G-RSV-Rev and pCAG-HIVgp using polyethylenimine. Cultured mouse NSCs were infected with Flag-tagged Egr1-expressing lentivirus constructs together with an M2rtTA-expressing lentivirus construct (#20342, Addgene) in NSC medium. After 16 to 20 h in a medium containing lentivirus, the medium was replaced with a medium for mouse NSCs. Doxycycline (2 mg/mL) was added only once to this medium to activate expression of Egr1. M2rtTA-transduced cells were used as control cells.
Real-Time qPCR Analysis.
Total RNA was isolated from cells using Sepasol-RNA I Super G (Nacalai Tesque) following the manufacturer’s instructions. RNA quality of all samples was checked by spectrophotometer. According to the kit protocol, reverse transcription reactions were carried out using a SuperScript VILO cDNA Synthesis Kit (Life Technologies). qRT-PCR was performed with SYBR green fluorescent dye using Step One Plus (Applied Biosystems) and Mx3000 (Stratagene). β-actin was used as an endogenous control to normalize samples.
Isolation of Nestin-EGFP+ Cells from DG.
Procedures for isolating Nestin-EGFP+ cells were modified from previously described dissociation and FACS protocols (48). Briefly, P10 and 8w Nestin-EGFP mice were euthanized, and each brain was promptly harvested. The DG was quickly microdissected under a dissection scope and minced with a scalpel. The dissociated DG was transferred into prewarmed papain (25 U/mL) solution and incubated at 33 °C for 45 min. The suspension was then washed with 2 mL Minimum Essential Medium α (Gibco) containing 5% BSA to stop enzyme activity, mechanically dissociated by gentle pipetting with a fire-polished Pasteur pipette, and centrifuged at 130 × g for 5 min. The cell pellet was then resuspended in 2 mL of Hanks’ balanced salt solution (HBSS) containing 250 U/mL DNase and centrifuged at 130 × g for 5 min. After centrifugation, the cells were suspended in HBSS. The cells were sorted on a FACS Aria II (BD Biosciences), with 7-AAD (559925, BD Pharmingen) added to exclude nonviable cells from the analysis. Cells dissociated from the DG of WT mice were used to draw gates for EGFP+ and EGFP− cell populations. The EGFP+ cells were sorted into N2 medium, centrifuged at 200 × g at 4 °C for 10 min, and stored in liquid nitrogen until use.
mRNA Isolation and cDNA Library Preparation.
We used 50,000 cells isolated from two mice at P10 and 30,000 cells isolated from three to five mice at 8w to construct a library for each sample for RNA-seq (n = 3 at each stage). mRNAs were isolated from the Nestin-EGFP+ cells with a Dynabeads mRNA DIRECT Micro Kit (Invitrogen), and the purified mRNAs were subjected to cDNA library construction using a NEB Next Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s protocols. Briefly, polyadenylated mRNA was isolated directly from the cells using Dynabeads Oligo (dT)25 (Invitrogen). mRNA was fragmented in NEB Next First Strand Synthesis Reaction Buffer by heating at 94 °C for 15 min. First-strand cDNA was reverse-transcribed from the fragmented mRNA and then used as template to synthesize second-strand cDNA. Deoxythymidine triphosphate was replaced with deoxyuridine triphosphate (dUTP) during the second-strand cDNA synthesis. The cDNA was end-repaired, dA-tailed, and ligated with NEB Next Adaptor. The second-strand cDNA containing dUTP was digested by USER enzyme, and a sequencing tag and barcode were introduced through a 12 to 15-cycle PCR amplification. The cDNA library was purified using AMPure XP beads (Beckman Coulter). The quality of the cDNA library was assessed using the Bioanalyzer High Sensitivity DNA Analysis kit (Agilent).
RNA-seq and Bioinformatics Analysis.
RNA-seq was performed with 50-bp single-end sequencing using an Illumina HiSEq 2500, described previously (51). Briefly, obtained reads were processed with the FASTX tool kit (52) to remove short (<30 bp) and low-quality (quality score < 20) reads, followed by trimming of the adaptor sequence. Processed reads were aligned to the mouse reference genome (mm10) using TopHat (53). Raw read counts were determined using Feature Counts software (54). The DESeq2 package (55) was used for differential gene expression analysis. Gene expression levels were quantified as transcripts per millions. GO analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery. We analyzed GO term enrichment in the Biological Process and Mouse Phenotype categories. GSEA was carried out using signal-to-noise as the ranking metric and with the “weighted” scoring scheme. For analyzing GSEA, we extracted a set of genes associated with NSC quiescence by a literature survey.
ATAC-seq and Data Analysis.
We used 50,000 cells isolated from two mice at P10 and 30,000 cells isolated from three to five mice at 8w to construct a library for each sample for ATAC-seq (n = 2 at each stage). ATAC-seq was performed as previously described (56) with minor modifications. Briefly, FACS-sorted fresh Nestin-EGFP+ cells from the DG were pelleted and incubated with cold lysis buffer (10 mM Tris⋅HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630). After removing the lysis buffer by centrifugation, samples were immediately subjected to a transposition reaction at 37 °C for 30 min with 2.5 μL transposase (Illumina Nextera DNA Preparation Kit). Transposed DNA was purified using a FastGene Gel/PCR Extraction Kit (Nippon Genetics). The purified DNA was amplified for 12 to 15 cycles using adapters from the Nextera index kit (Illumina) according to the manufacturer’s instructions, and PCR purification was performed using AMPure XP beads (Beckman Coulter) to remove large fragments and remaining primers. Quality of the cDNA library was assessed using the Bioanalyzer High Sensitivity DNA Analysis kit (Agilent). An Illumina HiSEq 2500 was used to perform 50-bp single-end sequencing. Sequencing reads were mapped to the mouse reference genome (mm10) from the University of California, Santa Cruz (UCSC) Genome Browser using Bowtie2. Open chromatin regions were analyzed using MACS software. The consensus sequence was identified by importing a 200-bp sequence surrounding the summit of the open chromatin regions into the MEME program. To confirm the signature of open chromatin regions, we used public ATAC-seq data obtained from NSCs and DG neurons (24, 25). To reveal the enrichment of factors in gained-open region, we used ChIP-Atlas (chip-atlas.org) and employed public ChIP-seq data for cell quiescence and/or chromatin remodeling–related factors from NSCs (26, 29, 30). Peak calling of these public data sets was performed using MACS version 1.4.1 with default parameters.
Chromatin Immunoprecipitation Assay.
Cells grown on 10-cm dishes were fixed in 1% formaldehyde-containing medium for 10 min and were then quenched by 125 mM glycine for 5 min. The cells were washed with ice-cold PBS twice, and cold PBS containing protease inhibitor was added. The cells were then harvested using a cell scraper and centrifuged at 8,000 × g at 4 °C for 10 min. The cell pellet was suspended in 1 mL of buffer 1 (140 mM NaCl, 1 mM ethylenediamine tetraacetic acid [EDTA], 50 mM Hepes-KOH [pH 7.5], 0.5% Nonidet P-40, 10% glycerol, 0.25% Triton X-100, protease inhibitor) and kept at 4 °C for 10 min. After centrifugation, as mentioned previously, the supernatant was discarded and the pellet was suspended in 1 mL of buffer 2 (20 mM Tris⋅HCl [pH 7.5], 200 mM NaCl, 1 mM EDTA, 0.5 mM ethylene glycol tetraacetic acid [EGTA], protease inhibitor). After a third centrifugation, the supernatant was again discarded and the pellet suspended in 400 μL of buffer 3 (20 mM Tris⋅HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, protease inhibitor). After incubation at 4 °C for 10 min, the lysate was sonicated with a Branson sonicator (Branson Ultrasonic) and then centrifuged at 20,000 × g at 4 °C for 10 min. The supernatant was collected, and 10% of its volume was used for input.
For each ChIP reaction, 50 μL of Dynabeads M-280 sheep anti-mouse IgG (Life Technologies) were washed twice with PBS. Beads were collected magnetically and conjugated to 2 μg of primary antibody in 1.2 mL of PBS with protease inhibitor. The suspensions were rotated at 4 °C overnight, after which the beads were collected by a magnet and washed twice with 1 mL of ice-cold PBS and once with 1 mL of buffer 3 and then suspended in 600 μL of buffer 3. Sheared chromatin was added to the beads and allowed to react overnight at 4 °C with rotation. The immunoprecipitated samples were washed once with buffer 3, twice with buffer 4 (20 mM Tris⋅HCl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS), three times with radioimmunoprecipitation assay buffer (1 mM EDTA, 50 mM Hepes-KOH [pH 7.4], 1% Nonidet P-40, 0.25 M LiCl, 0.5% Na-deoxycholate), and once with TE50 buffer (50 mM Tris⋅HCl [pH 8.0], 10 mM EDTA). Immunoprecipitated DNAs or input DNAs were eluted by adding 100 μL of elution buffer (TE50 buffer, 1% SDS) and incubated at 65 °C overnight. Samples were supplemented with 100 μL of Tris-EDTA buffer (10 mM Tris⋅HCl [pH 8.0], 1 mM EDTA) and treated with RNase A at 37 °C for 1 h and proteinase K at 50 °C for 2 h. DNA was purified using a FastGene Gel/PCR Extraction Kit (Nippon Genetics) according to the manufacturer’s protocol. Mouse anti-FLAG antibody (Sigma) was used for ChIP-seq.
ChIP-seq and Data Analysis.
Deep sequencing of immunoprecipitated chromatin was performed using the Illumina HiSEq 2500 system, as described previously (51). Briefly, sequencing libraries were made using the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England Biolabs). Single-end 50-bp sequence reads were trimmed based on read length and read quality using the FASTX tool kit and aligned to the mouse genome (UCSC mm10) using Bowtie v2.2.4 (57). All redundant reads were removed from further analysis. SAM files were converted to the BAM format using SAMtools (v0.1.19) (58). Peak calling to detect Egr1-occupied loci was performed using MACS version 1.4.1 with default parameters and the input library as control (59). Genomic annotation of the regions identified from the ChIP-seq data were performed using GREAT (60). For boxplot analysis, the mouse genome was partitioned into 500-bp bins, and signal enrichment was calculated as reads per kilobase per million mapped reads. The consensus sequence was identified by importing a 200-bp sequence surrounding the summit of the peak of Egr1 into the MEME program (61). Egr1 binding loci that overlapped with open chromatin regions were calculated using BEDTools.
Voluntary Running.
For the voluntary running experiments, an Igloo Fast-Tracs running wheel system (Animec, K-3250) was placed in the home cage. Animals were weaned at P28 and randomly divided into four groups: Ctrl (pups received distilled water), Ctrl + RW (pups received distilled water, with a running wheel), MDZ (pups received MDZ), and MDZ + RW (pups received MDZ, with a running wheel).
Behavioral Tests.
Before performing behavioral tests, mice were handled for 3 min in the laboratory animal housing room and then transferred to the testing room and acclimated for at least 1 h. All experiments were performed sequentially using the same set of male mice and were conducted between 13:30 and 16:30. The level of background noise during behavioral testing was about 50 dB. After each trial, the apparatus was wiped clean.
Open Field Test.
The open field test was conducted as previously described (62). In brief, locomotor activity was measured for 10 min using an open field apparatus made of white plastic (50 × 50 × 30 [H] cm). An LED light system was positioned 50 cm above the center of the field (100 lx at the center of the field). Total distance traveled (centimeters), average moving speed (centimeters/seconds), and time spent in the central area (30% of the field) (seconds) were measured by Image OF4 software (O’Hara & Co., Ltd.).
NLR Task.
NLR was conducted as previously described (21, 63). In brief, mice were habituated to a white plastic chamber (50 × 50 × 30 [H] cm) for 10 min on each of the 3 d prior to the NLR task. After the 3-d habituation, for a familiarization session, each mouse was left in the testing chamber, in which two identical objects were placed in adjacent corners of the testing arena, and allowed to freely explore the objects for 5 min and then taken back to its home cage for 5 min. Before a testing session, one object was shifted to a new location near the corners in the box. In a testing session, the mouse was again placed in the testing chamber and allowed to explore the familiar and displaced objects for 5 min. All sessions were recorded with an overhead video tracking system, and exploration behavior was defined as activities such as whisking, sniffing, and rearing against the object. Time spent exploring each object during the familiarization and testing sessions was manually scored. Exploration ratios for novel located objects were calculated using the equation t novel /(t novel + t familiar), as described previously (63). The testing chamber and objects were washed with 70% ethanol before the next mouse was tested.
FC Test.
Contextual and cued FC tests were conducted as previously described (62). In brief, on the conditioning trial (Day 1), each mouse was placed in a conditioning chamber (17 × 10 × 10 cm, 100 lx) with clear acrylic walls and a shock-grid floor made of stainless steel rods (2 mm in diameter, spaced 5 mm apart) and allowed to explore freely for 1 min. A 65-dB tone was then presented for 30 s. During the final 2 s of tone presentation, a 0.5-mA foot shock was delivered. The set of tone and foot shock was repeated 3 times automatically using a tone and shock generator controlled by Time FZ software (O’Hara & Co., Ltd.). The next day (Day 2), to evaluate contextual fear memory, mice were placed in the same context arena without tone or shock for 5 min. On the third day (Day 3), to evaluate cued fear memory, mice were placed in a novel context chamber with a dim light (20 lx) and solid white plastic walls without a shock-grid floor and allowed to explore freely for 3 min. The conditioning tone was then presented for 3 min. Overhead infrared cameras recorded behavior, and the duration of freezing (i.e., absence of movement except for breathing) was automatically calculated using Image FZ2 software.
Quantification and Statistical Analysis.
Statistical analysis was performed using GraphPad Prism software (GraphPad Software). Data distribution was assumed to be normal, but this was not formally tested. Statistical analysis was done using unpaired t tests and one-way ANOVA with post hoc analysis using Tukey’s multiple comparison test. Data are presented as mean ± SEM. Results were considered significant when P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank H. Nakashima, S. Katada, and T. Imamura for discussions and Y. Nakagawa for excellent secretarial assistance. We appreciate technical assistance from the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas JP16H06527 and JP16K21734 (to K.N.), a Grant-in-Aid for Young Scientists (B) JP18K14820 (to T.M.), a GSK Japan Research Grant 2017 (to T.M.), a Grant-in-Aid for Scientific Research (B) JP21H02808 (to T.M.), and a Research Grant for Public Health Science (to H.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107596118/-/DCSupplemental.
Data Availability
All data and materials have been made available within the submitted material or in a public repository. The RNA-seq, ATAC-seq, and ChIP-seq data obtained in this study have been uploaded to National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE166322.
References
- 1.Wilder R. T., et al., Early exposure to anesthesia and learning disabilities in a population-based birth cohort. Anesthesiology 110, 796–804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprung J., et al., Attention-deficit/hyperactivity disorder after early exposure to procedures requiring general anesthesia. Mayo Clin. Proc. 87, 120–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jevtovic-Todorovic V., et al., Early exposure to common anesthetic agents causes widespread neurodegeneration in the developing rat brain and persistent learning deficits. J. Neurosci. 23, 876–882 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vutskits L., Xie Z., Lasting impact of general anaesthesia on the brain: Mechanisms and relevance. Nat. Rev. Neurosci. 17, 705–717 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Creeley C. E., Olney J. W., The young: Neuroapoptosis induced by anesthetics and what to do about it. Anesth. Analg. 110, 442–448 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Zhao C., Deng W., Gage F. H., Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Aimone J. B., et al., Regulation and function of adult neurogenesis: From genes to cognition. Physiol. Rev. 94, 991–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J., et al., Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao H., et al., Long-range GABAergic inputs regulate neural stem cell quiescence and control adult hippocampal neurogenesis. Cell Stem Cell 21, 604–617.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weir C. J., Mitchell S. J., Lambert J. J., Role of GABAA receptor subtypes in the behavioural effects of intravenous general anaesthetics. Br. J. Anaesth. 119 (suppl. 1), i167–i175 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Kain Z. N., et al., Trends in the practice of parental presence during induction of anesthesia and the use of preoperative sedative premedication in the United States, 1995–2002: Results of a follow-up national survey. Anesth. Analg. 98, 1252–1259 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa H., Yamazaki S., Abe T., Oda Y., Midazolam as a first-line agent for status epilepticus in children. Brain Dev. 22, 239–242 (2000). [DOI] [PubMed] [Google Scholar]
- 13.DiMaggio C., Sun L. S., Li G., Early childhood exposure to anesthesia and risk of developmental and behavioral disorders in a sibling birth cohort. Anesth. Analg. 113, 1143–1151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman K., et al., Isoflurane anesthesia has long-term consequences on motor and behavioral development in infant rhesus macaques. Anesthesiology 126, 74–84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J., et al., Early developmental exposure to repetitive long duration of midazolam sedation causes behavioral and synaptic alterations in a rodent model of neurodevelopment. J. Neurosurg. Anesthesiol. 31, 151–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai S., Takagi Y., Kaneko S., Kurosawa T., Effect of three types of mixed anesthetic agents alternate to ketamine in mice. Exp. Anim. 60, 481–487 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Youssef M., et al., Ablation of proliferating neural stem cells during early life is sufficient to reduce adult hippocampal neurogenesis. Hippocampus 28, 586–601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giri P. K., et al., Pretreatment with minocycline improves neurogenesis and behavior performance after midazolam exposure in neonatal rats. Neuroreport 29, 153–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J. J., Fanselow M. S., Modality-specific retrograde amnesia of fear. Science 256, 675–677 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Goodman T., et al., Young hippocampal neurons are critical for recent and remote spatial memory in adult mice. Neuroscience 171, 769–778 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Manning E. E., Ransome M. I., Burrows E. L., Hannan A. J., Increased adult hippocampal neurogenesis and abnormal migration of adult-born granule neurons is associated with hippocampal-specific cognitive deficits in phospholipase C-β1 knockout mice. Hippocampus 22, 309–319 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Phillips R. G., LeDoux J. E., Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 106, 274–285 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Shin J., et al., Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Y., et al., Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 20, 476–483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toda T., et al., Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21, 618–634.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Fraser J., et al., Common regulatory targets of NFIA, NFIX and NFIB during postnatal cerebellar development. Cerebellum 19, 89–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min I. M., et al., The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell 2, 380–391 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Dixon J. R., et al., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beagan J. A., et al., YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27, 1139–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martynoga B., et al., Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769–1786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starks R. R., Biswas A., Jain A., Tuteja G., Combined analysis of dissimilar promoter accessibility and gene expression profiles identifies tissue-specific genes and actively repressed networks. Epigenetics Chromatin 12, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veyrac A., et al., Zif268/egr1 gene controls the selection, maturation and functional integration of adult hippocampal newborn neurons by learning. Proc. Natl. Acad. Sci. U.S.A. 110, 7062–7067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R., et al., Id4 downstream of Notch2 maintains neural stem cell quiescence in the adult hippocampus. Cell Rep. 28, 1485–1498.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Madabhushi R., et al., Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Z., et al., EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 10, 3892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodges H. C., et al., Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 25, 61–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C., et al., Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev. Cell 45, 753–768.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Praag H., Kempermann G., Gage F. H., Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266–270 (1999). [DOI] [PubMed] [Google Scholar]
- 39.van Praag H., Shubert T., Zhao C., Gage F. H., Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci. 25, 8680–8685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih J., et al., Delayed environmental enrichment reverses sevoflurane-induced memory impairment in rats. Anesthesiology 116, 586–602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinn G. A., Sasaki Russell J. M., Banh E. T., Lee S. C., Sall J. W., Voluntary exercise rescues the spatial memory deficit associated with early life isoflurane exposure in male rats. Anesth. Analg. 129, 1365–1373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adam R. C., et al., NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nat. Cell Biol. 22, 640–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutta S., Sengupta P., Men and mice: Relating their ages. Life Sci. 152, 244–248 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Sorrells S. F., et al., Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Jiménez E. P., et al., Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi M., Saito H., Suzuki M., Mori K., Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport 11, 1991–1996 (2000). [DOI] [PubMed] [Google Scholar]
- 47.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 48.Sakai A., et al., Ectopic neurogenesis induced by prenatal antiepileptic drug exposure augments seizure susceptibility in adult mice. Proc. Natl. Acad. Sci. U.S.A. 115, 4270–4275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh J., et al., IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J. Cell Biol. 164, 111–122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uezono N., et al., Prior treatment with anti-high mobility group box-1 antibody boosts human neural stem cell transplantation-mediated functional recovery after spinal cord injury. Stem Cells 36, 737–750 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Matsuda T., et al., Pioneer factor NeuroD1 rearranges transcriptional and epigenetic profiles to execute microglia-neuron conversion. Neuron 101, 472–485.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Patel R. K., Jain M., NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7, e30619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D., et al., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLean C. Y., et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey T. L., et al., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202-W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juliandi B., et al., Reduced adult hippocampal neurogenesis and cognitive impairments following prenatal treatment of the antiepileptic drug valproic acid. Stem Cell Reports 5, 996–1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mumby D. G., Gaskin S., Glenn M. J., Schramek T. E., Lehmann H., Hippocampal damage and exploratory preferences in rats: Memory for objects, places, and contexts. Learn. Mem. 9, 49–57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials have been made available within the submitted material or in a public repository. The RNA-seq, ATAC-seq, and ChIP-seq data obtained in this study have been uploaded to National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE166322.