Key Words: apoptosis, canonical Wnt pathway, differentiation, MiR-103-3p, Ndel1, neural stem cells, neurogenesis, proliferation
Abstract
The regulation of adult neural stem cells (NSCs) is critical for lifelong neurogenesis. MicroRNAs (miRNAs) are a type of small, endogenous RNAs that regulate gene expression post-transcriptionally and influence signaling networks responsible for several cellular processes. In this study, miR-103-3p was transfected into neural stem cells derived from embryonic hippocampal neural stem cells. The results showed that miR-103-3p suppressed neural stem cell proliferation and differentiation, and promoted apoptosis. In addition, miR-103-3p negatively regulated NudE neurodevelopment protein 1-like 1 (Ndel1) expression by binding to the 3′ untranslated region of Ndel1. Transduction of neural stem cells with a lentiviral vector overexpressing Ndel1 significantly increased cell proliferation and differentiation, decreased neural stem cell apoptosis, and decreased protein expression levels of Wnt3a, β-catenin, phosphor-GSK-3β, LEF1, c-myc, c-Jun, and cyclin D1, all members of the Wnt/β-catenin signaling pathway. These findings suggest that Ndel1 is a novel miR-103-3p target and that miR-103-3p acts by suppressing neural stem cell proliferation and promoting apoptosis and differentiation. This study was approved by the Animal Ethics Committee of Nantong University, China (approval No. 20200826-003) on August 26, 2020.
Chinese Library Classification No. R453; R363; Q421
Introduction
Adult hippocampal neurogenesis is a complicated process that involves proliferation, survival, differentiation, and integration of newborn neurons into pre-existing neuronal networks (de Miranda et al., 2017; Chen et al., 2018a; Rizk et al., 2021). It has been shown that adult neural stem cells (NSCs) contribute to the plastic nature of the mature central nervous system (CNS) (Bond et al., 2015; Bao and Song, 2018). NSCs can self-renew and differentiate into three major CNS cell types: neurons, astrocytes, and oligodendrocytes (Shoemaker and Kornblum, 2016; Zarco et al., 2019). NSCs are involved in the maintenance, development, repair, and regeneration of the CNS, and could potentially be used to treat neurodegenerative diseases by repairing damaged and diseased parts of the CNS (Taupin, 2011; Shoemaker and Kornblum, 2016; Sugaya and Vaidya, 2018). Numerous preclinical studies have found that transplantation of exogenous NSCs is an effective therapy that significantly complements or replaces damaged tissues damaged by ischemic stroke (Boese et al., 2018; Zhang et al., 2019b). Another report demonstrated that, in mouse models of Alzheimer's disease, exogenous NSCs can migrate to injury sites, where they reduce amyloid beta levels and attenuate plaque formation (Boese et al., 2020). Thus, cell therapy based on NSCs is a promising tool for the treatment of neurodegenerative diseases.
Most of the genome is transcribed, although not all transcripts are translated; indeed, transcription of the human genome is thought to produce thousands of unique noncoding RNAs (Hangauer et al., 2013). Noncoding RNAs are central to the intricate and dynamic regulation of various molecular processes in the CNS, such as synaptic plasticity, neuronal differentiation, and synaptic protein synthesis (Kanai et al., 1996; Satterlee et al., 2007; Daidone et al., 2021). MicroRNAs (miRNAs) are approximately 22-nt-long, small, regulatory, noncoding RNAs that can functionally silence genes in a post-transcriptional manner by complementary base-pairing with the target mRNA (Lu and Rothenberg, 2018; Matsuyama and Suzuki, 2019; Zhang et al., 2021). miRNAs play functional roles not only in cell proliferation, differentiation, and apoptosis, but also in tumorigenesis and host-pathogen interactions (Cai et al., 2009; Cao and Zhen, 2018). To date, a large number of miRNAs have been identified that are expressed in the mammalian brain (Kou et al., 2020). Multiple studies have shown that miRNAs are abundant in the CNS, where they are essential regulators of gene expression (Stoicea et al., 2016; Chandran et al., 2017). Previous studies have suggested that brain-enriched miRNAs are potential regulators of brain development and function that play roles in neurogenesis, synapse formation and plasticity, neuroinflammation, and neurodegeneration (Nguyen et al., 2018; Cho et al., 2019). For example, Li et al. (2019) reported that miR-223 deficiency significantly ameliorates CNS inflammation and demyelination, as well as the clinical symptoms of autoimmune encephalomyelitis, by targeting the autophagy-related 16-like 1 gene. Furthermore, Lehmann et al. (2012) found that the miRNA let-7 activates expression of RNA-sensing Toll-like receptor 7 and induces neurodegeneration. Although these findings are encouraging, miRNA-based regulation during neurogenesis still requires further elucidation, especially considering the large number of miRNAs expressed in the CNS.
To better understand the function of the miRNA regulatory network in NSCs, we overexpressed miR-103-3p in NSCs and evaluated cell proliferation, differentiation, and apoptosis.
Materials and Methods
Animals
Twenty pregnant Sprague-Dawley rats (gestational day 15) were obtained from the Laboratory Animal Center of Nantong University (license No. SYXK (Su) 2017-0046). Every effort was made to minimize the number and suffering of animals used in this study. The study was approved by the Animal Ethics Committee of Nantong University, China (approval No. 20200826-003) on August 26, 2020.
Cell culture
NSCs were obtained from the embryos on embryonic day (E)15, as described previously (He et al., 2018). In brief, after intraperitoneal anaesthetization with 2 mL/kg equithesin (pentobarbital sodium, Cat# Y0002194, Sigma-Aldrich, St. Louis, MO, USA; chloral hydrate, Cat# 15307, Sigma-Aldrich), the embryos were removed by cesarean section, and the embryonic hippocampi were isolated. The hippocampi were then combined and rapidly converted into a single-cell suspension by mechanical dissociation. The cell suspensions were maintained in cell culture flasks in Dulbecco's modified Eagle's medium/F12 (1:1; Gibco, Grand Island, NY, USA) containing 2% B27 (Gibco) supplemented with epidermal growth factor (20 ng/mL; Sigma-Aldrich) and basic fibroblast growth factor (20 ng/mL; Sigma-Aldrich). Cells were incubated at 37˚C 5% CO2 and passaged every 5 days by dissociating the newly formed neurospheres with trypsin (Gibco). The NSCs were then randomly divided into miR-NC, miR-103-3p, LV-NC, LV-Ndel1, and LV-Ndel1 + miR-103-3p groups.
MiRNA transfection and lentiviral transduction
Transfection and transduction were performed when NSCs were 60–80% confluent. Vector controls and miRNAs (miR-NC and miR-103-3p) were purchased from Ribobio (Guangzhou, China). Cells were transfected with miR-NC or miR-103-3p using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The lentiviruses (LV-NC and LV-Ndel1) were purchased from Genechem (Shanghai, China) and transduced into NSCs at a multiplicity of infection of 10. Briefly, the media were replaced with the infection complex medium, which was composed of lentiviral supernatant and fresh NSC growth medium (Dulbecco's modified Eagle's medium/F12 containing 2% B27 supplemented with epidermal growth factor and basic fibroblast growth factor). Neurosphere diameters were measured using an EVOS FL Auto fluorescence microscope (Invitrogen).
Flow cytometry assays
At 24 hours after transduction, cells were harvested for cell cycle, apoptosis, and differentiation analyses. For the cell cycle analysis, cells were fixed in ice-cold 75% ethanol at 4°C overnight, followed by staining with Propidium Iodide/RNase Staining Buffer (BD Biosciences, San Diego, CA, USA). For the apoptosis analysis, a PE Annexin V Apoptosis Detection Kit (BD Biosciences) was used to stain the harvested cells according to the manufacturer's instructions. For the differentiation analysis, cells were trypsinized, collected, washed twice in ice-cold phosphate-buffered saline, and fixed in 1× Fix/Perm Buffer at 4°C for 50 minutes. After washing twice with 1× Perm/Wash Buffer, the cells were incubated with an APC-conjugated anti-class III beta-tubulin (Tuj1) antibody (BD Biosciences) at 4°C for 50 minutes. The cells were then washed twice with 1× Perm/Wash Buffer, centrifuged, and resuspended in 350 µL of flow cytometry staining buffer. All the stained cells were assessed using a FACSCalibur Instrument (BD Biosciences).
5-Ethynyl-2′-deoxyuridine assay
At 24 hours after transduction, cell proliferation was measured using a Cell-Light 5-ethynyl-2?-deoxyuridine (EdU) Apollo567 In Vitro Kit (Ribobio) according to the manufacturer's protocol, as previously described (Zheng et al., 2020). Briefly, the cells were exposed to EdU for 2 hours, then washed and fixed with 4% paraformaldehyde for 30 minutes. Following washing with phosphate-buffered saline, the cells were blocked with phosphate-buffered saline containing 0.5% Triton X-100 for 10 minutes. Cells were incubated with 200 μL of 1× Apollo® staining reaction solution for 30 minutes, and the nuclei were stained with Hoechst 33342 (Abcam, Cambridge, MA, USA, Cat# ab228551, 1:1000).
Immunofluorescence assay
At 24 and 96 hours after transduction, cells were fixed with 4% paraformaldehyde for 15 minutes and then rinsed in phosphate-buffered saline three times. Cells were then blocked with 0.3% Triton X-100 containing 10% goat serum for 2 hours. Next, the cells were incubated with one of the following primary antibodies at 4°C overnight: mouse monoclonal anti-Nestin (Millipore, Billerica, MA, USA, Cat# MAB353, #RRID:AB_94911, 1:200), rabbit monoclonal anti-Ki67 (Abcam, Cat# ab16667, #RRID:AB_302459, 1:200), mouse monoclonal anti-microtubule associated protein 2 (Millipore, Cat# M1406, #RRID:AB_477171, 1:1000), or mouse monoclonal and anti-DCX (Abcam, Cat# ab18723, #RRID:AB_732011, 1:1000). Then, samples were incubated with an appropriate secondary antibody (Alexa Fluor488-conjugated goat anti-mouse IgG, Cat# A-11029, #RRID:AB_138404, 1:1000, or Alexa Fluor594-conjugated goat anti-rabbit IgG, Invitrogen, Cat# A-11037, #RRID:AB_2534095, 1:1000) for 2 hours at room temperature. Finally, the cell nuclei were stained with Hoechst 33342 (Abcam). All cells were examined using an EVOS FL Auto fluorescence microscope (Invitrogen). The results are expressed as percentages.
Immunohistochemistry
Cell apoptosis was measured using a Super-Sensitive Horseradish Peroxidase Immunohistochemistry Kit (rabbit, Sangon Biotech, Shanghai, China) according to the manufacturer's protocol. At 24 hours after transduction, cells were fixed with 4% paraformaldehyde for 30 minutes, then incubated with endogenous peroxidase blocking solution for 15 minutes. Samples were pre-incubated in blocking solution for 30 minutes and then incubated with a rabbit anti-Caspase 3 antibody (Cell Signaling Technology, Danvers, MA, USA, Cat# 9664, #RRID AB_2070042, 1:1000) overnight at 4°C. Next, the samples were subjected to color development with DAB and hematoxylin (Vector Laboratories, Burlingame, CA, USA) counterstaining. All cells were examined under the EVOS FL Auto fluorescence microscope, and the average optical density of the positive area was measured.
Real-time quantitative polymerase chain reaction analysis
At 24 hours after transduction, total RNA was extracted from cells using TRIzol (Vazyme Biotech, Nanjing, China). Next, 1 μg of total RNA was reverse-transcribed into cDNA using the HiScriptII Reverse Kit (Vazyme Biotech) according to the manufacturer's instructions. Amplification was tracked using an AceQ real-time-quantitative polymerase chain reaction (qPCR) Kit (Vazyme Biotech). Primers for qPCR were synthesized by Sangon Biotech and are listed in Additional Table 1.
Additional Table 1.
The sequences of primers for real-time quantitative polymerase chain reaction analysis
| Gene | Sequence |
|---|---|
| Bax | Forward: 5’-ACA GAT CAT GAA GAC AGG GGC-3’ |
| Reverse: 5’-CAA GGT CAG CTC AGG TGT CT-3’ | |
| Caspase3 | Forward: 5’-CAA CAA CGA AAC CTC CGT GG-3’ |
| Reverse: 5’-ACA CAA GCC CAT TTC AGG GT-3’ | |
| Gapdh | Forward: 5’-CCA CGG CAA GTT CAA CGG CAC AG-3’ |
| Reverse: 5’-GAC GCC AGT AGA CTC CAC GAC AT-3’ | |
| Map2 | Forward: 5’-CTT GAT TCT ATT GCC CTT GGG TTT A-3’ |
| Reverse: 5’-CAT CCA TCG TTC CGC TAG TGT TG-3’ | |
| Ndel1 | Forward: 5’-AGC ACC CGT TCA TCA CAT CT-3’ |
| Reverse: 5’-GAT GCT TGG CAG GAG CTT AGA-3’ | |
| Neurod1 | Forward: 5’-CAG GGT TAT GAG ATC GTC ACT ATT C-3’ |
| Reverse: 5’-CCT TCT TGT CTG CCT CGT GTT CC-3’; | |
| P53 | Forward: 5’-TAG CGG TGC TAG CCA GAA GT-3’ |
| Reverse: 5’-AAC CGG AAA TGC TCC TGC AA-3’ |
Gapdh: Glyceraldehyde-3-phosphate dehydrogenase; Map2: microtubule associated protein 2; Ndel1: NudE neurodevelopment protein 1 like 1; Neurod1: neuronal differentiation 1.
Western blot analysis
Total protein was extracted from cells with RIPA buffer (Solarbio, Beijing, China) 48 hours after transduction. The protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Millipore), which were blocked with 5% non-fat dry milk (Sangon Biotech) for 2 hours and incubated with one of the following primary antibodies overnight at 4°C: rabbit monoclonal anti-Ndel1 (Abcam, Cat# ab124895, #RRID:AB_10975781, 1:1000), rabbit monoclonal anti-Wnt3a (Abcam, Cat# ab219412, 1:1000), rabbit monoclonal anti-β-Catenin (Cell Signaling Technology, Cat# 8480, #RRID:AB_11127855, 1:1000), rabbit monoclonal anti-glycogen synthase kinase 3β (GSK-3β; Cell Signaling Technology, Cat# 12456, #RRID:AB_2636978, 1:1000), rabbit monoclonal anti-phospho-GSK-3β (Cell Signaling Technology, Cat# 5558, #RRID:AB_10013750, 1:1000), rabbit monoclonal anti-lymphoid enhancer binding factor 1 (Abcam, Cat# ab137872, 1:1000), rabbit monoclonal anti-MYC proto-oncogene, bHLH transcription factor (Abcam, Cat# ab168727, 1:1000), rabbit monoclonal anti-c-Jun (Abcam, Cat# ab218576, 1:1000), rabbit monoclonal anti-cyclin D1 (Abcam, Cat# ab156448, 1:1000), rabbit polyclonal anti-Bax (Abcam, Cat# ab32503, #RRID:AB_725631, 1:1000), rabbit monoclonal anti-p53 (Abcam, Cat# ab182733, 1:1000), rabbit monoclonal anti-Caspase3 (Cell Signaling Technology, Cat# 9664, #RRID:AB_2070042, 1:1000), or mouse monoclonal anti-β-actin (Abcam, Cat# ab8226, #RRID:AB_306371, 1:1000). This was followed by incubation with secondary antibodies (horseradish peroxidase-conjugated anti-rabbit IgG (Abcam, Cat# ab205718, #RRID:AB_2819160, 1:1000)and anti-mouse IgG (Abcam, Cat# ab205719, #RRID:AB_2755049, 1:1000)) for 2 hours at room temperature. Next, the protein bands were visualized using enhanced chemiluminescence reagent (Bio-Rad, Hercules, CA, USA). The intensity of each band was quantified using Software Quantity One (Bio-Rad). β-Actin was used as a loading control.
Target gene prediction and luciferase reporter assay
miRNA target genes were predicted using TargetScan (http://www.targetscan.org), miRDB (http://www.mirdb.org/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/); targets were considered authentic when they were predicted by all three databases. The tissue expression patterns and Gene Ontology annotations of the 131 selected predicted target genes were analyzed using DAVID 6.7 (https://david.ncifcrf.gov/).
Wild-type (WT) and mutant (Mut) versions of the miR-103-3p binding site in the NDE1 3′-UTR were synthesized by Genechem. 293T cells (Nantong University, Nantong, China) were transfected with miR-103-3p or miR-NC and with WT-Ndel1 or Mut-Ndel1 using Lipofectamine 3000 (Invitrogen). Relative luciferase activity was analyzed by dual-luciferase assay (Promega, Madison, WI, USA) 72 hours after transfection; firefly luciferase activity was normalized to Renilla luciferase activity for the calculations.
Statistical analysis
All quantitative data are expressed as the mean ± standard deviation (SD) of at least three separate experiments. GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA) was used to analyze the results. Two-tailed paired Student's t-test and one-way analysis of variance followed by Tukey's post hoc test were used to compare corresponding groups, as indicated. P < 0.05 was considered statistically significant.
Results
Overexpression of miR-103-3p in NSCs inhibits cell proliferation and promotes apoptosis
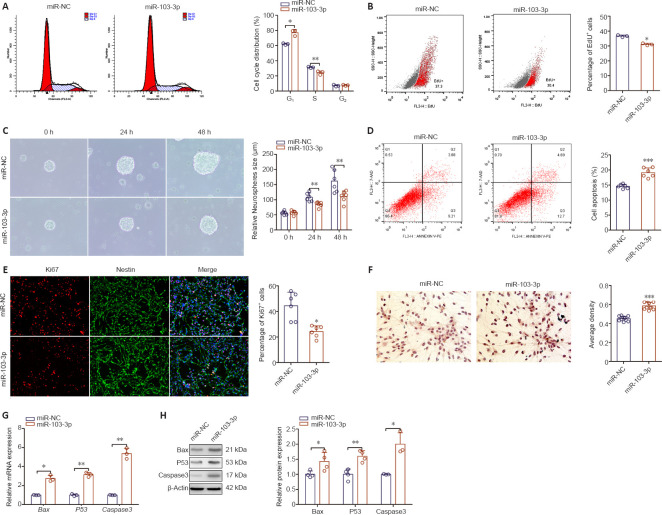
To determine the effect of miR-103-3p on NSC proliferation, we transfected NSCs with miR-103-3p or miR-NC. As shown in Figure 1A, the percentage of cells in G1 phase in the miR-103-3p group was increased (P < 0.05), whereas the percentage of cells in S phase was decreased (P < 0.01), compared with the miR-NC group. Transfection with miR-103-3p notably decreased the number of EdU-positive cells compared with the miR-NC–transfected group (P < 0.05; Figure 1B). In addition, the diameter of neurospheres that formed in the miR-103-3p group was dramatically decreased compared with the miR-NC group (P < 0.01; Figure 1C). Using immunofluorescent Ki67 staining, we found that miR-103-3p overexpression dramatically decreased the NSC growth rate compared with miR-NC overexpression (P < 0.001; Figure 1D). Next, we examined whether miR-103-3p affects apoptosis. Flow cytometry analysis showed that miR-103-3p overexpression significantly increased the ratio of apoptotic to non-apoptotic NSCs compared with miR-NC overexpression (P < 0.05; Figure 1E). Consistent with these results, immunohistochemistry showed that more cells were Caspase 3-positive in the miR-103-3p group compared with the control group (P < 0.001; Figure 1F). qPCR and western blot analyses showed that transfection with miR-103-3p significantly increased the mRNA (P < 0.05) and protein (P < 0.01) expression levels of three cell survival-related markers, Bax, P53, and Caspase 3 (Figure 1G and H), compared with the control group. Taken together, these results indicate that miR-103-3p overexpression suppresses NSC proliferation and promotes NSC apoptosis.
Figure 1.
Effects of miR-103-3p overexpression on neural stem cell (NSC) proliferation and apoptosis 24 hours after transduction.
(A) Cell cycle distribution of NSCs as detected by flow cytometry. (B) Assessment of NSC proliferation by EdU assay. (C) Representative images of neurospheres (arrows) transduced with miR-103-3p or miR-NC. The miR-103-3p group had a lower sphere-forming capacity compared with the miR-NC group. (D) NSC apoptosis was detected by flow cytometry. (E) Ki67 immunofluorescence staining (red, stained with Alexa Fluor594). The ratio of Ki67-positive to Ki67-negative cells in the miR-NC group was higher than in the miR-103-3p group. (F) Immunohistochemical detection of Caspase 3-positive cells. The ratio of Caspase 3-immunopositive to Caspase 3-immunonegative cells in the miR-NC group was lower than that in the miR-103-3p group. Scale bars: 400 μm in C, 200 μm in D, 100 μm in F. (G, H) The mRNA and protein expression levels of the cell survival-related gene, Bax, P53, and Caspase 3 were measured by quantitative polymerase chain reaction and Western blot analysis. Expression levels were normalized to miR-NC. Data are expressed as the mean ± SD. Each point represents one independent experiment performed in duplicate. *P< 0.05, **P< 0.01, ***P< 0.001, vs. miR-NC group (two-tailed paired Student's t-test). EdU: 5-Ethynyl-2?-deoxyuridine; NSC: neural stem cell.
miR-103-3p overexpression suppresses NSC differentiation into neurons
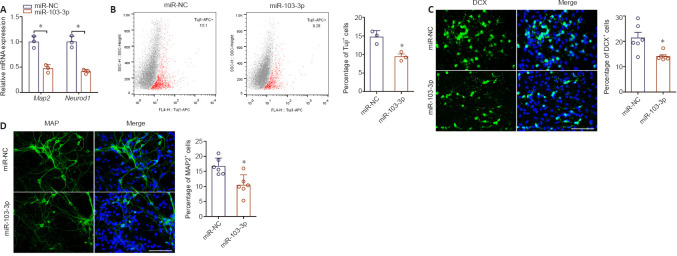
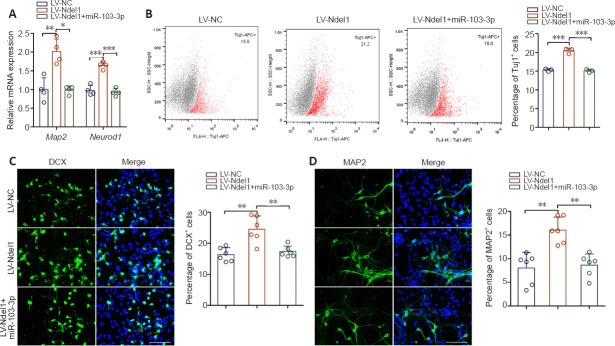
To characterize the role of miR-103-3p in NSCs differentiation, we performed qPCR analysis of the expression of two nerve-specific molecules, Map2 and Neurod1 (Sánchez et al., 2000; Lai et al., 2020), in cells overexpressing miR-103-3p. Both of these genes were downregulated significantly in the miR-103-3p group compared with the miR-NC group (P < 0.05; Figure 2A). Flow cytometry analysis showed that, compared with the miR-NC group, the percentage of Tuj1-positive cells was notably decreased in the miR-103-3p group (P < 0.05; Figure 2B). Similarly, an immunofluorescence assay showed that there were fewer DCX- and MAP2-positive cells in the miR-103-3p group than in the miR-NC group (P < 0.05; Figure 2C and D). These results demonstrate that miR-103-3p suppresses NSC differentiation.
Figure 2.
Effects of miR-103-3p overexpression on NSC differentiation 96 hours after transduction.
(A) Map2 and Neurod1 expression were measured by quantitative polymerase chain reaction. The relative mRNA expression was normalized to miR-NC. (B) Flow cytometry quantification of Tuj1-positive cells after transfection. (C, D) Representative images and quantification of DCX- (green, stained with Alexa Fluor488) and MAP2- (green, stained with Alexa Fluor594) positive cells. The miR-103-3p group had a lower ratio of DCX- and MAP2-positive cells compared with the other groups. Scale bars: 100 μm. Data are expressed as the mean ± SD. Each point represents one independent experiment performed in duplicate. *P< 0.05, vs. miR-NC group (two-tailed paired Student's t-test). DCX: Doublecortin; MAP2: microtubule-associated protein 2; NSC: neural stem cell; Tuj1: class III beta-tubulin.
miR-103-3p regulates Ndel1 expression in NSCs
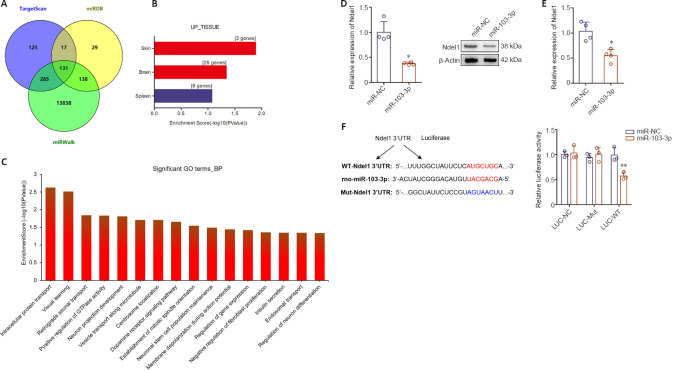
To explore the potential molecular mechanism by which miR-103-3p promotes apoptosis and inhibits proliferation and differentiation in NSC cells, miR-103-3p binding sites were predicted using TargetScan, miRDB, and miRWalk (Figure 3A). The 131 predicted target genes were further analyzed using DAVID 6.7. Of these 131 potential target genes, 25 were found to be significantly expressed in the brain (Figure 3B). Next, Gene Ontology analysis was conducted to identify biological processes that were enriched in these 25 predicted target genes expressed in the brain (Figure 3C). Among the 25 target genes, Ndel1 was enriched in retrograde axonal transport, neuron projection development, positive regulation of GTPase activity, vesicle transport along microtubules, centrosome localization, and establishment of mitotic spindle orientation. qPCR analysis showed that Ndel1 mRNA expression was downregulated in cells transfected with miR-103-3p compared with cells transfected with miR-NC (P < 0.05; Figure 3D). Western blot analysis further confirmed that Ndel1 was downregulated in the miR-103-3p group compared with the miR-NC group (P < 0.05; Figure 3E). Importantly, a luciferase reporter assay showed that miR-103-3p binds to the 3'-UTR of Ndel1 (P < 0.01; Figure 3F).
Figure 3.
miR-103-3p targets Ndel1 in NSCs.
(A) miR-103-3p target genes predicted by bioinformatic analysis. (B) The tissue expression patterns of the predicted miR-103-3p target genes were analyzed using DAVID 6.7. (C) Gene Ontology (GO) analysis of predicted target genes. Nde1 mRNA (D) and protein (E) expression were measured by quantitative polymerase chain reaction and western blot analyses, respectively. The relative mRNA and protein expression levels were normalized to miR-NC. (F) Luciferase reporter analyses of 293T cells co-transfected with Luciferase-Ndel1 and miR-103-3p or vector. Data are expressed as the mean ± SD. Each point represents one independent experiment performed in duplicate. *P< 0.05, **P< 0.01, vs. miR-NC group (two-tailed paired Student's t-test or one-way analysis of variance followed by Tukey's post hoc test). Ndel1: NudE neurodevelopment protein 1-like 1; NSC: neural stem cell.
miR-103-3p regulates NSC proliferation and apoptosis by targeting Ndel1
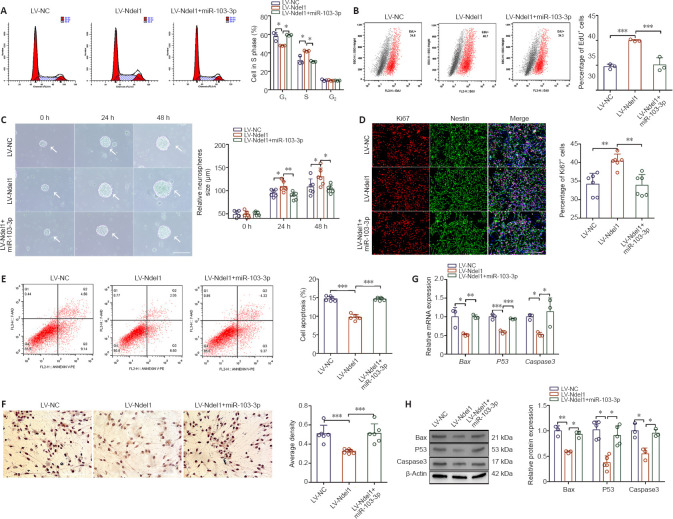
Since miR-103-3p regulates Ndel1 expression, we further examined Ndel1 function in NSCs. As shown in Figure 4A, Ndel1 overexpression decreased the number of cells in G1 phase and increased the number of cells in S phase compared with the LV-NC group (P < 0.05). miR-103-3p overexpression significantly rescued these phenotypes. An EdU assay demonstrated that Ndel1 overexpression increased the number of EdU-positive cells compared with the LV-NC group, and that transfection with miR-103-3p partially rescued this response (P < 0.001; Figure 4B). In addition, Ndel1 overexpression dramatically increased the sphere-forming capacity of NSCs compared with the LV-NC group (P < 0.05), while co-transfection with miR-103-3p reversed this effect (P < 0.01) (Figure 4C). Consistent with this, we found that Ndel1 overexpression dramatically increased the number of Ki67-positive cells compared with the LV-NC group, and concomitant transfection with miR-103-3p reversed this effect (P < 0.01; Figure 4D).
Figure 4.
miR-103-3p regulates NSC proliferation and apoptosis 24 hours after transduction by targeting Ndel1.
(A) Cell cycle distribution of NSCs as detected by flow cytometry. (B) Assessment of NSC proliferation by EdU assay. (C) Representative images of neurospheres (arrows) transfected with LV-Ndel1 or transduced with miR-103-3p. The LV-Ndel1 group had a higher sphere-forming capacity compared with the other two groups. (D) Proliferation was detected by Ki67 immunofluorescence staining (red, stained with Alexa Fluor594). The ratio of Ki67-positive to Ki67-negative cells in the LV-Ndel1 group was higher than in the LV-NC and LV-Ndel1 + miR-103-3p groups. (E) NSC apoptosis as detected by flow cytometry. (F) Immunohistochemical detection of Caspase 3 (brown). Markedly fewer cells in the LV-Ndel1 group were Caspase 3-immunopositive compared with the LV-NC and LV-Ndel1 + miR-103-3p groups. Scale bars: 400 μm in C, 200 μm in D, 100 μm in F. (G, H) The mRNA and protein expression levels of Bax, P53, and Caspase 3 were measured by quantitative polymerase chain reaction and western blot analyses, respectively. The relative mRNA and protein expression levels were normalized to LV-NC. Data are expressed as the mean ± SD. Each point represents one independent experiment performed in duplicate. *P< 0.05, **P< 0.01, ***P< 0.001 (one-way analysis of variance followed by Tukey's post hoc test). EdU: 5-Ethynyl-2?-deoxyuridine; Ndel1: NudE neurodevelopment protein 1-like 1; NSC: neural stem cell.
To characterize the role of Ndel1 in NSC apoptosis, we overexpressed Ndel1 in NSCs. Flow cytometry analysis showed that Ndel1 overexpression decreased the ratio of apoptotic to non-apoptotic NSCs compared with the LV-NC group, while co-overexpression of miR-103-3p increased apoptosis (P < 0.001; Figure 4E). Immunohistochemistry analysis showed that there were fewer Caspase 3-positive cells in the LV-Ndel1 group compared with the LV-NC group and that miR-103-3p partially rescued this effect (P < 0.001; Figure 4F). Similarly, qPCR and western blot analyses showed that Ndel1 overexpression significantly decreased the mRNA and protein expression levels of Bax, P53, and Caspase 3 compared with the LV-NC group, and that this effect was reversed by co-overexpression of miR-103-3p (P < 0.05, P < 0.01 or P < 0.001; Figure 4G and H). Taken together, these results indicate that miR-103-3p suppresses NSC proliferation and induces NSC apoptosis by targeting Ndel1.
miR-103-3p regulates NSC differentiation by targeting Ndel1
As shown in Figure 5A, Ndel1 overexpression substantially increased the expression of Map2 and Neurod1 compared with the LV-NC group, and co-transfection with miR-103-3p partially inhibited this effect (P < 0.05, P < 0.01 or P < 0.001). Flow cytometry analysis demonstrated that the percentage of Tuj1-positive cells notably increased after Ndel1 overexpression, whereas co-overexpression with miR-103-3p reversed this effect (P < 0.001; Figure 5B). Consistent with this, we found that Ndel1 overexpression dramatically increased the number of neurons, whereas miR-103-3p decreased the number of neurons (P < 0.01; Figure 5C and D). These data indicated that miR-103-3p suppresses NSC differentiation by regulating Nde1 concentrations.
Figure 5.
miR-103-3p regulates NSC differentiation by targeting Ndel1.
(A) Map2 and Neurod1 mRNA expression levels were detected by quantitative polymerase chain reaction. The relative mRNA expression levels were normalized to LV-NC. (B) Flow cytometry detection of Tuj1-positive cells after transfection. (C, D) Representative images and quantification of DCX- (green, stained with Alexa Fluor488) and MAP2- (green, stained with Alexa Fluor488) positive cells. The LV-Ndel1 group had a higher ratio of DCX- and Map2-positive cells to DCX- and Map2-negative cells than the other two groups. Scale bars: 100 μm. Data are expressed as the mean ± SD. Each point represents one independent experiment performed in duplicate. *P< 0.05, **P< 0.01, ***P< 0.001 (one-way analysis of variance followed by Tukey's post hoc test). DCX: Doublecortin; MAP2: microtubule-associated protein 2; Ndel1: NudE neurodevelopment protein 1-like 1; Neurod1: neuronal differentiation 1; NSC: neural stem cell; Tuj1: class III beta-tubulin.
miR-103-3p regulates the Wnt/β-catenin signaling pathway through Ndel1
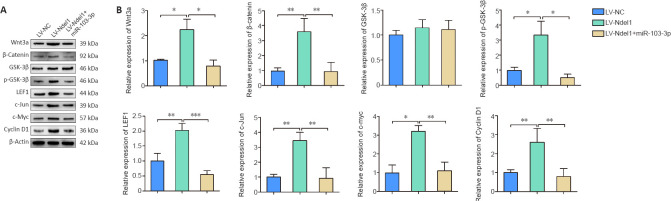
To investigate the regulatory effects of miR-103-3p on the Wnt/β-catenin pathway, we detected the expression of Wnt3a, β-catenin, GSK-3β, phospho-GSK-3β, LEF1, c-myc, c-Jun, and cyclin D1 by western blot. The results showed that the levels of Wnt3a, β-catenin, phospho-GSK-3β, LEF1, c-myc, c-Jun, and cyclin D1 expression were higher in the LV-Ndel1 group, but lower in the LV-Ndel1 + miR-103-3p group, than in the LV-NC group (P < 0.05, P < 0.01 or P < 0.001; Figure 6A and B). These results illustrate that miR-103-3p regulates the Wnt/β-catenin signaling pathway through Ndel1.
Figure 6.
Nde1 overexpression promotes activation of the Wnt/β-catenin pathway.
(A) Wnt3a, β-catenin, GSK-3β, p-GSK-3β, LEF1, c-myc, c-Jun, and cyclin D1 protein expression are induced by Ndel1 overexpression. (B) Quantification of Wnt3a, β-catenin, GSK-3β, p-GSK-3β, LEF1, c-myc, c-Jun, and cyclin D1 protein expression results. The data are presented as mean ± SD from three independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001 (one-way analysis of variance followed by Tukey's post hoc test). c-myc: MYC proto-oncogene; GSK-3β: glycogen synthase kinase 3β; LEF1: lymphoid enhancer binding factor 1; Ndel1: NudE neurodevelopment protein 1-like 1; p-GSK-3β: phospho-glycogen synthase kinase 3β.
Discussion
Evidence indicates that abnormal expression of miR-103 plays an important role in cancer. For example, Chen et al. (2018b) found that miR-103 post-transcriptionally downregulates the expression of Sal-like 4, inhibits glioma cell proliferation, migration, and invasion, and induces glioma cell apoptosis. Yu et al. (2018) showed that miR-103 regulates cell proliferation through PI3K/AKT signaling by directly targeting the phosphatase and tensin homologue gene in bladder cancer. Chen et al. (2019) found that miR-103 expression is negatively correlated with Axin2 expression and affects Wnt/β-catenin signaling activity, leading to poor overall survival in patients with colorectal cancer. To date, however, few studies have investigated the role of miR-103 in neurogenesis. One study by Yang et al. (2018) found that miR-103 promotes total neurite outgrowth and suppresses cells apoptosis in a PC12 cell model of Alzheimer's disease, suggesting that this miRNA plays a role in Alzheimer's disease progression. Furthermore, Croizier et al. (2018) demonstrated that inhibition of miR-103 reduces the number of pro-opiomelanocortin-positive cells and promotes the differentiation of Pomc progenitor cells into NPY neurons. In addition, Li et al. (2018) found that miR-103 protects against apoptosis and autophagy in lipopolysaccharide-injured PC12 cells and a rat model of spinal cord injury. Taken together, current evidence suggests that miR-103 is notable for being evolutionarily conserved and involved in regulating multiple cellular processes such as cell division, autophagy, apoptosis, angiogenesis (Zhou and Rigoutsos, 2014; Shi et al., 2018; Zhang et al., 2019a). In this study, we found that miR-103-3p overexpression significantly suppresses cell proliferation, induces apoptosis, and inhibits differentiation, suggesting that regulation of miR-103-3p could potentially be useful as a novel therapeutic approach in the treatment of neurodegenerative diseases.
miRNAs bind to the 3'-UTR of target genes to regulate their expression (Remsburg et al., 2019). Here, we investigated the molecular mechanisms regulating the effects of miR-103-3p on NSCs and found a possible interaction between miR-103-3p and the 3'-UTR of Ndel1. GO analysis showed that Ndel1 is significantly expressed in the brain and is enriched in pathways related to neuron development and cell division, which suggests that it may have multiple regulatory effects on cell growth and survival. Ndel1 was originally identified as a binding partner of lissencephaly 1, which interacts with dynein and modifies its activity (Inaba et al., 2016). Recent studies have reported that Ndel1 plays multifaceted roles in neurodevelopmental processes, such as mitosis, neuronal development, and neuronal migration (Okamoto et al., 2015; Inaba et al., 2016; Ye et al., 2017; Woo et al., 2019). In this study, we demonstrated that Ndel1 promotes NSC proliferation, inhibits NSC apoptosis, and promotes NSCs differentiation.
In this study, we found that miR-103-3p and Ndel1 expression were regulated by activation of the Wnt/β-catenin pathway. Furthermore, Ndel1 activated the canonical Wnt/β-catenin signaling pathway, whereas miR-103-3p had the opposite effect. The Wnt signal transduction cascade controls a variety of biological phenomena throughout development and during adulthood in all animals. Wnt signaling plays an important role in neuronal synapse formation and remodeling, dendritic growth and arborization, neurotransmission, neuroplasticity, neurogenesis, and neuroprotection (Maguschak and Ressler, 2012). However, there were some limitations to our study. Firstly, the upstream regulatory mechanism of miR-103-3p requires further investigation. Secondly, it remains unclear whether miR-103-3p also regulates other target genes.
In conclusion, in this study, we demonstrated that miR-103-3p significantly regulates NSC proliferation, apoptosis, and differentiation. Additionally, we showed that miR-103-3p expression is inversely correlated with Ndel1 expression. Ndel1 overexpression promoted NSC proliferation, inhibited NSC apoptosis, and promoted NSCs differentiation into neurons via the Wnt/β-catenin pathway. However, further studies are needed to explore the upstream regulatory mechanism of miR-103-3p, as well as to determine whether miR-103-3 regulates the expression of other target genes involved in neurogenesis.
Additional files:
Additional file 1: Open peer review reports 1 (96.2KB, pdf) and 2 (90.7KB, pdf) .
Additional Table 1: The sequences of primers for real-time quantitative polymerase chain reaction analysis.
Footnotes
P-Reviewers: Allen S, Barnstable C, Zhou Z; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Crow E, Yu J, Song LP; T-Editor: Jia Y
Conflicts of interest:The authors declare that they have no competing interests.
Financial support:The study was supported by Graduate Scientific Research Innovation Program of Jiangsu Province of China, No. KYCX19 2066 (to WL); and Project Funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education institutions China, No. 03081023 (to GHJ). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: The study was approved by the Animal Ethics Committee of Nantong University, China (approval No. 20200826-003) on August 26, 2020.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open peer reviewers: Scott Allen, The University of Sheffield, UK; Colin Barnstable, Penn State College of Medicine, USA; Zhibin Zhou, University of California, USA.
Funding:The study was supported by Graduate Scientific Research Innovation Program of Jiangsu Province of China, No. KYCX19 2066 (to WL); and Project Funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education institutions China, No. 03081023 (to GHJ).
References
- 1.Bao H, Song J. Treating brain disorders by targeting adult neural stem cells. Trends Mol Med. 2018;24:991–1006. doi: 10.1016/j.molmed.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boese AC, Hamblin MH, Lee JP. Neural stem cell therapy for neurovascular injury in Alzheimer's disease. Exp Neurol. 2020;324:113112. doi: 10.1016/j.expneurol.2019.113112. [DOI] [PubMed] [Google Scholar]
- 3.Boese AC, Le QE, Pham D, Hamblin MH, Lee JP. Neural stem cell therapy for subacute and chronic ischemic stroke. Stem Cell Res Ther. 2018;9:154. doi: 10.1186/s13287-018-0913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–395. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao T, Zhen XC. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018;24:586–597. doi: 10.1111/cns.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12–22. doi: 10.1016/j.neuint.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Hu S, Wu Z, Liu J, Li S. The role of miR-132 in regulating neural stem cell proliferation, differentiation and neuronal maturation. Cell Physiol Biochem. 2018a;47:2319–2330. doi: 10.1159/000491543. [DOI] [PubMed] [Google Scholar]
- 9.Chen HY, Lang YD, Lin HN, Liu YR, Liao CC, Nana AW, Yen Y, Chen RH. miR-103/107 prolong Wnt/β-catenin signaling and colorectal cancer stemness by targeting Axin2. Sci Rep. 2019;9:9687. doi: 10.1038/s41598-019-41053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LP, Zhang NN, Ren XQ, He J, Li Y. miR-103/miR-195/miR-15b regulate SALL4 and inhibit proliferation and migration in glioma. Molecules. 2018b;23:2938. doi: 10.3390/molecules23112938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho KHT, Xu B, Blenkiron C, Fraser M. Emerging roles of miRNAs in brain development and perinatal brain injury. Front Physiol. 2019;10:227. doi: 10.3389/fphys.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croizier S, Park S, Maillard J, Bouret SG. Central Dicer-miR-103/107 controls developmental switch of POMC progenitors into NPY neurons and impacts glucose homeostasis. Elife. 2018;7:e40429. doi: 10.7554/eLife.40429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daidone M, Cataldi M, Pinto A, Tuttolomondo A. Non-coding RNAs and other determinants of neuroinflammation and endothelial dysfunction: regulation of gene expression in the acute phase of ischemic stroke and possible therapeutic applications. Neural Regen Res. 2021;16:2154–2158. doi: 10.4103/1673-5374.310607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Miranda AS, Zhang CJ, Katsumoto A, Teixeira AL. Hippocampal adult neurogenesis: does the immune system matter? J Neurol Sci. 2017;372:482–495. doi: 10.1016/j.jns.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 15.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He H, Li W, Peng M, Qin J, Shi J, Li H, Tian M, Zhang X, Lv G, Jin G. MicroRNA expression profiles of neural stem cells following valproate inducement. J Cell Biochem. 2018;119:6204–6215. doi: 10.1002/jcb.26831. [DOI] [PubMed] [Google Scholar]
- 17.Inaba H, Goto H, Kasahara K, Kumamoto K, Yonemura S, Inoko A, Yamano S, Wanibuchi H, He D, Goshima N, Kiyono T, Hirotsune S, Inagaki M. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol. 2016;212:409–423. doi: 10.1083/jcb.201507046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanai K, Kako M, Aikawa T, Hino K, Tsubouchi H, Takehira Y, Iwabuchi S, Kawasaki T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Core promoter mutations of hepatitis B virus for the response to interferon in e antigen-positive chronic hepatitis B. Am J Gastroenterol. 1996;91:2150–2156. [PubMed] [Google Scholar]
- 19.Kou X, Chen D, Chen N. The regulation of microRNAs in Alzheimer's disease. Front Neurol. 2020;11:288. doi: 10.3389/fneur.2020.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M, Pan M, Ge L, Liu J, Deng J, Wang X, Li L, Wen J, Tan D, Zhang H, Hu X, Fu L, Xu Y, Li Z, Qiu X, Chen G, Guo J. NeuroD1 overexpression in spinal neurons accelerates axonal regeneration after sciatic nerve injury. Exp Neurol. 2020;327:113215. doi: 10.1016/j.expneurol.2020.113215. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kälin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Chen T, Zhu Y, Xiao X, Bu J, Huang Z. MiR-103 alleviates autophagy and apoptosis by regulating SOX2 in LPS-injured PC12 cells and SCI rats. Iran J Basic Med Sci. 2018;21:292–300. doi: 10.22038/ijbms.2018.25980.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Zhou D, Ren Y, Zhang Z, Guo X, Ma M, Xue Z, Lv J, Liu H, Xi Q, Jia L, Zhang L, Liu Y, Zhang Q, Yan J, Da Y, Gao F, Yue J, Yao Z, Zhang R. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy. 2019;15:478–492. doi: 10.1080/15548627.2018.1522467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maguschak KA, Ressler KJ. A role for WNT/β-catenin signaling in the neural mechanisms of behavior. J Neuroimmune Pharmacol. 2012;7:763–773. doi: 10.1007/s11481-012-9350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama H, Suzuki HI. Systems and synthetic microRNA biology: from biogenesis to disease pathogenesis. Int J Mol Sci. 2019;21:132. doi: 10.3390/ijms21010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen LS, Fregeac J, Bole-Feysot C, Cagnard N, Iyer A, Anink J, Aronica E, Alibeu O, Nitschke P, Colleaux L. Role of miR-146a in neural stem cell differentiation and neural lineage determination: relevance for neurodevelopmental disorders. Mol Autism. 2018;9:38. doi: 10.1186/s13229-018-0219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto M, Iguchi T, Hattori T, Matsuzaki S, Koyama Y, Taniguchi M, Komada M, Xie MJ, Yagi H, Shimizu S, Konishi Y, Omi M, Yoshimi T, Tachibana T, Fujieda S, Katayama T, Ito A, Hirotsune S, Tohyama M, Sato M. DBZ regulates cortical cell positioning and neurite development by sustaining the anterograde transport of Lis1 and DISC1 through control of Ndel1 dual-phosphorylation. J Neurosci. 2015;35:2942–2958. doi: 10.1523/JNEUROSCI.5029-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remsburg C, Konrad K, Sampilo NF, Song JL. Analysis of microRNA functions. Methods Cell Biol. 2019;151:323–334. doi: 10.1016/bs.mcb.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Rizk M, Vu J, Zhang Z. Impact of pediatric traumatic brain injury on hippocampal neurogenesis. Neural Regen Res. 2021;16:926–933. doi: 10.4103/1673-5374.297057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez C, Díaz-Nido J, Avila J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog Neurobiol. 2000;61:133–168. doi: 10.1016/s0301-0082(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 32.Satterlee JS, Barbee S, Jin P, Krichevsky A, Salama S, Schratt G, Wu DY. Noncoding RNAs in the brain. J Neurosci. 2007;27:11856–11859. doi: 10.1523/JNEUROSCI.3624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi FP, Wang XH, Zhang HX, Shang MM, Liu XX, Sun HM, Song YP. MiR-103 regulates the angiogenesis of ischemic stroke rats by targeting vascular endothelial growth factor (VEGF) Iran J Basic Med Sci. 2018;21:318–324. doi: 10.22038/IJBMS.2018.27267.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker LD, Kornblum HI. Neural stem cells (NSCs) and proteomics. Mol Cell Proteomics. 2016;15:344–354. doi: 10.1074/mcp.O115.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoicea N, Du A, Lakis DC, Tipton C, Arias-Morales CE, Bergese SD. The miRNA journey from theory to practice as a CNS biomarker. Front Genet. 2016;7:11. doi: 10.3389/fgene.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugaya K, Vaidya M. Stem cell therapies for neurodegenerative diseases. Adv Exp Med Biol. 2018;1056:61–84. doi: 10.1007/978-3-319-74470-4_5. [DOI] [PubMed] [Google Scholar]
- 37.Taupin P. Neurogenesis, NSCs, pathogenesis and therapies for Alzheimer's disease. Front Biosci (Schol Ed) 2011;3:178–190. doi: 10.2741/s143. [DOI] [PubMed] [Google Scholar]
- 38.Woo Y, Kim SJ, Suh BK, Kwak Y, Jung HJ, Nhung TTM, Mun DJ, Hong JH, Noh SJ, Kim S, Lee A, Baek ST, Nguyen MD, Choe Y, Park SK. Sequential phosphorylation of NDEL1 by the DYRK2-GSK3β complex is critical for neuronal morphogenesis. Elife. 2019;8:e50850. doi: 10.7554/eLife.50850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, Wang H, Shu Y, Li X. miR-103 promotes neurite outgrowth and suppresses cells apoptosis by targeting prostaglandin-endoperoxide synthase 2 in cellular models of Alzheimer's disease. Front Cell Neurosci. 2018;12:91. doi: 10.3389/fncel.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RYC, Kim J, Song H, Ming GL, Zhang M. DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis. Neuron. 2017;96:1041–1054.e5. doi: 10.1016/j.neuron.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu QF, Liu P, Li ZY, Zhang CF, Chen SQ, Li ZH, Zhang GY, Li JC. MiR-103/107 induces tumorigenicity in bladder cancer cell by suppressing PTEN. Eur Rev Med Pharmacol Sci. 2018;22:8616–8623. doi: 10.26355/eurrev_201812_16625. [DOI] [PubMed] [Google Scholar]
- 42.Zarco N, Norton E, Quiñones-Hinojosa A, Guerrero-Cázares H. Overlapping migratory mechanisms between neural progenitor cells and brain tumor stem cells. Cell Mol Life Sci. 2019;76:3553–3570. doi: 10.1007/s00018-019-03149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Zhang C, Wang H, Qi Y, Kan Y, Ge Z. Effects of miR 103a 3p on the autophagy and apoptosis of cardiomyocytes by regulating Atg5. Int J Mol Med. 2019a;43:1951–1960. doi: 10.3892/ijmm.2019.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang GL, Zhu ZH, Wang YZ. Neural stem cell transplantation therapy for brain ischemic stroke: Review and perspectives. World J Stem Cells. 2019b;11:817–830. doi: 10.4252/wjsc.v11.i10.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Cui SS, Zhou ZC, Hu XH, Yang XH. MicroRNA regulates histone deacetylase in the treatment of bone-related diseases. Zhongguo Zuzhi Gongcheng Yanjiu. 2021;25:2767–2774. [Google Scholar]
- 46.Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, Zhang L, Chen J. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73. doi: 10.1186/s12943-020-01183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Rigoutsos I. MiR-103a-3p targets the 5’ UTR of GPRC5A in pancreatic cells. RNA. 2014;20:1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.