Abstract
DRH-3 is critically involved in germline development and RNA interference (RNAi) facilitated chromosome segregation via the 22G-siRNA pathway in Caenorhabditis elegans. DRH-3 has similar domain architecture to RIG-I-like receptors (RLRs) and belongs to the RIG-I-like RNA helicase family. The molecular understanding of DRH-3 and its function in endogenous RNAi pathways remains elusive. In this study, we solved the crystal structures of the DRH-3 N-terminal domain (NTD) and the C-terminal domains (CTDs) in complex with 5′-triphosphorylated RNAs. The NTD of DRH-3 adopts a distinct fold of tandem caspase activation and recruitment domains (CARDs) structurally similar to the CARDs of RIG-I and MDA5, suggesting a signaling function in the endogenous RNAi biogenesis. The CTD preferentially recognizes 5′-triphosphorylated double-stranded RNAs bearing the typical features of secondary siRNA transcripts. The full-length DRH-3 displays unique structural dynamics upon binding to RNA duplexes that differ from RIG-I or MDA5. These features of DRH-3 showcase the evolutionary divergence of the Dicer and RLR family of helicases.
INTRODUCTION
RNA interference (RNAi) is important for the viability and growth of Caenorhabditis elegans (C. elegans) larvae (1–4). RNAi refers to sequence-specific gene silencing that is triggered by small duplex RNAs. These non-coding RNA molecules are key players in RNAi. The two pathways of generating small RNAs in C. elegans are cleavage of double-stranded RNA (dsRNA) by Dicer and de novo synthesis of secondary siRNAs by worm RNA-dependent RNA polymerase (RdRp). Worm Dicer (DCR-1) is required to produce microRNAs (miRNAs) and small interfering RNAs (siRNAs), which are short (21–23 nt) dsRNAs bearing 5-phosphate (5′-p) ends and 3′ overhangs (5–7). Worm-specific RdRps can transcribe endogenous siRNAs using messenger RNA (mRNA) templates (2,8,9). These secondary siRNAs exhibit a distinct polarity (5′ to 3′ on the antisense strand) and bear triphosphate moieties at the 5′ end (5′-ppp). Secondary siRNAs can be produced through multiple amplification rounds which may result in more effective mRNA silencing in worms (10,11)
DRH-3 was first reported as a Dicer interacting protein and required for germline RNAi (12). This protein is important for germline development and chromosome segregation (13). The drh-3 mutants are sterile and exhibit defects in germline proliferation and gametogenesis. Aoki et al. reported immunoprecipitation data showing that DRH-3 instead of worm Dicer (DCR-1), directly interacts with worm RdRps, such as RRF-1, indicating DRH-3 could be involved in the RdRp activities (2). DRH-3 is required for the biogenesis of secondary siRNAs, including endogenous 26G and 22G siRNAs (3,14). Deep sequencing data suggest that both 22 and 21 nt 5′ G small RNAs are dramatically reduced in the drh-3 mutants, indicating that DRH-3 plays a role in the biogenesis of 22G RNAs, in C. elegans (15). Vasale et al. have performed a qRT-PCR analysis of 26G target genes expression and shown the 26G-RNA targets could be up-regulated in the drh-3 mutant (14).
DRH-3 is orthologous to Dicer and most closely related to the RIG-I-like receptors (RLRs), which include RIG-I (retinoic acid inducible gene 1), MDA5 (melanoma differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology 2) (16,17). The RLRs play an essential role in sensing viral infection and initiating an interferon-mediated antiviral immune response. RLRs share similar a domain architecture that supports specific detection of viral RNA. Upon binding of a short double-stranded RNA (dsRNA) bearing a 5′ triphosphate, RIG-I undergoes conformational changes that switch it from an auto-inhibitory state to an active state that exposes the N-terminal caspase activation and recruitment domains (CARDs), which then interact with the adaptor protein MAVS to activate type-I interferon expression. MDA5 adopts an open but inactive conformation that oligomerizes on long RNA duplexes, which brings its CARDs into proximity for activation of MAVS (16,18–21). DRH-3 has a domain architecture that is similar to RLRs, including an N-terminal domain (NTD), a DExH/D helicase core and a C-terminal domain (CTD) (22–25). Intriguingly, the DRH-3 NTD shares low amino acid sequence identity with the N-terminal tandem CARDs of RIG-I (11%) and MDA5 (12%) (23,24), but it may serve a different function in the case of DRH3. For example, direct RNA binding and enzymatic analyses have shown that the NTD of DRH3 contributes to cooperative, ATP-dependent dimerization on dsRNA molecules that are 22–25 bp in length (26,27). These studies indicated that the DRH-3 NTD contributes to RNA length discrimination and RNA 5′-terminus selection. Without the NTD, length-dependent RNA binding by DRH-3 is abolished. In addition, the N-terminal truncation variant develops an affinity for 5′-ppp over 5′-OH, suggesting a possible regulatory role for the NTD in dsRNA recognition (27). In vivo experiments have shown that the NTD of DRH-1 helps to suppress viral propagation, thereby playing an important role in the antiviral RNAi pathway (28). However, the biological function of DRH-3 NTD in RNAi is currently not yet characterized, making it particularly important to obtain structural information on this enigmatic domain.
The helicase core of DRH-3, which contains two Rec-A-like domains and a unique insertion domain (Hel2i), is highly related to the helicase domains in Dicer and the other RLRs (16,29–31). The sequential and structural conserved motifs for the binding of RNA and ATP are well characterized: motifs Q, I, II and III bind to ATP; motif VI in Hel2 facilitates ATP binding and hydrolysis; and motifs Ia-Ib and IV-V interact with RNA backbone in a sequence-independent manner (16,22). Previous studies of RIG-I Hel2i showed that this insertion domain ‘walks’ along dsRNA like a scanner and this movement may relate to ATP hydrolysis (32). The DRH-3 helicase domain is essential for RNA binding and ATP hydrolysis (27).
The CTD of DRH-3 is also highly homologous to the CTD of RLRs (33,34). Despite the apparent conservation in the overall CTD structure within this family of proteins, the RNA recognition specificity is different, which has important functional implications for viral RNA sensing by RLRs (27,35–38). For example, the RIG-I CTD shows high binding affinity to the 5′-triphosphate/diphosphate- (5′-ppp/5′-pp) terminus of short dsRNA species (32,34–36,39–41), whereas the MDA5 CTD preferentially recognizes the internal stem region of long dsRNA and interacts with weaker affinity (38,42,43). The LGP2 CTD can bind both the stem and termini of dsRNA (37). Similarly, the CTD of DRH-3 may recognize specific RNA features in a way that contributes to its function in the endogenous RNAi pathway.
Structural and functional studies of Dicer proteins and RLRs have greatly enhanced our understanding of their roles in RNAi and antiviral defense mechanisms. Unfortunately, the molecular mechanisms of endogenous RNAi pathways and the role played by DRH-3 are still poorly understood. Although full-length DRH-3 (DRH-3FL) can bind to both ssRNA and dsRNA, its ATPase activity can only be stimulated by dsRNA (with a kcat of 500/min). For optimal enzymatic function, DRH-3 requires RNAs of ∼22 bp, which is the typical length of a microRNA. The footprint or minimum dsRNA length for binding two molecules of any RLR is about 20–24 bp. Given that DRH-3 is structurally similar to RLRs, one ∼22 bp dsRNA is likely to be occupied by two DRH-3 monomers (27).
Here, we report crystal structures of the DRH-3 NTD and the CTD in complex with 5′-triphosphorylated RNA. In addition, we monitored the overall conformation and dynamic behavior of the DRH-3FL protein in solution using hydrogen/deuterium exchange mass spectrometry (HDX-MS). Together, our data provide new insights into the RNA binding specificity and structural dynamics of DRH-3, which will guide functional analyses of the endogenous RNAi pathway in worms.
MATERIALS AND METHODS
Protein expression and purification
The cDNA encoding the C. elegans DRH-3FL (UniProt Q93413) was inserted into the bacterial expression vector Champion pET-SUMO (Invitrogen). The resulting plasmid encodes DRH-3FL with an N-terminal hexahistidine (His6)-SUMO tag. The DRH-3FL gene was also used as a template for generating recombinant constructs of individual domains of DRH-3. Constructs were designed based on bioinformatics analysis and secondary structure predictions carried out using BLAST (44), GlobPlot (45) and JPred (46). The His6-SUMO-tagged NTD of DRH-3 (residues 1–335) was cloned into the expression vector pNIC28-Bsa4 (47). The CTD of DRH-3 (residues 940–1108) was cloned into the expression vector pET15b (Novagen), and the resulting clone carries an N-terminal His6 tag followed by a thrombin cleavage site. All cloned DNA sequences were confirmed by DNA sequencing. The recombinant constructs were transfected into chemically competent Escherichia coli cells [BL21(DE3) Rosetta II (Novagen), BL21(DE3) Rosetta T1R (Novagen) and BL21-CodonPlus (DE3)-RIL cells (Agilent Technologies)] for protein expression.
The transformed cells were grown in Luria broth (LB) with 50 μg ml−1 kanamycin and 35 μg ml−1 chloramphenicol at 37°C, and protein expression was induced with a final concentration of 500 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) once the OD600 reached 0.6–0.8. The incubation temperature was then lowered to 16°C, and the bacterial cell culture was further grown for an additional 20 h for protein expression. The cell pellet was harvested by centrifugation at 4000 ×g for 15 min at 4°C. The cells were resuspended in a buffer containing 25 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol and 5 mM β-mercaptoethanol and lysed by a Panda Plus 2000 homogenizer (GEA Niro Soavi). The lysate was clarified by high-speed centrifugation at 20 000 × g at 4°C for 40 min. The supernatant was then incubated with Ni-NTA resin (BioRad) that had been equilibrated with lysis buffer for 1 h at 4°C. After binding, the resin was washed with lysis buffer containing up to 20 mM imidazole, and the bound proteins were eluted with lysis buffer containing 200 mM imidazole. For purification of DRH-3FL and DRH-3 CTD, an additional step of heparin affinity chromatography was done to remove nucleic acid contamination. Briefly, the protein solution was allowed to bind to a 5 ml HiTrap Heparin HP column (GE Healthcare), and a linear gradient of increasing NaCl concentration was used to elute the protein fractions. Pure target protein normally elutes at approximately 400–500 mM NaCl. The eluted protein was then dialyzed overnight with SUMO protease or thrombin to remove the tag. The cleaved DRH-3FL was then further purified using a Superdex 200 HiLoad 16/600 size-exclusion column (GE Healthcare), while the cleaved DRH-3 CTD and DRH-3 NTD were purified using a Superdex 75 HiLoad 16/600 size-exclusion column (GE Healthcare). Peak fractions were pooled and concentrated to 5–15 mg ml–1 in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol and 2 mM DTT. The protein concentration was determined by absorbance spectroscopy at 280 nm, and the purity was assessed by SDS-PAGE.
The N-terminal His6-SUMO-tagged human RIG-I CTD (residues 802–925) was cloned into the expression vector pNIC28-Bsa4 (47) and was expressed and purified in the same way as the DRH-3 CTD.
Crystallization, data collection and structure determination
All crystals were grown by the hanging-drop vapor diffusion method. The native DRH-3 NTD crystals were grown at 0.2 M ammonium fluoride (pH 6.2), 20% w/v polyethylene glycol 3350 and 5 mM DTT at 4°C. The selenomethionine (SeMet)-incorporated DRH-3 NTD crystals were grown in 0.2 M potassium sodium tartrate tetrahydrate (pH 7.4), 20% w/v polyethylene glycol 3350 and 5 mM DTT at 4°C. The DRH-3 CTD was mixed with single-strand 5′-ppp 8-nt RNA at a 1:1 ratio. Crystals were grown from 0.2 M ammonium acetate, 0.1 M Tris pH 8.5, 25% w/v polyethylene glycol 3350 at 18°C. The DRH-3 CTD was mixed with self-complementary 5′-ppp 12-mer GC-rich RNA at a 2:1 molar ratio. Crystals were grown from 0.1 M Tris pH 7.5, 30% w/v polyethylene glycol 6000 at 18°C. Crystals of CTD-RNA complexes were flash-frozen with the reservoir solution and 25% (v/v) glycerol as cryoprotectant. Diffraction intensities for native DRH-3 NTD and SeMet DRH-3 NTD were collected at SLS-PXIII using the PILATUS 2 M-F detector and the multiaxis PIRGo goniometer (Paul Scherrer Institute, Villigen, Switzerland). Integration, scaling and merging of intensities were carried out using the programs iMOSFLM (48) and SCALA (49) from the CCP4 suite (50). Diffraction intensities for CTD-RNA complexes were recorded at NE-CAT beamline ID-24 at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL, USA). Integration, scaling and merging of the intensities were carried out with the programs XDS (51) and SCALA (52). The structure of the DRH-3 NTD was determined using the single-wavelength anomalous dispersion (SAD) method with the online server AutoRickshaw (53,54). The structure of the DRH-3 CTD in complex with the single-strand 5′-ppp 8-nt RNA was solved by the SAD method using SHELX (55). The structure of the DRH-3 CTD in complex with the 5′ppp dsRNA12 was solved by molecular replacement using Phaser (56). The models were further developed using rounds of manual building in COOT (57) and refined using Phenix (58). Statistics on the data collection and structure refinement for all three structures are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | ||||
|---|---|---|---|---|
| Macromolecule | DRH-3 NTD (Native) | DRH-3 NTD (SeMet) | DRH-3 CTD: 5′ppp 8-nt ssRNA | [DRH-3 CTD: 5′ppp 12-bp dsRNA]2 |
| Wavelength (Å) | 0.9998 | 0.9794 | 1.2819 | 0.9792 |
| Resolution (Å)a | 85.56–2.80 (2.95– 2.80) | 47.63–3.11(3.28–3.11) | 43.04–1.89 (1.93–1.89) | 43.96–1.60 (1.63–1.60) |
| Space group | P 41 21 2 | P 41 21 2 | P 43 21 2 | P 2 21 21 |
| Unit cell a/b/c(Å) | 93.18/93.18/215.96 | 89.93/89.93/215.68 | 56.843/56.843/131.75 | 33.775/93.090/133.823 |
| α/β/γ (°) | 90/90/90 | 90/90/90 | 90/90/90 | 90/90/90 |
| Unique reflections | 24012 (3401) | 16670(2324) | 17289 (984) | 56358 (2845) |
| Multiplicity | 11.1 | 12.8 | 9.2 | 3.1 |
| Completeness (%) | 99.3 (99.0) | 99.7(98.3) | 96.3 (88.4) | 99.4 (99.9) |
| I/sigma(I) | 10.9 (2.2) | 11.0(2.1) | 12.0 (1.7) | 14.9(1.6) |
| R merge b | 0.134 (0.896) | 0.241(1.352) | 0.142 (1.715) | 0.042 (0.761) |
| Wilson B-factor | 66.9 | 59.3 | 26.7 | 20.1 |
| CC1/2 | 1 (0.727) | 0.996(0.664) | 0.996 (0.378) | 0.998 (0.586) |
| Refinement statistics | ||||
| Reflections used in refinement | 22582 | 17263 | 56281 | |
| Number of atoms | 5390 | 1678 | 3650 | |
| macromolecules | 5252 | 1521 | 3188 | |
| ligands | 0 | 33 | 130 | |
| water | 138 | 124 | 332 | |
| Protein residues | 659 | 167 | 322 | |
| R work | 0.2337 | 0.1777 | 0.1692 | |
| R free c | 0.286 | 0.2213 | 0.1988 | |
| Rmsd from ideal bond length (Å) | 0.008 | 0.008 | 0.017 | |
| Rmsd from ideal bond angle (°) | 1.19 | 0.991 | 1.88 | |
| Ramachandran statistics | ||||
| Ramachandran favored (%) | 97.6 | 95.73 | 98.2 | |
| Ramachandran allowed (%) | 100 | 100 | 100 | |
| Ramachandran outliers (%) | 0 | 0 | 0 | |
| Clashscore | 6.53 | 2.66 | 5.53 | |
| Average B-factor | 86.3 | 37.06 | 29.77 | |
| macromolecules | 86.8 | 36.32 | 28.96 | |
| solvent | 67.43 | 38.98 | 36.60 | |
aValues in parentheses indicated values in the highest resolution shell.
bRmerge = Σ|Iobs − Iavg|/ΣIavg.
cRfree = Σ|Fobs − Fcalc|/ΣIcalc.Rfree was calculated with 5% of reflections.
RNA preparations
The 5′-ppp RNAs used for crystallization and HDX-MS were synthesized using T7 polymerase in vitro transcription. Transcription was carried out using template DNA with a T7 promoter (5′-G TAATACGACTCACTATA…), and the sequence was initiated by GTP. The reaction was then phenol-chloroform extracted, precipitated with 80% ethanol and purified on a 20% denaturing gel. The transcribed RNAs were run on a 20% denaturing gel to test for purity and size. All palindromic dsRNA oligonucleotides used for assays were synthesized on a MerMade 12 DNA-RNA synthesizer (BioAutomation) as previously described (59,60). Synthesized RNAs were deprotected (59) and gel purified. The synthetic RNAs were further analyzed for purity by mass spectrometry (Novatia LLC). FAM-labeled RNA was purchased from Integrated DNA Technologies. All RNA sequences are listed in Supplementary Table S1.
Limited proteolysis
A 1 mg ml–1 trypsin stock solution was prepared in trypsin digestion reaction buffer (25 mM HEPES, 5% glycerol, 150 mM NaCl and 1 mM DTT). DRH-3 CTD proteins and DRH-3 CTD-RNA complexes were incubated with trypsin at 1:2000 w/w for 0–2 h at 25°C. The reactions were quenched by the addition of SDS loading dye and boiling for 5 min. Samples were analyzed by SDS-PAGE.
Thermal shift assay
The thermal shift assay involves monitoring changes in the fluorescence signal of SYPRO Orange dye as it interacts with a protein undergoing thermal unfolding. The SYPRO Orange dye was supplied by Invitrogen (catalog number S6650) at 5000× concentration in DMSO and diluted in assay buffer for use. Each test sample (50 μl total volume) in a 96-well PCR plate (BioRad) contained 25 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM DTT, 5% glycerol, 5× SYPRO Orange dye, 5 μM enzyme and 6 μM RNA. In negative control samples, the buffer was added instead of test RNA(s). The plate was sealed with optical quality sealing tape and heated up from 25 to 95°C in increments of 1°C. Fluorescence changes in the wells were monitored simultaneously with a charge-coupled (CCD) camera. The wavelengths for excitation and emission were 485 and 575 nm, respectively. The temperature midpoint for the protein unfolding transition, the melting temperature (Tm), was calculated using BioRad iQ5 software.
Fluorescence polarization (FP) competition assay
Fluorescence polarization experiments were performed on a Synergy H1 plate reader (BioTek, Vermont) in 25 mM HEPES, 2.5% glycerol, 75 mM NaCl, 0.5 mM DTT and 0.01% Triton-X. The excitation and emission wavelengths were set to 485 and 510 nm, respectively. The 5′-OH hairpin RNA was purchased from IDT, and fluorescein amidite (FAM) was labeled at the hairpin region of the RNA.
Polarization was calculated using the equation:
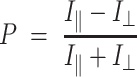 |
Where P is the fluorescence polarization, 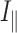 is the intensity with polarizer parallel and
is the intensity with polarizer parallel and 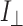 is the intensity with polarizer perpendicular.
is the intensity with polarizer perpendicular.
For the equilibrium binding using the FAM-labeled RNA as the ligand, the binding data were fit using the Hill equation with a Ymin:
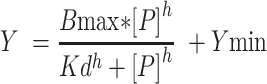 |
Where Y is the concentration of the Protein: RNA complex; [P] is the total protein concentration; 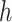 is the Hill coefficient, Kd is the equilibrium dissociation constant, Bmax is the intercept on the x-axis which represents the maximum specific binding, and Ymin is the biological activity of Fam-labeled RNA without protein binding.
is the Hill coefficient, Kd is the equilibrium dissociation constant, Bmax is the intercept on the x-axis which represents the maximum specific binding, and Ymin is the biological activity of Fam-labeled RNA without protein binding.
For competition assays, 400 nM DRH-3 CTD and 5 nM FAM-labeled RNA (R0, Supplementary Table S1) were mixed and incubated in 25 mM HEPES, 2.5% glycerol, 75 mM NaCl, 0.5 mM DTT, 0.01% Triton-X in a Greiner Bio-One 384-well Black Polystyrene Microplate for 1 h at room temperature. RNA competitors were serially diluted over a concentration range from 10 μM to 2.4 nM. RNA competitors (R1, R1P, R1OH, R2 and R3, Supplementary Table S1), which were serially diluted over a concentration range from 10 μM to 2.4 nM, were then added and distributed equally.
The titration curve was fitted with log (inhibitor) versus response–variable slope (four parameters) using GraphPad Prism 5 (GraphPad software, San Diego California USA, www.graphpad.com).
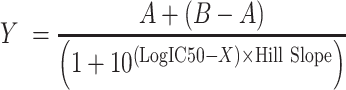 |
Where X is the concentration of competitor RNAs, A is the polarization of free FAM-RNA, B is the polarization of free FAM-RNA bound to protein, and Hill Slope is the slope factor.
The inhibition (%) or IC50 was calculated using the equation below.
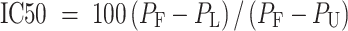 |
where PF is the polarity of fully bound 5′-OH hairpin RNA; PU is the polarity of unbound 5′-OH hairpin RNA, and PL is the polarity of 5′-OH hairpin RNA in the presence of RNA competitors.
Hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS)
DRH-3FL proteins were exposed to deuterated water for 1 h followed by a quenching step (low pH on ice) to stop hydrogen/deuterium exchange. Proteins were digested into peptide fragments by low-temperature and low-pH compatible pepsin. The resulting peptides were subsequently separated by capillary-flow liquid chromatography and measured by mass spectrometry. The level of deuterium incorporation was then determined by examining the change in the mass centroid. HDX Workbench (61) was utilized to analyze the data and generate graphs. HDX-MS results for DRH-3 proteins are shown as a graphical heatmap that revealed the level of deuterium incorporation (%D) for detected peptides. The heatmap of HDX was subsequently projected onto the crystal structure of DRH-3 CARDs and DRH-3FL models with and without RNA ligand to represent the molecular structure and protein dynamics of DRH-3.
Structure modeling
Homology models of DRH-3 proteins were generated using SWISS-MODEL (62). DRH-3 CARD (PDB code: 6JPX), RIG-I helicase domain (PDB code: 5E3H) and DRH-3 CTD (PDB code: 6JPY) crystal structures were used as input reference models. For illustration purposes, the HDX-MS data were taken into account to model the relative positions of the domains.
Negative staining transmission electron microscopy
The samples were prepared as follows: 50 μg/ml of DRH-3FL recombinant protein were mixed with 3 μg/ml of dsRNA of different lengths in buffer containing 25 mM HEPES pH 7.5, 0.1 M KCl, 5 mM MgCl2, 1.5 mM ADP-AlF4. RNAs used in this experiment are listed in Supplementary Table S1. The mixture was then incubated on ice for 30 min. After incubation, 4 μl of each sample were applied to glow-discharged carbon-coated grids. The grids were then negatively stained with uranyl acetate [1% (wt/vol)] and imaged with an FEI Tecnai 12 transmission electron microscope at an accelerating voltage of 200 kV. Micrographs were taken at -1 μm defocus, 49 000 × magnification and 3.061Å per pixel.
Analytical size-exclusion chromatography
Purified DRH-3FL and gel purified RNA sample was diluted to 27 and 29 μM, respectively, in the size-exclusion chromatography (SEC) running buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM DTT and 5% glycerol. The protein–RNA complex was prepared by mixing 140 μl of diluted DRH-3FL with 140 μl of diluted RNA for an almost equimolar protein to RNA ratio. The mixture was then incubated for 30 min on ice before injection into Superdex 200 Increase 10/300 GL column. Fractions containing protein and RNA were identified by measuring their absorbance at 280 and 260 nm. The absorbance against elution volume was plotted using the scatter plot function in R.
RESULTS
DRH-3 NTD contains unique tandem CARDs in a parallel arrangement
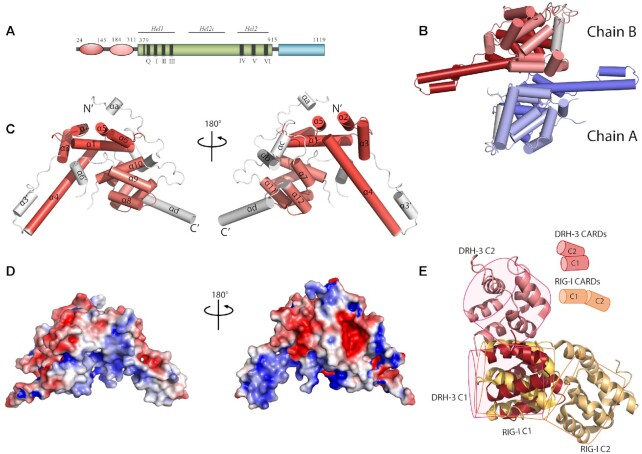
As observed previously for RLR proteins, the DRH-3 NTD is connected to the helicase-CTD RNA binding module via a long linker (Figure 1A). The DRH-3 NTD crystal structure was solved at 2.8 Å resolution (Figure 1B), revealing a domain that is composed primarily of α helices arranged in two tandem helical bundles (Figure 1C). The two bundles adopt Greek-key-like folds linked by loops and small helices (63,64). We compared the structures of the α-helical bundles with their respective top 5 hits from the DALI server (65) (Supplementary Figure S1 and Supplementary Table S2). All hits fell into the CARD domain family of proteins that participate either in apoptosis [nucleolar protein 3 (NOL3), procaspase-9 (pc-9) and apoptotic protease activating factor 1 (Apaf-1)] or inflammation [caspase recruitment domain family member 11 (CARD11), ICEBERG, RIG-I, B Cell CLL/Lymphoma 10 (Bcl10) and Bcl10-interacting CARD protein (BinCARD)]. Despite the low sequence identities shared by the two α-helical bundles of the top hits (the sequence identity is <10%), the relative orientation of the helices is similar. We, therefore, refer to the two helical bundles in the DRH-3 NTD as CARD1 (helix α1–6) and CARD2 (helix α7–12). The first helix in DRH-3 CARD1 has a kink, which is a common feature for CARDs (66,67). The third and fourth helices in DRH-3 CARD1 and CARD2 are longer than canonical CARDs, resulting in a different overall shape (68).
Figure 1.
The overall structure of the DRH-3 N-terminal domain. (A) Schematic representation of the domain architecture of DRH-3. NTD, worm-specific N-terminal domain; HEL, the helicase domain; CTD, the C-terminal RNA-binding domain. The residues numbers corresponding to the predicted domain boundaries were labeled accordingly. (B) Crystal structure of dimeric DRH-3 NTD. Two DRH-3 NTD molecules in the asymmetric unit are shown as cartoons. Chain A and Chain B are indicated in different colors. (C) Cylinder depiction of DRH-3 NTD structure with helices labeled. DRH-3 NTD contains two CARD-like α-helical bundles, highlighted in red (NTD_C1) and pink (NTD_C2) colors, respectively. The topology of a 6-α-helical bundle is that of a Greek Key. Cylinders represent the helices, and the arrow represents the rotation direction of the polypeptide chain. (D) The surface of DRH-3 NTD colored according to the electrostatic surface charges where blue is for positive charges, red for negative charges, and white for the neutral surface. (E) Superposition of DRH-3 NTD1 with RIG-I CARD1. The relative position of the two CARD/CARD-like domain in DRH-3 and RIG-I are compared and illustrated in simplified cartoon models.
The two CARDs in DRH-3 adopt a side-by-side arrangement. In contrast, the two CARD domains of RIG-I are more similar to each other (20% amino acid sequence identity) and are oriented in a head-to-tail arrangement (Figure 1E). CARD1 and CARD2 interact with each other through electrostatic interactions between a positively charged Arginine residue (R30, α1, CARD1) and a negatively charged Aspartic acid residue (D188, α7, CARD2) (Supplementary Figure S2). The CARD1–CARD2 interface is further stabilized by hydrogen bonds between helices α1-α10 (Y28-L270) and helices α4-α7 (N125-Y184) (Supplementary Figure S2). We further analyzed the electrostatic potential at the solvent-accessible surface of DRH-3 CARDs (Figure 1D). The surface of DRH-3 CARDs contains several highly charged patches. The positive patches fall onto the helix stem composed of helices α4 and α3′; the negative patches were on the upper helix stem region (helices α2 and α3). These surfaces may contribute to the functions of DRH-3 because death-fold proteins signal via electrostatic interactions (67).
There are two DRH-3 NTD molecules (rmsd 0.367 Å for 305 superimposed Cα atoms) in one asymmetric unit. To examine whether the dimeric interface was biologically relevant, we analyzed the structure using PDBePISA (69) (Supplementary Figure S3). The interface area (540 Ų) is approximately 10% of the total solvent-accessible area (5273 Ų). Although this dimer interface is not extensive, it may provide a hint of a physiological function for the DRH-3 NTD. DRH-3 NTD alone and RNA-free DRH-3FL exist as monomers in solution (Supplementary Figure S4), but dsRNA-associated DRH-3 can dimers. Therefore, the observed interface may represent a possible dimerization interface of DRH-3 NTD in the context of the DRH-3FL protein upon binding to certain RNA species. It is a common strategy for CARDs to interact through complimentary charged surfaces (64). Due to high sequence divergence, the interaction interfaces among CARDs can be quite variable (67). It has been reported that CARDs from RIG-I and MDA5 are capable of forming three types of intermolecular interactions (type Ia/b, IIa/b and IIIa/b) (43,70,71). The interface of DRH-3 CARD1–CARD2 belongs to none of these groupings, perhaps because the two CARDs in DRH-3 are not identical. The two CARDs in DRH-3 are distinct in sequence (only 11% amino acid sequence identity) and structure (high rmsd value of 4.9 Å for all Cα atoms). CARDs usually form oligomers via homotypic interactions (64), as observed for the Apaf-1 CARD and caspase-9 CARD which is the first reported structure of a CARD-CARD interaction (72). We speculate that the DRH-3 CARD1 and CARD2 interact with different inter- or intra- molecular targets, to regulate specific signaling events. Overall, the NTD of DRH-3 folds into a set of novel tandem CARDs, and these may function by interacting with other CARD-containing proteins in C. elegans RNAi pathways.
DRH-3 CTD recognizes 5′ tri-phosphorylated dsRNA.
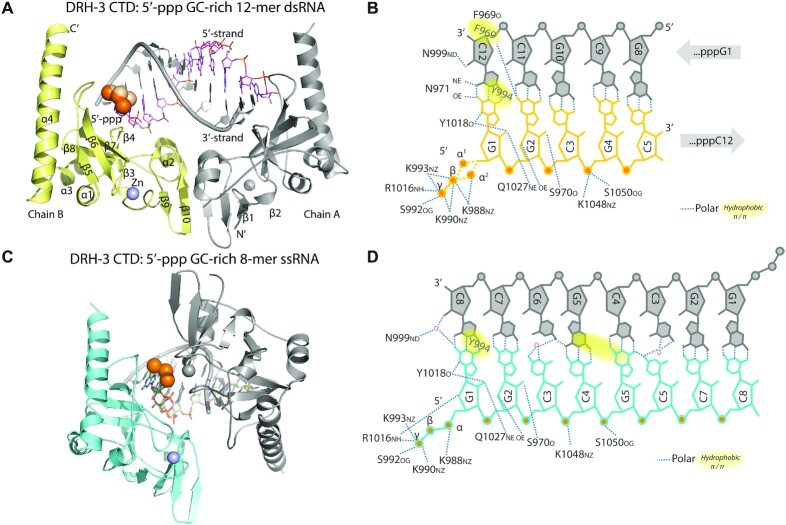
To elucidate the structural basis for secondary siRNA recognition, we obtained two crystal structures of the DRH-3 CTD in complex with 5′-ppp RNAs (Figure 2A, C and Table 1). The overall conformation of the DRH-3 CTD is similar to that of the RLR CTDs: they all contain a saddle-like shape that consists of two antiparallel β-sheets (β-sheet1: β1–2 and β9–10, β-sheet2: β5–8) and exhibits a high degree of shape complementarity to their RNA ligands. The long helix (α4) at the C-terminus, which stacks on the backside of the 5′-ppp binding site, is a unique feature of DRH-3. A C4-type zinc finger binding motif (strands β1–2 and β6–7) is found in the DRH-3 and RLR CTDs, and it captures a zinc ion that plays a role in maintaining the overall fold of the CTD (35–38). Despite the structural similarity, the CTD of DRH-3 has a low sequence identity to the CTDs of RIG-I (22%), MDA5 (21%) and LGP2 (22%).
Figure 2.
5′-ppp RNA recognition by DRH-3 CTD. (A) The overall structure of DRH-3 in complex with 5′-ppp 12-mer dsRNA. The crystallographic asymmetric unit contains a 2:1 complex between DRH-3 CTD and the dsRNA. Chain B and the 5′ stand are colored. The 5′-ppp is shown as orange spheres and the zinc molecule is shown as a purple sphere. (B) The schematic diagram of contacts between DRH-3 CTD and 5′-ppp 12-mer dsRNA. (C) The overall structure of DRH-3 in complex with 5′-ppp 8-mer ssRNA. The crystallographic asymmetric unit contains a 1:1 complex between DRH-3 CTD and the ssRNA. The symmetry mate structure is shown in gray. The 5′-ppp is shown as orange spheres and the zinc molecule is shown as a purple sphere. (D) The schematic diagram of contacts between DRH-3 CTD and 5′-ppp 8-mer ssRNA.
We observe that the RNA 5′-ppp participates in an extensive network of electrostatic interactions with the DRH-3 CTD (Figure 2B, D and Supplementary Figure S5). For example, K992 and R1016 interact with γ-phosphate; K990 and K993 interact with γ- and β-phosphates; and K988 interacts with α- and β-phosphates. Similarly charged residues within the CTD of RIG-I are important for binding 5′-ppp RNAs (K858-K861) (35,36,40). In addition, residues K1048 and S1050 in the conserved KWK motif (28,73) mediate interactions between the DRH-3 CTD and the RNA sugar-phosphate backbone. The 3′-stranded RNA contributes to RNA binding via a hydrogen bond between C12 and residues F969. Notably, the DRH-3 CTD binds to 5′-ppp dsRNA and 5′-ppp ssRNA in a similar manner (Figure 2 and Supplementary Figure S5).
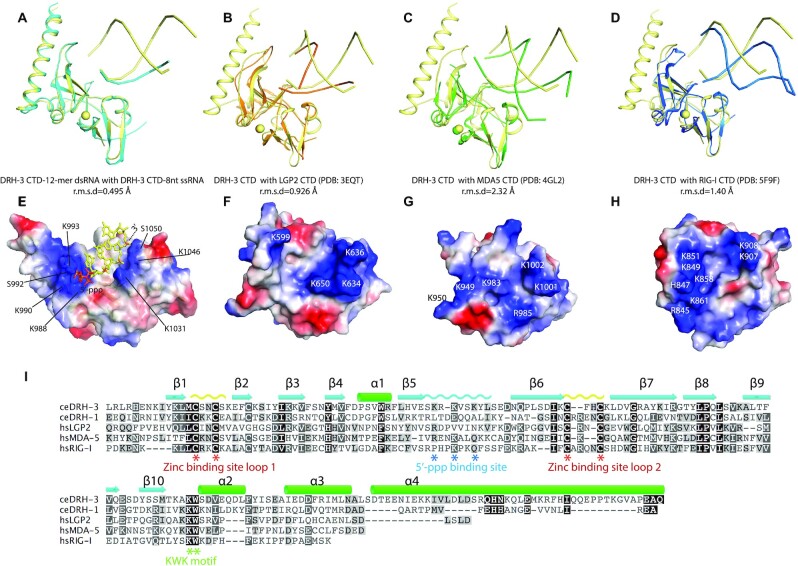
We then compared the DRH-3 CTD with the available human RLR structures (Figure 3A–D). Superposition of the DRH-3 CTD with the RLR CTDs resulted in rmsd values ranging from 0.9 to 2.3 Å for all superimposed Cα atoms, indicating a high level of structural similarity despite a relatively low sequence similarity. Intriguingly, the 5′-ppp binding loop between β-strands 5 and 6 is similar in DRH-3, RIG-I, and LGP2, which is consistent with their preferences for dsRNA 5′-ppp or pp (Figure 3A–D). To further interpret these observations, the electrostatic potential at the solvent-accessible surface of DRH-3 CTD was analyzed. A continuous positively charged surface area was identified, and it corresponds to the 5′-ppp recognition and the RNA phosphate backbone binding sites (Figure 3E). This positively charged patch is also observed in LGP2, MDA-5 and RIG-I CTD (Figure 3F–H).
Figure 3.
Comparison of C-terminal domain of DRH3 and RLRs. (A) Superposition of DRH-3 CTD structures bound to the 5′-ppp 12-mer dsRNA and the 5′-ppp 8-mer ssRNA. (B) Superposition of the RNA-bound DRH-3 CTD (yellow) and LGP2 CTD (orange, PDB code: 3EQT). (C) Superposition of the RNA-bound DRH-3 CTD (yellow) and MDA5 CTD (green, PDB code: 4GL2). (D) Superposition of the RNA-bound DRH-3 CTD (yellow) and RIG-I CTD (orange, PDB code: 3EQT). Polypeptide chains are shown as cartoons. Structural superimposition was done by pairwise alignment and represented by Pymol (98). (E) Electrostatic charge at solvent-accessible surfaces of dsRNA-bound DRH-3 CTD. Positively charged surfaces are colored blue and negatively charged surfaces are red. Key residues involved in the RNA binding were labeled. (F–H) Electrostatic charge at solvent-accessible surfaces of LGP2 CTD, MDA5 CTD and RIG-I CTD. Each CTD is shown in the same orientation as in Figure 3A–C. Positively charged surfaces are colored in blue and negatively charged surfaces are colored in red. The key charged residues are indicated. (I) Alignment of amino acid sequences of DRH and RLR CTDs. Sequences were aligned using the program ClustalW and visualized using Geneious software. The conservation of the amino acids is indicated by colors.
To determine the RNA binding characteristics of DRH-3, we tested how 5′- terminal modifications (such as 5′-ppp, 5′-OH and 5′-p) and 5′ or 3′ overhanging regions on the 5′-ppp dsRNA affect RNA binding to the DRH-3 CTD (Supplementary Table S2). First, we examined whether the bound RNA could protect the DRH-3 CTD from trypsin digestion (Supplementary Figure S6A). Trypsin cleavage site prediction suggested that the CTD in DRH-3, DRH-1 and RIG-I share at least 2 similar cleavage sites at RNA binding regions, including the RNA-Cap binding site and the KWK motif. Therefore, we set out to determine whether RNA binding would affect the trypsin digestion rate. Unbound DRH-3 CTD was digested into two major fragments after 5 min of treatment with trypsin. When the DRH-3 CTD was incubated with different dsRNA species before treatment with trypsin for 5 min, the digestion patterns were very similar, suggesting that the RNA did not protect the DRH-3 CTD from trypsin digestion (Supplementary Figure S6A). In contrast, all the 12-mer dsRNAs protected RIG-I CTD and DRH-1 CTD from trypsin digestion (Supplementary Figure S6B and C), especially blunt-ended 5′-ppp and 5′-pp and 5′-ppp with a 3′-overhang, suggesting that RIG-I CTD prefers binding to 5′-ppp RNAs. We then conducted thermal stability assays to determine the effect of RNA binding on protein stability (Supplementary Figure S6D and E). The assay was done with RNA ligands bearing various modifications at their 5′ or 3′ end to determine if DRH-3 has a preference for a particular RNA. No significant difference in the melting temperature (Tm) was observed in the presence and absence of the RNA ligands, indicating that RNA binding failed to change the thermal stability of DRH-3 CTD (Supplementary Figure S6D). In contrast, all 12-bp RNAs used in this study increased the melting temperature (Tm) of DRH-3 CTD from 1°C for 5′-ppp + 5′G to a maximum of 9°C for 5′-ppp + 3′G, suggesting that binding of short dsRNA stabilizes the DRH-3 CTD, particularly 5′-ppp dsRNAs (Supplementary Figure S6E). Specifically, the observed increase in the melting temperature suggests that 5′-ppp dsRNA molecules containing a 3′ overhang have higher binding affinity to the DRH-3 CTD, consistent with the trypsin digestion data. Similarly, all dsRNAs, especially the 5′-ppp with a 3′-overhang, increased the thermal stability of the RIG-I CTD (Supplementary Figure S6D). Together, results from the limited proteolysis and thermal stability assays suggest that the DRH-3 CTD preferentially binds to dsRNA molecules containing a 5′-ppp.
To further characterize this finding, we measured RNA binding using a fluorescence polarization (FP)-based competition assay. The binding affinity of the DRH-3 CTD to a FAM-labeled 5′-OH RNA hairpin probe was determined to be 217.0 nM (Supplementary Figure S7A). We then titrated each competitor RNAs through a range of concentrations that resulted in a maximum polarity of 50% (IC50 values) (Supplementary Figure S7B-C). Consistent with the crystallographic findings showing apparent interactions between 5′-ppp RNA ligands and specific amino acids of the DRH-3 CTD, 5′-ppp RNAs showed the highest degree of competition with the probe (with IC50 values ranging from 80 to 234 nM), indicating that the DRH-3 CTD preferentially recognizes 5′-ppp RNAs. In contrast, the 5′-OH and 5′-p RNAs competed poorly with the RNA probe with relatively high IC50 values of 631 to 752 nM. Overall, DRH-3 CTD preferentially recognizes 5′ppp blunt-ended dsRNAs.
DRH-3FL displays an open conformation.
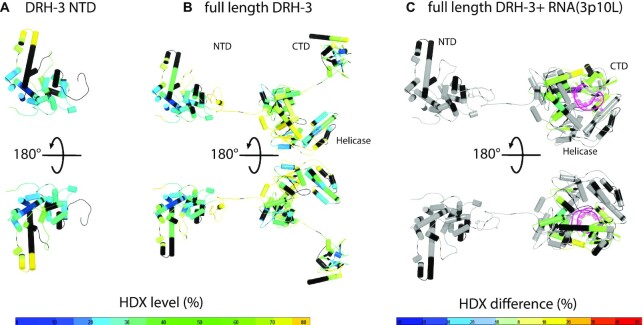
HDX-MS was conducted to examine the overall architecture of DRH-3FL in response to different ligands. HDX-MS was previously used to dissect the overall conformational changes of RIG-I and MDA5 upon ligand binding (74–76). The exchange rates of amide hydrogens to deuterium in the target protein in the presence of deuterated solvent were mapped against the amino acid sequence and structural models of DRH-3 conformational states. The NTD CARDs in isolation and CARDs of the DRH-3FL free protein showed similar deuteration profiles (Figure 4A, B; Supplementary Figure S8A and S8B), suggesting a lack of intramolecular interactions between the CARDs and the core central domains of DRH-3FL. This finding is different from what is observed with RIG-I, in which the CARDs are sequestered in an autoinhibitory conformation (20,29,75). The majority of peptides on DRH-3 CARDs (86%) exhibited a magnitude of deuterium uptake that was <50%. Low H/D exchange level suggests that the DRH-3 CARDs are well-folded and thus, less accessible to solvent. This contrasts with the less ordered linker region between the CARDs and the helicase domain (HEL). As expected, the linker region (I351-E368) was more solvent-exposed (with 70% deuteriation) in DRH-3FL. Therefore, the overall conformation of DRH-3FL appears to be similar to that of free MDA5, in which the various protein domains are relatively unconstrained (75).
Figure 4.
Conformational dynamics of DRH-3. Deuterium uptake profiles of the DRH-3 NTD (A) and DRH-3FL (B) are presented in a front and back view. The structure of DRH-3 NTD was obtained from this study and the DRH-3FL model was generated by assembling all three domains via fusing peptide bonds. The deuterium exchange data were mapped onto the structural models of DRH-3 NTD and DRH-3FL. The % deuterium exchange in the deuterium exchange of each peptide is colored according to the scale bar. Blue indicates the lowest and orange indicates the highest exchange rate. Black indicates that the region has no amide hydrogen exchange activities. (C) HDX-MS differential map for DRH-3FL with a short hairpin RNA in a front and back view. Differential single amino acid consolidation HDX data were mapped on to full-length RNA-bound DRH-3 structural model. The percentages of deuterium differences are color-coded according to the scale bar.
To map the RNA binding interface and RNA-induced conformational dynamics of DRH-3FL in solution, we performed HDX-MS experiments for DRH-3FL in the presence of the short 5′-ppp hairpin RNA 3p10L [5′-ppp GGACGUACGUUUCGACGUACGUCC, also known as SLR10 (77)], which was shown to bind and activate RIG-I to initiate the type I interferon-mediated immune response (32,75,77). In the presence of 3p10L, several conserved motifs within the helicase domain (motifs Ia, II, IVa, Vb and VI) and the CTD (the 5′-ppp binding site and the KWK motif) displayed a significant reduction in deuteration, suggesting their involvement in RNA binding. A helix of Hel2i and the Pincer region were also affected by 3p10L binding (Figure 4C and Supplementary Figure S8C). Notably, the H/D exchange rate of the CARDs in the DRH-3FL with or without 3p10L remained unchanged, suggesting that the binding of 3p10L had no allosteric influence on the dynamics of the CARDs, unlike the RIG-I CARDs. It’s known that RIG-I is autoinhibited by the formation of an intra-molecular CARD2-Hel2i interaction (20,78). The 3p10L can only be occupied by one DRH-3 molecule, and therefore dsRNA binding may not be sufficient to induce a conformational change in DRH-3. These results may suggest that DRH-3 activation may require its cooperative dimerization (22,27).
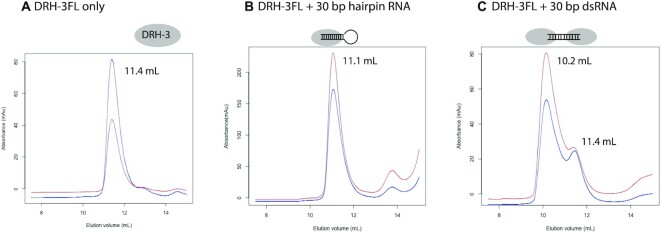
The molecular state of DRH-3FL in the presence of 30-bp RNAs was evaluated by analytical size exclusion chromatography. Monomeric DRH-3FL forms a 1:1 stable complex with the 30 bp hairpin RNA carrying only one 5′ppp end and a 2:1 stable complex with the 30 bp dsRNA carrying two 5′ppp ends (Figure 5B and C). We also used negative staining electron microscopy to directly visualize the ability of DRH-3FL to oligomerize upon binding longer dsRNAs. The recombinant DRH-3FL protein was incubated with dsRNA of 100 bp (Supplementary Table S1). Samples were then applied on carbon-coated grids and negatively stained with uranyl acetate. Particles of the free DRH-3FL are largely monomeric (Supplementary Figure S9A). The observed DRH-3 forms rounded particles in the presence of 100 bp dsRNA, with no sign of RNA-dependent filaments (Supplementary Figure S9B). This is in contrast to MDA5 which can form RNA-mediated helical oligomers (43,79–82).
Figure 5.
DRH-3 forms stable complexes with short dsRNAs with 5′-ppp. (A) Size exclusion chromatography (SEC) profile of DRH-3FL. Blue and red lines represent the absorbance at 280 nm (A280) and 260 nm (A260), respectively. The elution volume of the monomeric DRH-3FL in Superdex 200 Increase 10/300 GL is 11.4 ml. The A260/A280 ratio is typical for proteins. (B) SEC profile of DRH-3FL mixed with the 30-bp hairpin RNA. DRH-3FL forms a 1:1 complex with a 30-bp hairpin RNA. A higher A260/A280 ratio indicates that DRH-3FL forms a complex with the hairpin RNA. The elution volume of the DRH-3FL RNA complex is 11.1 ml. (C) SEC profile of DRH-3FL with a 30-bp dsRNA. DRH-3FL proteins form a dimer upon binding to the 30-bp dsRNA. The elution volume of the major DRH-3FL RNA complex is 10.2 ml, suggesting that more than one monomer is bound to the RNA.
In summary, the DRH-3FL protein adopts an extended conformation where the N-terminal CARDs, HEL and CTD behave as solvent-exposed domains connected by flexible linkers. DRH-3 behaves somewhat differently from both RIG-I and MDA5, as there are no apparent intra-molecular auto-inhibitory interactions nor the formation of dsRNA-mediated filaments (19–21,79,82,83).
DISCUSSION
In this study, we characterized the structure and function of DRH-3 protein from C. elegans. From its crystal structure, the DRH-3 NTD is observed to be structurally similar to death-fold proteins, especially CARDs. In mammals, CARDs play important roles in apoptosis, immune responses and cytokine processing signaling pathways, while in worms, CARDs are restricted to apoptotic proteins (64). DRH-3 is the first CARD-containing protein involved in the RNAi pathway that has been discovered thus far. Different from classical CARD-containing proteins, the two CARDs of DRH-3 arrange themselves in parallel and interact at a native interface, indicating their distinctive physiological roles. CARD–CARD interaction is required for CARD-containing proteins to perform their signaling function (67). Therefore, if DRH-3 CARDs are not bound to each other, they may interact with other CARD-containing proteins via intermolecular CARD–CARD interactions during the RNAi process. Alternatively, they may bind to a completely different type of interaction partner. Oligomer formation among CARDs primarily depends on interactions between charged surfaces and is relatively independent of amino acid sequences. As such, the DRH-3 CARDs and their prospective binding partners are expected to exhibit complementarity of charged interfaces. Given the involvement of DRH-3 in 22G siRNA biogenesis, it is likely to sense the 5′ triphosphorylated ∼22 bp dsRNA along with other partner proteins, such as worm-specific RdRp and Agos (2,3,13,14).
Oligomerization of CARDs is thought to enhance translocation rates and provide a basis for cooperativity in ATPase hydrolysis and/or translocation (16,29,53,71,79). The activation of CARD oligomerization may require specific activation mechanisms. For example, RIG-I signaling is regulated by ubiquitin ligases through K63-linked ubiquitin ligation at the CARD domain (50,84,85). In particular, TRIM25 is suggested to interact with the CARDs and assists tetramer formation (86). Notably, the residues T170 and K172 in CARD2 are critical for RIG-I ubiquitination (87). The phosphorylation of residue Thr170 affects the interface between CARD1–CARD2, while residue K172 is the ubiquitination site. However, noncovalent ubiquitination is dispensable for MDA5 oligomerization and signaling. In contrast, MDA5 CARD2 has a lower affinity for K63-Ubn and forms oligomers in a concentration-dependent but not ubiquitin-dependent manner (43,88). The previous qRT-PCR analysis shown drh-1, the worm homolog of drh-3, mediates the intracellular pathogen response upon viral infection and is required induction of ubiquitin ligase complex component skr-5 (89). The NTD or the CARDs of DRH-1 and DRH-3 may contain ubiquitination sites for their oligomerization thus signaling. Additionally, the linker between CARD2 and the helicase domain is ∼30 amino acids in DRH-3, ∼50 amino acids long in RIG-I and ∼100 aa long in MDA5 (43,88). Instead of forming a tetramer like RIG-I CARDs or a larger oligomer like MDA5 CARDs, the shorter linker probably only allows DRH-3 to form a dimer. Another indication is that the cooperativity of DRH-3 on dsRNA is weaker than MDA5 but stronger than RIG-I (27).
Our structural studies show a positively charged binding site on DRH-3 CTD for 5′-ppp. A similar type of 5′-ppp binding site is also observed within the RIG-I CTD (Figure 3). In humans, the 5′-ppp dsRNA is a viral pathogen-associated molecular pattern (PAMP) that is specifically recognized by RIG-I, whose activation triggers downstream Type I interferon responses. The binding affinities of 5′-OH dsRNA and 5′-ppp ssRNA are particularly low compared to that of 5′-ppp dsRNA (35,40,41). ATP hydrolysis facilitates the removal of lower-affinity self-RNAs from the RIG-I binding site (32,34,41,90–92). Similarly, the DRH-3 CTD can discriminate different types of RNA terminus and this may contribute to target RNA selectivity. The FP-based competition assays carried out in this study show that DRH-3 prefers RNAs terminated by a 5′-ppp (a signature of RdRp-generated RNAs) in comparison with 5′-p and 5′-OH, Dicer-cleaved RNAs, or other exogenous RNAs, suggesting that it plays a role in secondary siRNA recognition.
The HDX-MS study suggests that the DRH-3FL is an allosteric enzyme that is regulated by RNA binding. The conformational changes during ATP hydrolysis of DRH-3 remain unclear. The domains of DRH-3 may cooperate to interact with RNA and substrate ATP for enzymatic activity. Unlike the CARDs in RIG-I, which are locked against the helicase domain in an auto-inhibitory conformation in the absence of RNA ligand, the NTD in DRH-3 does not show any intramolecular interaction with the helicase domain. The crystallographic dimeric form of the NTD suggests a possible biological relevance, as many CARD domains reported so far oligomerize for function (93–96). In C. elegans, RdRps transcribe secondary small regulatory RNAs de novo from mRNA-derived templates (2,8,9). Together with a dual Tudor domain protein, DRH-3 is reported to be involved in the biogenesis of secondary small RNAs, including endogenous 22G and 26G RNAs and exogenous secondary virus-derived siRNAs (2,3,12,97). It is hypothesized that DRH-3 may facilitate the translocation of RdRp along with the template RNA and that the RNA-dependent ATPase function of DRH-3 is essential for this role (2), although direct observation of DRH3 motion has not been reported. The functional activation of DRH-3 is largely dependent on its RNA binding and ATP hydrolysis status and its protein partners. Full understanding of the mechanism for DRH-3 activation and its function requires future structural and functional studies using additional components, along with more biologically relevant systems such as a worm or its tissue extracts containing the active endo-siRNA biogenesis complex.
DATA AVAILABILITY
The atomic coordinates and structure factors for the DRH-3 N-terminal CARDs have been deposited with the Protein Data bank under accession code 6M6Q. The atomic coordinates and structure factors for the DRH-3 CTD domains in complex with 5′pppRNAs have been deposited to the Protein Data bank under accession codes 6M6R and 6M6S.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the beamline staff at TPS 05A beamline in National Synchrotron Radiation Research Center, Hsinchu, Taiwan, MXII beamline in Australian Light Source, Melbourne, Australia and PSIII beamline in Swiss Light Source (SLS) Paul Scherrer Institute, Switzerland and NECAT 24-ID at Argonne National Laboratory, Argonne, IL, the US for providing us outstanding support during the data collection. The authors wish to thank Drs. El Sahili Abbas and Chong Wai Liew for their help on X-ray data collection at synchrotron X-ray beamlines. The acknowledgments extend to the Protein Production Platform (PPP) at Nanyang Technological University School of Biological Sciences for assistance with high-throughput screening of protein overexpression. O.F. is a research scientist supported by the Howard Hughes Medical Institute. D.L. was a research fellow supported by the Howard Hughes Medical Institute. A.M.P. is an Investigator in the Howard Hughes Medical Institute.
Author Contributions: K.L., A.M.P. and D.L. designed the study. K.L., J.Z., M.W., N.M.T., O.F. and D.L. performed the experiments. All authors contributed to the results analysis and critical evaluation and preparation of the manuscript.
Notes
Present address: Jie Zheng, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Contributor Information
Kuohan Li, Lee Kong Chian School of Medicine, Nanyang Technological University, EMB 03-07, 59 Nanyang Drive 636921, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive 637551, Singapore; NTU Institute of Structural Biology, Nanyang Technological University, EMB 06-01, 59 Nanyang Drive 636921, Singapore.
Jie Zheng, The Scripps Research Institute, Jupiter, FL 33458, USA.
Melissa Wirawan, Lee Kong Chian School of Medicine, Nanyang Technological University, EMB 03-07, 59 Nanyang Drive 636921, Singapore; NTU Institute of Structural Biology, Nanyang Technological University, EMB 06-01, 59 Nanyang Drive 636921, Singapore.
Nguyen Mai Trinh, Lee Kong Chian School of Medicine, Nanyang Technological University, EMB 03-07, 59 Nanyang Drive 636921, Singapore; NTU Institute of Structural Biology, Nanyang Technological University, EMB 06-01, 59 Nanyang Drive 636921, Singapore.
Olga Fedorova, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520, USA; Howard Hughes Medical Institute, Chevy Chase, MD 20815, USA.
Patrick R Griffin, The Scripps Research Institute, Jupiter, FL 33458, USA.
Anna M Pyle, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520, USA; Howard Hughes Medical Institute, Chevy Chase, MD 20815, USA.
Dahai Luo, Lee Kong Chian School of Medicine, Nanyang Technological University, EMB 03-07, 59 Nanyang Drive 636921, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive 637551, Singapore; NTU Institute of Structural Biology, Nanyang Technological University, EMB 06-01, 59 Nanyang Drive 636921, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Health’s National Medical Research Council, Individual Research Grant (OF-IRG) [NMRC/OFIRG/0075/2018]; Singapore Ministry of Education, Education Academic Research Fund Tier 1 [2018-T1-002-010]; Howard Hughes Medical Institute. Funding for open access charge: Singapore Ministry of Health’s National Medical Research Council, Individual Research Grant (OF-IRG) [NMRC/OFIRG/0075/2018].
Conflict of interest statement. None declared.
REFERENCES
- 1.Tabara H., Yigit E., Siomi H., Mello C.C.. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002; 109:861–871. [DOI] [PubMed] [Google Scholar]
- 2.Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H.. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007; 26:5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu W., Shirayama M., Conte D. Jr, Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J.et al.. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009; 36:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Yigit E., Li W.X., Ding S.W.. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009; 5:e1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czech B., Hannon G.J.. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011; 12:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghildiyal M., Zamore P.D.. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009; 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman E.J., Carrington J.C.. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007; 8:884–896. [DOI] [PubMed] [Google Scholar]
- 8.Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A.. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001; 107:465–476. [DOI] [PubMed] [Google Scholar]
- 9.Smardon A., Spoerke J.M., Stacey S.C., Klein M.E., Mackin N., Maine E.M.. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000; 10:169–178. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Montgomery T.A., Fischer S.E., Garcia S.M., Riedel C.G., Fahlgren N., Sullivan C.M., Carrington J.C., Ruvkun G.. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 2012; 22:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai H.Y., Chen C.C., Conte D. Jr, Moresco J.J., Chaves D.A., Mitani S., Yates J.R. 3rd, Tsai M.D., Mello C.C. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell. 2015; 160:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D. Jr, Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G.et al.. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006; 124:343–354. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M., Ando R., Nakazawa T., Yudazono T., Tsutsumi N., Hatanaka N., Ohgake T., Hanaoka F., Eki T.. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells Dev. Mol. Cell. Mech. 2007; 12:997–1010. [DOI] [PubMed] [Google Scholar]
- 14.Vasale J.J., Gu W., Thivierge C., Batista P.J., Claycomb J.M., Youngman E.M., Duchaine T.F., Mello C.C., Conte D. Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA. 2010; 107:3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W., Shirayama M., Conte D., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E., Keys J., Stoltz M.J.et al.. Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009; 36:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo D., Kohlway A., Pyle A.M.. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 2013; 10:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou J., Chang M., Nie P., Secombes C.J.. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 2009; 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita T., Onoguchi K., Onomoto K., Hirai R., Yoneyama M.. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 2007; 89:754–760. [DOI] [PubMed] [Google Scholar]
- 19.Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M. Jr, Patel S.S., Marcotrigiano J.. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011; 479:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S.. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011; 147:423–435. [DOI] [PubMed] [Google Scholar]
- 21.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M.. Structural insights into RNA recognition by RIG-I. Cell. 2011; 147:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matranga C., Pyle A.M.. Double-stranded RNA-dependent ATPase DRH-3: insight into its role in RNAsilencing in Caenorhabditis elegans. J. Biol. Chem. 2010; 285:25363–25371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M. Jr, Akira S.et al.. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005; 175:2851–2858. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T.. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004; 5:730–737. [DOI] [PubMed] [Google Scholar]
- 25.Kang D.C., Gopalkrishnan R.V., Wu Q., Jankowsky E., Pyle A.M., Fisher P.B.. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA. 2002; 99:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T., Murakami T., Hirose Y., Eki T.. Purification and Characterization of Double-Stranded Nucleic Acid-Dependent ATPase Activities of Tagged Dicer-Related Helicase 1 and its Short Isoform in Caenorhabditis elegans. Genes (Basel). 2020; 11:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald M.E., Vela A., Pyle A.M.. Dicer-related helicase 3 forms an obligate dimer for recognizing 22G-RNA. Nucleic Acids Res. 2014; 42:3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., Zhang R., Wang J., Ding S.W., Lu R.. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc. Natl. Acad. Sci. 2013; 110:16085–16090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo D.Toward a crystal-clear view of the viral RNA sensing and response by RIG-I-like receptors. RNA Biol. 2014; 11:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolakofsky D., Kowalinski E., Cusack S.. A structure-based model of RIG-I activation. RNA. 2012; 18:2118–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hur S.Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019; 37:349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M.. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013; 14:772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J.. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. 2016; 113:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M. Jr, Inagaki F., Fujita T.. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008; 29:428–440. [DOI] [PubMed] [Google Scholar]
- 35.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P.. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010; 18:1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P.. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011; 39:1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Ranjith-Kumar C.T., Brooks M.T., Dharmaiah S., Herr A.B., Kao C., Li P.. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 2009; 284:13881–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pothlichet J., Burtey A., Kubarenko A.V., Caignard G., Solhonne B., Tangy F., Ben-Ali M., Quintana-Murci L., Heinzmann A., Chiche J.D.et al.. Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One. 2009; 4:e7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P.. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell. 2008; 29:169–179. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T.et al.. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010; 17:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vela A., Fedorova O., Ding S.C., Pyle A.M.. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J. Biol. Chem. 2012; 287:42564–42573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang D.-c., Gopalkrishnan R.V., Wu Q., Jankowsky E., Pyle A.M., Fisher P.B.. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. 2002; 99:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S.. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013; 152:276–289. [DOI] [PubMed] [Google Scholar]
- 44.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linding R., Russell R.B., Neduva V., Gibson T.J.. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003; 31:3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cole C., Barber J.D., Barton G.J.. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008; 36:W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savitsky P., Bray J., Cooper C.D., Marsden B.D., Mahajan P., Burgess-Brown N.A., Gileadi O.. High-throughput production of human proteins for crystallization: the SGC experience. J. Struct. Biol. 2010; 172:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welker N.C., Maity T.S., Ye X., Aruscavage P.J., Krauchuk A.A., Liu Q., Bass B.L.. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell. 2011; 41:589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W.Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P.Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 53.Panjikar S., Parthasarathy V., Lamzin V.S., Weiss M.S., Tucker P.A.. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005; 61:449–457. [DOI] [PubMed] [Google Scholar]
- 54.Panjikar S., Parthasarathy V., Lamzin V.S., Weiss M.S., Tucker P.A.. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009; 65:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheldrick G.M.Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thillier Y., Decroly E., Morvan F., Canard B., Vasseur J.J., Debart F.. Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N(7))-methyl transferase. RNA. 2012; 18:856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zlatev I., Manoharan M., Vasseur J.J., Morvan F.. Solid-phase chemical synthesis of 5′-triphosphate DNA, RNA, and chemically modified oligonucleotides. Curr. Protoc. Nucleic Acid Chem. 2012; Chapter 1:Unit1.28. [DOI] [PubMed] [Google Scholar]
- 61.Pascal B.D., Willis S., Lauer J.L., Landgraf R.R., West G.M., Marciano D., Novick S., Goswami D., Chalmers M.J., Griffin P.R.. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 2012; 23:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L.et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steward A., McDowell G.S., Clarke J.. Topology is the principal determinant in the folding of a complex all-alpha Greek key death domain from human FADD. J. Mol. Biol. 2009; 389:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kersse K., Verspurten J., Vanden Berghe T., Vandenabeele P.. The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 2011; 36:541–552. [DOI] [PubMed] [Google Scholar]
- 65.Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouchier-Hayes L., Martin S.J.. CARD games in apoptosis and immunity. EMBO Rep. 2002; 3:616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kao W.P., Yang C.Y., Su T.W., Wang Y.T., Lo Y.C., Lin S.C.. The versatile roles of CARDs in regulating apoptosis, inflammation, and NF-kappaB signaling. Apoptosis Internat. J. Program. Cell Death. 2015; 20:174–195. [DOI] [PubMed] [Google Scholar]
- 68.Ver Heul A.M., Gakhar L., Piper R.C., Ramaswamy S. Crystal structure of a complex of NOD1 CARD and ubiquitin. PLoS One. 2014; 9:e104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 70.Ferrao R., Wu H.. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012; 22:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu B., Peisley A., Tetrault D., Li Z., Egelman E.H., Magor K.E., Walz T., Penczek P.A., Hur S.. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell. 2014; 55:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin H., Srinivasula S.M., Wu G., Fernandes-Alnemri T., Alnemri E.S., Shi Y.. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999; 399:549–557. [DOI] [PubMed] [Google Scholar]
- 73.Takahasi K., Kumeta H., Tsuduki N., Narita R., Shigemoto T., Hirai R., Yoneyama M., Horiuchi M., Ogura K., Fujita T.et al.. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J. Biol. Chem. 2009; 284:17465–17474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yong H.Y., Luo D.. RIG-I-like receptors (RLRs) as novel targets for pan-antiviral and novel vaccination strategies against emerging and re-emerging viral infections. Front. Immunol. 2018; 9:1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng J., Yong H.Y., Panutdaporn N., Liu C., Tang K., Luo D.. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015; 43:1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ho V., Yong H.Y., Chevrier M., Narang V., Lum J., Toh Y.X., Lee B., Chen J., Tan E.Y., Luo D.et al.. RIG-I activation by a designer short RNA ligand protects human immune cells against dengue virus infection without causing cytotoxicity. J. Virol. 2019; 93:e00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Linehan M.M., Dickey T.H., Molinari E.S., Fitzgerald M.E., Potapova O., Iwasaki A., Pyle A.M.. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018; 4:e1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramanathan A., Devarkar S.C., Jiang F., Miller M.T., Khan A.G., Marcotrigiano J., Patel S.S.. The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection. Nucleic Acids Res. 2016; 44:896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu Q., Qu K., Modis Y.. Cryo-EM structures of MDA5-dsRNA filaments at different stages of ATP hydrolysis. Mol. Cell. 2018; 72:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T., Hohng S., Hur S.. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. USA. 2012; 109:E3340–E3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berke I.C., Yu X., Modis Y., Egelman E.H.. MDA5 assembles into a polar helical filament on dsRNA. Proc. Natl. Acad. Sci. USA. 2012; 109:18437–18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berke I.C., Modis Y.. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012; 31:1714–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T., Hur S.. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. USA. 2011; 108:21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao D., Yang Y.K., Wang R.P., Zhou X., Diao F.C., Li M.D., Zhai Z.H., Jiang Z.F., Chen D.Y.. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One. 2009; 4:e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oshiumi H., Miyashita M., Inoue N., Okabe M., Matsumoto M., Seya T.. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010; 8:496–509. [DOI] [PubMed] [Google Scholar]
- 86.D’Cruz A.A., Kershaw N.J., Hayman T.J., Linossi E.M., Chiang J.J., Wang M.K., Dagley L.F., Kolesnik T.B., Zhang J.G., Masters S.L.et al.. Identification of a second binding site on the TRIM25 B30.2 domain. Biochem. J. 2018; 475:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferrage F., Dutta K., Nistal-Villan E., Patel J.R., Sanchez-Aparicio M.T., De Ioannes P., Buku A., Aseguinolaza G.G., Garcia-Sastre A., Aggarwal A.K.. Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure. 2012; 20:2048–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu B., Hur S.. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015; 12:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sowa J.N., Jiang H., Somasundaram L., Tecle E., Xu G., Wang D., Troemel E.R.. The Caenorhabditis elegans RIG-I homolog DRH-1 mediates the intracellular pathogen response upon viral infection. J. Virol. 2020; 94:e01173-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lassig C., Matheisl S., Sparrer K.M., de Oliveira Mann C.C., Moldt M., Patel J.R., Goldeck M., Hartmann G., Garcia-Sastre A., Hornung V.et al.. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. eLife. 2015; 4:e10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fitzgerald M.E., Rawling D.C., Potapova O., Ren X., Kohlway A., Pyle A.M.. Selective RNA targeting and regulated signaling by RIG-I is controlled by coordination of RNA and ATP binding. Nucleic Acids Res. 2017; 45:1442–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rawling D.C., Fitzgerald M.E., Pyle A.M.. Establishing the role of ATP for the function of the RIG-I innate immune sensor. eLife. 2015; 4:e09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin S.C., Lo Y.C., Wu H.. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010; 465:885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park H.H., Logette E., Raunser S., Cuenin S., Walz T., Tschopp J., Wu H.. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007; 128:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scott F.L., Stec B., Pop C., Dobaczewska M.K., Lee J.J., Monosov E., Robinson H., Salvesen G.S., Schwarzenbacher R., Riedl S.J.. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009; 457:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao T., Towb P., Wasserman S.A., Sprang S.R.. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999; 99:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F.et al.. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009; 139:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DeLano W.L.The PyMOL Molecular Graphics System. 2002; San Carlos, CA: DeLano Scientific. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for the DRH-3 N-terminal CARDs have been deposited with the Protein Data bank under accession code 6M6Q. The atomic coordinates and structure factors for the DRH-3 CTD domains in complex with 5′pppRNAs have been deposited to the Protein Data bank under accession codes 6M6R and 6M6S.