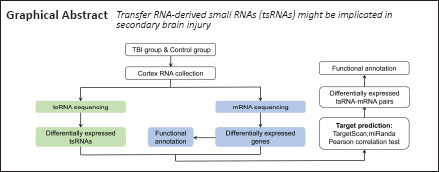
Key Words: gene set enrichment analysis, inflammation, integrated analysis, neurodegenerative disease, next-generation sequencing, secondary injury, synaptic function, transfer RNA-derived small RNAs, transfer RNAs, traumatic brain injury
Abstract
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are a recently established family of regulatory small non-coding RNAs that modulate diverse biological processes. Growing evidence indicates that tsRNAs are involved in neurological disorders and play a role in the pathogenesis of neurodegenerative disease. However, whether tsRNAs are involved in traumatic brain injury-induced secondary injury remains poorly understood. In this study, a mouse controlled cortical impact model of traumatic brain injury was established, and integrated tsRNA and messenger RNA (mRNA) transcriptome sequencing were used. The results revealed that 103 tsRNAs were differentially expressed in the mouse model of traumatic brain injury at 72 hours, of which 56 tsRNAs were upregulated and 47 tsRNAs were downregulated. Based on microRNA-like seed matching and Pearson correlation analysis, 57 differentially expressed tsRNA-mRNA interaction pairs were identified, including 29 tsRNAs and 26 mRNAs. Moreover, Gene Ontology annotation of target genes revealed that the significantly enriched terms were primarily associated with inflammation and synaptic function. Collectively, our findings suggest that tsRNAs may be associated with traumatic brain injury-induced secondary brain injury, and are thus a potential therapeutic target for traumatic brain injury. The study was approved by the Beijing Neurosurgical Institute Animal Care and Use Committee (approval No. 20190411) on April 11, 2019.
Chinese Library Classification No. R446; R742; Q344+.13
Introduction
Traumatic brain injury (TBI) pathogenesis is a complex process that results from primary and secondary insults (Galgano et al., 2017). Primary injury is caused by mechanical force and occurs at the moment of injury; it is followed by delayed and protracted secondary injury (Bae et al., 2018). Secondary injury occurs as a consequence of diverse pathological mechanisms, including excitotoxicity (Fujikawa, 2015; Tehse and Taghibiglou, 2019), oxidative stress (Cornelius et al., 2013; Greco et al., 2016; Chen et al., 2017), cerebral metabolic dysfunction (Glenn et al., 2003; Soustiel et al., 2005), cerebrovascular pathology (Len and Neary, 2011; Ramos-Cejudo et al., 2018; Sun et al., 2021), chronic inflammatory events (Kumar et al., 2015; Corrigan et al., 2016; Clark et al., 2019), and mitochondrial dysfunction (Hiebert et al., 2015; Pandya et al., 2019). Because of the heterogeneous nature of its complicated pathogenesis, no effective therapy is available to improve clinical outcomes for patients with TBI. A better understanding of the precise molecular mediators underlying TBI pathogenesis, especially in secondary injury-associated processes, is critical for developing effective therapeutic approaches for TBI patients.
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are a recently established family of regulatory small non-coding RNAs that modulate diverse biological processes, including sperm maturation (Peng et al., 2012; Sharma et al., 2016), the onset and progression of multiple types of cancers (Balatti et al., 2017), stress responses (Fu et al., 2009; Thompson and Parker, 2009; Yamasaki et al., 2009; Kumar et al., 2016), transposon control (Martinez et al., 2017; Schorn et al., 2017; Zhang et al., 2017), intergenerational epigenetic inheritance (Chen et al., 2016; Shi et al., 2019; Zhang et al., 2019b), and neuronal function (Karaiskos and Grigoriev, 2016). tsRNAs are derived from tRNAs, which are highly conserved and essential components of translation machinery. Pre-tRNAs are transcribed by RNA polymerase III and generate mature tRNA after endonucleolytic cleavage by RNases P and Z and the subsequent addition of a CCA tail (Phizicky and Hopper, 2010). Mature tRNAs generally resemble a cloverleaf secondary structure, composed of the dihydrouridine loop, anticodon loop, variable loop, TψC loop, and amino acid acceptor stem (Schimmel, 2018). Based on the mapped region of mature tRNA or pre-tRNA, tsRNA are classified into four major categories: 5′ or 3′ tRNA halves, tRNA-derived RNA fragment (tRF)-5, tRF-3, and tRF-1 (Kumar et al., 2016). With the rapid advances of next-generation sequencing coupled with its plummeting price, the numbers and types of identified tsRNA are increasing. However, the functional roles of tsRNA in physiological and pathological states are far less understood.
The endonucleolytic cleavage of tRNA is a conserved response to genetic and environmental stress, including inflammation, oxidative stress, and metabolic perturbation (Thompson and Parker, 2009; Anderson and Ivanov, 2014; Li et al., 2018; Hogg et al., 2019). Increasing evidence indicates that tsRNA abnormalities are involved in neurological dysfunction and may contribute to the development of neurodegenerative diseases (Schaffer et al., 2014; Karaiskos and Grigoriev, 2016; Hogg et al., 2019; Qin et al., 2020). These findings prompted us to speculate that tsRNA might be implicated in TBI-associated pathophysiological changes. To address this question, a combination of genome-wide tsRNA and messenger RNA (mRNA) sequencing was adopted to investigate the underlying role of tsRNA in TBI.
Materials and Methods
Animals
Adult male C57BL/6 mice (n = 30, 8–10 weeks, 25–30 g) were obtained from Beijing Vital River Experimental Animals Technology Co., Ltd. (Beijing, China; license No. SCXK-(Jing) 2016-0006). All animals were housed individually in temperature- (22 ± 2°C) and humidity- (50–60%) controlled animal quarters with food and water ad libitum, and were maintained on a 12-hour light/dark cycle. All animal procedures were approved by the Beijing Neurosurgical Institute Animal Care and Use Committee (approval No. 20190411) on April 11, 2019. All experiments were designed and reported according to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Controlled cortical impact models
Mice were randomly divided into two groups (n = 15 per group): control and TBI. As in our previously published work (Zhang et al., 2020), mice were anesthetized with 2% isoflurane (RWD Life Science Co., Shenzhen, China) inhalation and positioned in a stereotaxic frame (RWD Life Science Co.). The temperature of each mouse was maintained at (37.0 ± 0.5)°C during surgery using a thermal plate. A midline scalp incision was made to expose the skull. Next, a 4.0-mm craniotomy was made over the right parietal bone using an electric drill (RWD Life Science Co.), without damaging the dura mater. Mice were then subjected to controlled cortical impact with a 3-mm diameter flat tip using an electromagnetic controlled cortical impact device (Pinpoint PCI3000 Precision Cortical Impactor, Hatteras Instruments, Cary, NC, USA) at a velocity of 3 m/s, with a 20 ms dwell time and a depth of 1.5 mm. The controlled cortical impact model was considered successful when cerebral cortical contusion was noticeable (Song et al., 2019). Following the impact, the burr hole was sealed with bone wax and the scalp incision was sutured closed. Animals in the control group underwent the same process, but without the cortical impact.
RNA extraction
Mice were deeply anesthetized and transcardially perfused with 100 mL ice-cold 0.9% saline at 72 hours post-injury. After removal of the brain, cerebral perilesional cortex tissue around 1 mm from the margin of the contusion site was acquired on a chilled stainless-steel plate overlying crushed ice. Subsequently, the RNA was extracted from the cortices using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Pooled samples can reduce variability and compensate for losses in the number of replicates (Takele Assefa et al., 2020). Therefore, following RNA isolation, the cortex lysates from three mice were pooled into one sample, and one group contained three biological replicates. The RNA purity and integrity were evaluated using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and an RNA 6000 Nano LabChip Kit (Agilent Technologies). Finally, the resulting RNA from each sample was split into two halves and used for mRNA sequencing (mRNA-seq) or tsRNA sequencing.
mRNA library construction and sequencing
Poly(A) RNA was purified from total RNA with RNA integrity number > 7.0 using poly-T oligo-attached magnetic beads (New England Biolabs, Ipswich, MA, USA) with two rounds of purification. Subsequently, RNA was fragmented into small pieces and reverse-transcribed to construct a cDNA library using the manufacturer's protocol for the mRNA-Seq sample preparation kit (Illumina, San Diego, CA, USA). The average insert size for the paired-end libraries was 300 bp (± 50 bp) and the quality of the sequencing library was determined using an Agilent 2100 Bioanalyzer with an Agilent DNA 1000 chip kit (Agilent). Next, paired-end sequencing was performed using an Illumina HiSeq 4000 (Illumina).
tsRNA library construction and sequencing
tsRNA undergo a large number of post-transcriptional modifications, which can block reverse transcriptase-mediated extension during cDNA synthesis (Cozen et al., 2015). Therefore, Escherichia coli alpha-ketoglutarate-dependent dioxygenase (AlkB) was used to demethylate N1-methyladenosine, N3-methylcytidine, and N1-methylguanosine prior to reverse transcription. Additionally, the following treatments were performed to remove terminal modifications that interfere with adaptor ligation to the RNA ends using the rtStar™ tRF&tiRNA Pretreatment Kit (Arraystar Inc., Rockville, MD, USA): 3′-aminoacyl (charged) deacylation to 3′-OH, 3′-cP (2′,3′-cyclic phosphate) removal to 3′-OH, and 5′-OH (hydroxyl group) phosphorylation to 5′-P (Qin et al., 2019). The libraries with 134–160 bp polymerase chain reaction-amplified fragments (corresponding to an RNA size range of 14–40 nt) were sequenced for 50 cycles using an Illumina NextSeq 500 as per the manufacturer's instructions, thus generating 50 bp single-end read datasets.
Data processing and analysis
For mRNA-seq, clean reads were obtained from raw reads by removing reads that contained adaptors, primers, or Q nucleotide quality scores lower than 20. The clean reads were then aligned to the mouse genome using the HISAT (2.0) package (http://www.ccb.jhu.edu/software/hisat/index.shtml). Next, StringTie (version 1.3.0, http://ccb.jhu.edu/software/stringtie/) was used to assemble transcripts with mapped reads and perform gene- and transcript-level quantifications as fragments per kilobase of transcript per million RNA-sequencing mapped reads.
For tsRNA sequencing, cytoplasmic tRNA sequences were downloaded from GtRNAdb (http://gtrnadb.ucsc.edu/) and mitochondrial tRNA sequences were predicted using tRNAscan-SE (http://trna.ucsc.edu/tRNAscan-SE/). The mature tRNA library was generated through the removal of intronic sequences and the addition of 3′-terminal CCA tails. In addition, to generate the precursor tRNA library,40 bp flanking genomic sequences were added to each side of the tRNA sequences. The reads were first subjected to adaptor removal, and reads with a length shorter than 14 nt or longer than 40 nt were then discarded with Cutadapt (version 1.17, https://cutadapt.readthedocs.io/en/stable/). Subsequently, trimmed reads were aligned to the mature tRNA sequences, which accepted only single mismatches with Bowtie software (version 1.2.2, http://bowtie-bio.sourceforge.net/index.shtml). Reads that were not mapped to the mature tRNA sequences were then aligned to the precursor tRNA sequences. The abundance of tsRNA was calculated by the read counts and normalized as the counts per million of total aligned reads. Known tRNA fragments were downloaded from tRFdb (http://genome.bioch.virginia.edu/trfdb/).
Differential expression analysis
Differential expression analysis was performed on raw counts to compare gene expression between the TBI and control groups using the edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html) and the DESeq2 package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html), which is based on a negative binomial regression model. To correct for multiple testing, the false discovery rate (FDR) was calculated using the Benjamini–Hochberg procedure. For mRNA-seq, differentially expressed genes were defined as those having |log2 (fold change)| > 1 and FDR < 0.05. For tsRNA sequencing, differentially expressed tsRNAs were defined as those having a fold change > 1.5 and P-value < 0.05.
tsRNA target prediction and correlation analysis
Increasing evidence indicates that tsRNA can regulate gene expression through microRNA (miRNA)-like seed matching mechanisms (Kumar et al., 2014). Therefore, two commonly used miRNA target prediction algorithms, TargetScan and miRanda, were used for the tsRNA target analysis. In addition to a context score percentile > 50 in TargetScan and max energy < –10 in miRanda, putative targets needed to be simultaneously predicted by both algorithms. Furthermore, to reduce false positives produced by miRNA prediction algorithms, the Pearson correlation test was used to analyze pair-wise correlations between tsRNAs and putative target genes using the expression data of tsRNA and mRNA. If r < –0.5 and P < 0.05, there was a significant negative correlation between tsRNA and gene expression. On the basis of regulatory relationships between tsRNAs and predicted targets genes, a network of tsRNAs and their target genes was constructed using Cytoscape software (version 3.7.2, https://cytoscape.org/).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis
To depict the biological functions of differentially expressed genes in mRNA-seq and tsRNA target genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (https://www.genome.jp/kegg/) and Gene Ontology (GO) analysis (http://geneontology.org/), respectively, were used. For the GO and KEGG analyses, a list of genes were imported to the web-based tool Metascape (https://metascape.org/gp/index.html#/main/step1), which is a biologist-oriented resource for the analysis of systems-level databases (Zhou et al., 2019). The FDR was calculated using the Benjamini–Hochberg procedure to account for multiple testing. Enriched terms with FDR < 0.05 were considered to be significantly enriched terms.
Protein-protein interaction network and key gene analysis
The protein–protein interaction network was obtained from the online STRING database (Search Tool for Recurring Instances of Neighbouring Genes; https://string-db.org/), which integrates both known and predicted protein–protein interactions (Szklarczyk et al., 2019). The potential interaction network was then imported into Cytoscape, which is a public source software for visualizing and analyzing molecular interaction networks. The application Cytohubba in Cytoscape was used to identify key genes based on the node degree method.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is a robust threshold-free computational method that determines whether a priori defined sets of genes are differentially expressed in different phenotypes (Subramanian et al., 2005). First, mouse genes were assigned to the corresponding human orthologs using the GSEA “Collapse dataset to gene symbols” feature with respective symbol remapping chip (mouse_Gene_Symbol_Remapping_MSigDB.v7.0.chip). GSEA software (version 4.0.3, https://www.gsea-msigdb.org/gsea/index.jsp) was used to determine canonical pathway enrichment (C2 KEGG subset of canonical pathways). The parameter of permutation was 1000. The normalized enrichment score reflected the enrichment degree in the gene expression data. The FDR was the estimated probability that the enriched gene set with a given normalized enrichment score represented a false-positive finding. Gene sets with FDR < 0.25 were defined as significantly enriched.
Results
Genome-wide profiling of tsRNAs in the mouse cortex after TBI
To dissect the transcriptomic landscape of tsRNAs in the peri-injured cortex of a mouse model of TBI, six cDNA libraries were generated and sequenced. On average, 11.0 million raw reads per cerebral cortex sample were obtained. After the removal of adaptors and read length filtering, approximately 8.3 million clean reads were acquired per sample, of which an average of 18.8% and 1.04% were mapped to mature tRNA and pre-tRNA, respectively.
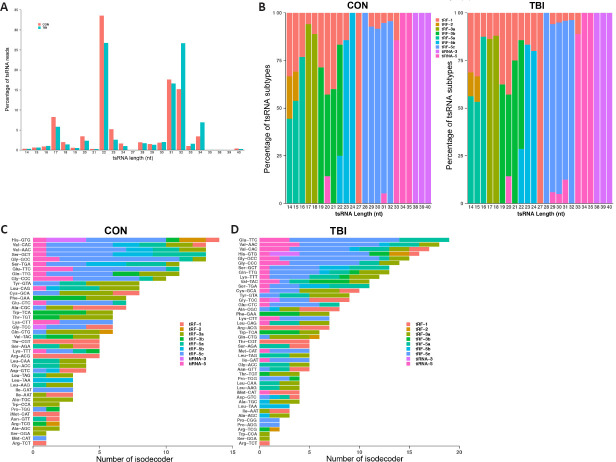
The sequencing results revealed that the most abundant tsRNAs were 22 nt in length, which accounted for 33.57% and 26.76% of the total clean reads in the control and TBI groups, respectively, and predominantly consisted of tRF-3b and tRF-5b (Figure 1A and B). Notably, the percentage of tsRNAs with 32 nt in length was significantly increased from 15.21% to 26.68% post-TBI; these were mainly comprised of tRF-5c (Figure 1A and B).
Figure 1.
Length and subtype distribution of tsRNAs in the cortex of a TBI mouse model.
(A) Read length distribution for tsRNAs. (B) The percentage of tsRNA subtypes against the tsRNA length. (C, D) The number of tsRNA subtypes against tRNA isodecoders in the control (C) and TBI groups (D). The x-axis indicates the number of tRNA isodecoders and the y-axis represents the tRNA isodecoders. The colors represent the subtypes of tsRNAs. CON: Control; TBI: traumatic brain injury; tiRNA: transfer RNA-derived stress-induced RNA; tRF: transfer RNA-derived RNA fragment; tsRNA: transfer RNA-derived small RNA.
tRNA isodecoders share the same anticodon but differ in their body sequences (Geslain and Pan, 2010, 2011; Rudinger-Thirion et al., 2011). Mounting evidence indicates that tRNA isodecoders undergo post-transcriptional regulation beyond translation (Geslain and Pan, 2011). In the present study, 254 and 324 isodecoders were detected in the control and TBI groups, respectively. In the control group, His-GTG had the most subtypes, with 14 isodecoders. In the TBI group, Glu-TTC had the most subtypes, with 19 isodecoders after injury (Figure 1C and D).
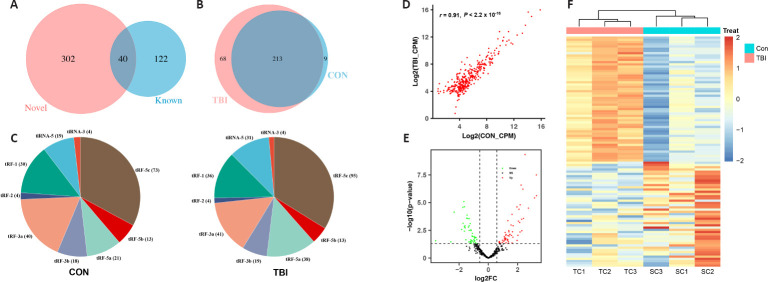
The sequencing results demonstrated that 342 tsRNAs were acquired in all libraries, of which 302 tsRNAs were specifically detected in our work compared with the known tRNA fragments from the tRFdb (Figure 2A). Furthermore, there were 68 and nine tsRNAs specifically expressed in the TBI and control groups, respectively, while 213 tsRNAs appeared in both groups (Figure 2B). Notably, the number of 5′ ends of tRNA-derived fragments, particularly tRF-5c, was markedly increased after TBI (Figure 2C).
Figure 2.
Expression patterns of tsRNA and the identification of differentially expressed tsRNAs in the cortex of a TBI mouse model.
(A) Venn diagram comparing the tsRNAs detected in the cortex of the TBI mouse model and the tsRNAs from the tRFdb database. (B) Venn diagram showing the commonly and specifically expressed tsRNAs in the control and TBI groups. (C) Pie chart illustrating the number of each subtype of tsRNAs in the control and TBI groups. (D) Scatter plot indicating the correlation of tsRNA expression between the control and TBI groups, analyzed using Pearson's correlation coefficient. r = 0.91, P < 2.2 × 10–16. (E) Volcano plot displaying differentially expressed tsRNAs. Upregulated and downregulated differentially expressed tsRNAs are highlighted in red and green, respectively. (F) Heatmap showing the hierarchical clustering of differentially expressed tsRNAs between the control and TBI groups. Blue indicates low expression levels and red indicates high expression levels. CON: Control; CPM: counts per million of total aligned reads; FC: fold change; SC: control cortex; TBI: traumatic brain injury; TC: TBI cortex; tiRNA: transfer RNA-derived stress-induced RNA; tRF: transfer RNA-derived RNA fragment; tsRNA: transfer RNA-derived small RNA.
Identification of differentially expressed tsRNAs in a mouse model of TBI
To investigate the potential role of tsRNAs in secondary injury post-TBI, differentially expressed tsRNAs were determined. The expression correlation of tsRNAs was first performed in the control and TBI groups, which indicated a strong correlation of tsRNA expression between the two groups (r = 0.91, P < 2.2 × 10–16; Figure 2D). Subsequently, 103 tsRNAs were observed to be differentially expressed in the mouse model of TBI at 72 hours, of which 56 tsRNAs were upregulated and 47 tsRNAs were downregulated (Figure 2E). Furthermore, differentially expressed tsRNAs were able to distinguish fairly well between the two groups, which implies a potential biological significance of tsRNAs in secondary injury after TBI (Figure 2F).
Interrogation of protein-coding gene alterations and biological functions
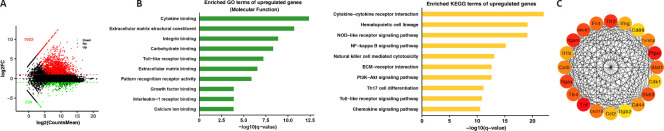
To explore the biological processes that occur in response to TBI injury, differentially expressed protein-coding genes were determined. Consequently, 1922 genes were upregulated relative to the control group upon TBI injury, and 294 genes were downregulated (Figure 3A). To characterize the biological functions of the upregulated genes, GO and KEGG analyses were conducted using the web-based tool Metascape. GO annotation results revealed that molecular functions of upregulated genes were mainly focused on interactions between biomolecules, such as cytokine binding, pattern recognition receptor activity, and calcium ion binding (Figure 3B). Moreover, the KEGG enrichment analysis revealed that significantly enriched pathways were largely implicated in inflammatory processes, including cytokine–cytokine receptor interaction, the nucleotide-binding oligomerization domain-like receptor signaling pathway, and the nuclear factor-κB pathway (Figure 3B). To determine which of the 1922 upregulated genes had critical roles in secondary injury post-TBI, a protein–protein interaction network was constructed using STRING and the CytoHubba application of Cytoscape. The hub genes (e.g., Tnfa, Tlr2, Tlr4, Il-1b, and Stat3) were primarily involved in inflammatory processes, which was consistent with the functional analysis by KEGG and GO (Figure 3C).
Figure 3.
Identification of differentially expressed genes (DEGs) in the cortex of a TBI mouse model and their functional annotation.
(A) Log-intensity ratios (M-values) versus log-intensity averages (A-values) plot showing the log2 fold changes (FC) of gene levels and the average counts. The red and green dots represent upregulated and downregulated DEGs, respectively. The values in red and green indicates the number of upregulated and downregulated DEGs, respectively. (B) GO molecular function term enrichment (left) and KEGG term enrichment (right) of upregulated DEGs. (C) Interaction network analysis of common DEGs. The color intensity indicates the degree of the node. The yellow and red colors represent low and high node degrees, respectively. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; TBI: traumatic brain injury.
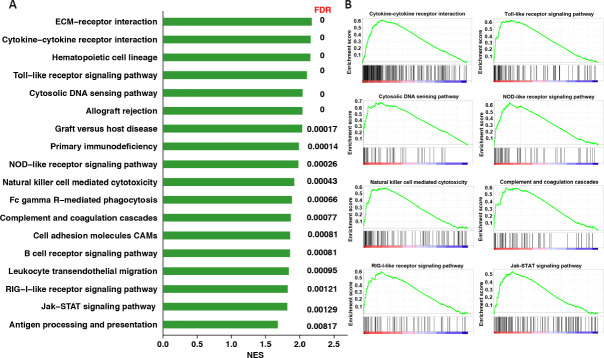
Given the bias caused by the arbitrary cutoff to identify significant genes, GSEA was adopted to identify the enriched biological processes. This analysis method uses information about all genes in the experiment to investigate the underlying biological mechanisms. After conversion to the corresponding human orthologs, an enrichment analysis was performed against the C2 KEGG subset of canonical pathways gene sets. This revealed that the enriched pathways by GSEA were also predominantly associated with inflammation (Figure 4), which further corroborated the functional identification of the KEGG and GO analyses.
Figure 4.
Gene set enrichment analysis of differentially expressed genes (DEGs) in the cortex of a TBI mouse model based on the KEGG gene set.
(A) Bar plots indicating enriched KEGG pathways. (B) GSEA enrichment plots of representative gene sets that were significantly enriched. The enrichment score reflects the degree to which the genes in a gene set were overrepresented at the extremes (top or bottom) of a ranked gene list. FDR: False discovery rate; GSEA: gene set enrichment analysis; KEGG: Kyoto Encyclopedia of Genes and Genomes; NES: normalized enrichment score; TBI: traumatic brain injury.
Correlation analysis of tsRNAs and their target genes
In light of the miRNA-like regulatory roles of tsRNAs in gene expression, two commonly used miRNA target prediction algorithms, TargetScan and miRanda, were used for the interaction analysis of tsRNAs and genes, both of which were differentially expressed after TBI. The correlation analysis of tsRNAs and their target genes identified 57 tsRNA–mRNA interaction pairs, which included 29 tsRNAs and 26 mRNAs (Figure 5A and B). Out of 57 pairs, 47 displayed a pattern of tsRNA upregulation while their corresponding targets were downregulated. GO analysis was then performed to obtain global insights into the physiological roles of the target genes. The GO annotation results demonstrated that the significantly enriched terms were primarily involved in inflammation and synaptic communication (Figure 5C). To further depict the interaction of tsRNAs and their target genes, the network between tsRNAs and their target genes was constructed using Cytoscape. This revealed that multiple tsRNAs can target the same gene, while a single tsRNA can also regulate multiple genes (Figure 6).
Figure 5.
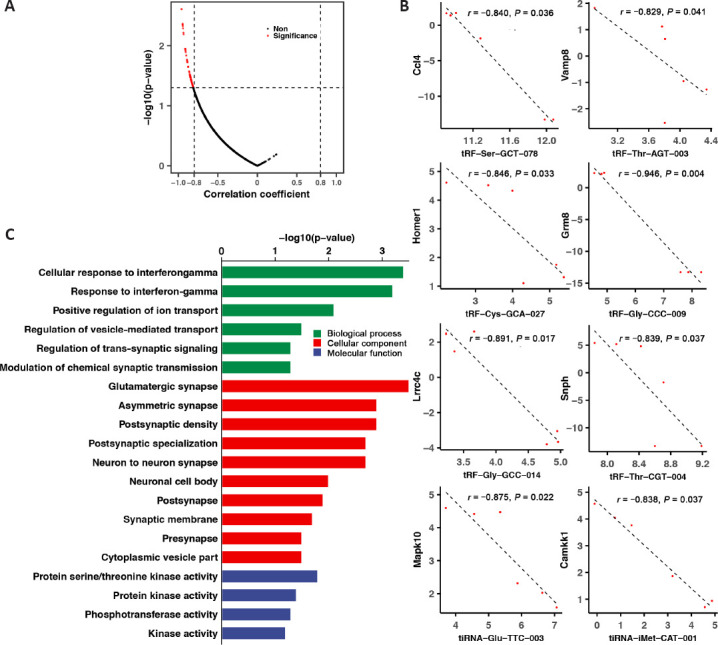
Integrated analysis of tsRNAs and their target mRNAs.
(A) Volcano plot showing the correlation between tsRNAs and their targets, both of which were differentially expressed after TBI. The red dots represent significantly correlated pairs. (B) Representative significantly correlated tsRNA–mRNA pairs. (C) GO enrichment analysis of predicted mRNA targets for tsRNAs. GO: Gene Ontology; TBI: traumatic brain injury; tsRNA: transfer RNA-derived small RNA.
Figure 6.
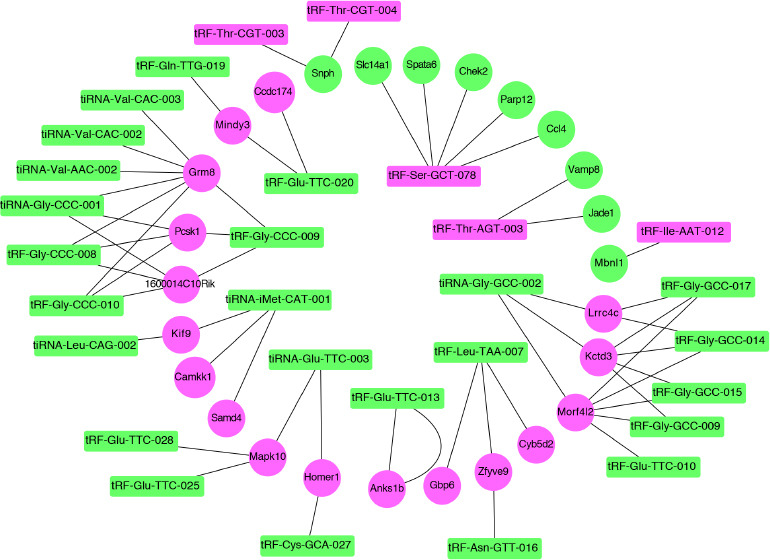
Interaction network of significantly correlated tsRNA-mRNA pairs.
The green and rose/violet dots represent upregulation and downregulation, respectively. tsRNA: Transfer RNA-derived small RNA.
Discussion
Although the mechanisms underlying TBI-associated secondary injury have been intensively investigated, there are no effective treatments available to date, which implies that alternative mechanisms are not yet fully understood. Given the pleiotropic role of tRNA-derived fragments in the central nervous system under physiological and pathophysiological conditions, the present work aimed to investigate whether tsRNAs are involved in secondary injury after TBI. To test this hypothesis, integrated next-generation sequencing analysis of tsRNA and mRNA was performed. The findings indicated that TBI exerted a large influence on tsRNA expression in the peri-injured cortex. Moreover, the mRNA targets of dysregulated tsRNA, identified by miRNA-like seed matching and expression correlation analysis, were primarily implicated in inflammation and synaptic functions.
tsRNAs can fine-tune gene expression at both the transcriptional and post-transcriptional levels in prokaryotes and eukaryotes, thereby playing key regulatory roles in various physiological and pathological events (Kim et al., 2017; Zhu et al., 2018). Recently, tsRNAs have gained more attention in neurological diseases (Qin et al., 2020). The interaction between cleavage, polyadenylation factor I subunit (CLP1), and the tRNA splicing endonuclease (TSEN) complex is necessary for tRNA splicing and maturation. Mutations in genes of the CLP1 and TSEN subunits result in neurological disorders, such as pontocerebellar hypoplasia (Abbott et al., 2014; Weitzer et al., 2015; Hayne et al., 2020). Hanada et al. (2013) reported that the absence of CLP1 activity in mice causes the accumulation of tyrosine pre-tRNA-derived small RNA fragments, which in turn sensitizes cells to oxidative stress-induced p53-dependent cell death, ultimately leading to a progressive loss of spinal motor neurons in the peripheral nerves, and thus muscle paralysis. Notably, in five unrelated families of the same ethnic group, a homozygous missense mutation (p.R140H) of CLP1 disturbed CLP1–TSEN complex integrity and caused the accumulation of linear tRNA introns. Abnormalities in tRNA-derived fragments led to severe sensory-motor deficits, cortical dysgenesis, and microcephaly in the affected individuals (Karaca et al., 2014). Concomitantly, in four independent consanguineous Turkish families, Schaffer et al. (2014) observed that the p.R140H mutation of CLP1 also results in cerebellar neurodegeneration because of accumulated pre-tRNA and reduced mature tRNA through p53-mediated neuronal loss. Together, these findings highlight that aberrant expression of tRNA-derived fragments is intimately associated with peripheral and central nervous system dysfunction.
In addition to genetic alterations, tsRNAs are often elevated during a variety of stress conditions, including neurological diseases (Thompson and Parker, 2009; Blanco et al., 2014; Hogg et al., 2019; Wang et al., 2019). Elkordy et al. (2018) demonstrated that oxidative stress, such as by the application of arsenite and hydrogen peroxide, induces tRNA cleavage and increases angiogenin-mediated generation of tRNA halves in a rat neuronal cell line (PC12). Moreover, the amount of tRNA halves is related to the degree of cell damage. Based on the observation that the generation of tRNA halves responds rapidly to oxygen–glucose deprivation, tRNA halves may be a novel biomarker for ischemia–reperfusion in PC12 cells (Elkordy et al., 2019). Intriguingly, aging can also greatly affect tsRNA expression, and 3′ tsRNAs display monotonic increases with age (Karaiskos and Grigoriev, 2016). Furthermore, Zhang et al. (2019a) identified eight differentially expressed tsRNAs in the brains of senescence-accelerated mouse-prone 8 mice, whose potential target genes were mainly implicated in neuronal function, including synapse formation and synaptic vesicle cycle pathways. In the present work, we observed that 103 tsRNAs were differentially expressed in a mouse model of TBI at 72 hours. Functional interrogation of the potential target genes of dysregulated tsRNA demonstrated that synapse-related functions, such as the regulation of vesicle-mediated transport and trans-synaptic signaling, were significantly enriched. Recently, it has been suggested that tsRNAs might be associated with the pathological processes of traumatic spinal cord injury through mitogen-activated protein kinase and brain-derived neurotrophic factor (Qin et al., 2019).
Accumulating evidence suggests that tsRNAs are involved in the pathophysiology of the inflammatory response (Dhahbi, 2015; Zhu et al., 2020), such as in cancer (Shan et al., 2020), alcoholic fatty liver disease (Zhong et al., 2019), and neurological disorders in particular (Su et al., 2020; Winek et al., 2020). The balance between the inflammatory response and immune suppression largely determines the post-stroke prognosis, and involves the cholinergic blockade of immune reactions (Hoover, 2017; Martín et al., 2018). Winek et al. (2020) reported that tsRNAs undergo significant changes in the peripheral blood of ischemic stroke patients at 2 days post-stroke; 143 tsRNAs were differentially expressed, of which 87% were upregulated. Notably, tsRNAs predominantly originate from lymphocytes and monocytes, particularly CD14+ monocytes. Further investigations have shown that tsRNAs might target cholinergic-associated monocytic transcription factors and regulate acetylcholine-mediated post-stroke immune suppression. Intriguingly, tsRNAs have been implicated in lifelong neuropathology in offspring (Su et al., 2020). Su et al. (2020) demonstrated that tsRNAs respond rapidly to maternal immune activation at the maternal–fetal interface, which can induce autism-related phenotypes in the offspring. Moreover, maternal immune activation triggers a decrease in 5′ tRNA halves and an increase in 3′ tsRNAs. In the mouse model of TBI in the current study, 54% of the differentially expressed tsRNAs were upregulated, similar to the pattern observed in the peripheral blood of ischemic stroke patients. In contrast to the changing trend of tsRNAs in the maternal immune activation autism model, 5′ tsRNAs were significantly upregulated post-TBI, which indicates that the biological function of disease-induced tsRNAs might be context specific. More importantly, TBI-associated tsRNAs were also involved in inflammation, such as tRF-Ser-GCT-078 and tRF-Thr-AGT-003, which target Ccl4 and Vamp8, respectively. Collectively, these data suggest that tsRNAs are potential contributors to regulating neuroinflammatory responses.
tsRNAs exert biological functions through multiple mechanisms, such as transcriptional silencing and repression and translational inhibition and enhancement (Xie et al., 2020; Yu et al., 2020). tsRNAs may interact with Argonaute proteins and function in a manner similar to miRNAs to regulate gene expression (Kumar et al., 2014). Maute et al. (2013) reported that tRNA-derived fragments, designated CU1276, repressed endogenous replication protein A1 in a 3′ untranslated region in a sequence-specific manner, thereby modulating proliferation and the DNA damage response. Interestingly, unlike conventional miRNA seed locations, Karaiskos and Grigoriev (2016) demonstrated that the potential seed regions of tsRNAs might exist at both ends of different tsRNAs. In particular, Jehn et al. (2020) reported that 5′ tRNA halves, which are highly expressed in the primate hippocampus, receive the most efficient gene silencing through the interactions between mid-regions of 5′ tRNA halves and coding DNA sequences or 3′ untranslated regions of target mRNAs. In addition to their direct binding to the 3′ untranslated region of mRNA transcripts, tsRNAs can interact with the RNA-binding protein Y-box binding protein-1 in a motif-specific manner and consequently suppress the stability of corresponding transcripts (Goodarzi et al., 2015). Furthermore, tsRNAs can fine-tune gene expression in terms of translational levels. Ivanov et al. (2011) revealed that angiogenin-induced tRNA fragments are able to displace eukaryotic initiation factor 4G/A from mRNA, and particularly uncapped mRNA, to inhibit translation in stressed cells. In contrast, Kim et al. (2017) demonstrated that LeuCAG-derived tsRNAs enhance the translation of two ribosomal protein mRNAs, RPS28 and RPS15, and ultimately regulate ribosome biogenesis. Together, these findings indicate that complex processes are associated with tsRNA-mediated biological functions. Based on potential miRNA-like actions, we identified 57 tsRNA–mRNA interaction pairs, of which 47 displayed the pattern of tsRNAs being upregulated while their corresponding targets were downregulated. Whether tsRNAs function in TBI through alternative mechanisms, such as translational regulation, remains unknown and requires further investigation.
To date, clinical trials for TBI have failed to show effective neuroprotection (Stein, 2015). Given the inherently heterogeneous nature of TBI and the multiplicity of its pathophysiological processes, the targeting of single events or molecules that are proposed to induce secondary injury is considered to be the most important factor underlying the failure of clinical trials (Loane and Faden, 2010). tsRNAs can act like miRNAs, and individual tsRNA can simultaneously fine-tune multiple genes, thereby influencing numerous biological processes. The manipulation of tsRNAs therefore provides a novel multipotential treatment for TBI. Our present and previous works have highlighted the intimate involvement of inflammatory mechanisms in the pathophysiological processes of TBI (Yang et al., 2021); thus, targeting inflammation-related tsRNAs might offer a potential strategy for the treatment of TBI. Moreover, the advancement of small RNA-based therapeutics, especially therapeutic small interfering RNA (siRNA), paves the way for the clinical translation of tsRNAs (Hu et al., 2020).
In summary, a greater understanding of the mechanisms underlying secondary injury will benefit the development of effective treatment modalities for TBI. The present study used integrated next-generation sequencing analysis of tsRNA and mRNA to provide the first evidence that tRNA-derived fragments might be involved in TBI-induced secondary injury. However, much remains to be elucidated regarding the detailed functions of tsRNA in secondary brain injury. For example, tRF-Ser-GCT-078 and tRF-Thr-AGT-003 have been implicated in inflammation, and the involvement of tRNA-derived stress-induced RNA (tiRNA)-Val-CAC-003 and tRF-Gly-Gcc-017 in synaptic function has been observed. However, more experimental evidence is needed to interrogate their functions in TBI using genetic approaches, such as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) and siRNA. In addition, TBI-induced spatiotemporal changes of tsRNAs and their relationships, such as causality or synergism, require further investigation.
Additional file: Open peer review report 1 (93.2KB, pdf) and 2 (89.2KB, pdf) .
Footnotes
P-Reviewers: Andrews CJ, Andrews MR; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Gardner B, Yu J, Song LP; T-Editor: Jia Y
Conflicts of interest:All authors in this manuscript have declared no conflict of interest.
Financial support:This work was supported by grants from the National Natural Science Foundation of China, Nos. 81471238, 81771327; and Construction of Central Nervous System Injury Basic Science and Clinical Translational Research Platform, Budget of Beijing Municipal Health Commission 2020, No. PXM2020_026280_000002 (all to BYL). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: The study was approved by the Beijing Neurosurgical Institute Animal Care and Use Committee (approval No. 20190411) on April 11, 2019.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open peer reviewers: Christopher J Andrews, University of Queensland, Australia; Melissa Renee Andrews, University of Southampton, UK.
Funding:This work was supported by grants from the National Natural Science Foundation of China, Nos. 81471238, 81771327; and Construction of Central Nervous System Injury Basic Science and Clinical Translational Research Platform, Budget of Beijing Municipal Health Commission 2020, No. PXM2020_026280_000002 (all to BYL).
References
- 1.Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet. 2014;5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae YH, Joo H, Bae J, Hyeon SJ, Her S, Ko E, Choi HG, Ryu H, Hur EM, Bu Y, Lee BD. Brain injury induces HIF-1α-dependent transcriptional activation of LRRK2 that exacerbates brain damage. Cell Death Dis. 2018;9:1125. doi: 10.1038/s41419-018-1180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG, Palamarchuk A, Hart JR, Bell C, Carosi M, Pescarmona E, Perracchio L, Diodoro M, Russo A, Antenucci A, Visca P, et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114:8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Hölter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus-Durner V, Hrab , de Angelis M, Káradóttir RT, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Guo Y, Yang W, Zheng P, Zeng J, Tong W. Connexin40 correlates with oxidative stress in brains of traumatic brain injury rats. Restor Neurol Neurosci. 2017;35:217–224. doi: 10.3233/RNN-160705. [DOI] [PubMed] [Google Scholar]
- 8.Clark DPQ, Perreau VM, Shultz SR, Brady RD, Lei E, Dixit S, Taylor JM, Beart PM, Boon WC. Inflammation in traumatic brain injury: roles for toxic A1 astrocytes and microglial-astrocytic crosstalk. Neurochem Res. 2019;44:1410–1424. doi: 10.1007/s11064-019-02721-8. [DOI] [PubMed] [Google Scholar]
- 9.Cornelius C, Crupi R, Calabrese V, Graziano A, Milone P, Pennisi G, Radak Z, Calabrese EJ, Cuzzocrea S. Traumatic brain injury: oxidative stress and neuroprotection. Antioxid Redox Signal. 2013;19:836–853. doi: 10.1089/ars.2012.4981. [DOI] [PubMed] [Google Scholar]
- 10.Corrigan F, Mander KA, Leonard AV, Vink R. Neurogenic inflammation after traumatic brain injury and its potentiation of classical inflammation. J Neuroinflammation. 2016;13:264. doi: 10.1186/s12974-016-0738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhahbi JM. 5’ tRNA halves: the next generation of immune signaling molecules. Front Immunol. 2015;6:74. doi: 10.3389/fimmu.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkordy A, Mishima E, Niizuma K, Akiyama Y, Fujimura M, Tominaga T, Abe T. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J Neurochem. 2018;146:560–569. doi: 10.1111/jnc.14321. [DOI] [PubMed] [Google Scholar]
- 14.Elkordy A, Rashad S, Shehabeldeen H, Mishima E, Niizuma K, Abe T, Tominaga T. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res. 2019;1714:8–17. doi: 10.1016/j.brainres.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 16.Fujikawa DG. The role of excitotoxic programmed necrosis in acute brain injury. Comput Struct Biotechnol J. 2015;13:212–221. doi: 10.1016/j.csbj.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galgano M, Toshkezi G, Qiu X, Russell T, Chin L, Zhao LR. Traumatic brain injury: current treatment strategies and future endeavors. Cell Transplant. 2017;26:1118–1130. doi: 10.1177/0963689717714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geslain R, Pan T. Functional analysis of human tRNA isodecoders. J Mol Biol. 2010;396:821–831. doi: 10.1016/j.jmb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geslain R, Pan T. tRNA: Vast reservoir of RNA molecules with unexpected regulatory function. Proc Natl Acad Sci U S A. 2011;108:16489–16490. doi: 10.1073/pnas.1113715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glenn TC, Kelly DF, Boscardin WJ, McArthur DL, Vespa P, Oertel M, Hovda DA, Bergsneider M, Hillered L, Martin NA. Energy dysfunction as a predictor of outcome after moderate or severe head injury: indices of oxygen, glucose, and lactate metabolism. J Cereb Blood Flow Metab. 2003;23:1239–1250. doi: 10.1097/01.WCB.0000089833.23606.7F. [DOI] [PubMed] [Google Scholar]
- 21.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greco T, Glenn TC, Hovda DA, Prins ML. Ketogenic diet decreases oxidative stress and improves mitochondrial respiratory complex activity. J Cereb Blood Flow Metab. 2016;36:1603–1613. doi: 10.1177/0271678X15610584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, Minis A, Sato F, Mimata H, Yoshimura A, Tamir I, Rainer J, Kofler R, Yaron A, Eggan KC, Woolf CJ, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayne CK, Schmidt CA, Haque MI, Matera AG, Stanley RE. Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Res. 2020;48:7609–7622. doi: 10.1093/nar/gkaa438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiebert JB, Shen Q, Thimmesch AR, Pierce JD. Traumatic brain injury and mitochondrial dysfunction. Am J Med Sci. 2015;350:132–138. doi: 10.1097/MAJ.0000000000000506. [DOI] [PubMed] [Google Scholar]
- 26.Hogg MC, Raoof R, El Naggar H, Monsefi N, Delanty N, O’Brien DF, Bauer S, Rosenow F, Henshall DC, Prehn JH. Elevation in plasma tRNA fragments precede seizures in human epilepsy. J Clin Invest. 2019;129:2946–2951. doi: 10.1172/JCI126346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoover DB. Cholinergic modulation of the immune system presents new approaches for treating inflammation. Pharmacol Ther. 2017;179:1–16. doi: 10.1016/j.pharmthera.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jehn J, Treml J, Wulsch S, Ottum B, Erb V, Hewel C, Kooijmans RN, Wester L, Fast I, Rosenkranz D. 5’ tRNA halves are highly expressed in the primate hippocampus and might sequence-specifically regulate gene expression. RNA. 2020;26:694–707. doi: 10.1261/rna.073395.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, Erdin S, Isikay S, Franco LM, Gonzaga-Jauregui C, Gambin T, Gelowani V, Hunter JV, Yesil G, Koparir E, Yilmaz S, et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res 5:ISCB Comm J-2758. 2016 doi: 10.12688/f1000research.10116.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, Zhang F, Chua MS, So S, Zhang QC, Sarnow P, Kay MA. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar RG, Boles JA, Wagner AK. Chronic inflammation after severe traumatic brain injury: characterization and associations with outcome at 6 and 12 months postinjury. J Head Trauma Rehabil. 2015;30:369–381. doi: 10.1097/HTR.0000000000000067. [DOI] [PubMed] [Google Scholar]
- 37.Len TK, Neary JP. Cerebrovascular pathophysiology following mild traumatic brain injury. Clin Physiol Funct Imaging. 2011;31:85–93. doi: 10.1111/j.1475-097X.2010.00990.x. [DOI] [PubMed] [Google Scholar]
- 38.Li S, Xu Z, Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes. 2018;9:246. doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loane DJ, Faden AI. Neuroprotection for traumatic brain injury: translational challenges and emerging therapeutic strategies. Trends Pharmacol Sci. 2010;31:596–604. doi: 10.1016/j.tips.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martín A, Domercq M, Matute C. Inflammation in stroke: the role of cholinergic, purinergic and glutamatergic signaling. Ther Adv Neurol Disord. 2018;11:1756286418774267. doi: 10.1177/1756286418774267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45:5142–5152. doi: 10.1093/nar/gkx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandya JD, Leung LY, Yang X, Flerlage WJ, Gilsdorf JS, Deng-Bryant Y, Shear DA. Comprehensive profile of acute mitochondrial dysfunction in a preclinical model of severe penetrating TBI. Front Neurol. 2019;10:605. doi: 10.3389/fneur.2019.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, Duan E. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin C, Xu PP, Zhang X, Zhang C, Liu CB, Yang DG, Gao F, Yang ML, Du LJ, Li JJ. Pathological significance of tRNA-derived small RNAs in neurological disorders. Neural Regen Res. 2020;15:212–221. doi: 10.4103/1673-5374.265560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin C, Feng H, Zhang C, Zhang X, Liu Y, Yang DG, Du LJ, Sun YC, Yang ML, Gao F, Li JJ. Differential expression profiles and functional prediction of tRNA-derived small RNAs in rats after traumatic spinal cord injury. Front Mol Neurosci. 2019;12:326. doi: 10.3389/fnmol.2019.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos-Cejudo J, Wisniewski T, Marmar C, Zetterberg H, Blennow K, de Leon MJ, Fossati S. Traumatic brain injury and Alzheimer's disease: the cerebrovascular link. EBioMedicine. 2018;28:21–30. doi: 10.1016/j.ebiom.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3’ UTR-embedded Alu element. Proc Natl Acad Sci U S A. 2011;108:E794–802. doi: 10.1073/pnas.1103698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, Rosti RO, Abdellateef M, Caglar C, Kasher PR, Cazemier JL, Weterman MA, Cantagrel V, Cai N, Zweier C, Altunoglu U, et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 52.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shan N, Li N, Dai Q, Hou L, Yan X, Amei A, Lu L, Wang Z. Interplay of tRNA-derived fragments and T cell activation in breast cancer patient survival. Cancers (Basel) 2020;12:2230. doi: 10.3390/cancers12082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J, Zhang Y, Zhou T, Chen Q. tsRNAs: The Swiss army knife for translational regulation. Trends Biochem Sci. 2019;44:185–189. doi: 10.1016/j.tibs.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song YM, Qian Y, Su WQ, Liu XH, Huang JH, Gong ZT, Luo HL, Gao C, Jiang RC. Differences in pathological changes between two rat models of severe traumatic brain injury. Neural Regen Res. 2019;14:1796–1804. doi: 10.4103/1673-5374.257534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soustiel JF, Glenn TC, Shik V, Boscardin J, Mahamid E, Zaaroor M. Monitoring of cerebral blood flow and metabolism in traumatic brain injury. J Neurotrauma. 2005;22:955–965. doi: 10.1089/neu.2005.22.955. [DOI] [PubMed] [Google Scholar]
- 58.Stein DG. Embracing failure: What the Phase III progesterone studies can teach about TBI clinical trials. Brain Inj. 2015;29:1259–1272. doi: 10.3109/02699052.2015.1065344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su Z, Frost EL, Lammert CR, Przanowska RK, Lukens JR, Dutta A. tRNA-derived fragments and microRNAs in the maternal-fetal interface of a mouse maternal-immune-activation autism model. RNA Biol. 2020;17:1183–1195. doi: 10.1080/15476286.2020.1721047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun TJ, Liu SJ, Xie FK, Huang XF, Zhang J, Jiang XH, Feng H, Yu AY. Role and hotspots of stem cell-derived exosome in the repair of traumatic brain injury. Zhongguo Zuzhi Gongcheng Yanjiu. 2021;25:123–127. [Google Scholar]
- 62.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takele Assefa A, Vandesompele J, Thas O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genomics. 2020;21:312. doi: 10.1186/s12864-020-6721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tehse J, Taghibiglou C. The overlooked aspect of excitotoxicity: Glutamate-independent excitotoxicity in traumatic brain injuries. Eur J Neurosci. 2019;49:1157–1170. doi: 10.1111/ejn.14307. [DOI] [PubMed] [Google Scholar]
- 65.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 66.Wang C, Zhao M, Wang J, Zhang D, Wang S, Zhao J. Expression analysis of transfer RNA derived fragments in the blood of patients with moyamoya disease: A preliminary study. Mol Med Rep. 2019;19:3564–3574. doi: 10.3892/mmr.2019.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weitzer S, Hanada T, Penninger JM, Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley interdisciplinary reviews RNA. 2015;6:47–63. doi: 10.1002/wrna.1255. [DOI] [PubMed] [Google Scholar]
- 68.Winek K, Lobentanzer S, Nadorp B, Dubnov S, Dames C, Jagdmann S, Moshitzky G, Hotter B, Meisel C, Greenberg DS, Shifman S, Klein J, Shenhar-Tsarfaty S, Meisel A, Soreq H. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc Natl Acad Sci U S A. 2020;117:32606–32616. doi: 10.1073/pnas.2013542117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Y, Yao L, Yu X, Ruan Y, Li Z, Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther. 2020;5:109. doi: 10.1038/s41392-020-00217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang MS, Xu XJ, Zhang B, Niu F, Liu BY. Comparative transcriptomic analysis of rat versus mouse cerebral cortex after traumatic brain injury. Neural Regen Res. 2021;16:1235–1243. doi: 10.4103/1673-5374.301028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu M, Lu B, Zhang J, Ding J, Liu P, Lu Y. tRNA-derived RNA fragments in cancer: current status and future perspectives. J Hematol Oncol. 2020;13:121. doi: 10.1186/s13045-020-00955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, Xu X, Niu F, Mao X, Dong J, Yang M, Gao F, Liu B. Corticosterone replacement alleviates hippocampal neuronal apoptosis and spatial memory impairment induced by dexamethasone via promoting brain corticosteroid receptor rebalance after traumatic brain injury. J Neurotrauma. 2020;37:262–272. doi: 10.1089/neu.2019.6556. [DOI] [PubMed] [Google Scholar]
- 74.Zhang S, Li H, Zheng L, Li H, Feng C, Zhang W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging (Albany NY) 2019a;11:10485–10498. doi: 10.18632/aging.102471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Shi J, Chen Q. tsRNAs: new players in mammalian retrotransposon control. Cell Res. 2017;27:1307–1308. doi: 10.1038/cr.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Shi J, Rassoulzadegan M, Tuorto F, Chen Q. Sperm RNA code programmes the metabolic health of offspring. Nat Rev Endocrinol. 2019b;15:489–498. doi: 10.1038/s41574-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong F, Hu Z, Jiang K, Lei B, Wu Z, Yuan G, Luo H, Dong C, Tang B, Zheng C, Yang S, Zeng Y, Guo Z, Yu S, Su H, Zhang G, Qiu X, Tomlinson S, He S. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res. 2019;29:548–561. doi: 10.1038/s41422-019-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu C, Sun B, Nie A, Zhou Z. The tRNA-associated dysregulation in immune responses and immune diseases. Acta Physiol (Oxf) 2020;228:e13391. doi: 10.1111/apha.13391. [DOI] [PubMed] [Google Scholar]
- 80.Zhu L, Liu X, Pu W, Peng Y. tRNA-derived small non-coding RNAs in human disease. Cancer Lett. 2018;419:1–7. doi: 10.1016/j.canlet.2018.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.