Abstract
Prokaryotic Argonautes (pAgos) have been proposed as more flexible tools for gene-editing as they do not require sequence motifs adjacent to their targets for function, unlike popular CRISPR/Cas systems. One promising pAgo candidate, from the halophilic archaeon Natronobacterium gregoryi (NgAgo), has been the subject of debate regarding its potential in eukaryotic systems. Here, we revisit this enzyme and characterize its function in prokaryotes. NgAgo expresses poorly in non-halophilic hosts with most of the protein being insoluble and inactive even after refolding. However, we report that the soluble fraction does indeed act as a nicking DNA endonuclease. NgAgo shares canonical domains with other catalytically active pAgos but also contains a previously unrecognized single-stranded DNA binding domain (repA). Both repA and the canonical PIWI domains participate in DNA cleavage activities of NgAgo. NgAgo can be programmed with guides to nick targeted DNA in Escherichia coli and in vitro 1 nt outside the 3′ end of the guide sequence. We also found that these endonuclease activities are essential for enhanced NgAgo-guided homologous recombination, or gene-editing, in E. coli. Collectively, our results demonstrate the potential of NgAgo for gene-editing and provide new insight into seemingly contradictory reports.
INTRODUCTION
Long prokaryotic Argonaute proteins (pAgos) are programmable endonucleases that have recently been proposed as flexible tools for genome editing (1). These enzymes bind single-stranded DNA and/or RNA molecules as guides, which then prime the enzyme for nicking of complementary target DNA, RNA, or both (1). Double stranded DNA cleavage requires two complementary guides, which may induce DNA repair and editing. Unlike Cas9-based gene editing strategies, however, pAgos have the distinct advantage of not requiring a protospacer adjacent motif (PAM) for function (2–5). Thus, pAgos are not limited to targets flanked by PAM sites and can potentially cut any DNA target regardless of composition. Despite this potential, no pAgo has been developed that rivals the simplicity and functionality of Cas9-based strategies.
Target recognition and cleavage is enabled by four canonical domains (3): N (N-terminal), PAZ (PIWI-Argonaute-Zwille), MID (middle) and PIWI (P element-induced wimpy testis) domains. The N-terminal domain is essential for target cleavage (6,7) and dissociation of cleaved strands (7,8), although the detailed mechanism remains poorly understood. The MID domain interacts with the 5′-end of the guide (9) and promotes binding to its target (10). The PAZ domain interacts with the 3′ end of the guide (11–14), protecting it from degradation (15). Finally, the PIWI domain plays a pivotal role in nucleic acid cleavage via the conserved catalytic tetrad, DEDX (D: aspartate, E: glutamate, X: histidine, aspartate or asparagine) (16).
Recent emerging evidence also suggests a role for accessory proteins in pAgo activity. Within prokaryote genomes, pAgos are often organized in operons with ssDNA binding proteins and helicases among other DNA modifying proteins (17) hinting at concerted function in vivo. Supplementing a pAgo with these proteins in vitro enhances reaction rates and target specificity, reduces biases in substrate composition preferences, and enables activity on more topologically diverse substrates (18). These effects are observed with several homologs of these accessory proteins for multiple pAgos. Moreover, pAgos also copurify with helicases, ssDNA binding proteins, and recombinases from both native and heterologous hosts (19,20) indicating conserved physical interactions in different prokaryotes. Given the need for these and potentially other unrecognized accessory proteins, in vivo evaluation of pAgos may more accurately reflect their activity.
Despite the potential for programmable cleavage activities by long pAgos, currently crystallized pAgos including TtAgo (2), MpAgo (5), PfAgo (21) and MjAgo (3,22) work at very high temperatures (>55°C). Thus, their use for gene editing and in vivo testing in common mesophilic organisms is currently infeasible. The halophilic Argonaute from the archaeon Natronobacterium gregoryi (NgAgo) was proposed as a promising candidate for pAgo-mediated gene editing, as it was believed to be active at mesophilic (∼37°C) temperatures (23). However, these claims have since been refuted due to an inability to demonstrate in vitro DNA cleavage or to replicate these findings in a number of eukaryotic hosts (24–28). NgAgo expression is poor, presumably due to its halophilic characteristics that make low salt expression challenging (29,30). Thus, all published in vitro cleavage assays have relied on refolded protein (18,28), which may be non-functional, resulting in the inconclusive results. Nonetheless, recent work by Fu and colleagues demonstrated that NgAgo may still have potential as a gene editor for prokaryotic hosts (19). While the authors were able to confirm that gene-editing was mediated by homologous recombination via RecA (19), which physically associated with NgAgo in an unanticipated manner, the specific role of NgAgo remained unclear. Here, we demonstrate that NgAgo is indeed a nicking DNA endonuclease by identifying residues that are required for DNA cleavage and its cleavage site. While the mechanism remains to be elucidated, we provide evidence that this activity is essential for NgAgo-mediated gene editing via homologous recombination repair.
MATERIALS AND METHODS
Strains and plasmids
Escherichia coli strains and plasmids used in this study are listed in Table 1. Cloning was carried out according to standard practices (31) with primers, template, and purpose listed in Supplementary Table S5. Plasmids were maintained in E. coli DH5α. NgAgo variants (wildtype, D663A/D738A, N-del, and repA with GST or His tag) that were used for in vitro activity assays were cloned into an IPTG-inducible T7 plasmid, pET32a-GST-ELP64. MG1655 (DE3) atpI::KanR-mNeonGreen was generated using recombineering (32) via donor plasmid pTKDP-KanR-mNeonGreen-hph. For gene-editing/recombination studies (33), p15-KanR-PtetRed was used as a donor plasmid (Table 1).
Table 1.
Strains and plasmids
| Name | Relevant genotype | Vector backbone | Plasmid origin | Source |
|---|---|---|---|---|
| Strains | ||||
| BL21 (DE3) | F– ompT gal dcm lon hsdSB (rB–mB–) λ (DE3) [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12 (λS) | (57) | ||
| MG1655 (DE3) | K-12 F– λ– ilvG– rfb-50 rph-1 (DE3) | (58) | ||
| MG1655 (DE3) atpI::KanR-mNeonGreen | K-12 F– λ– ilvG– rfb-50 rph-1 (DE3) atpI::KanR-mNeonGreen | This study | ||
| Plasmids | ||||
| pBSI-SceI (E/H) | bla | ColE1 derivative | (59) | |
| pTXTL-p70a-T7RNAP | Bla, P70-T7RNAP | unknown | Arbor Biosciences | |
| pET32a-GST-ELP64 | bla, lacI, PT7-GST-ELP64 | pBR322 | Professor Xin Ge (University of California, Riverside) | |
| pTKDP-hph | bla, hph, sacB | pMB1 | (32) | |
| pCas9-CR4 | cat, PTet-Cas9 | p15A | (60) | |
| pET-GST-Ago-His | bla, lacI, PT7-GST-NgAgo-His | pET32a-GST-ELP64 | pBR322 | This study |
| pET32a-His-Ago | bla, lacI, PT7-GST-NgAgo-His | pET32a-GST-ELP64 | pBR322 | This study |
| pET32a-His-repA | bla, lacI, PT7-His-repA | pET32a-GST-ELP64 | pBR322 | This study |
| pET-GST-N-del-His | bla, lacI, PT7-GST-N-del-His | pET32a-GST-ELP64 | pBR322 | This study |
| pET-GST-N-del/D663A/D738A-His | bla, lacI, PT7-GST- N-del/D663A/D738A -His | pET32a-GST-ELP64 | pBR322 | This study |
| pTKDP-KanR-mNeonGreen-hph | bla, hph, KanR-mNeonGreen | pTKDP-hph | pMB1 | This study |
| p15-KanR-PtetRed | cat, KanR-mNeonGreen, PTet-gam-beta-exo | pCas9-CR4 | p15A | This study |
| pET32-BFP | Amp, lacI, PT7-BFP | pET32a-GST-ELP64 and pBAD-mTagBFP2 | pBR322 | This study |
| pIncw-mNeonGreen | cat | pN565 (61) (origin of replication); pCas9-CR4 (60) (cat) | pIncW | This study |
NgAgo expression and purification
GST-NgAgo or His-NgAgo variants were expressed in BL21 (DE3) grown with 100 μg/ml ampicillin. 5 ml cultures started from single colonies were grown for 16 h before subculturing in 100 ml of LB Miller containing ampicillin. Expression was induced with 0.1 mM IPTG at OD600 = 0.5 for either 4 h at 37°C or overnight at 22°C overnight before harvesting the cells at 7500 rpm (11 500 g) at 4°C for 5 min. The cell pellet was resuspended in TN buffer (10 mM Tris and 100 mM NaCl, pH 7.5) and lysed via sonication at a medium power setting (∼50 W) in 10 s intervals, with intervening 10 s incubations on ice to reduce heat denaturation. Cell lysates were then clarified at 12 000 rpm at 4°C for 30 min. The supernatant was collected as a soluble protein fraction. Both soluble and insoluble (cell pellet) fractions were purified via His-IDA nickel column (Clontech Laboratories, Mountain View, CA. Cat. No. 635657) according to the manufacturer instructions. Insoluble NgAgo protein was refolded on the column after denaturation with guanidium chloride as follows. The bacterial lysate was resuspended with equilibration buffer (50 mM sodium phosphate, 6 M guanidine–HCl, 300 mM NaCl, 20 mM imidazole, pH 7.4) and incubated with a pre-equilibrated column at 4°C for an hour. Then the column was washed with equilibration buffer and wash buffer (50 mM sodium phosphate, 6 M guanidine-HCl, 300 mM NaCl, 40 mM imidazole, pH 7.4). The protein was then refolded with refolding buffer (100 mM NaH2PO4, 10 mM Tris–Cl, 2 M NaCl, pH 8) at 4°C for 20 min before elution with elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 300 mM imidazole, pH 7.4). GST-tagged NgAgo variants were purified by glutathione agarose (Thermo Fisher Scientific, Waltham, MA. Cat. No. 16100) according to the manufacturer protocol.
Cell-free expression of NgAgo and activity assay
Cell-free TXTL reactions contained 5′ phosphorylated DNA guides targeting mNeonGreen (sequences in Supplementary Table S6), Chi6 oligos, IPTG, plasmids encoding T7RNA polymerase (pTXTL-p70a-T7RNAP) and NgAgo variants, including wildtype, D663A/D738A, repA, N-del and N-del/D663A/D738A (Table 2). Reactions were incubated at 29°C for 20 h to promote NgAgo expression before being supplemented to 125 mM NaCl and incubated at 37°C for folding for 24 h. Control TXTL reactions with mNeonGreen as a reporter were used to validate successful protein production by the cell-free system (Supplementary Figure S13). MgCl2 to a final concentration of 62.5 μM was then added along with target or non-target plasmid for reaction at 37°C for an hour. RNase A (70 ng or >490 units) (Millipore Sigma, Burlington, MA. Cat. No. R6513-10MG) was then added to each reaction to remove transcribed RNA at 37°C for 10 min. The reaction mixtures were then mixed with 0.5% SDS to dissociate any proteins and 6X loading dye before gel electrophoresis. The gel was visualized under a blue light with SYBR Safe DNA gel stain (ThermoFisher Scientific, Waltham, MA).
Table 2.
Materials for NgAgo variants production by cell-free system
| Volume (μl) | Final concentration | Remarks | |
|---|---|---|---|
| Cell-free system mixture | 4.5 | - | |
| 5′ phosphorylated DNA guides | 0.5 | 1 μM | |
| Chi6 oligos | 0.5 | 1 μM | Protect linear DNA from recBCD degradation (62) |
| IPTG | 0.5 | 0.5 mM | Induce NgAgo variants expression |
| pTXTL-p70a-T7RNAP | 0.5 | 2.4 nM | Encodes T7RNA polymerase for induction of NgAgo variants |
| Plasmids encoding NgAgo variants or mNeonGreen control | 0.5 | 6 nM |
NgAgo cut site identification
The shifted bands (nicked and/or linearized plasmid DNA) from the cell-free-based activity assay were gel-extracted and subjected to primer extension with 0.2 mmol of each single oligonucleotide over 45 rounds (oligonucleotides PEP1 or PEP12 with 60°C annealing temperature; Supplementary Table S5). Primer-extended single-stranded DNAs were then ligated at the 3′ end with a 5′ phosphorylated adapter sequences (adapter P-BacF) before amplification by PCR with the extension and adaptor primers. Amplified DNA were then A-tailed and cloned to an intermediate plasmid via the pGEMT-Easy vector system according to manufacturer instructions (Promega, Madison, WI). All ligation products were transformed and grown in liquid culture. The mixed population in liquid culture, representing all amplified nicked products, was then miniprepped and sequenced via next generation sequencing. Assuming equal amplification efficiency for each nicked product, abundance within the sequenced library represents the preference for a given nicking site. Libraries were generated using the Illumina DNA prep kit and run on a MiSeq 500-cycle kit (Illumina, San Diego, CA). Demultiplexing was achieved using Illumina's bcl2fastq program and mapped against the pNCS-mNeonGreen using BBMap (34). The nicked sites of the target DNA were identified based on the terminating bases of the primer-extended sequences. All the primers and adapters are listed in Supplementary Table S5.
Survival assay
BL21 (DE3) was transformed with target plasmid pIncw-mNeonGreen and NgAgo expression plasmid before being made electrocompetent. Electrocompetent cells were transformed with either no guides or 1 μg total of FW, RV, or both guides for targets as indicated (Supplementary Table S6) and plated on ampicillin and chloramphenicol selective LB Miller agar plate with 0.1 mM IPTG before 16–20 h incubation at 37°C. Colonies were counted to measure survival rate of transformants. The unguided control was normalized to 100% and guided-treatments were normalized to the unguided control. Availability of guide for NgAgo binding post transformation was validated in control experiments with labeled oligonucleotides (sequence in Supplementary Table S6; Supplementary Figure S12; see Supplemental Notes).
Gene-editing assay
MG1655 (DE3) atpI::KanR-mNeonGreen was transformed with pET-GST-NgAgo-His (to induce DNA cleavage) and p15-KanR-PtetRed (for lambda-red recombinase expression and to provide donor DNA for repair) and made electrocompetent. Electrocompetent cells were transformed with either no guides or 1.2 μl of 100 μM guides (FW, RV, or both; sequences in Supplementary Table S6) targeting mNeonGreen and incubated in LB Miller with ampicillin, chloramphenicol, and IPTG for an hour. These cultures were then diluted ten-fold in LB Miller containing ampicillin (working concentration: 100 μg/ml), chloramphenicol (working concentration: 25 μg/ml), IPTG (working concentration: 0.1mM), and anhydrotetracycline (aTc) (working concentration: 50 μg/ml), incubated until OD600 = 0.2 before plating with and without kanamycin (working concentration: 50 μg/ml). Colony forming units (CFU) were counted after 16–20 h incubation at 37°C. The unguided control was normalized to 100% and guided-treatments were normalized to the unguided control.
Phyre 2 and HHpred analysis
NgAgo protein (IMG/M Gene ID: 2510572918) was analyzed via Phyre 2 (35) with normal mode on 2018 November 19. The normal mode pipeline involves detecting sequence homologues, predicting secondary structure and disorder, constructing a hidden Markov model (HMM), scanning produced HMM against library of HMMs of proteins with experimentally solved structures, constructing 3D models of NgAgo, modelling insertions/deletions, modelling of amino acid sidechains, submission of the top model, and transmembrane helix and topology prediction (32). NgAgo was analyzed via HHpred (36,37) (https://toolkit.tuebingen.mpg.de/#/tools/hhpred) on 2018 November 27. The parameters for HHpred were HHblits = >uniclust30_2018_08 for multiple sequence alignment (MSA) generation method, three for maximal number of MSA generation steps, 1e–3 for E-value incl. threshold for MSA generation, 0% for minimum sequence identity of MSA hits with query, 20% for minimum coverage of MSA hits, during_alignment for secondary structure scoring, local for alignment mode, off for realign with MAC, 0.3 for MAC realignment threshold, 250 for number of target sequences and 20% for minimum probability in hit list.
Phylogenetic analysis
BLAST was used to compare NgAgo protein sequence with all the isolates in the database via the IMG/M server (https://img.jgi.doe.gov/). Representative full-length Argonautes with a repA domain were used to represent each species. Selected pAgos with repA domains and some well-characterized pAgos (SeAgo from Synechococcus elongatus (38), TtAgo from Thermus thermophilus (2), CbAgo from Clostridium butyricum (39), CbcAgo from CWBI 1009 of C. butyricum (40), CpAgo from Clostridium perfringens (41), KmAgo from Kurthia massiliensis (42), IbAgo from Intestinibacter bartlettii (41), LrAgo from Limnothrix rosea (43), RsAgo from Rhodobacter sphaeroides (44), MjAgo from Methanocaldococcus jannaschii (3), PfAgo from Pyrococcus furiosus (21), MkAgo from Methanopyrus kandleri (16), MpAgo from Marinitoga piezophile (5), and AaAgo from Aquifex aeolicus (45)) were compared, and the midpoint rooted tree was generated via the server http://www.genome.jp/tools-bin/ete with unaligned input type, mafft_default aligner, no alignment cleaner, no model tester, and fasttree_default Tree builder parameters. The nwk output file was then used for phylogenetic tree generation using Iroki (46).
RESULTS
NgAgo has canonical N-terminal, PIWI, MID and PAZ domains, and a putative single stranded DNA binding (repA) domain
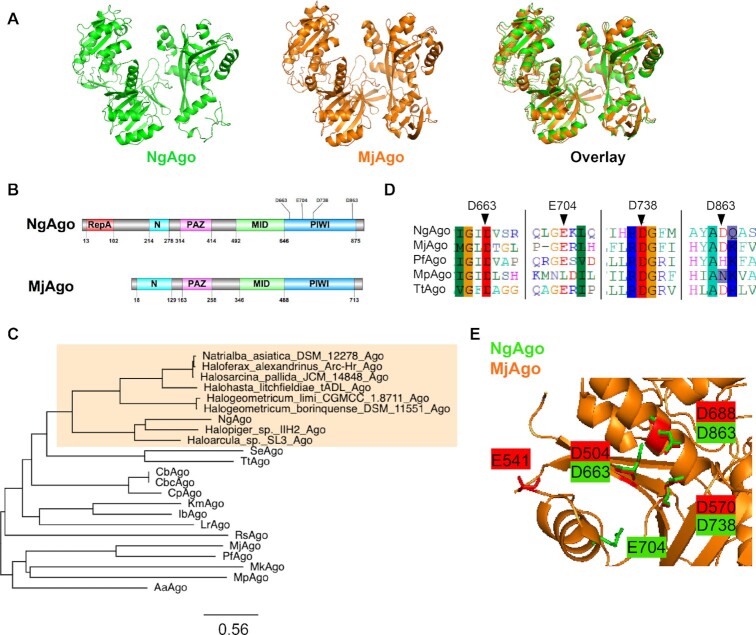
Given the ongoing debate of the function of NgAgo, we analyzed its sequence (IMG/M Gene ID: 2510572918) with Phyre 2 (35) and HHpred (36,37) to predict its structure based on characterized structural homologs. Phyre 2 and HHpred analyses found with high confidence (probability = 100%) that NgAgo shares structural features with catalytically active pAgos and eukaryotic Agos (eAgos) including archaeal MjAgo, bacterial TtAgo, and eukaryotic hAgo2 (Supplementary Tables S1 and S2). Since MjAgo is the most similar archaeal pAgo, and thus likely shares a common archaeal ancestor with NgAgo, we used it as a template for comparative modelling. The predicted NgAgo structure is similar to the crystal structure of MjAgo, consisting of canonical N-terminal, PAZ, MID and PIWI domains (Figure 1A and B). However, the N-terminal domain of NgAgo, which plays a key role in targeted cleavage, is truncated, relative to MjAgo. This may suggest a novel mechanism for strand displacement and binding.
Figure 1.
NgAgo belongs to a distinct clade of pAgos with a catalytic DEDX tetrad and novel repA domain. (A) Phyre 2 simulation 3D structure based on MjAgo structure (PDB: 5G5T). NgAgo structure is similar to MjAgo structure except for at the N-terminal domain. (B) Domain architecture analysis of NgAgo based on Phyre2 and HHpred reveals that NgAgo has an uncharacterized repA domain, a truncated N-terminal domain, a MID domain, and a PIWI domain. (C) Phylogenetic analysis of repA-containing pAgos (orange shaded) found from BLASTP against all isolates via JGI-IMG portal and other characterized pAgos. (D) The catalytic tetrad of NgAgo is conserved with catalytically active pAgos including MjAgo, PfAgo, MpAgo, and TtAgo in sequence alignment. (E) All residues of the catalytic tetrad (D663, E704, D738, and D863) DEDD, except E704 are structurally colocalized with the catalytic tetrad of MjAgo (D504, E541, D570 and D688).
Structural analysis also identified an uncharacterized oligonucleotide/oligosaccharide-binding (OB) fold domain between residues 13 and 102 of NgAgo that commonly binds single-stranded DNA in eukaryotes and prokaryotes (47) (Figure 1B). This OB domain has recently been identified as a new feature of pAgos (17). As repA proteins were the most common matches on both Phyre 2 and HHpred, we will refer to this OB domain as repA (Supplementary Tables S3 and S4). While the repA domain is absent in all characterized pAgos, at least 12 sequenced pAgo homologs share this domain. Phylogenetic analysis showed that all the repA-containing pAgos were from halophilic Archaea forming a clade that is distinct from that of the current well-characterized thermophilic and mesophilic pAgos (Figure 1C). This monophyletic group of repA-containing pAgos may represent a distinct class of pAgos that is currently unrecognized in the literature (see Supplemental Notes) (17).
Our analysis of NgAgo also confirmed the presence of a conserved catalytic tetrad, DEDX (X: H, D or N) (16), which is critical for nucleic acid cleavage by the PIWI domain of Argonautes. The catalytic tetrad (D663, E704, D738 and D863) of NgAgo aligns well with those from other catalytically active pAgos, including MjAgo (3), PfAgo (21), MpAgo (5) and TtAgo (2) (Figure 1D). Moreover, structural alignment of NgAgo and MjAgo display good colocalization of D663, D738, and D863 within the catalytic tetrad suggesting that NgAgo may have similar nucleic acid cleavage activity (Figure 1E).
Soluble, but not refolded, NgAgo exhibits DNA cleavage activity in vitro
As halophilic proteins tend to be insoluble in low-salt environments due to their sequence adaptations (29,30,48), we first optimized expression conditions to obtain more soluble NgAgo protein (Supplementary Figure S1). NgAgo was still unstable in optimal expression conditions, as evidenced by truncated peptide products (Supplementary Figure S1B). We purified wildtype NgAgo from both the soluble and insoluble fractions to test for 5′P-ssDNA guide-dependent DNA cleavage (Supplementary Figure S2). Insoluble NgAgo was refolded during purification using established methods (28). Purified NgAgo from the soluble fraction (sNgAgo) nicks plasmid DNA and genomic DNA, independent of a guide, as evidenced by the presence of the nicked and linearized plasmid (Supplementary Figure S3A; see Supplemental Notes). However, refolded NgAgo from the insoluble lysate fraction (rNgAgo) has little or no DNAse activity (Supplementary Figure S3B), consistent with established literature (24,26).
RepA and PIWI domains of NgAgo participate in DNA cleavage
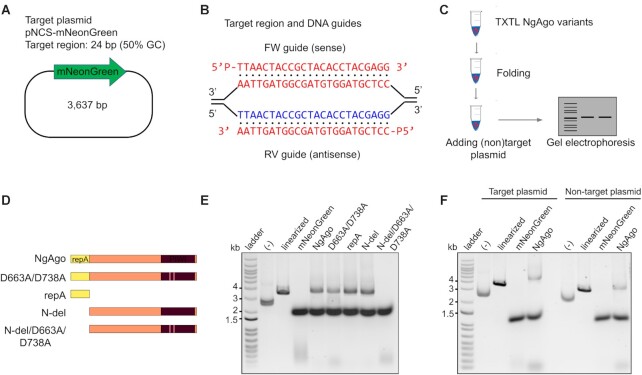
To rule out the possibility of non-specific host nuclease impurities (Supplementary Figure S4), we pursued cell-free expression of NgAgo. This approach has successfully been used to rapidly prototype other endonucleases including CRISPR-Cas endonucleases (49). NgAgo expression was induced in the presence of 5′ phosphorylated guides that targeted a plasmid substrate, pNCS-mNeonGreen (Figure 2A, B). NaCl was supplemented after expression to promote proper folding of the halophilic enzyme (Figure 2C). To identify regions critical for DNA cleavage, we constructed and expressed the repA domain of NgAgo (residues 1–102), a truncated NgAgo without the repA domain (residues 105–887, referred to as N-del) and D663A/D738A point mutations in the full-length protein and N-del variant (Figure 2D). D663A/D738A is a double mutant within the catalytic tetrad that corresponds to the inactive catalytic double mutant D478A/D546A of TtAgo (2,50).
Figure 2.
NgAgo variants degrade plasmid DNA in vitro via the repA domain and D663/D738 residues in the PIWI domain. (A) Target plasmid pNCS-mNeonGreen contains a 24-base pair target site with 50% GC content. (B) 5′ phosphorylated DNA guides binds to target sequence in pNCS-mNeonGreen. (C) Procedure for bacterial cell-free-system production of NgAgo and DNA degradation assessment. (D) NgAgo variants used in the in vitro assay to identify which domain is essential for nicking and cleaving activity. (E) Plasmids were treated with NgAgo variants or mNeonGreen as an endonuclease negative control for an hour before analysis on an agarose gel. Wildtype and D663A/D738A degrades plasmids DNA while N-del degrades plasmid DNA with compromised activity. N-del/D663A/D738A loses the ability to degrade plasmid DNA. (F) NgAgo degrades both target plasmid pNCS-mNeonGreen and non-target plasmid pBSI-SceI (E/H). Negative controls (–) are plasmids without any treatments. TXTL treated supercoiled plasmid reproducibly migrates more rapidly due to unknown mechanisms (Supplementary Figure S14).
Mutations to NgAgo abolished observed DNA cleavage activity, suggesting that cleavage was NgAgo dependent (Figure 2E). Both wildtype NgAgo and D663A/D738A linearized substrate DNA suggesting catalytic activity beyond the PIWI domain (51) or rescue of functionality by other domains even in the presence of a PIWI mutation. Both repA and PIWI domains participate in DNA cleavage with each being sufficient for activity as cleavage was retained in both repA and N-del mutants. It is unclear how the repA domain might lead to DNA damage although it does possess single-stranded DNA binding activity (Supplementary Figure S3C). Nonetheless, only in the presence of both a repA deletion and PIWI mutation, N-del/D663A/D738A, is DNA degradation completely lost. When a non-target plasmid with no complementarity to the supplied guides was incubated with the enzymes, a linearized product was still observed (Figure 2F). That is, NgAgo-induced DNA degradation may be both target specific and non-specific, consistent with proposed pAgo models of non-specific DNA ‘chopping’ for guide acquisition of unloaded pAgos and enhanced specific cleavage of complementary sequences (50).
NgAgo specifically nicks DNA complementary to guide
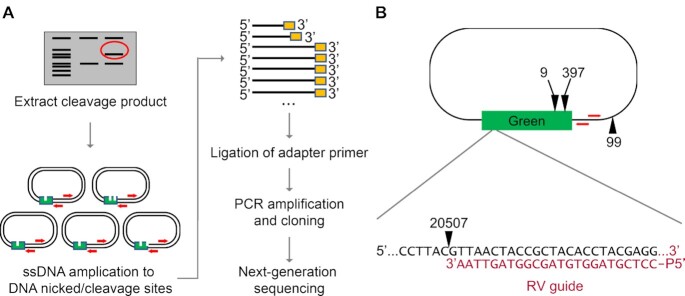
To validate guided sequence specific-cleavage via NgAgo, we sequenced the linearized products from our cell-free expressed cleavage studies (the high MW bands in Figure 2E). These digestion products were amplified and cloned before Illumina sequencing of the cleavage products (Figure 3A). The predominant product observed (20,507 reads) for target plasmid treated with wildtype NgAgo was cleaved 1 bp downstream from the 3′ end of the reverse (RV) guide (Figure 3B). Other cleavage products were far less abundant (397, 99, and 9 reads, respectively) (Figure 3B and Supplementary Table S7) and shared low similarity with the intended target (16.67–50%; Supplementary Table S7). The abundance of these products is similar to reads for random environmental contamination (e.g. non-E. coli rRNA sequences) and is thus likely to be the result of low levels of off-target nicking due to guide mismatch or chemical damage. The overwhelming abundance of a nicked product associated with the guide sequence (>97% of wildtype sequenced products) suggests that NgAgo is indeed targeted by a guide. However, we did not observe any targeted cleavage site associated with the forward (FW) guide; this result was robust and was observed from multiple primer extensions that primed at either end of the target gene in the plasmid (Supplementary Table S8). Other pAgos such as CbAgo exhibit an in vitro AT sequence preference at the 5′ end of the target sequence, presumably due to its low duplex stability that promotes DNA melting for pAgo recognition in the absence of supplemented helicase or other factors (39). The cell-free performance of our guides is consistent with a similar 5′ AT preference for NgAgo; the RV guide targets 5′TTAA while the FW guide targets 5′CCTC. Nonetheless, NgAgo exhibits unique cleavage properties as it does not cleave between nucleotides complementary to position 10 and 11 in the guide like other pAgos (43).
Figure 3.
NgAgo specifically nicks DNA complementary to the guide. (A) Experimental workflow for cut site identification of NgAgo is illustrated here (see method section). (B) The cut sites of wildtype NgAgo and their corresponding number of reads from next-generation sequencing are illustrated on the relative location on the target plasmid, pNCS-mNeonGreen. Red arrows indicate primer extension sites.
While D663A/D738A, repA, and N-del NgAgo mutants appeared to have some cleavage activity, their products were not guide dependent. Cleavage products shared less than 41.6% similarity with supplied guides (Supplementary Table S7). Nonetheless, for repA and N-del mutants, two mNeonGreen cleavage products dominated (24 618 and 7453 reads, respectively). The D663A/D738A PIWI mutants displayed weak bias towards a single site; however no cleavage product exceeded 577 reads. Moreover, the reduction in total reads and band intensity (Figure 2E) suggest that the PIWI domain is the primary driver of DNA nicking. Counterintuitively, detected cleavage products were unique for each NgAgo mutant (Supplementary Table S7). However, our data indicate that both PIWI and rep domains of wildtype NgAgo are needed for guide-dependent cleavage of complementary DNA. Furthermore, the data also suggest nonspecific nicking/cleavage of NgAgo is possible in the absence of ssDNA/guide interacting domains.
NgAgo has specific in vivo activity at plasmid and genomic loci in bacteria
Next, we tested whether NgAgo can be programmed to target DNA in vivo. We chose E.coli instead of mammalian cells as our model because NgAgo, like most pAgos, lacks helicase activity needed to separate DNA strands for pAgo recognition and nicking of complementary sequences (18). The rapid rate of bacterial DNA replication increases the abundance of accessible unpaired DNA targets for NgAgo activity. Additionally, E. coli lack histones, which are known to inhibit pAgo activity (22).
Studies have reproducibly demonstrated an ability of NgAgo to reduce gene expression (26,28) and have suggested RNA cleavage as a possible mechanism (51). However, two alternative hypotheses could also explain this phenomenon: (i) NgAgo cuts DNA leading to poor expression and (ii) NgAgo inhibits transcription by tightly binding DNA. To distinguish between these three hypotheses, we created a two-plasmid system that harbors an inducible NgAgo expression cassette on one plasmid and another that serves as a target harboring a transcriptionally inactive pseudogene target, mNeonGreen, and a selectable marker or essential gene under selective conditions, cat (Figure 4A). NgAgo was expressed in cells with both these plasmids and transformed with phosphorylated guide ssDNA (P-ssDNA) targeting different strands of mNeonGreen, including forward (FW, sense/coding), reverse (RV, antisense/non-coding), both FW and RV, or without a guide. After transformation, these cells were streaked on selective media (Figure 4B). When guides were targeted to the transcriptionally silent mNeonGreen (Supplementary Figure S5), fewer than half the colony forming units were observed relative to unguided controls (Figure 4C). Control studies with either guides alone or NgAgo alone did not identify any cell toxicity, suggesting that the reduction in survival was due to NgAgo activity (Supplementary Figures S6 and S7). As similar results were obtained regardless of the strand targeted and the target produced no RNA, NgAgo must interact at the DNA level. One possible mechanism is plasmid curing and loss of the selective marker through cleavage of the test plasmid, in agreement with our in vitro (Supplementary Figures S3 and S8) and cell-free studies (Figure 2). Using BFP controls in place of NgAgo does not reduce survival when incubated with guides complementary to the pseudogene mNeonGreen (Figure 4C), confirming the survival reduction effect requires NgAgo expression. Finally, this effect is target specific. When targeted to an absent locus (tetA), there were no significant changes in the number of surviving colonies relative to unguided controls (Figure 4C). This assay only quantifies activity relative to an unguided control and as such cannot measure off-target activity present in unguided controls. However, the reduction of survival in a guide- and target-dependent manner suggests that NgAgo has the capacity for targeted DNA endonuclease activity in vivo in E. coli, consistent with our cell-free studies (Figure 3).
Figure 4.
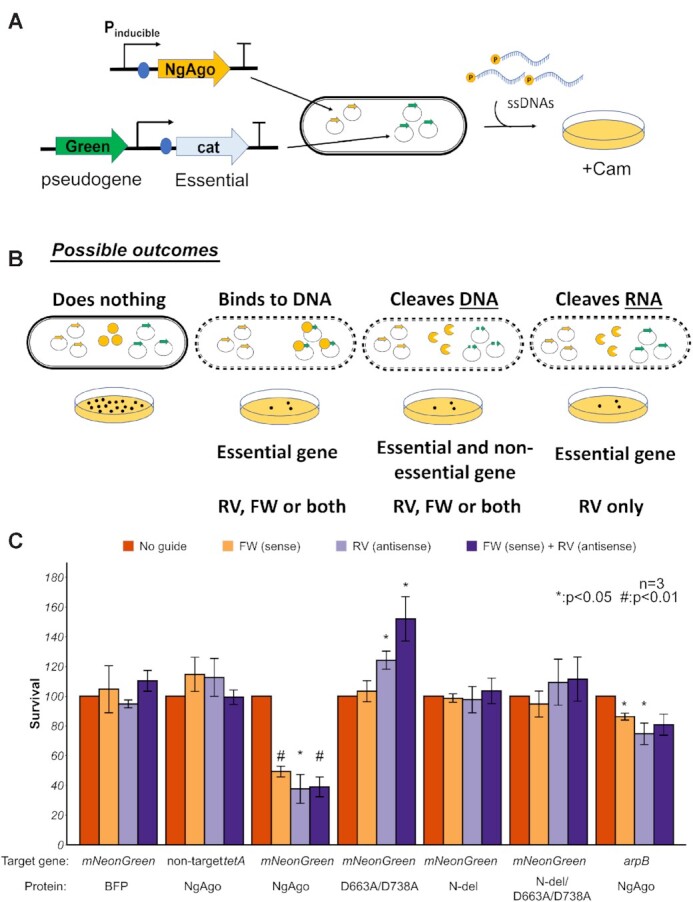
NgAgo can be programmed to target DNA in E. coli. (A) Workflow of testing NgAgo function in E. coli. Two plasmids system used to test the function of NgAgo. One plasmid harbors NgAgo driven by T7 inducible promoter while the other low-copy plasmid serves as the target of NgAgo, including an untranscribed pseudogene, mNeonGreen. (B) Four possible outcomes relative to an unguided control including no interaction, DNA binding, DNA cleaving and RNA binding/cleaving, reveal the function of NgAgo. (C) Survival rate targeting a pseudogene (mNeonGreen) on the plasmid or targeting a nonessential gene (arpB) in the genome with NgAgo variants or BFP control.
To confirm that the reduced survival is not limited to targets on the plasmid, we also targeted a genomic locus, arpB. arpB is a non-essential pseudogene that is interrupted by a stop codon (52). Since arpB RNA is not required for survival (i.e. the arpB mutant is nonlethal), RNA cleavage would not reduce survival. However, double stranded DNA breaks in E. coli are lethal due to inhibited genome replication (53). As targeting arpB did reduce survival (Figure 4C), this suggests NgAgo also cleaves genomic DNA, consistent with our plasmid cleavage results.
Additional control experiments targeting a plasmid-borne chloramphenicol resistance gene (cat) further validate our inference of NgAgo dependence (Supplementary Figure S9). As before, all guides that target the cat locus reduce survival in the presence of NgAgo. However, when a protein BFP control is used, survival is only reduced in the presence of reverse guide. Unlike the forward guide, reverse guide may hybridize with transcribed cat mRNA to form an RNA/DNA duplex that may be rapidly destroyed by RNAse H, removing chloramphenicol resistance. However, the use of forward guide alone with BFP does not induce this effect
Next, we asked if repA and PIWI domains are required for targeting in E.coli by evaluating the ability of different variants to target mNeonGreen. Our results showed that the PIWI mutant (D663A/D738A) and truncated repA deletion (N-del) lost the ability to reduce survival (Figure 4C), suggesting disruption of targeted DNA cleavage in agreement with our sequencing of cell-free cleavage products (Supplementary Tables S7 and S8). The PIWI mutation, however, enhanced survival activity (Figure 4C). While the mechanisms remain to be fully elucidated, single stranded DNA binding proteins (SSBs) have been implicated in initiating plasmid replication (54,55). We hypothesize that the intact SSB domain (repA) and guide recognition domains (N, MID and PAZ) allow for targeted plasmid interactions in the presence of guide. These interactions coupled with SSB activity of the mutant might enhance plasmid replication in the absence of nicking via the PIWI domain resulting in increased numbers of cat resistant colonies. Nonetheless, both intact repA and PIWI domains were required for targeted NgAgo activity, consistent with our cell-free studies.
DNA-cleaving domains are needed for NgAgo programmable genome editing in bacteria
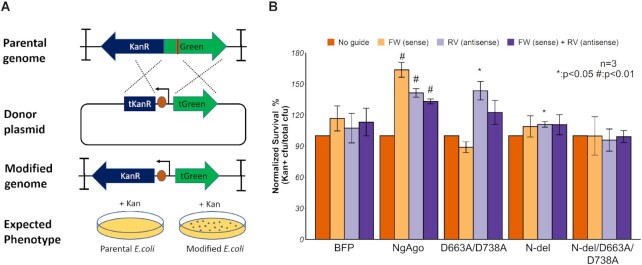
Since we have shown that NgAgo can cleave DNA in vitro and in E. coli, we asked whether this activity was essential for reproducible gene editing by NgAgo observed in other prokaryotes (19). To test for NgAgo gene editing activity, we created a kanamycin sensitive MG1655 (DE3) strain harboring a cassette composed of a kanR resistance gene lacking an RBS and promoter and a mNeonGreen gene flanked by two double terminators (Figure 5A). This arrangement prevented any KanR/mNeonGreen expression from transcription read-through and translation from upstream and downstream genes. We then provided a donor plasmid with a truncated mNeonGreen, a constitutive promoter, an RBS, and a truncated kanR, which is also KanR– but can recombine with our locus to create a KanR+ phenotype (Figure 5A). As DNA breaks in E. coli are lethal, repair via recombination should increase the number of KanR+ transformants if NgAgo induces DNA cleavage. We validated this system with CRISPR/Cas9, which showed a 4-fold enhancement in recombination efficiency (Supplementary Figure S10).
Figure 5.
NgAgo enhances gene-editing via λ-red-mediated homologous recombination in E.coli. (A) Design of gene-editing assay in MG1655 (DE3). KanR and mNeonGreen (Green) cassette without promoter and RBS, flanked by two double terminators, is integrated in MG1655 (DE3). Donor plasmid with truncated mNeonGreen (tGreen) encodes a nonfunctional truncated KanR (tKanR). Guide was transformed to target the mNeonGreen (red line). After successful gene editing, modified genome has a functional KanR cassette, enabling survival in Kan selective plate. (B) NgAgo variants enhance gene editing efficiency with ∼1 microgram of guide (s) relative to an unguided control while blue fluorescent protein (BFP) control has no enhancement with guides. Error bars are the standard errors generated from three replicates. Statistically significant results are indicated with * (P-value < 0.05, paired t-test).
Wildtype NgAgo increased homologous recombination efficiency when provided with FW, RV and both guides compared to an unguided control (Figure 5B), demonstrating that guide-dependent NgAgo activity can enhance gene editing. Subsequent sequencing of the kanamycin-resistant colonies confirmed the inserted promoter and RBS (Supplementary Figure S11). In contrast, a BFP protein control showed no statistically significant enhancement in recombination compared to the unguided control (Figure 5B). The PIWI mutant, D663A/D738A, displayed no significant enhancement of recombination with FW or both guides (Figure 5B), consistent with its weak cleavage activity (Figure 2E). However, the reverse guide displayed reduced but some statistically significant enhancement in homologous recombination. While the mechanism behind this pattern is unclear, these data suggest that the catalytic tetrad within the PIWI domain is not necessary for enhanced homologous recombination under some conditions. This observation has been independently confirmed in other published studies and attributed to interactions with host accessory proteins such as recA to enhance strand invasion and recombination (19). The N-del mutant of NgAgo lacking the repA domain displayed even weaker statistically significant enhancement in homologous recombination above unguided controls (11%) in the presence of the RV guide only (Figure 5B). The N-del/D663A/D738A catalytic mutant showed no increase in gene editing activity in the presence of FW, RV, or both guides compared to an unguided control. This trend in homologous recombination enhancement is consistent with our observed DNA endonuclease activities (Figures 2E and 4C) suggesting that the DNA endonuclease activity mediated by the repA and PIWI domains enhances homologous recombination and gene editing.
DISCUSSION
NgAgo has been subject to intense debate in the literature in recent years (23,24,26,27,56). Although previous studies suggested that refolded NgAgo does not cut DNA in vitro (18,51), consistent with our findings, we establish that soluble NgAgo can, in fact, nick DNA in vitro. That is, refolded NgAgo, which has been historically studied due to the poor soluble expression of this halophilic enzyme, may not be an accurate assessment of NgAgo activities. However, when soluble protein is concentrated and isolated, there is indeed some capacity for nonspecific or guide-independent DNA cleavage as we have demonstrated in vitro. Moreover, this behavior may be salt dependent, reflecting the halophilic lifestyle of the native host; NgAgo expressed from cells grown with LB Lennox showed no activity in our hands (data not shown) relative to that produced from cells grown on LB Miller (this work). Our parallel studies in cell-free expression systems that allow for control of salt conditions and lack potentially contaminating endonuclease expression confirm this observation. Our sequencing results validate that this cleavage behavior is predominantly guide dependent and that cleavage occurs 1 bp outside of the 3′ end of guide (5′ of the complementary target sequence). Although we did not find a cleavage site corresponding to the forward guide, this failure might be due to guide preference, as seen in CbAgo (39). Most importantly, we generated a catalytically dead N-del/D663A/D738A mutant making it unlikely that the detected activity is the result of sample contamination.
NgAgo activity is mediated not only by the PIWI domain, like canonical pAgos, but also an uncharacterized and previously unrecognized accessory repA or single-stranded DNA binding domain fused to the N-terminus that appears common among halophilic pAgos (Figure 1C). Our work is the first report to suggest a role for this domain in NgAgo function and identify it as a confounding variable in the ongoing conversation regarding NgAgo activity. Previously studied ‘catalytic’ mutants left this domain intact and were unable to detect a change in NgAgo function, which was attributed to sample contamination or inactivity (51). However, this and growing evidence from the literature (18–20) suggest that accessory proteins and domains may be essential for pAgo function. As homologous accessory proteins from heterologous hosts can mediate function (18,19), we investigated whether in vivo cleavage, as observed via cell survival and DNA recombination efficiency, would be induced by NgAgo and its mutants. Not only were these assay results consistent with DNA cleavage, but they also importantly suggested an ability to target specific gene loci via single-stranded 5′P DNA guides. However, there were slight differences in performance of the forward guide between in vivo and cell-free studies, potentially due to the presence of accessory proteins from the host. Host-supplied helicase, for example, may unwind target DNA, eliminating potential sequence preferences observed in cell-free based validation. Our work here underscores the role of unrecognized accessory proteins, supplied via the expression host, and a need to characterize these proteins to more accurately assess pAgo activity.
Finally, our results provide supporting evidence to encourage the development of NgAgo for gene-editing. When provided with homologous target and donor sequences, NgAgo can enhance homologous recombination. Much like other pAgos, the PIWI domain participates in DNA editing in prokaryotes as shown here and by Fu et al. (19). Moreover, without repA, PIWI mutants of NgAgo exhibit reduced cleavage activity with a concomitant reduction in homologous recombination efficiency. Both the repA deletion and the PIWI mutation (N-del/D663A/D738) are needed to fully abolish catalytic and gene-editing functions. In the presence of both functional domains, NgAgo can effectively enhance homologous recombination by inducing a double stranded break at a targeted region. Despite the programmable DNA-cleaving ability of NgAgo, there remain several challenges to its development as a robust tool for gene-editing applications: an elucidation of its mechanism, the potential for guide-independent or off-target cleavage, unknown accessory proteins needed for function, poor expression, salt dependence, and potentially low activity in eukaryotic hosts. Nonetheless, further insight may lead to protein engineering strategies to overcome these hurdles and develop NgAgo as a robust tool for gene-editing.
CONCLUSIONS
Based on the above findings, we conclude that NgAgo is a novel DNA endonuclease that belongs to an unrecognized class of pAgos defined by a characteristic repA domain. NgAgo uses both a well-conserved catalytic tetrad in PIWI and a novel uncharacterised repA domain to cleave DNA. This cleavage mediates the efficiency of gene-editing via NgAgo in prokaryotes. Despite the challenges of NgAgo, our work establishes innovative approaches to probe NgAgo activity (and that of other pAgos) and identifies critical protein features for its development as a next generation synthetic biology tool.
DATA AVAILABILITY
Additional notes and data are available in the Supplemental materials. Next generation sequencing reads used to determine NgAgo cleavage targets can be found under BioProject PRJNA748512.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Xin Ge (University of California, Riverside) and Dr Kristala J. Prather (Massachusetts Institute of Technology) for providing pET32a-GST-ELP64 and MG1655 (DE3), respectively. We also thank Dr Mathew Tantama (Purdue University) for providing pBAD-mTagBFP2.
Author contributions: K.V.S., F.G. and K.Z.L. designed the experiments. K.Z.L., M.A.M., A.K., A.L., K.F. and P.P. conducted and analyzed the experiments. K.V.S., F.G. and K.Z.L. supervised research and experimental design. K.V.S., K.Z.L., M.A.M. and F.G. wrote the manuscript.
Notes
Present address: Kevin V. Solomon, Department of Chemical & Biomolecular Engineering, University of Delaware, Newark, DE 19706, USA.
Contributor Information
Kok Zhi Lee, Department of Agricultural and Biological Engineering, Purdue University, West Lafayette, IN 47906, USA.
Michael A Mechikoff, Department of Agricultural and Biological Engineering, Purdue University, West Lafayette, IN 47906, USA.
Archana Kikla, Department of Biological Sciences, Purdue University, West Lafayette, IN 47906, USA.
Arren Liu, Department of Biological Sciences, Purdue University, West Lafayette, IN 47906, USA.
Paula Pandolfi, Department of Biological Sciences, Purdue University, West Lafayette, IN 47906, USA.
Kevin Fitzgerald, Department of Agricultural and Biological Engineering, Purdue University, West Lafayette, IN 47906, USA.
Frederick S Gimble, Purdue University Interdisciplinary Life Science Program (PULSe), Purdue University, West Lafayette, IN 47906, USA; Department of Biochemistry, Purdue University, West Lafayette, IN, 47906, USA.
Kevin V Solomon, Department of Agricultural and Biological Engineering, Purdue University, West Lafayette, IN 47906, USA; Purdue University Interdisciplinary Life Science Program (PULSe), Purdue University, West Lafayette, IN 47906, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ralph W. and Grace M. Showalter Research Trust [Award 41000622]; USDA National Institute of Food and Agriculture [Hatch Multistate Project S1041]; Purdue Research Foundation Fellowships [60000025, 60000029]; startup funds from the Colleges of Engineering and Agriculture. Funding for open access charge: Startup Funds.
Conflict of interest statement. K.V.S. and K.Z.L. have filed a patent related to this work.
REFERENCES
- 1.Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools. Nat. Rev. Microbiol. 2018; 16:5. [DOI] [PubMed] [Google Scholar]
- 2.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 4.Enghiad B., Zhao H.. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017; 6:752–757. [DOI] [PubMed] [Google Scholar]
- 5.Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauptmann J., Dueck A., Harlander S., Pfaff J., Merkl R., Meister G.. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 2013; 20:814. [DOI] [PubMed] [Google Scholar]
- 7.Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L.. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013; 3:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak P.B., Tomari Y.. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012; 19:145. [DOI] [PubMed] [Google Scholar]
- 9.Ma J.-B., Yuan Y.-R., Meister G., Pei Y., Tuschl T., Patel D.J.. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005; 434:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Künne T., Swarts D.C., Brouns S.J.J.. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014; 22:74–83. [DOI] [PubMed] [Google Scholar]
- 11.Lingel A., Simon B., Izaurralde E., Sattler M.. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004; 11:576. [DOI] [PubMed] [Google Scholar]
- 12.Ma J.-B., Ye K., Patel D.J.. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004; 429:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hur J.K., Zinchenko M.K., Djuranovic S., Green R.. Regulation of Argonaute slicer activity by guide RNA 3′end interactions with the N-terminal lobe. J. Biol. Chem. 2013; 288:7829–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., Van Der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryazansky S., Kulbachinskiy A., Aravin A.A.. The expanded universe of prokaryotic Argonaute proteins. mBio. 2018; 9:e01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt E.A., Evans T.C. Jr, Tanner N.A.. Single-stranded binding proteins and helicase enhance the activity of prokaryotic argonautes in vitro. PLoS One. 2018; 13:e0203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L., Xie C., Jin Z., Tu Z., Han L., Jin M., Xiang Y., Zhang A.. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019; 47:3568–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D.. Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell. 2020; 182:1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., Van Der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stöckl S., Tinnefeld P., Schneider S., Klingl A.. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cyranoski D.Authors retract controversial NgAgo gene-editing study. Nat. News. 2017; 10.1038/nature.2017.22412. [DOI] [Google Scholar]
- 24.Javidi-Parsijani P., Niu G.G., Davis M., Lu P., Atala A., Lu B.S.. No evidence of genome editing activity from Natronobacterium gregoryi Argonaute (NgAgo) in human cells. PLoS One. 2017; 12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z., Tan S., Xu L., Gao L., Zhu H., Ma C., Liang X.. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017; 145:20–23. [DOI] [PubMed] [Google Scholar]
- 26.Burgess S., Cheng L.Z., Gu F., Huang J.J., Huang Z.W., Lin S., Li J.S., Li W., Qin W., Sun Y.J.et al.. Questions about NgAgo. Protein & Cell. 2016; 7:913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khin N.C., Lowe J.L., Jensen L.M., Burgio G.. No evidence for genome editing in mouse zygotes and HEK293T human cell line using the DNA-guided Natronobacterium gregoryi Argonaute (NgAgo). PLoS One. 2017; 12:e0178768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin Y.Y., Wang Y.M., Liu D.. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 2016; 26:1349–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elcock A.H., McCammon J.A.. Electrostatic contributions to the stability of halophilic proteins. J. Mol. Biol. 1998; 280:731–748. [DOI] [PubMed] [Google Scholar]
- 30.Tadeo X., López-Méndez B., Trigueros T., Laín A., Castaño D., Millet O.. Structural basis for the amino acid composition of proteins from halophilic archea. PLoS Biol. 2009; 7:e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch E.F., Maniatis T.. Molecular Cloning: A Laboratory Manual. 1989; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 32.Tas H., Nguyen C.T., Patel R., Kim N.H., Kuhlman T.E.. An integrated system for precise genome modification in Escherichia coli. PLoS One. 2015; 10:e0136963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S.. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015; 81:2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bushnell B.BBMap: A Fast, Accurate, Splice-Aware Aligner Lawrence Berkeley National Lab. 2014; Berkeley, CA: LBNL. [Google Scholar]
- 35.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann L., Stephens A., Nam S.-Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V.. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018; 430:2237–2243. [DOI] [PubMed] [Google Scholar]
- 37.Söding J., Biegert A., Lupas A.N.. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005; 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olina A., Kuzmenko A., Ninova M., Aravin A.A., Kulbachinskiy A., Esyunina D.. Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 2020; 17:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J.. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019; 47:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Quintans N., Bowden L., Berenguer J., Mencía M.. DNA interference by a mesophilic Argonaute protein, CbcAgo. F1000Res. 2019; 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T., Shi W., Li C., Wang Y., Zhao F.et al.. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37°C. Cell Discov. 2019; 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Li W., Jiang X., Wang Y., Zhang Z., Liu Q., He R., Chen Q., Yang J., Wang L.et al.. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021; 49:1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A.. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019; 47:5822–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyoshi T., Ito K., Murakami R., Uchiumi T.. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016; 7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashid U.J., Paterok D., Koglin A., Gohlke H., Piehler J., Chen J.C.-H.. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007; 282:13824–13832. [DOI] [PubMed] [Google Scholar]
- 46.Moore R.M., Harrison A.O., McAllister S.M., Polson S.W., Wommack K.E.. Iroki: automatic customization and visualization of phylogenetic trees. PeerJ. 2020; 8:e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynn R.L., Zou L.. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010; 45:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller-Santos M., de Souza E.M., Pedrosa F. de O., Mitchell D.A., Longhi S., Carrière F., Canaan S., Krieger N.. First evidence for the salt-dependent folding and activity of an esterase from the halophilic archaea Haloarcula marismortui. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids. 2009; 1791:719–729. [DOI] [PubMed] [Google Scholar]
- 49.Marshall R., Maxwell C.S., Collins S.P., Jacobsen T., Luo M.L., Begemann M.B., Gray B.N., January E., Singer A., He Y.et al.. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell. 2018; 69:146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M., Zhang Y., Zhao H., Lou J., Wang Y.. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell. 2017; 65:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sunghyeok Y., Taegeun B., Kyoungmi K., Omer H., Hwan L.S., Young K.Y., Kang-In L., Seokjoong K., Jin-Soo K.. DNA-dependent RNA cleavage by the Natronobacterium gregoryi Argonaute. 2017; bioRxiv doi:20 January 2017, preprint: not peer reviewed 10.1101/101923. [DOI]
- 52.Goodall E.C.A., Robinson A., Johnston I.G., Jabbari S., Turner K.A., Cunningham A.F., Lund P.A., Cole J.A., Henderson I.R.. The essential genome of Escherichia coli K-12. mBio. 2018; 9:e02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons L.A., Goranov A.I., Kobayashi H., Davies B.W., Yuan D.S., Grossman A.D., Walker G.C.. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 2009; 191:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatelier E.L., Jannière L., Ehrlich S.D., Canceill D.. The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMβ1 from Gram-positive bacteria. J. Biol. Chem. 2001; 276:10234–10246. [DOI] [PubMed] [Google Scholar]
- 55.Jovanovic O.S., Ayres E.K., Figurski D.H.. The replication initiator operon of promiscuous plasmid RK2 encodes a gene that complements an Escherichia coli mutant defective in single-stranded DNA-binding protein. J. Bacteriol. 1992; 174:4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z., Tan S., Xu L., Gao L., Zhu H., Ma C., Liang X.. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017; 145:20–23. [DOI] [PubMed] [Google Scholar]
- 57.Wood W.B.Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 1966; 16:118–133. [DOI] [PubMed] [Google Scholar]
- 58.Tseng H.-C., Martin C.H., Nielsen D.R., Prather K.L.J.. Metabolic engineering of Escherichia coli for enhanced production of (R)-and (S)-3-hydroxybutyrate. Appl. Environ. Microbiol. 2009; 75:3137–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu Y., Tenney K., Li H., Gimble F.S.. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J. Mol. Biol. 2008; 382:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reisch C.R., Prather K.L.J.. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci. Rep. 2015; 5:15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhodius V.A., Segall-Shapiro T.H., Sharon B.D., Ghodasara A., Orlova E., Tabakh H., Burkhardt D.H., Clancy K., Peterson T.C., Gross C.A.et al.. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013; 9:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall R., Maxwell C.S., Collins S.P., Beisel C.L., Noireaux V.. Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems. Biotechnol. Bioeng. 2017; 114:2137–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional notes and data are available in the Supplemental materials. Next generation sequencing reads used to determine NgAgo cleavage targets can be found under BioProject PRJNA748512.