Graphical abstract
Keywords: Mitochondrial haplogroups, Alzheimer’s disease risk, Heterogeneity, Subgroup
Highlights
-
•
There is heterogeneity among subgroups of haplogroup J which influences AD risk.
-
•
The heterogeneity among haplogroup J influences the MCI-to-AD conversion risk.
-
•
The heterogeneity among subgroups of haplogroup J is independent of Aβ and p-tau.
Abstract
Introduction
The impact of mitochondrial haplogroups on Alzheimer’s disease (AD) risk has not been fully elucidated and warrants further investigation at the subgroup level.
Objectives
The aim of this research is to evaluate the association between mitochondrial haplogroups and AD risk in subgroups level.
Methods
In total, 809 AD Neuroimaging Initiative subjects were assessed using mtDNA sequencing, the AD Assessment Scale-Cognitive Subscale (ADAS-cog), hippocampal volume measurements, the hypometabolic convergence index (HCI), and MCI-to-AD conversion proportion measurements.
Results
The frequency of haplogroup J was significantly higher than that of other haplogroups in the AD group (p = 0.013). According to the correlation between haplogroup J-specific SNPs and ADAS-cog, haplogroup J was divided into four subgroups harboring exacerbating SNPs, protective SNPs, both exacerbating and protective SNPs, or irrelevant SNPs. The subgroups harboring exacerbating SNPs exhibited higher AD risk represented by the levels of ADAS-cog, hippocampal volume, HCI, and MCI-to-AD conversion proportion than other subgroups.
Conclusion
Heterogeneity existed among the subgroups of haplogroup J, which suggested that different subgroups exhibited different levels of AD risk. This study provides novel insights into the correlation between mitochondrial haplogroups and AD risk.
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by the progressive impairment of cognitive function [1], [2], [3]. The pathogenesis of AD has not been fully elucidated [2]. However, previous studies have highlighted the genetic heritability of AD [4], [5].
Several AD genetic risk factors have been identified, including the apolipoprotein E gene, which is a strong genetic risk factor for AD. Three rare mutations in amyloid beta precursor protein (APP), Presenilin 1 (PSEN1), and PSEN2 are usually observed and result in an early onset of AD [5], [6]. However, previous studies have suggested that the vast majority of AD cases are probably driven by several genomic variants [7]. Therefore, genome-wide association studies (GWAS) that aim to identify massive potential genomic risk factors have been conducted [7]. Presently, more than 30 genetic risk factors have been identified based on these GWAS studies [7], [8].
Most of these GWAS studies have focused on the association between AD and nuclear genetic risk factors. However, few studies have reported that certain nuclear genetic risk factors that encode mitochondria-related proteins may impact mitochondrial function directly or indirectly [8], [9], [10], [11]. According to the findings of these studies, mitochondrial dysfunction may play an important role in AD pathogenesis. Mitochondrial dysfunction increases ROS generation, leading to oxidative damage, a key feature that precedes widespread plaque pathology in the brains of patients with AD [10]. As thirteen mitochondrial electron transport chain proteins are transcribed from the circular mitochondrial genome (mtDNA), which additionally contributes two ribosomal RNAs (12S and 16S rRNAs, backbone of the mitoribosome) and twenty-two tRNAs to the organelle’s own translational system, mitochondrial dysfunction in AD may arise, at least in part, from the existence of somatic or inherited mtDNA variants [8], [10], [12], [13]. However, further studies are warranted to provide more evidence for this theory. Edland et al. found a high mother-to-father ratio among affected parents of the subjects with AD [14]. Moreover, mtDNA polymorphisms occur more frequently in subjects with AD [15], [16], [17], [18]. Therefore, the association between mtDNA variants and AD risk should be further investigated.
Variations in mtDNA are often described by established haplotype groups [19]. A human mitochondrial haplogroup refers to a unique set of mtDNA polymorphisms, reflecting mutations accumulated by a discrete maternal lineage [20]. Mitochondrial haplogroups are represented by a single letter (e.g., H, V, and L), while mitochondrial subgroups are further defined by additional numbers/letters (e.g., H51A1 and L3) [19]. Several previous studies have investigated the association between the mitochondrial haplogroup and AD risk and suggested that mitochondrial haplogroups U, H, J, and so on may be significant AD risk factors [8], [19], [21]. However, the conclusions from mitochondrial haplogroup-related AD risk factor studies may be discrepant. Maruszak et al. reported that cluster JT seems to be a protective factor for patients with AD, while Tranah et al. suggested that individuals with haplogroup J were more likely to perform worse on the MMSE test [22], [23]. The reasons for mitochondrial haplogroup-related studies lacking inter-study agreement are currently unknown. However, these discrepancies contribute towards mitochondrial genetic variation to AD risk inconclusively. To clarify the association between mitochondrial haplogroups and AD risk, the reasons for mitochondrial haplogroup-related studies lacking inter-study agreement should be investigated. We then aimed to investigate whether the heterogeneity among subgroups of haplogroups, which influences AD risk, was explored in previous studies lacking inter-study agreement. Investigation at the subgroup level is beneficial to clarify the association between mitochondrial haplogroups and AD risk.
To test this hypothesis, a more comprehensive analysis of the association between the mitochondrial haplogroup and AD risk is performed at the subgroup level. In this study, we not only analyzed mitochondrial haplogroup frequencies from mtDNA-sequenced members of the AD Neuroimaging Initiative (ADNI) cohort, but also investigated the association between AD-related biomarkers through the AD Assessment Scale (ADAS), hippocampal volume measurements, and hypometabolic convergence index (HCI). This study provides greater insight into the contribution of mitochondrial haplogroups to AD risk.
Methods
Ethics statement
All experiments involving human patients were conducted according to the ethical policies and procedures approved by the Resource Allocation Review Committee (RARC), consisting of members independent of ADNI, approved by the NIA, and chaired by Dr. Tom Montine at the University of Washington, Seattle, Washington, USA (NO. U19 AG024904).
ADNI cohort
ADNI was launched in 2003 as a public-private partnership led by Michael W. Weiner, MD [8]. The primary goal of ADNI is the investigation of the possibility of combining serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment for determination of the progression of MCI and early onset of AD [8]. Descriptive and demographic characteristics of the ADNI participants were recorded and made accessible. ADNI is a longitudinal multicenter clinical study [8]. ADNI participants are classified as AD, MCI, or norm aging, and the group assignment can change over time. In this study, a subset of ADNI participants who underwent complete mtDNA sequencing (n = 809) were analyzed.
Cognitive function test
In this study, AD Assessment Scale-Cognitive Subscale (ADAS-Cog) was used to assess the cognitive function of ADNI participants. The ADAS is a brief cognitive test battery that is used to assess learning and memory, language production, language comprehension, constructional praxis, ideational praxis, and orientation [24]. The test items related to the cognitive test of the ADAS are provided in the order indicated. The word recall test is conducted first and the word recognition task test is conducted subsequently, which enables the subjects to distinguish between the words provided in the two tasks without considerable confusion [24]. Following completion of the objective testing, subjective clinical ratings of language ability and the ability to remember test instructions are performed by the examiner. ADAS-cog was administered at baseline and at 6, 12, 18, 24, and 36 months.
MRI scanning
All study subjects were subjected to 1.5 Tesla MRI scans conducted at regular intervals throughout the study [24]. The MRI scans for the ADNI participants were conducted at baseline and at 6, 12, 18, 24, and 36 months. The raw MRI imaging data were processed and analyzed using FreeSurfer 4.3 by the UCSF team. This processing includes multiple steps as follows: (1) motion correction and averaging of multiple volumetric T1-weighted images (when more than one is available) [25]; (2) removal of non-brain tissue by performing a hybrid watershed/surface deformation procedure; (3) automated Talairach transformation [25]; (4) segmentation and normalization of the intensity of the subcortical white matter and deep gray matter volumetric structures (including hippocampus, amygdala, caudate, putamen, and ventricles) [25]; (5) characterization of the tessellation of the gray matter-white matter boundary [25]; (6) automated topology correction and surface deformation following intensity gradients for optimal placement of the gray/white and gray/cerebrospinal fluid borders where the greatest shift in intensity defines the transition to the other tissue class [25].
Fluorodeoxyglucose (FDG)-PET scanning
In total, 50% of the subjects underwent fluorodeoxyglucose (FDG) PET scans completed at regular intervals throughout the study in addition to 1.5 Tesla MRI scans [24]. The FDG-PET scan was conducted at baseline and at 6, 12, 24, and 36 months. In this study, a global index for FDG-PET, termed the HCI, which summarizes voxel-wise information into a single measurement, was used. HCI represents the level of hypometabolism in an FDG-PET image [26]. HCI was calculated based on FDG-PET imaging by Dr. Koeppe’s team at the University of Michigan. Briefly, this process contains several steps as follows: first, to generate the t-score, the normalized FDG-PET scan images acquired by SPM (Wellcome Trust Center for Neuroimaging, UCL, UK) in probable AD patients were compared with those of NCs; second, the t-score map was transformed to the z-score map through the above-mentioned method; finally, to calculate HCI, all voxels whose z-scores were negative in both maps were summed, and the results were divided by 10,000 [26].
Cerebral spinal fluid (CSF) biomarkers
CSF samples were collected from all subjects who provided consent at the Baseline and M12 Visit [24]. All biomarker samples were collected in the morning before breakfast and after an overnight fast [24]. Only consumption of water was permitted until blood was drawn and the lumbar puncture was completed. In this study, three widely accepted AD biomarkers, namely , total tau (t-tau), and phosphorylated tau (p-tau), were analyzed. All CSF , t-tau, and p-tau181 concentrations were measured using a micro-bead-based multiplex immunoassay, the INNO-BIA AlzBio3 RUO test (Fujirebio, Ghent, Belgium) using the Luminex platform [27].
mtDNA sequencing and annotation
Whole genome sequencing conducted with support from the Brin-Wojcicki Foundation and the Alzheimer’s Association was performed using samples obtained from 818 subjects from the ADNI study by Illumina’s non-CLIA laboratory at roughly 30–40 x coverage in 2012 and 2013. The mtDNA sequence was obtained by remapping the original whole-genome sequencing data. Using the original mappings, all reads that mapped to the mitochondrial genome or those that were unmapped were extracted using SAMTools and mapped to NC_01292 using Burrows-Wheeler Aligner [28], [29]. Next, local realignments were performed around indels and base recalibration with GATK to refine the mappings. Finally, FreeBayes (-p 1 –F 0.6, and removed variants with quality less than 20) were used to join call variants and convert the resulting VCF file to FASTA with vcf2fasta (vcflib, https://github.com/vcflib/vcflib) [24], [30]. Mitochondrial variants for each sample were annotated through 9228 mitochondrial DNA coding and RNA sequence variants and 2792 control region variants downloaded from MITOMAP [31]. Mitochondrial haplotypes were annotated with Phy-Mer [32]. Phy-Mer is used to report the five most likely mitochondrial haplotypes and a score, where 1 is a perfect score. For each of the samples, the top hit was selected [24]. All samples presented with scores > 0.99, except one sample that presented with a score of 0.988 [24].
Statistical analysis
Mean and standard deviation values were calculated for the descriptive statistics of the continuous measures. Frequencies and relative frequencies comprising haplogroup frequencies, the proportion of AD subjects, and the proportion of MCI-to-AD conversion were determined for the descriptive statistics of categorical measures. For continuous measures, including ADAS-cog, hippocampal volumes, HCI, Aβ levels, and p-tau/tau ratio, the ANONA test was performed to compare the mean values among different groups. The objective of performing the ANOVA test was to estimate the differences among the subject groups based on the degree of variation of a particular measurement [33]. Haplogroup frequencies, the proportion of AD subjects, and MCI-to-AD conversion were compared among different groups using the Fisher’s exact test, which is a commonly used and conceptually attractive approach for evaluating the difference in observed frequencies among different groups. Forest plots were generated to represent the differences in proportions of subjects with AD and MCI-to-AD conversion among different groups, which display the odds ratios (ORs) with corresponding 95% confidence intervals. Furthermore, the association between subgroups of different haplogroups and cognitive function was evaluated. For this evaluation, LASSO regression was used to select subgroups related to ADAS-cog. LASSO regression is a popular technique for feature selection that can exclude irrelevant features by shrinking coefficients to zeros [34]. A least angle regression algorithm is used to solve LASSO regression [35]. We analyzed the association between subgroups of haplogroups related to AD risk and ADAS-cog using LASSO regression, which shrunk the coefficients of irrelevant subgroups to zero and removed the irrelevant subgroups.
Results
ADNI cohort demography
Samples obtained from 818 subjects in the ADNI cohort were analyzed by performing whole genome sequencing. However, samples from nine subjects were removed due to failure in quality control. Therefore, 809 subjects were included in this study. The demography information of this subset of ADNI is presented in Table A1 (supplementary materials). The subjects used in this study were divided into three groups as follows: AD (n = 48), MCI (n = 480), and control (n = 281) groups. The proportion of males in the MCI (58%) and control (48%) groups was not significantly different (p > 0.05). The proportion of males in the MCI (58%) and control (48%) groups exceeded that of the AD group (38%). The age of the subjects in these three groups was similar.
The AD group has higher haplogroup J frequency
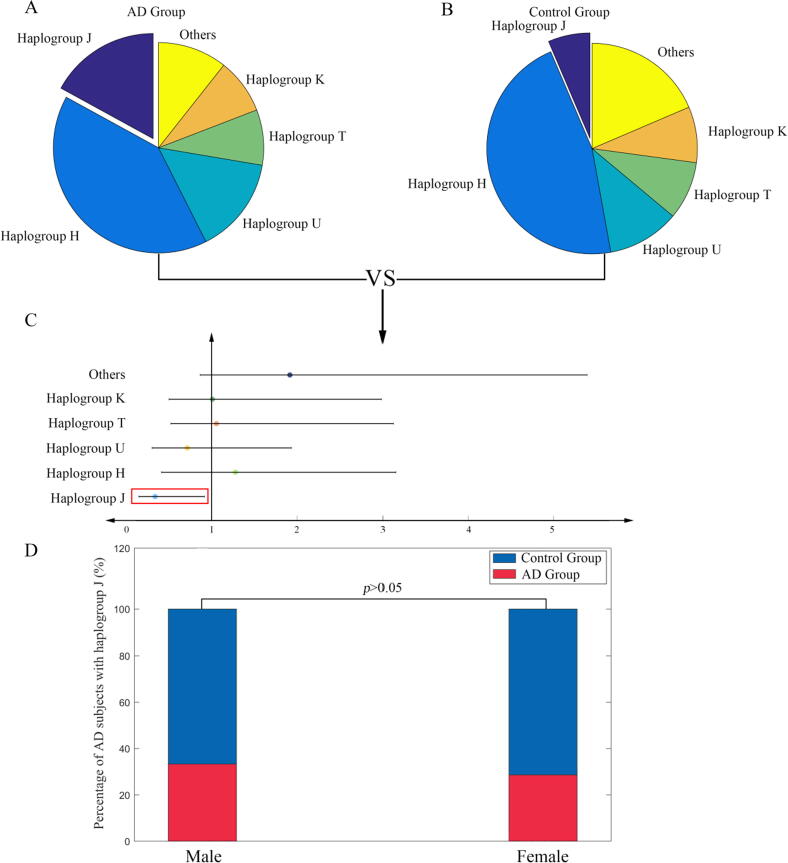
First, we analyzed the different frequencies of mitochondrial haplogroups in the ADNI cohort. In this subset of the ADNI cohort (n = 809), 15 different types of mitochondrial haplogroups were observed. The frequencies of five mitochondrial haplogroups (haplogroups J, H, U, T, and K) exceeded 5%. The frequencies of 10 mitochondrial haplogroups (haplogroups R, I, L, A, N, B, V, W, X, and M) were less than 5%, which were merged into the other category in the following statistical analysis. To test whether there was a difference in the frequencies of mitochondrial haplogroups between the AD (n = 48) and control (n = 281) groups, the Fisher’s exact test was performed and a forest plot was generated (shown in Fig. 1). Our results showed that the frequency of haplogroup J in the AD group exceeded that in the control group (OR = 0.33, 95% CI 0.19–0.58, p = 0.013), suggesting that haplogroup J might be related to AD risk. Furthermore, to investigate the gender specificity of AD risk among subjects with haplogroup J, the proportion of subjects with AD carrying haplogroup J among males and females was compared. The results showed that the proportion of subjects with AD carrying haplogroup J was not significantly different between male and female subjects (p > 0.05, compared by the Fisher’s exact test). This suggests that no gender specificity exists in subjects with AD carrying haplogroup J.
Fig. 1.
A: The distribution of haplogroups in the Alzheimer’s disease (AD) group. B: The distribution of haplogroups in the control group. C: Forest plots showing odds ratios (control group vs AD group) and 95th percentile confidence intervals for haplogroups. The differences in subjects with AD among different haplogroups were compared by the Fisher’s exact test. D: The proportion of subjects with AD carrying haplogroup J in males and females. The difference of frequencies in males and females were compared by the Fisher’s exact test.
The cognitive function of subjects with haplogroup J
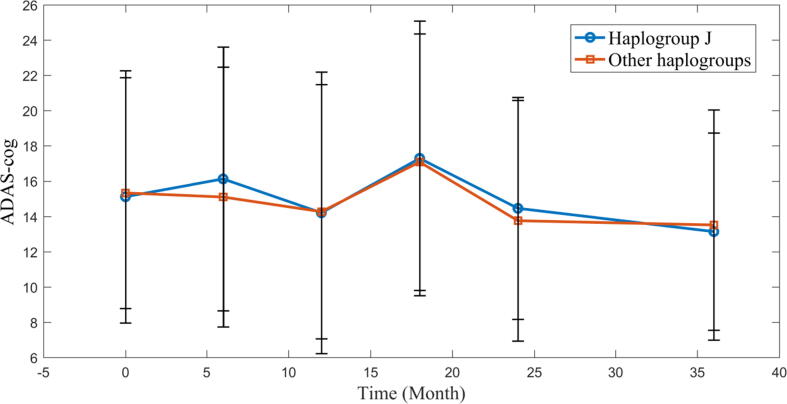
The above-mentioned results suggest that presence of the haplogroup J is related to AD risk. Furthermore, we hypothesized that patients with AD or MCI with haplogroup J might present with worse cognitive function than those with other haplogroups. To test our hypothesis, the cognitive function of patients with AD and MCI with haplogroup J and other haplogroups were compared (n = 528). The results (Fig. 2) showed that the ADAS-cog levels in these two groups were not significantly different. Haplogroup J, an AD risk factor, did not influence cognitive function, which seemed counterintuitive. To investigate this aspect, a new hypothesis was proposed wherein presence of heterogeneity among the subgroups of haplogroup J was considered, which might explain the cognitive function difference observed between patients with haplogroup J and patients with other haplogroups.
Fig. 2.
The time course of AD Assessment Scale-Cognitive Subscale (ADAS-cog) in the haplogroup J and other haplogroups. The ADAS-cog between haplogroup J and other haplogroups were compared by ANOVA. There is no significant difference between these two groups.
The heterogeneity in subgroups of haplogroup J
To test the hypothesis of heterogeneity in subgroups of haplogroup J, we investigated the association between different subgroups of haplogroup J and ADAS-cog in AD/MCI subjects (n = 69). However, in the ADNI cohort, the subjects with haplogroup J were distributed into multiple subgroups dispersedly and each subgroup contained few samples, thus rendering impossible direct analysis of their association with ADAS-cog. Therefore, a statistical strategy was considered in this study (Fig. 3). As the mitochondrial haplogroups are determined by analysis of mtDNA SNPs, data on the haplogroup J-specific mtDNA SNPs were extracted from the WGS dataset. Our aim was to investigate the heterogeneity in subgroups of haplogroup J. Hence, SNPs that were commonly observed in subjects with haplogroup J (frequency > 90%) were excluded. To avoid potential statistical bias, the SNPs that were rare in subjects with haplogroup J (frequency < 5%) were also excluded. Thereafter, data on 66 haplogroup J-specific mtDNA SNPs were extracted (presented in Supplementary Materials Table A2). Second, the correlation between these SNPs and ADAS-cog was evaluated by performing LASSO regression. The results showed that 16 SNPs were positively associated with ADAS-cog (termed as exacerbating SNPs), 13 were negatively associated with ADAS-cog (termed as protective SNPs), and 37 were irrelevant to ADAS-cog (termed as irrelevant SNPs). According to the results of LASSO regression, the subgroups were regrouped into the four following clusters: subgroups harboring exacerbating SNPs (38%, n = 26), subgroups harboring protective SNPs (22%, n = 15), subgroups harboring both exacerbating and protective SNPs (16%, n = 11), and subgroups harboring irrelevant SNPs (25%, n = 17) (Supplementary Materials Table A3). We then compared the ADAS-cog among these four clusters (Fig. 3B). Subgroups harboring exacerbating SNPs had significantly higher ADAS-cog levels than those in the other three clusters. The ADAS-cog levels in subgroups harboring both exacerbating and protective SNPs and those harboring irrelevant SNPs were relatively similar. The subgroups harboring protective SNPs had lower ADAS-cog levels than those in the other three clusters.
Fig. 3.
A: The LASSO regression coefficient between mtDNA SNPs and ADAS-cog. B: The time course of ADAS-cog among four subgroup clusters of haplogroup J. C: Comparison of ADAS-cog levels among four subgroup clusters of haplogroup J. D: Comparison of ADAS-cog levels between subgroups harboring exacerbating SNPs. E: Comparison of ADAS-cog levels between subgroups harboring irrelevant SNPs. F: Comparison of ADAS-cog levels between subgroups harboring protective SNPs. G: Comparison of ADAS-cog levels between subgroups harboring both exacerbating and protective SNPs. H: Comparison of ADAS-cog levels among subgroup clusters in haplogroup J and other haplogroups. The levels of ADAS-cog in different groups were compared by ANOVA. ** p < 0.01, * p < 0.05.
To gain further insight into the heterogeneity in subgroups of haplogroup J, we compared the above-mentioned four clusters with the other haplogroups. The results are shown in Fig. 3C-H. The ADAS-cog level was significantly higher in subgroups harboring exacerbating SNPs than that in the other haplogroups. Subgroups harboring both exacerbating and protective SNPs and those harboring irrelevant SNPs exhibited similar ADAS-cog levels to the other haplogroups. The subgroups harboring protective SNPs had lower ADAS-cog levels than those in the other haplogroups.
These results suggest that heterogeneity exists in subgroups of haplogroup J. Different subgroups exhibit different levels of AD risk, and subgroups harboring exacerbating SNPs play a more important role in AD risk.
Image evidence for heterogeneity in subgroups of haplogroup J
To provide further evidence to validate the hypothesis of heterogeneity in subgroups of haplogroup J, we analyzed the MRI image and FDG-PET data (n = 69). The hippocampal volume was selected as the MRI image biomarker. First, the hippocampal volumes in the four clusters were compared with each other (Fig. 4A-F). Subgroups harboring exacerbating SNPs exhibited more decreased hippocampal volumes than the other three clusters. The subgroups harboring both exacerbating and protective SNPs had similar hippocampal volumes to subgroups harboring irrelevant SNPs. The hippocampal volumes in subgroups harboring protective SNPs were higher than those in the other three clusters. To gain more conclusive evidence, the hippocampal volumes of the above-mentioned four clusters were compared with those of other haplogroups. The hippocampal volumes in the subgroups harboring exacerbating SNPs decreased significantly compared to those of other haplogroups. The subgroups harboring protective SNPs showed remarkably increased hippocampal volumes compared with those of other haplogroups. The subgroups harboring both exacerbating and protective SNPs and those harboring irrelevant SNPs had similar hippocampal volumes with other haplogroups.
Fig. 4.
A: Comparison of the hippocampal volume levels among four subgroup clusters of haplogroup J. B: Comparison of hippocampal volume levels between subgroups harboring exacerbating SNPs. C: Comparison of hippocampal volume levels between subgroups harboring irrelevant SNPs. D: Comparison of hippocampal volume levels between subgroups harboring protective SNPs. E: Comparison of hippocampal volume levels between subgroups harboring both exacerbating and protective SNPs. F: Comparison of hippocampal volume levels among subgroup clusters in haplogroup J and other haplogroups. G: Comparison of hypometabolic convergence index (HCI) levels among four subgroup clusters of haplogroup J. H: Comparison of HCI levels between subgroups harboring exacerbating SNPs. I: Comparison of HCI levels between subgroups harboring irrelevant SNPs. J: Comparison of HCI levels between subgroups harboring protective SNPs. K: Comparison of HCI levels between subgroups harboring both exacerbating and protective SNPs. L: Comparison of HCI levels among subgroup clusters in haplogroup J and other haplogroups. The levels of hippocampal volume and HCI in different groups were compared by ANOVA. ** p < 0.01, * p < 0.05.
Furthermore, we analyzed the FDG-PET results to gain more insights into the heterogeneity in subgroups of haplogroup J. In this study, a global index for FDG-PET, termed as HCI, was selected to quantify the cerebral metabolic variation in subjects with ADNI. With higher HCI, the subjects may develop more severe hypometabolism throughout the brain. Although ADNI provides HCI data at multiple time points, data at these time points (except baseline) contain markedly few samples with haplogroup J (less than five samples). Therefore, to avoid potential statistical bias, HCI data at any time point that were obtained after analysis of less than five samples with haplogroup J were excluded. Finally, only HCI data at baseline were included in this study. First, the HCIs in the four clusters of haplogroup J were compared with each other (Fig. 4G-L). The subgroups harboring protective SNPs had significantly lower HCI levels compared with those in the other three clusters. Second, the four clusters of haplogroup J were also compared with other haplogroups. The subgroups harboring both exacerbating and protective SNPs, those harboring irrelevant SNPs, and those harboring exacerbating SNPs had similar HCI levels to other haplogroups. The HCI levels in subgroups harboring protective SNPs decreased significantly compared with those of other haplogroups.
In summary, our results suggest that the levels of hippocampal volumes and HCI among different subgroups of haplogroup J and other haplogroups were significantly different. These results revealed the physiological basis for the difference among different subgroups, which provided further evidence for validation of the hypothesis on the heterogeneity in subgroups of haplogroup J.
Different MCI-to-AD conversion proportions in different subgroups in haplogroup J
To obtain further insights into the impact of different subgroups in haplogroup J on AD risk, the MCI-to-AD conversion proportions in different subgroups were investigated (n = 480). First, the different MCI-to-AD conversion proportions in the four clusters of haplogroup J were compared. The results (Fig. 5A-G) showed that subgroups harboring both exacerbating and protective SNPs and subgroups harboring exacerbating SNPs had a significantly higher MCI-to-AD conversion proportion than that in other subgroups. Second, the four clusters of haplogroup J were compared to those of other haplogroups. The results showed that the MCI-to-AD conversion proportion in the subgroups harboring both exacerbating and protective SNPs and subgroups harboring exacerbating SNPs increased significantly compared with that of other haplogroups. These results show that the MCI-to-AD conversion proportion varies in different subgroups of haplogroup J, suggesting that the heterogeneity of haplogroup J is also associated with MCI-to-AD conversion risk.
Fig. 5.
A: Comparison of the MCI-to-AD conversion proportion among four subgroup clusters of haplogroup J. B: Comparison of the MCI-to-AD conversion proportion between subgroups harboring exacerbating SNPs. C: Comparison of the MCI-to-AD conversion proportion between subgroups harboring irrelevant SNPs. D: Comparison of the MCI-to-AD conversion proportion between subgroups harboring protective SNPs. E: Comparison of the MCI-to-AD conversion proportion between subgroups harboring both exacerbating and protective SNPs. F: Comparison of the MCI-to-AD conversion proportion among subgroup clusters in haplogroup J and other haplogroups. G: Forest plots showing odds ratios (subgroup clusters in haplogroup J vs. other haplogroups) and 95th percentile confidence intervals for haplogroups. The frequencies of MCI-to-AD conversion in different groups were compared by the Fisher’s exact test. ** p < 0.01, * p < 0.05.
The independence of heterogeneity in haplogroup J to Aβ and tau
We explored whether the heterogeneity in subgroups of haplogroup J was Aβ- or tau-dependent. To investigate this, the levels of CSF Aβ and the p-tau/tau ratio in different subgroups of haplogroup J were compared (n = 528). As the number of samples in the subgroups harboring protective SNPs and those harboring irrelevant SNPs was less than five, only those harboring both exacerbating and protective SNPs and exacerbating SNPs alone as well as other haplogroups were included. The results (Fig. 6) showed that the levels of CSF Aβ and the p-tau/tau ratio had no significant difference in different groups, suggesting that heterogeneity in haplogroup J was independent of Aβ and tau.
Fig. 6.
A: Cerebral spinal fluid (CSF) Aβ levels in different subgroup clusters and other haplogroups. B: CSF p-tau and total tau ratio levels in different subgroups clusters and other haplogroups. The levels of Aβ and the p-tau/tau ratio in different groups were compared by ANOVA.
Discussion
In this study, the heterogeneity of subgroups of haplogroup J was investigated, which suggested that different subgroups exhibited different levels of AD risk. This study is beneficial to elucidate the association between mitochondrial haplogroups and AD risk. As heterogeneity exists among subgroups of haplogroups, the association between the mitochondrial haplogroup and AD risk may be ambiguous. The discovery of heterogeneity necessitates conduction of subgroup-level research that can recognize the risk factor accurately from data comprising a mixture of factors analyzed. Additionally, the heterogeneity hypothesis provides a novel explanation for AD risk research involving the discovery of mitochondria haplogroups lacking inter-study agreement.
This study highlights the impact of mitochondrial haplogroup J on AD risk and the heterogeneity among subgroups of haplogroup J. Several previous studies have reported an association between haplogroup J and AD risk. However, the results of these studies seem to be inconsistent. Tranah et al. and Swerdlow et al. have suggested that haplogroup J is a risk factor for AD [8], [23]. Maruszak et al. have reported that haplogroup J is a protective factor for AD [22]. Andrews et al. have reported that a significant association between haplogroup J and AD risk does not exist [36]. The discrepancy in these results may be explained by the heterogeneity among subgroups of haplogroup J. J1B1-related subgroups J1B1A1, J1B1A1B, and J1B1B2 are protective subgroups. This finding is consistent with that of a study conducted by Maruszak et al., which highlights that subgroup J1B1 plays a protective role against AD [22]. Otherwise, the cohort containing mixed types of subgroups may counteract the differences among haplogroups. The same phenomenon has been reported in several previous studies that could not indicate an association between haplogroup J and AD [37], [38]. Nevertheless, Tranah et al. observed a more severe cognitive decline in the haplogroup J cohort, which might comprise a high proportion of subgroups harboring exacerbating SNPs [23]. In summary, the discrepancy in the results reported by previous studies on the association between mitochondrial haplogroups and AD risk may be attributed to the heterogeneity of subgroups in haplogroups. The cohort used in studies containing different proportions of different subgroups may report different conclusions. Therefore, it is essential to analyze the association between mitochondrial haplogroups and AD risk at the subgroup level to ascertain the existence of heterogeneity among subgroups.
As the bias of haplogroups/subgroups is attributed to a set of mtDNA SNPs, the biological basis of heterogeneity among subgroups may rely on the different mtDNA SNPs harbored by different subgroups. Therefore, studies conducted to investigate heterogeneity among subgroups also provide novel insights into the association of the single mtDNA SNP with AD risk. For example, the mtDNA SNP C242T, one of the defining SNPs for the subgroup J1B1A, is recognized as a protective SNP [39]. If a study is conducted to investigate whether a single mtDNA SNP C242T protects against AD, the cohort, including J1B1A and its subgroups (e.g., J1B1A1, J1B1A1A, J1B1A1B, and J1B1A1C) may be analyzed. However, heterogeneity among these subgroups may affect the results. For example, the SNP C5463T, one of the defining SNPs for subgroup J1B1A1A, is an exacerbating SNP, suggesting that subjects in subgroup J1B1A1A harbor both exacerbating and protective SNPs [39]. The cognitive function of subjects in these subgroups was significantly different compared with those of the other subgroups, such as J1B1A1 and J1B1A1B, which harbored protective SNPs only. Therefore, studies using cohorts with different proportions of different subgroups may result in reporting of controversial conclusions. A similar issue might have been encountered in other previous studies. Hutchin et al. reported that the mtDNA 4336G mutation was related to AD risk in a North American cohort [16]. However, two other studies using different cohorts did not confirm these results [40], [41]. The bias of heterogeneity among subgroups is attributed to a set of mtDNA SNPs, and studies involving single mtDNA SNPs may inevitably result in the formation of different types of subgroups. Therefore, even for studies at the single SNP level, subgroup analysis is essential.
A distinct statistical strategy has been developed for conducting subgroup-level research. Statistical analysis at the subgroup level is extremely challenging for samples distributed in multiple subgroups dispersedly. The dispersed distribution of samples renders it impossible to analyze a single subgroup using traditional statistical analysis. Even if the sample size is extended, this issue cannot be addressed satisfactorily because to ensure that each subgroup comprises sufficient samples, the total sample size needs to be increased by multiple folds, and this may be difficult. To address this issue, a distinct statistical strategy was developed to enrich the samples at the subgroup level by merging multiple subgroups into clusters. This merging process was performed based on haplogroup-specific SNPs. These SNPs were divided into three categories (exacerbating SNPs, protective SNPs, and irrelevant SNPs) by performing LASSO regression. According to the above-mentioned classification, different subgroups can be merged into four clusters (subgroups harboring exacerbating SNPs, subgroups harboring protective SNPs, subgroups harboring both exacerbating and protective SNPs, and subgroups harboring irrelevant SNPs), which can ensure that each cluster contains sufficient samples for statistical analysis with a relatively small total sample size. Our statistical strategy can be adjusted for application in other subgroup level studies, which may facilitate the investigation of the association between mitochondrial haplogroups and AD at the subgroup level.
There are two limitations of this study. First, few subgroups of haplogroup J were not observed in the ADNI cohort. The characteristics of the remaining subgroups remain to be further investigated. Second, this study focused on the heterogeneity among subgroups of haplogroup J. The heterogeneity among subgroups of other haplogroups remains to be elucidated.
Conclusions
This study highlighted the association between haplogroup J and AD risk at the subgroup level. The heterogeneity among subgroups of haplogroup J was revealed, which suggested that different subgroups exhibited different levels of AD risk. The heterogeneity and association between haplogroup J and AD risk are independent of Aβ/p-tau levels. This study provides novel insights into the association between mitochondrial haplogroups and AD risk.
Compliance with ethics requirements
All experiments involving human patients were conducted according to the ethical policies and procedures approved by the Resource Allocation Review Committee (RARC), consisting of members independent of ADNI, approved by the NIA, and chaired by Dr. Tom Montine at the University of Washington, Seattle, Washington, USA (U19 AG024904).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study was supported by the Natural Science Foundation of China (nos. 81903703 and 81773806), “Double First-Class” University project (no. CPU2018GY19).
Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Rev December 5, 2013, Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego, USA. ADNI data were disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.02.003.
Contributor Information
HaoChen Liu, Email: haochenliu@cpu.edu.cn.
Yixuan Zhang, Email: yxzhang@stu.cpu.edu.cn.
XiaoQuan Liu, Email: lxq@cpu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Asadbegi M., Yaghmaei P., Salehi I., Komaki A., Ebrahim-Habibi A. Investigation of thymol effect on learning and memory impairment induced by intrahippocampal injection of amyloid beta peptide in high fat diet- fed rats. Metab Brain Dis. 2017;32(3):827–839. doi: 10.1007/s11011-017-9960-0. [DOI] [PubMed] [Google Scholar]
- 2.Ramezani M., Komaki A., Hashemi-Firouzi N., Mortezaee K., Faraji N., Golipoor Z. Therapeutic effects of melatonin-treated bone marrow mesenchymal stem cells (BMSC) in a rat model of Alzheimer's disease. J Chem Neuroanat. 2020;108:101804. doi: 10.1016/j.jchemneu.2020.101804. [DOI] [PubMed] [Google Scholar]
- 3.Shahidi S., Zargooshnia S., Asl S.S., Komaki A., Sarihi A. Influence of N-acetyl cysteine on beta-amyloid-induced Alzheimer's disease in a rat model: a behavioral and electrophysiological study. Brain Res Bull. 2017;131:142–149. doi: 10.1016/j.brainresbull.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Bellou E., Baker E., Leonenko G., Bracher-Smith M., Daunt P., Menzies G. Age-dependent effect of APOE and polygenic component on Alzheimer's disease. Neurobiol Aging. 2020;93:69–77. doi: 10.1016/j.neurobiolaging.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Yang J., Schneider J.A., De Jager P.L., Bennett D.A., Zhang H.-Y. Genome-wide interaction analysis of pathological hallmarks in Alzheimer's disease. Neurobiol Aging. 2020;93:61–68. doi: 10.1016/j.neurobiolaging.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strittmatter W.J., Saunders A.M., Schmechel D., Pericakvance M., Enghild J., Salvesen G.S. APOLIPOPROTEIN-E - HIGH-AVIDITY BINDING TO BETA-AMYLOID AND INCREASED FREQUENCY OF TYPE-4 ALLELE IN LATE-ONSET FAMILIAL ALZHEIMER-DISEASE. PNAS. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram L., Tanzi R.E. Alzheimer disease risk genes: 29 and counting. Nat Rev Neurol. 2019;15(4):191–192. doi: 10.1038/s41582-019-0158-4. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow R.H., Hui D., Chalise P., Sharma P., Wang X., Andrews S.J. Exploratory analysis of mtDNA haplogroups in two Alzheimer's longitudinal cohorts. Alzheimer's & dementia: J Alzheimer's Assoc. 2020;16(8):1164–1172. doi: 10.1002/alz.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang S.J., Ma T.R., Miranda R.D., Balestra M.E., Mahley R.W., Huang Y.D. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. PNAS. 2005;102(51):18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleck D., Phu L., Verschueren E., Hinkle T., Reichelt M., Bhangale T. PTCD1 is required for mitochondrial oxidative-phosphorylation: possible genetic association with Alzheimer's Disease. J Neurosci. 2019;39(24):4636–4656. doi: 10.1523/JNEUROSCI.0116-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pa J., Andrews S.J., Swerdlow R.H. Mitochondria and Alzheimer's: Is PTCD1 the smoking gun? Trends Neurosci. 2019;42(11):759–762. doi: 10.1016/j.tins.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallberg B.M., Larsson N.G. Making proteins in the powerhouse. Cell Metab. 2014;20(2):226–240. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Rackham O., Mercer T.R., Filipovska A. The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley Interdisciplinary Reviews-Rna. 2012;3(5):675–695. doi: 10.1002/wrna.1128. [DOI] [PubMed] [Google Scholar]
- 14.Edland S.D., Silverman J.M., Peskind E.R., Tsuang D., Wijsman E., Morris J.C. Increased risk of dementia in mothers of Alzheimer's disease cases: evidence for maternal inheritance. Neurology. 1996;47(1):254–256. doi: 10.1212/wnl.47.1.254. [DOI] [PubMed] [Google Scholar]
- 15.Brown M.D., Shoffner J.M., Kim Y.L., Jun A.S., Graham B.H., Cabell M.F. Mitochondrial DNA sequence analysis of four Alzheimer's and Parkinson's disease patients. Am J Med Genet. 1996;61(3):283–289. doi: 10.1002/(SICI)1096-8628(19960122)61:3<283::AID-AJMG15>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Hutchin T., Cortopassi G. A MITOCHONDRIAL-DNA CLONE IS ASSOCIATED WITH INCREASED RISK FOR ALZHEIMER-DISEASE. PNAS. 1995;92(15):6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin F.H., Lin R., Wisniewski H.M., Hwang Y.W., Grundkeiqbal I., Healylouie G. DETECTION OF POINT MUTATIONS IN CODON-331 OF MITOCHONDRIAL NADH DEHYDROGENASE SUBUNIT-2 IN ALZHEIMER BRAINS. Biochem Biophys Res Commun. 1992;182(1):238–246. doi: 10.1016/s0006-291x(05)80136-6. [DOI] [PubMed] [Google Scholar]
- 18.Shoffner J.M., Brown M.D., Torroni A., Lott M.T., Cabell M.F., Mirra S.S. Mitochondrial DNA variants observed in alzheimer disease and parkinson disease patients. Genomics. 1993;17(1):171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 19.Ridge P.G., Kauwe J.S.K. Mitochondria and Alzheimer's disease: the role of mitochondrial genetic variation. Curr Genetic Med Rep. 2018;6(1):1–10. doi: 10.1007/s40142-018-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li HZ, Liu DH, Lu JX and Bai YD. Physiology and Pathophysiology of Mitochondrial DNA. In: Scatena R, Bottoni P, Giardina B, editors. Advances In Mitochondrial Medicine; 2012. pp. 39-51. [DOI] [PMC free article] [PubMed]
- 21.van der Walt J.M., Dementieva Y.A., Martin E.R., Scott W.K., Nicodemus K.K., Kroner C.C. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365(1):28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 22.Maruszak A., Safranow K., Branicki W., Gaweda-Walerych K., Pospiech E., Gabryelewicz T. The impact of mitochondrial and nuclear DNA variants on late-onset alzheimer's disease risk. J Alzheimers Dis. 2011;27(1):197–210. doi: 10.3233/JAD-2011-110710. [DOI] [PubMed] [Google Scholar]
- 23.Tranah G.J., Nalls M.A., Katzman S.M., Yokoyama J.S., Lam E.T., Zhao Y.Q. Mitochondrial DNA sequence variation associated with dementia and cognitive function in the elderly. J Alzheimers Dis. 2012;32(2):357–372. doi: 10.3233/JAD-2012-120466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ADNI. ADNI General Procedures Manual; 2010.
- 25.Hu Z, Pan Z, Lu H and Li W. Classification of Alzheimer’s Disease Based on Cortical Thickness Using AdaBoost and Combination Feature Selection Method. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. pp. 392-401.
- 26.Chen K., Ayutyanont N., Langbaum J.B.S., Fleisher A.S., Reschke C., Lee W. Characterizing Alzheimer's disease using a hypometabolic convergence index. NeuroImage. 2011;56(1):52–60. doi: 10.1016/j.neuroimage.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsson A., Vanderstichele H., Andreasen N., De Meyer G., Wallin A., Holmberg B. Simultaneous measurement of beta-amyloid((1–42)), total tau, and phosphorylated tau (Thr(181)) in cerebrospinal fluid by the xMAP technology. Clin Chem. 2005;51(2):336–345. doi: 10.1373/clinchem.2004.039347. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. Durbin R and Genome Project Data P. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrison E and Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint. 2012; 2012(1):arXiv:1207.3907.
- 31.Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M. Procaccio V and Wallace DC. mtDNA variation and analysis using mitomap and mitomaster. Curr Protocols Bioinformatics. 2013;44(1) doi: 10.1002/0471250953.bi0123s44. 23.21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro-Gomez D., Leipzig J., Shen L.S., Lott M., Stassen A.P.M., Wallace D.C. Phy-Mer: a novel alignment-free and reference-independent mitochondrial haplogroup classifier. Bioinformatics. 2015;31(8):1310–1312. doi: 10.1093/bioinformatics/btu825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armstrong R.A., Eperjesi F., Gilmartin B. The application of analysis of variance (ANOVA) to different experimental designs in optometry. Ophthalmic Physiol Opt. 2002;22(3):248–256. doi: 10.1046/j.1475-1313.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the Lasso. J Royal Statistical Soc Ser B-Methodol. 1996;58(1):267–288. [Google Scholar]
- 35.Efron B., Hastie T., Johnstone I., Tibshirani R. Least angle regression. Ann Stat. 2004;32(2):407–451. [Google Scholar]
- 36.Andrews S.J., Fulton-Howard B., Patterson C., McFall G.P., Gross A., Michaelis E.K. Pa J and Alzheimers Dis Neuroimaging I. Mitonuclear interactions influence Alzheimer's disease risk. Neurobiology Of. Aging. 2020;87 doi: 10.1016/j.neurobiolaging.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chinnery P.F., Taylor G.A., Howell N., Andrews R.M., Morris C.M., Taylor R.W. Mitochondrial DNA haplogroups and susceptibility to AD and dementia with Lewy bodies. Neurology. 2000;55(2):302. doi: 10.1212/wnl.55.2.302. [DOI] [PubMed] [Google Scholar]
- 38.Elson J.L., Herrnstadt C., Preston G., Thal L., Morris C.M., Edwardson J.A. Does the mitochondrial genome play a role in the etiology of Alzheimer's disease? Hum Genet. 2006;119(3):241–254. doi: 10.1007/s00439-005-0123-8. [DOI] [PubMed] [Google Scholar]
- 39.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30(2):E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 40.Wragg M.A., Talbot C.J., Morris J.C., Lendon C.L., Goate A.M. No association found between Alzheimer's disease and a mitochondrial tRNA glutamine gene variant. Neurosci Lett. 1995;201(2):107–110. doi: 10.1016/0304-3940(95)12146-3. [DOI] [PubMed] [Google Scholar]
- 41.Zsurka G., Kálmán J., Juhász A., Császár A., Raskó I., Janka Z. No mitochondrial haplotype was found to increase risk for alzheimer’s disease. Biol Psychiatry. 1998;44(5):371–373. doi: 10.1016/s0006-3223(97)00461-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.