Abstract
The activation of eukaryotic DNA replication origins needs to be strictly controlled at multiple steps in order to faithfully duplicate the genome and to maintain its stability. How the checkpoint recovery and adaptation protein Polo-like kinase 1 (Plk1) regulates the firing of replication origins during non-challenged S phase remained an open question. Using DNA fiber analysis, we show that immunodepletion of Plk1 in the Xenopus in vitro system decreases replication fork density and initiation frequency. Numerical analyses suggest that Plk1 reduces the overall probability and synchrony of origin firing. We used quantitative chromatin proteomics and co-immunoprecipitations to demonstrate that Plk1 interacts with firing factors MTBP/Treslin/TopBP1 as well as with Rif1, a known regulator of replication timing. Phosphopeptide analysis by LC/MS/MS shows that the C-terminal domain of Rif1, which is necessary for its repressive action on origins through protein phosphatase 1 (PP1), can be phosphorylated in vitro by Plk1 on S2058 in its PP1 binding site. The phosphomimetic S2058D mutant interrupts the Rif1-PP1 interaction and modulates DNA replication. Collectively, our study provides molecular insights into how Plk1 regulates the spatio-temporal replication program and suggests that Plk1 controls origin activation at the level of large chromatin domains in vertebrates.
Graphical Abstract
Graphical Abstract.
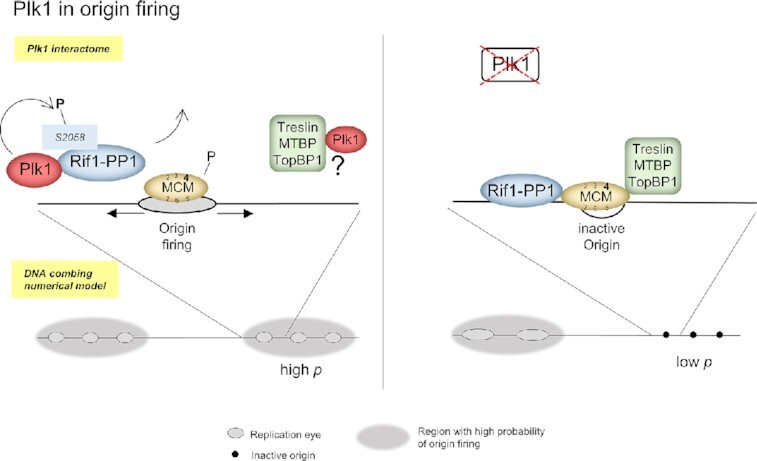
Plk1 promotes DNA replication in regions with high probabilty of origin firing via a negative regulation of Rif1/PP1.
INTRODUCTION
To maintain genome stability, eukaryotic DNA replication must be strictly controlled in space and time during S phase (1). In higher eukaryotes, DNA replication starts from several thousand replication origins, grouped in clusters, which are activated at different times during S phase. Genome-wide studies have shown that large adjacent genomic segments have similar replication timing, which are called replication domains (2). However, only few factors that regulate such a multiscale spatio-temporal organization are known.
Initiation of DNA replication is divided into two steps, origin licensing and origin activation or firing. During G1 phase, the assembly of the pre-replicative complex (pre-RC) at origins is initiated by the loading of the origin recognition complex (ORC) to DNA sequences, which then recruits Cdc6, Cdt1 and the inactive double hexamer helicase, the MCM 2–7 complex (origin licensing step). For origins to fire, the helicase needs to be activated, which converts the pre-RC into two active bi-directional replication forks or replisomes. This requires the coordinated action of Cyclin- and Dbf4-dependent kinases (respectively, CDKs and DDKs), which leads to the recruitment of Cdc45 and the GINS complex to the pre-RC to form the core of the replicative helicase, the CMG complex (Cdc45-MCM-GINS). The activation of the CMG complex is a critical, multistep process. In metazoans, Treslin/TICRR (3,4) mediates CDK-dependency of origin firing by a phosphorylation-mediated interaction with TopBP1 (5,6). MTBP binds Treslin and forms a stable protein complex in human and Xenopus (7,8) and together with TopBP1 build the Treslin/MTBP/TopBP1 complex, which serves as a platform to regulate the pre-initiation complex (pre-IC) formation, composed of CMG, Treslin/MTBP/TopBP1 and pol ϵ. After CMG formation, active replisomes assemble before bi-directional DNA synthesis can start. The competition between origins for limiting factors such as Sld3/Treslin and Dbp11/TopBP1, contribute to set their firing time in budding yeast and Xenopus respectively (9,10). Another factor regulating replication timing is Rif1 (Rap1-interacting factor), which modulates late replication in yeast (11,12), mice (13) and human (14). In higher eukaryotes, Rif1 binds protein phosphatase 1 (PP1) via the SILK and RVSF motifs within its C-terminal part. This targets PP1 to pre-RC phospho-sites, acting both negatively on origin activation by counteracting the phosphorylation of MCM by DDK (15–17), and positively on the licensing step (18). In metazoans, Rif1 also modulates cellular responses and suppresses DNA replication after the inhibition of Cdc7 (19), and Rif1 bound to the nuclear lamina regulates replication firing time as a possible organiser of chromatin architecture (20).
The spatio-temporal DNA replication program is additionally regulated by the ATR/Chk1 dependent replication checkpoint during normal S phase, even in the absence of fork stalling or DNA damage both in Xenopus (21–24) and in mammalian cells (25,26). In the Xenopus in vitro system, activated replication origins are spaced 5–15 kb apart and clustered in early- and late-firing groups of origins (21,27,28), the late clusters being inhibited by ATR and Chk1 (21,24). By modelling either the unchallenged or the checkpoint-inhibited replication pattern, we recently proposed that, as in other eukaryotes, the genome would be segmented into regions of low and high probability of origin firing. We showed that the latter escapes intra-S checkpoint regulation (29). The necessity for the segmentation of the genome into high and low probability of origin firing to model the experimental observations implies that, in the regions with high probability of origin firing, the temporal difference between two firing events would be smaller than in other regions of the genome. This leads to an observed synchrony of origin firing and therefore to an effective observed clustering of replication eyes on a single DNA fiber. One open remaining question is why origin firing is under the control of the replication checkpoint in regions with low probability of origin firing (supposedly late firing (30)), while the regions with high probability of origin firing (supposedly early firing) escape its regulation. It has been proposed that the checkpoint adaptation and recovery protein Polo-like kinase 1 (Plk1) would act close to forks to suppress the checkpoint action (31–33) when forks stall. Plk1 contains a polo-box domain (PBD) that interacts with phosphorylated sequences in substrates (34). During prolonged fork stalling, Plk1 targets the checkpoint adaptor Claspin, primed by ATR phosphorylation, leading to the inactivation of Chk1 in Xenopus (35). Plk1 also interacts with TopBP1 during homologous recombination (36). Several studies identified, after stress induction, that Plk1 interacts with MCM2 and MCM7 (31,37), Hbo1 (38) and Orc2 (39), which underlined the role of Plk1 in regulating pre-RC assembly. During normal S phase, we have recently shown that Plk1 accumulates on chromatin and moreover, that Plk1 depletion from Xenopus replicating extracts leads to a decreased DNA replication, an activation of Chk1 and a decrease in Cdk2 activity (40). We did not detect any changes in pre-RC proteins bound on chromatin after Plk1 depletion, suggesting that Plk1 does not target the licensing step in the absence of external stress in this system. However, the spatio-temporal DNA replication program upon modulation of Plk1 has not been investigated to better understand the role of this kinase in origin activation.
Here, we used DNA fiber analysis to demonstrate that Plk1 depletion decreased fork density and initiation frequency of replication origins. Numerical analyses suggest that Plk1 depletion reduced the overall probability of origin firing by decreasing the availability of limiting origin firing factors and the synchrony of origin firing by lowering the fraction of early firing regions. However, Plk1 did not relieve origin inhibition locally as it has been proposed. Quantitative proteomics of chromatin protein dynamics after Plk1 depletion and ChIP-mass spectroscopy revealed multiple Plk1 interacting partners, including MTBP/Treslin/TopBP1 and Rif1, all of them confirmed by co-immunoprecipitation experiments. In vitro, Plk1 could phosphorylate C-terminal Rif1 on the consensus sites for PP1 binding. The depletion of Plk1 increased Rif1–PP1 binding and decreased the phosphorylation of MCM4 on chromatin, whereas the addition of the C-terminal domain of Rif1 showed the opposite effects together with an acceleration of DNA synthesis. Mutations of one of the phosphorylated residues within the PP1 binding site revealed that this phosphorylation not only modulates Rif1–PP1 interaction but also DNA replication. Altogether, our study provides molecular insights into how Plk1 regulates the DNA replication program in vertebrates and suggests that Plk1 promotes initiation of DNA replication at the level of large genomic regions.
MATERIALS AND METHODS
Reagents and antibodies
Antibodies and reagents used in this study are described in Supplementary Methods S1 and Supplementary Methods S2, unlisted basic chemicals were from Sigma, Saint-Quentin-Fallavier, France. Recombinant functional His-tagged Xenopus Plk1 was expressed and purified as described in (40). C-terminal Xenopus Rif1 (Rif-CTD, 296 amino acids, MW: 32.4 kDa), cloned in pET30a vector, a kind gift from B. Dunphy and A. Kumagai (41), was expressed in Escherichia coli C41(DE3) cells, purified by Nickel-Sepharose chromatography (GE Healthcare) and used as an antigen to raise antibodies in rabbits at a commercial facility (Covalab, Villeurbanne, France). Mutant Xenopus Rif1–CTD S2058A and S2058D DNA sequences were ordered from Twist Bioscience (San Francisco, USA), subcloned in a pET30a vector, expressed and purified as the wild type Rif1–CTD.
Replication of sperm nuclei in Xenopus egg extracts
Replication competent extracts from unfertilized Xenopus eggs were prepared as described (42) and used freshly made unless stated otherwise. When sperm chromatin is incubated in Xenopus egg extract, it is assembled into normal interphase nuclei, which synchronously start replication after a typical 20 to 30 min lag phase; replication is completed 40–60 min later. Sperm nuclei were incubated in untreated mock or Plk1 depleted extracts, in the presence of cycloheximide (250 μg/ml) and energy mix (7.5 mM creatine phosphate, 1 mM ATP, 0.1 mM EGTA, pH 7.7, 1 mM MgCl2). Replication was allowed to continue for indicated time. For DNA combing experiments, Biotin-dUTP (20 μM) was added at the start of the reaction and labelled DNA was purified at different times during S phase. For nascent strand analysis by alkaline electrophoresis, 0.2 μCi [α32P]-dCTP (3000 Ci/mmol) (Hartmann Analytic, Braunschweig, Germany) was added and DNA purified, separated and incorporation was analyzed as described (28). When indicated, Rif1–CTD recombinant protein was added at a final concentration of 640 nM or 1 μM for overexpression or pull-down experiments, respectively. Our institutional Animal Care and Use Committee (IACUC) namely Paris Center and South number 59 approved the study and the protocols herein (approvals number 2012-0062 and 2012-0063), following the French and the European laws on animal experimentation.
Immunodepletions and immunoprecipitations
For immunodepletion, anti-Xenopus Plk1 or pre-immune sera were incubated overnight at 4°C with protein A Sepharose beads. Beads were washed with extraction buffer EB (50 mM HEPES, 50 mM KCl, 5 mM MgCl2) and incubated in extracts 1 h at 4°C (volume ratio 1:3). For immunoprecipitations, Dynabeads protein G were saturated with 1 mg/ml BSA for 1 h in 0.1 M acetate buffer pH 5, 0.1% NP40, washed and bound to pre-immune, Plk1 or Rif1 sera. Coupled beads were washed in EB buffer, then incubated for 1 h with crude LSS extract or nucleoplasmic extract. Beads were washed three times in 10 mM HEPES (pH 7.5), 150 mM NaCl, 0.5% NP40, 2.5 mM EGTA, and twice in the same buffer without NP40, then eluted in Laemmli sample buffer. Nucleoplasmic extracts were prepared by lysing nuclei isolated as described in the ChIP section (see below) in NIBS buffer 0.5% Triton X100.
Molecular combing and detection by fluorescent antibodies
DNA was extracted and combed as described (43). Biotin was detected with streptavidin-conjugated AlexaFluor594 followed by biotinylated anti-avidin antibody (repeated twice). DNA was labeled by anti-DNA antibody (mouse), followed by secondaries; AlexaFluor488-labeled rabbit anti-mouse, then goat anti-rabbit antibodies for enhancement (24).
Measurements and data analysis
Images of the combed DNA molecules were acquired and measured as described (43). For each combing experiment, a total of 9–60 Mb DNA was measured. The fields of view were chosen at random. The means of fibers lengths were comparable inside individual experiments to avoid biases in eye-to-eye distances. Images of DNA combed molecules were visualized and acquired using a 100× objective on a Zeiss Evolve inverted microscope, associated with the Axiovision software. Measurements in pixel on each fiber were made in ImageJ and then converted into kilobases and analyzed by using Matlab (vR2013a). Replication eyes were defined as the incorporation tracks of biotin–dUTP of at least 1kb. Replication eyes were considered to be the products of two replication forks, incorporation tracks at the extremities of DNA fibers were considered to be the products of one replication fork. When the label was discontinuous, the tract of unlabeled DNA needed to be at least 1 kb to be considered as a real gap. The replication extent was determined as the sum of eye lengths divided by the total DNA length. Fork density was calculated as the total DNA divided by the total number of forks. The midpoints of replication eyes were assumed as the origins of replication. Eye-to-eye distances (ETED), also named inter-origin distances, were measured between the midpoints of adjacent replication eyes. The tracks of biotin-dUTP at the extremities of the fibers were not considered in the measurement of eye-to-eye distances and replication eyes length, but were considered in the calculation of the replication extent and the fork density, as products of a single replication fork. The frequency of initiation was obtained as the number of new initiations defined as replication eyes smaller than 3 kb divided by the length of unreplicated DNA and by the interval in which a detectable initiation event can occur, that is Δt = 180 s considering a replication fork speed of ∼0.5 kb/min (21).
Chromatin purification and western blot analysis
For analysis of chromatin-bound proteins, proteins were recovered using a protocol slightly modified from (44). Replication reactions were diluted into a 13-fold volume of ELB buffer (10 mM HEPES pH 7.5, 50 mM KCl, 2.5 mM MgCl2) containing 1 mM DTT, 0.2% Triton X100, protease inhibitors and phosphatase inhibitors (Supplementary Methods S2); chromatin was recovered through a 500 mM sucrose cushion in ELB buffer at 6780 g, 50 s, 4°C. Interphase was washed twice with 200 μl ELB, 250 mM sucrose. The pellet was resuspended in Laemmli sample buffer. For western blots, proteins were subjected to SDS-PAGE (Polyacrylamide gel electrophoresis) and transferred to PVDF membranes subsequently incubated with the indicated primary antibody followed by incubation with the appropriate peroxidase coupled secondary or horseradish peroxidase-labeled protein A (1/20 000). Immunodetection was performed according to the manufacturer and peroxidase activity was revealed using Super Signal West Femto Chemiluminescence Kit. Imaging of western blots were performed on Biorad ChemiDoc system and optical densities were obtained using Biorad ImageLab software. To increase the band separation of high molecular weight proteins (>48 kDa), SDS-PAGE were performed on 8.5% acrylamide gels prepared with a 120:1 acrylamide:bisacrylamide ratio. For quantitative analysis of chromatin-bound proteins by mass spectrometry (Chromass), sperm nuclei (4000 nuclei/μl) were incubated in mock or Plk1 depleted egg extracts and were allowed to replicate for 40 min. Experiments were carried out in three replicates incubated in parallel. Samples were prepared as previously described (45) for mass spectrometry analysis.
Kinase assays
In vitro phosphorylation of Rif1–CTD by Plk1 was performed by adding 590 nM recombinant Plk1, 0.1 μCi [γ32P]-ATP (Hartmann Analytic, Braunschweig, Germany), 50 μM ATP, 6 μM of recombinant Rif1–CTD or Casein as control substrate for 30 min at 23°C in kinase buffer (20 mM HEPES, pH 7.2, 10 mM MgCl2, 2 mM DTT, 0.1 mM EGTA) with or without 100 nM Plk1 inhibitor BI2536. Reactions were stopped in Laemmli sample buffer, proteins were separated by SDS-PAGE. Gels were dried and bands were quantified on a Phosphorimager Typhoon Trio (GE Healthcare). An aliquot of each reaction was withdrawn before the addition of [γ32P]-ATP and loaded on a Stain-Free gel (BioRad) for visualization. For phosphorylation assay in extracts, Rif1–CTD (1 μM) was incubated in mock or Plk1 depleted egg extract for 1 h in the presence of 0.1 μCi [γ32P]-ATP and energy mix, at 22°C. Reactions were stopped by dilution in three volume of slurry Nickel Sepharose beads in EB, binding was allowed for 20 min under shaking at 4°C, beads were washed twice with EB/NP-40 (0.1%) and four times in EB, bound proteins were eluted by Laemmli sample buffer, followed by 12% SDS-PAGE and quantification. Rif1–CTD pulldown assays for PP1 binding were performed using magnetic Ni-NTA beads (CubeBiotech, Monheim, Germany).
Chromatin immunoprecipitation for Plk1 interactions analysis (ChIP)
Sperm nuclei (4000 nuclei/μl) were incubated in fresh extracts and were allowed to replicate for 15 and 60 min; reactions were stopped by a 1/5 dilution in nuclear isolation buffer (NIBS) (50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 10% sucrose). Nuclei were isolated by centrifugation through 150 μl of a 20% sucrose cushion in NIBS buffer for 5 min at 5000 g (4°C). The purification was repeated once and the pellet resuspended in 100 μl ChIP buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA). Crosslinking was performed using 1% formaldehyde for 8 min, reaction was quenched by 250 mM Glycine and nuclei were pelleted for 5 min at 5000 g (4°C), then lysed in 200 μl ChIP buffer containing 0.5% Triton X100 (ChIP lysis buffer). The DNA was sheared into fragments of 100–300 bp using Bioruptor Plus sonicator (Diagenode, Liège, Belgium) in 1.5 ml TPX tubes; 30 cycles 30 s/30 s ON/OFF (high power) then the soluble DNA was clarified by centrifugation for 15 min at 19 000 g (4°C). Sample was divided threefold; One third was reversed cross-linked in 50 mM Tris pH 8, 10 mM EDTA, 1% SDS at 65°C overnight, treated with 1 μg/μl RNAse A for 1h at 37°C, then with 1 μg/μl proteinase K for 2h at 55°C, extracted with phenol–chloroform, ethanol precipitated for DNA shredding control on agarose gel electrophoresis. The two other thirds were diluted 2-fold and incubated overnight at 4°C with either anti-Plk1 or control IgG DMP-crosslinked to Protein A Sepharose. Beads were washed twice with 200 μl ChIP lysis buffer, once in 200 μl ChIP buffer containing 500 mM NaCl and then once in 200 μl ChIP buffer. Finally, the beads were resuspended in 20 μl 2× Laemmli sample buffer, samples were separated by SDS PAGE and analyzed by western blot or by mass spectrometry.
Statistical analysis
Plots and statistical analysis were made using GraphPad version 6.0 (La Jolla, CA, USA) or Matlab (vR2013a), means or medians with standard error of mean (SEM), tests and P-values are indicated in the legends of figures or in figures.
Chromass and ChIP LC–MS/MS analysis and data processing
Trypsin-generated peptides of the different samples, ChIP and Chromass, were analyzed with two different instruments. Nano-liquid chromatography elution conditions were similar for both systems, with a flow rate of 300–400 nl/min and an acetonitrile gradient of 5–35% (v/v) acetonitrile over 100 or 180 min for nanoElute (Bruker) and nanoRSLC (Thermo) systems, respectively.
For the ChIP samples, proteins were analyzed with a Triple-TOF 4600 mass spectrometer (ABSciex) coupled to the nanoRSLC system (ThermoFisher, Illkirch, France) equipped with a trap column (Acclaim PepMap100C18, 75 μm i.d. × 2 cm, 3 μm) and an analytical column (Acclaim PepMapRSLCC18, 75 μm i.d. × 75 cm, 2 μm, 100 Å). MS/MS spectra were acquired by a data-dependent acquisition method involving selection of the 20 precursors giving the most intense signals, CID fragmentation with the Q1 quadrupole set at low resolution to improve sensitivity. MS raw data were processed with MS Data Converter and PeakView softwares (ABSciex). Protein identifications were performed using the MASCOT search engine (Matrix science, London, UK) against the Xenbase V9.2 database (46) with carbamidomethylation of cysteines set as fixed modification and oxidation of methionines as variable modification. Peptide and fragment tolerances were set at 25 ppm and 0.05 Da, Proteins were validated when identified with at least two unique peptides. Only ions with a score higher than the identity threshold and a false-positive discovery rate of less than 1% (Mascot decoy option) were considered. Mass spectrometry based-quantification was performed using spectral count. MS/MS spectral count values were obtained from Scaffold software. Missing values occurring in spectral count datasets at protein-level were imputed by a constant value fixed at 0.5 and 0.4.
For the Chromass triplicates samples, proteins were analyzed with a Tims-TOF Pro mass spectrometer (Bruker) coupled to the nanoElute system (Bruker) equipped with a trap column (nanoEase C18, 100 Å, 5 μm, 180 μm × 20 mm) and an analytical column (Aurora, 25 cm × 75 μm, C18 1.6 μm). MS/MS spectra were acquired with the PASEF ion mobility-based acquisition mode and MS raw data were processed with DataAnalysis software (Bruker). The MS files were processed with the MaxQuant software version 1.6.6.0 and searched with Andromeda search engine against the Xenbase V9.2 database. To search parent mass and fragment ions, we set a mass deviation of 10 and 20 ppm, respectively. The minimum peptide length was set to 7 amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (Met) and N-term acetylation were set as variable modifications. The false discovery rates (FDRs) at the protein and peptide level were set to 1%. Scores were calculated in MaxQuant as described previously (47). The reverse and common contaminants hits were removed from MaxQuant output. Proteins were quantified according to the MaxQuant label-free algorithm using LFQ intensities; protein quantification was obtained using at least two peptides per protein. Match between runs was allowed. Proteins functions and groups were attributed using GO-term function retrieved from Uniprot database (48), manually curated.
Statistical and bioinformatic analysis, including heatmaps, profile plots and clustering, were performed with Perseus software (version 1.6.12) freely available at www.perseus-framework.org (49). For statistical comparison, we set two groups, each containing up to three biological replicates. We then filtered the data to keep only proteins with at least two valid values in at least one group. Next, the data were imputed to fill missing data points by creating a Gaussian distribution of random numbers with a standard deviation of 33% relative to the standard deviation of the measured values and 1.8 standard deviation downshift of the mean to simulate the distribution of low signal values. We performed a t-test, P-value <0.05, S0 = 0.1. The heatmap and hierarchical clustering of proteins that survived the test was performed in Perseus (1.6.12.0) on logarithmic LFQ intensities after z-score normalization of the data, using Euclidean distances (49).
Identification of phospho-sites in Rif1 by LC/MS/MS
Phosphosite detection was performed after in vitro phosphorylation reactions with recombinant XPlk1 and XRif1–CTD, samples were further processed as followed: S-Trap™ micro spin column (Protifi, Hutington, USA) digestion was performed according to manufacturer's instructions. Briefly, 10 μl of each sample was adjusted to 25 μl with 5% SDS/50 mM TEAB buffer supplemented with 20 mM TCEP and 50 mM chloracetamide for reduction and alkylation, respectively. Aqueous phosphoric acid was then added to a final concentration of 1.2% following by the addition of S-Trap binding buffer (90% aqueous methanol, 100 mM TEAB, pH 7.1). Mixtures were then loaded on S-Trap columns. Two extra washing steps were performed for thorough SDS elimination. Samples were digested with 1 μg of trypsin (Promega) at 47°C for 1 h. After elution, peptides were vacuum dried and resuspended in 40 μl of 10% ACN, 0.1% TFA in HPLC-grade water prior to MS analysis. For each run, 1 μl was injected in a nanoRSLC-Q Exactive PLUS (RSLC Ultimate 3000) (Thermo Scientific, Waltham MA, USA). Peptides were loaded onto a μ-precolumn (Acclaim PepMap 100 C18, cartridge, 300 μm i.d. × 5 mm, 5 μm) (Thermo Scientific, Waltham MA, USA), and were separated on a 50 cm reversed-phase liquid chromatographic column (0.075 mm ID, Acclaim PepMap 100, C18, 2 μm) (Thermo Scientific, Waltham, MA, USA). Chromatography solvents were (A) 0.1% formic acid in water, and (B) 80% acetonitrile, 0.08% formic acid. Peptides were eluted from the column with the following gradient 5–40% B (38 min), 40% to 80% (1 min). At 39 min, the gradient stayed at 80% for 4 min and, at 43 min, it returned to 5% to re-equilibrate the column for 16 min before the next injection. One blank was run between samples to prevent sample carryover. Peptides eluting from the column were analyzed by data dependent MS/MS, using top-10 acquisition method. Peptides were fragmented using higher-energy collisional dissociation (HCD). Briefly, the instrument settings were as follows: resolution was set to 70 000 for MS scans and 17 500 for the data dependent MS/MS scans in order to increase speed. The MS AGC target was set to 3.106 counts with maximum injection time set to 200 ms while MS/MS AGC target was set to 1.105 with maximum injection time set to 120 ms. The MS scan range was from 400 to 2000 m/z.
The MS files were processed with the MaxQuant software version 1.6.6.0 and searched with Andromeda search engine against the Xenopus laevis database from XENLA_9.2_Xenbase.pep.fa. To search parent mass and fragment ions, we set an initial mass deviation of 4.5 and 20 ppm, respectively. The minimum peptide length was set to 7 amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Carbamidomethylation (C) was set as fixed modification, whereas phosphorylation (STY), oxidation (M) and N-term acetylation were set as variable modifications. Match between runs was not allowed. The false discovery rates (FDRs) at the protein and peptide level were set to 1%. Scores were calculated in MaxQuant as described previously (47). The reverse and common contaminants hits were removed from MaxQuant output. The phosphopeptides output table and the corresponding logarithmic intensities were used for phosphopeptide analysis. The phosphopeptide table was expanded to separate individual phosphosites (47).
Numerical simulations
A Monte Carlo method was used to simulate the DNA replication program and the simulation results were compared to the DNA combing data for mock and Plk1 depleted samples by using a genetic optimization algorithm as described previously (29). In the simulation, the genome is constituted as a vector of 106 blocks of value 0 for the unreplicated blocks and 1 for the replicated blocks. Each block represents 1 kb. Potential origins are distributed randomly along the genome with an average density of one origin per 2.3kb (50). Origin initiation is regulated by the encounter between a not activated origin and a limiting factor, which number 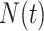 increases during S phase as
increases during S phase as 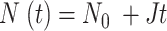 . The probability of successful origin firing after an encounter between the limiting factor and a potential origin is heterogeneous over the genome: it corresponds to
. The probability of successful origin firing after an encounter between the limiting factor and a potential origin is heterogeneous over the genome: it corresponds to 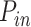 for a fraction θ of the genome and to
for a fraction θ of the genome and to 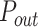 for the remining 1 – θ fraction. Moreover, we consider that the probability within a distance
for the remining 1 – θ fraction. Moreover, we consider that the probability within a distance 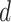 from an active replication fork is
from an active replication fork is 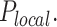 Once an origin is activated, the two replication forks elongate in opposite direction of one block for each step of calculation. The limiting factor is sequestrated by the forks and is made available for further origin activation once two converging forks merge.
Once an origin is activated, the two replication forks elongate in opposite direction of one block for each step of calculation. The limiting factor is sequestrated by the forks and is made available for further origin activation once two converging forks merge.
RESULTS
Plk1 depletion decreases initiation frequency and reduces the synchrony of origin firing
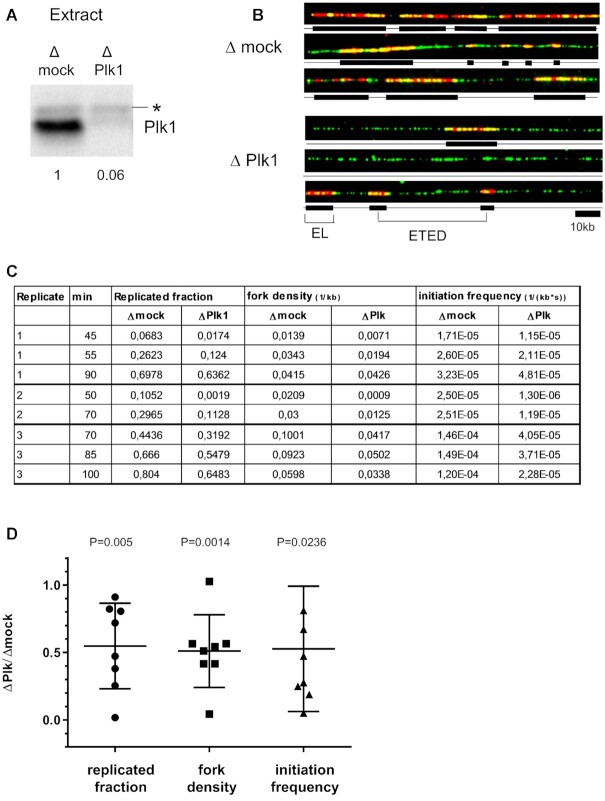
Plk1 depletion induces a decrease in the rate of DNA synthesis, while adding back recombinant Plk1 rescues replication (40). The decrease could either result from a reduction of the frequency of origin firing or from a decrease in fork speed or from both. In order to directly monitor origin firing, we performed DNA combing experiments in the Xenopus in vitro system. This model system has the advantage that all nuclei synchronously enter S phase, which easily allows to analyze origin activation all over S phase. Plk1 was efficiently depleted from egg extracts by a specific anti-Xenopus Plk1 antibody (Figure 1A). Sperm nuclei (2000 nuclei/μl) were incubated in mock or Plk1 depleted egg extract in presence of Biotin-dUTP. The reactions were stopped at different times during S phase and the DNA was purified, combed and labelled to be visualized by fluorescence microscopy (Figure 1B). In three independent experiments, 4000 DNA fibers (467 Mb) were measured and analyzed for replicated fraction, fork density and initiation frequency, eye-to eye distances, eye lengths (Figure 1C and Supplementary Table S1). In all experiments, replication fork density and initiation frequency decreased after Plk1 depletion, with a more pronounced effect during early and mid S phase (Figure 1C and Supplementary Figure S1). Considering all early to late S phase time points, mean replicated fractions significantly decreased to 54.8% (Figure 1D) after Plk1 depletion. Likewise, we also observed a significant mean decrease of fork density (51.1%) and of initiation frequency (52.7%) in the absence of Plk1. Altogether, these observations demonstrate that the decrease in DNA synthesis after Plk1 depletion is due to a reduction of the number of activated origins.
Figure 1.
Plk1 depletion decreases replication fork density and initiation frequency during unchallenged S phase. (A) Xenopus egg extracts were mock- or Plk1-depleted and remaining Plk1 was quantified on western blot using anti-Plk1 antibody. * marks a non-specific band. (B) Sperm nuclei (2000 nuclei/μl) were incubated in mock- or Plk1-depleted Xenopus egg extracts in the presence of Biotin-dUTP for different times and analyzed by DNA combing. Selection of three representative DNA fibers of early S phase from mock- (top three) or Plk1-depleted (bottom three) extract; in red, replication eyes labeled by Biotin-dUTP, in green, whole DNA fibers. (C) Replicated fraction (%), fork density (kb–1) and frequency of initiation (kb–1s–1) for three independent experiments at 2 or 3 time points were calculated and reported in the table. (D) scatter blot of ratios Δ Plk/Δmock of replication parameters for each line shown in C, with mean (n = 8) and standard deviation, P values, one sample t-test, compared to theoretical mean 1.
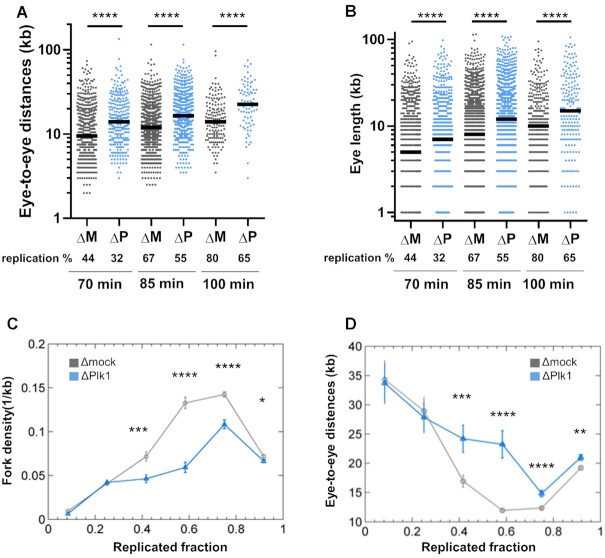
After considering global, averaged replication parameters described above, we precisely analyzed local parameters on individual fibers. Eye-to-eye distances were significantly increased in the absence of Plk1 (median increase 1.4- and 1.6-fold) during mid and late S phase (Figure 2A and Supplementary Figure S2), consistent with the global decrease in initiation frequency (Figure 1D). Further, we observed that eye lengths were also significantly larger during mid and late S phase after Plk1 depletion (Figure 2B, 1.4–1.5-fold increase of medians and Supplementary Figure S2). This result could indicate that after Plk1 depletion, replication fork speed was increased. However, during very early S phase, mean eye lengths were not larger but actually smaller after Plk1 depletion (Supplementary Table S1), arguing against a direct, positive effect on fork speed. Alternatively, this average increase in eye lengths could result from fewer small eye lengths, which means a decrease in new initiation events after Plk1 depletion (Figure 1C, D, Supplementary Figure S3A).
Figure 2.
Plk1 depletion increases eye-to-eye distances, independently of replicated fraction of DNA fibers. Sperm nuclei (2000 nuclei/μl) were incubated for different times in mock- (ΔM, dark grey) or Plk1-depleted (ΔP, blue) Xenopus egg extracts in the presence of Biotin-dUTP. (A) Eye-to-eye distance distribution (n = 77–1151) and (B) eye length distribution (n = 196–2507) (kb) (scatter dot plots with median) of one combing experiment representative of middle and late S phase. All DNA fibers were then grouped together in different bins according to their replicated fraction. Means (n = 51–1189) with SEM of (C) fork density (kb–1) and (D) eye-to-eye distances (kb), calculated for the different bins of replicated fractions of all fibers. * indicates significant difference (Mann–Whitney U test, two-sided, P< 0.05: P-values: * 0.01–0.05; ** 0.001–0.01; *** 0.0001–0.001; ****<0.0001).
In order to exclude that the changes in replication parameters after Plk1 depletion are not simply a consequence of a delay in entering S phase or a delay of the replication program, we grouped together all fibers from the three experiments and sorted within six bins corresponding to the replicated fraction of each single fiber. We observed a significant decrease of the mean frequency of initiation and fork density and, at the same time, a significant increase in the mean eye-to-eye distances and eyes lengths after Plk1 depletion (Figure 2C, D and Supplementary Figure S3B, C). Altogether, this shows that Plk1 depletion modifies replication parameters, independently of the replicated fraction of individual fibers, and does not just induce a general delay of the replication program. We further noticed that, after Plk1 depletion, the number of unreplicated DNA fibers and fibers with zero to four replication eyes were overrepresented, whereas fibers with more than five eyes were underrepresented. Fibers with replication extent of 85–99% were overrepresented (Supplementary Figure S4A–C). This observation suggests that Plk1 depletion affects the distribution of active origins in a heterogeneous manner along the genome.
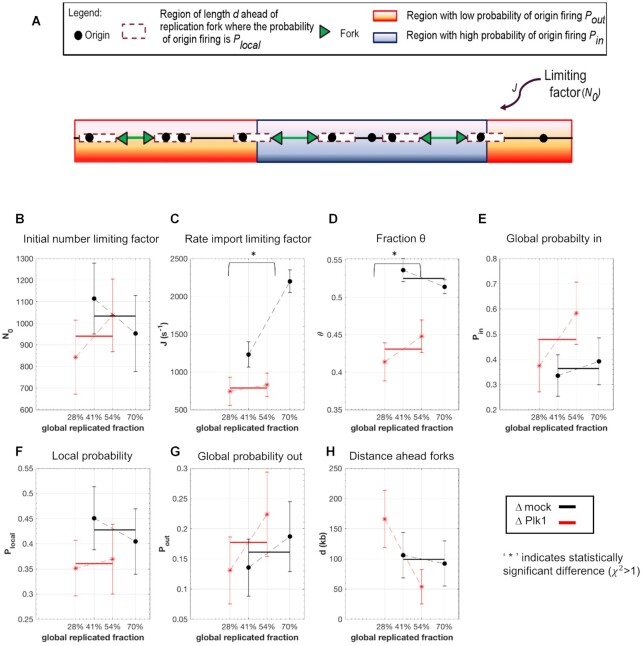
Next, we fitted these experimental combing data by our recently developed minimal model of DNA replication (29). This model assumed that origin firing is stochastic and given by the encounter of limiting replication factors with potential origins and the import/activation rate of limiting replication factors, J. The probability of origin firing is heterogeneous along the genome, meaning replication origins located in a fraction θ of the genome have a probability of firing 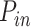 higher than the probability of firing
higher than the probability of firing 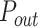 of those located in the remaining fraction 1- θ (Figure 3A). The fraction θ of the genome corresponds to regions of efficient and synchronous firing. Finally, the presence of replication forks promotes other origin firing with a probability
of those located in the remaining fraction 1- θ (Figure 3A). The fraction θ of the genome corresponds to regions of efficient and synchronous firing. Finally, the presence of replication forks promotes other origin firing with a probability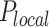 within a distance d from the fork.
within a distance d from the fork.
Figure 3.
Numerical simulations: Plk1 depletion decreases the fraction of the genome with high probability origin firing, θ, and the rate of import of limiting factors, J. (A) Diagram of numerical model from (29): Potential replication origins located in a fraction θ of the genome have a probability of firing 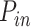 higher than probability of firing
higher than probability of firing 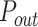 of potential origins located in the complementary genome fraction 1 - θ. The firing of a potential origin requires its encounter with limiting factors whose number
of potential origins located in the complementary genome fraction 1 - θ. The firing of a potential origin requires its encounter with limiting factors whose number 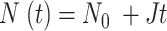 increases as S phase progresses. The origin firing probability over a distance d ahead of a replication fork is enhanced of a probability
increases as S phase progresses. The origin firing probability over a distance d ahead of a replication fork is enhanced of a probability  . (B–H) Inferred model parameters by fitting Δmock and ΔPlk1 depleted combing data for two different time points as indicated with replicated fraction: circles indicate the mean value of the parameters over 100 different runs of the genetic algorithm and the error bar are standard deviations, solid lines are the mean (n = 2) values of consecutive parameters whose differences are not statistically different, (*) indicates significant difference between the Δmock and ΔPlk samples, student t-test as described in Appendix 2 of (29), (
. (B–H) Inferred model parameters by fitting Δmock and ΔPlk1 depleted combing data for two different time points as indicated with replicated fraction: circles indicate the mean value of the parameters over 100 different runs of the genetic algorithm and the error bar are standard deviations, solid lines are the mean (n = 2) values of consecutive parameters whose differences are not statistically different, (*) indicates significant difference between the Δmock and ΔPlk samples, student t-test as described in Appendix 2 of (29), ( P values < 10–7).
P values < 10–7).
For both our experimental conditions, mock and Plk1 depleted samples (70 and 85 min time point of Figure 2A and B), the model well reproduced all replication parameters (Supplementary Figure S5). By comparing pairwise values of model parameters for the Δmock and ΔPlk1 samples, only the reduction of θ and J values were statistically significant (Figure 3B–H). This is consistent with the observed decrease of initiation frequency, fork density and increase of eye-to-eye distances by DNA combing. According to the model, this demonstrates that Plk1 depletion could reduce the fraction θ of the genome that presents an efficient activation of origins, thereby reducing the extent of regions where replication origins seem to fire synchronously. In addition, Plk1 depletion would reduce the overall firing probability via the reduction of the rate of available limiting replication factors.
Dynamics of the chromatin proteome after Plk1 depletion
Since our results above suggest that Plk1 depletion affects the availability of replication factors, we analyzed the dynamics of the abundance of chromatin-bound proteins after Plk1 depletion using an unbiased global and quantitative label-free proteomic approach, i.e. Chromass (45). In triplicate reactions, sperm nuclei were allowed to initiate replication in either mock or Plk1 depleted egg extracts, chromatin was purified after 40 min and subjected to LC/MS/MS. We could quantify 868 proteins across samples and found 62 proteins significantly modulated by Plk1 depletion in the chromatin fraction compared to the control: 27 increased and 35 decreased (Figure 4 and Supplementary Table S2). Amongst the proteins decreasing, we found several replication proteins associated with the active replication fork such as Pol δ and ϵ, GINS3, Rfc2 and Rfc4, consistent with the decrease in the fork density observed by DNA combing experiments. No increase but in fact a decrease of factors like ATM, Mre11 and MSH6 was also found to be consistent with our observation that Plk1 depletion itself does not induce DNA damage in the absence of exogenous replication stress (40). We were intrigued by the increase of three important replication factors involved in the regulation of origin firing: MTBP, TopBP1 and Rif1. MTBP together with Treslin and TopBP1 are necessary for the assembly of the pre-initiation complex (Pre-IC), but also for the checkpoint response.
Figure 4.
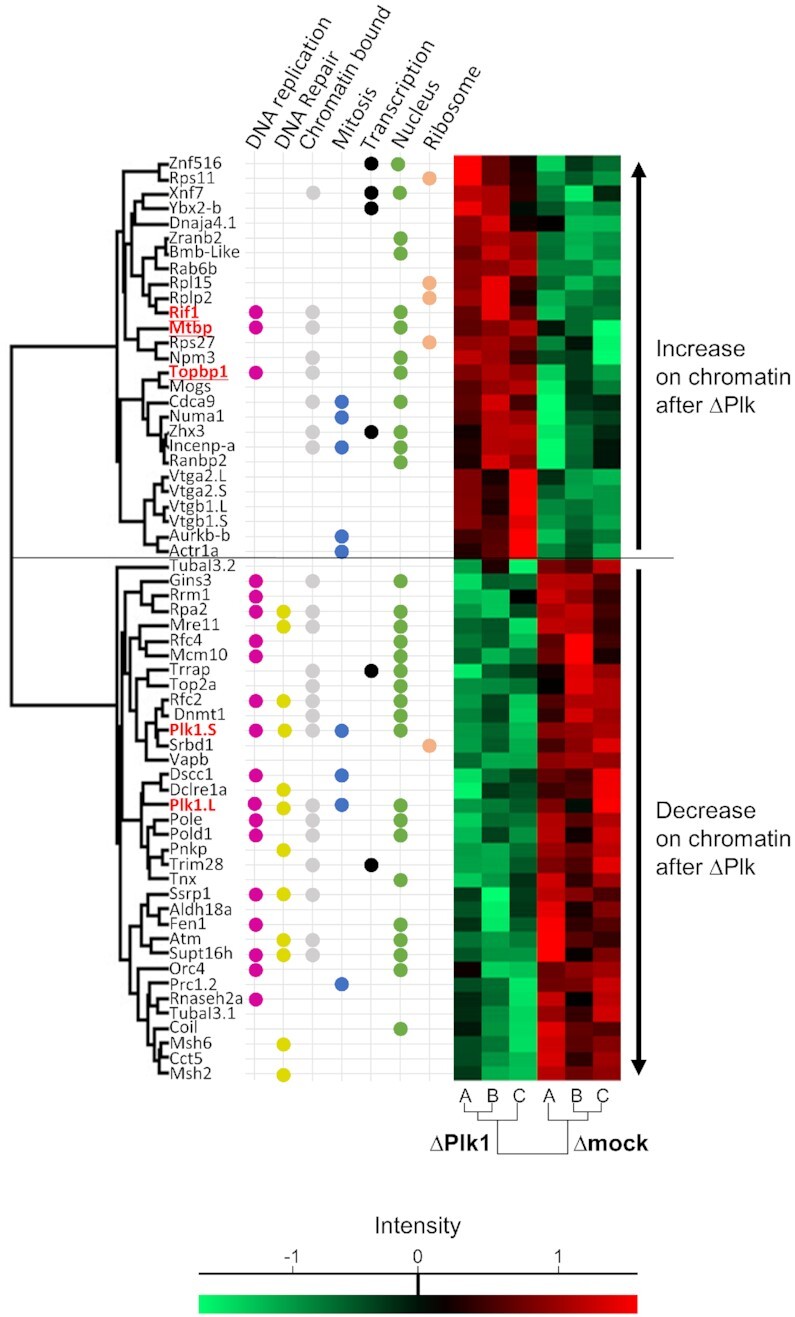
Dynamics of chromatin bound proteins after Plk1 depletion by LC/MS/MS. Sperm nuclei (4000 nuclei/μl) were allowed to replicate in Xenopus egg extracts, chromatin was isolated and analyzed according the Chromass protocol (Materials and Methods). Heat map of significant (P< 0.05) increased or decreased proteins in Plk1 depleted samples versus control samples (A, B, C are triplicates). Protein MS1 intensities are normalized across samples using z score: red indicates intensity above average, bright green below. For each protein, GO-term functions retrieved from Uniprot database manually curated. Plk1.L and Plk1.S signifies proteins encoded by the two different subgenomes L and S of X. laevis.
Plk1 depletion leads to a slower release of Treslin/MTBP/TopBP1 from chromatin during S phase
First, we confirmed in six immunodetections experiments that the levels of MTBP, TopBP1 and, to a lesser extent, Treslin increased significantly on chromatin after Plk1 depletion compared to mock depletion (Figure 5A and B). To ascertain the specificity of Plk1 depletion, we added back recombinant XPlk1 to the extracts, which rescued chromatin Treslin/MTBP/TopBP1 levels comparable to those of mock. Since it was surprising to observe higher levels of these firing factors along with a decrease of origin firing, we followed the dynamics of Treslin/MTBP/TopBP1 on chromatin in a time course experiment along S phase in the presence or absence of Plk1 (Figure 5C and quantification in Supplementary Figure S6). Consistent with Treslin/MTBP/TopBP1 being necessary for Cdc45 recruitment, we observed that all three proteins preceded Cdc45 loading on chromatin, which indicated the start of S phase at 30 min. Treslin/MTBP/TopBP1 levels on chromatin peaked during early S phase, preceding maximal fork density (compare with parallel experiment, Figure 1 C, replicate 3), then declined afterwards, in agreement with what is observed for their yeast homologues Sld3/Sld7 and Dbp11, which do not travel with replication forks (51–54). However, MTBP, TopBP1 and Treslin remained bound to chromatin for a longer period in the absence of Plk1. In budding yeast, the MTBP homologue Sld7 needs to be released from chromatin for CMG activation to allow elongation (53). Therefore, Plk1 depletion induced a longer persistence of the Treslin/MTBP/TopBP1 complex on chromatin, possibly slowing down the recycling frequency of these proteins.
Figure 5.
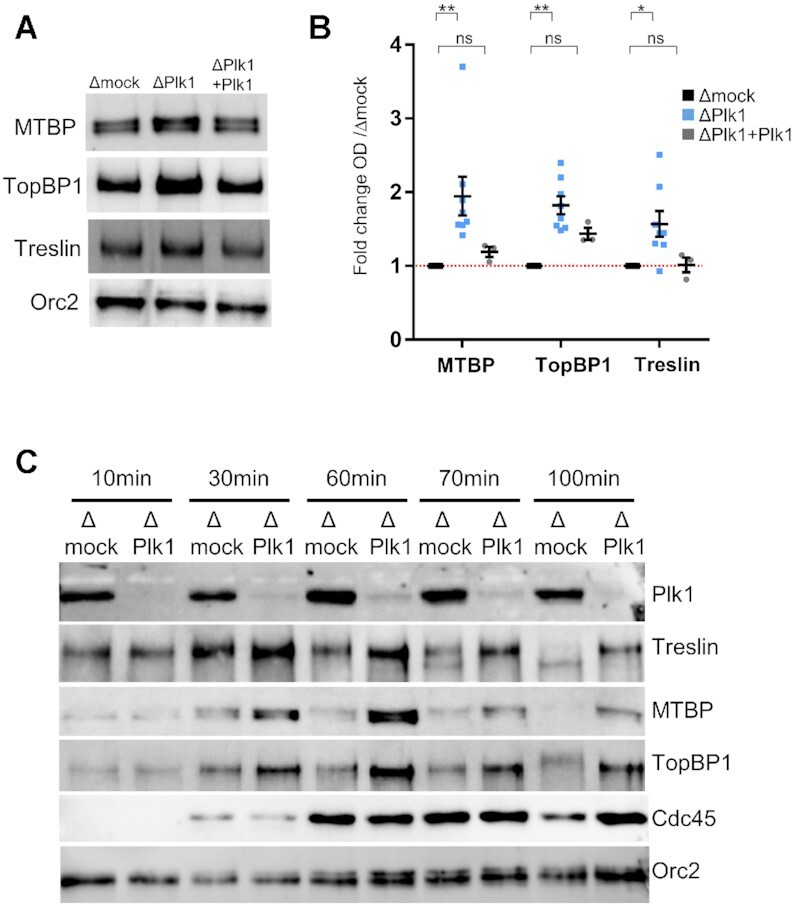
Plk1 depletion stabilizes the binding of Treslin, MTBP and TopBP1 on chromatin. Sperm nuclei (2000 nuclei/μl) were added to mock- and Plk1-depleted egg extracts. Isolated chromatin was subjected to gel electrophoresis and western blot analysis using the indicated antibodies. (A) Representative western blot at 70 min for mock-depleted extracts, Plk1-depleted extracts and Plk1-depleted extracts replenished with recombinant Plk1 (300 nM). (B) Mean fold increase with SEM of MTBP, TopBP1, and Treslin optical density of bands in western blots (OD) normalized to Orc2 in six independent experiments at mid S phase (60–70 min) for depletions (n = 8), three experiments for add back with recombinant Plk1 (n = 3). * indicates significant differences, ns not significant, two-sided, unpaired t-test. (C) As in (A) but chromatin fractions were isolated at indicated times, in parallel with combing experiment from Figure 2.
MTBP is phosphorylated in an ATR/Chk1 and DDK dependent manner
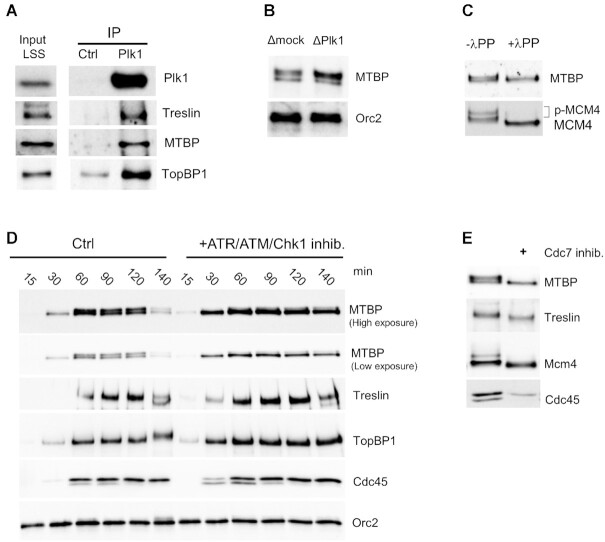
In order to investigate whether Plk1 could directly interact with Treslin/MTBP and TopBP1, we performed immunoprecipitations from interphase egg extracts using mock or Plk1 antibody coupled to protein A coated beads. We observed that Treslin, MTBP and TopBP1 were specifically enriched on Plk1 beads compared to mock beads (Figure 6A), supporting a specific interaction between Plk1 and the Treslin/MTBP/TopBP1 complex. On high resolution SDS-PAGE gels, MTBP appeared as a doublet on western blots; moreover, after Plk1 depletion the upper band was increased in comparison to the lower band (Figure 6B). This suggested that MTBP could be post-translationally modified, possibly by phosphorylation. Digestion with Lambda-phosphatase demonstrated that the upper MTBP band was indeed a phosphorylated form (Figure 6C), which increased following S phase progression (Figure 6D). It was previously shown that Treslin is phosphorylated by Chk1 (23) in Xenopus. We therefore asked whether the phosphorylation of MTBP could also be dependent on the replication checkpoint kinases ATR/ATM and Chk1. Upon inhibition of ATR/ATM by caffeine together with the Chk1 inhibitor AZD 7762, we found that almost all MTBP remained unphosphorylated (Figure 6 D). Inhibiting ATR/ATM alone had a smaller effect on MTBP dephosphorylation whereas Chk1 inhibition alone had no visible effect on MTBP phosphorylation (Supplementary Figure S7). This observation could be explained by a cooperative effect of the checkpoint kinases as concluded in a recent study, published while our work was in revision (55). In yeast, the Treslin homologue Sld3 shifts in a DDK dependent manner in G1 phase (56). We therefore tested whether DDK inhibition by the Cdc7 inhibitor PHA767491 (17,57) would also cause shifts of both MTBP and Treslin (Figure 6E and Supplementary Figure S8). Both proteins indeed shifted down upon Cdc7 inhibition in chromatin fractions. Altogether, these data strongly suggest that the phosphorylation of Treslin and MTBP is regulated by the ATR/Chk1 dependent checkpoint and by DDK. To conclude, Plk1 interacts with Treslin/MTBP and TopBP1 and, after Plk1 depletion, the Treslin/MTBP/TopBP1 complex showed an increased and prolonged association to chromatin. Consequently, a slower recycling of these rate-limiting initiation factors could explain a decrease in origin firing, possibly via a direct interaction between Plk1 and the phospho-Treslin/MTBP/TopBP1 proteins.
Figure 6.
Plk1 co-immunoprecipitates with MTBP, Treslin and TopBP1. (A) Egg extracts were incubated with control (IP Ctrl) or Plk1 (IP Plk1) antibody coupled Sepharose beads. Immunoprecipitates and input extract were subjected to gel electrophoresis and western blot analysis using the indicated antibodies. (B) Chromatin fractions, 60 min after the addition of 2000 nuclei/μl in mock- or Plk1-depleted extracts, analyzed by western blot with indicated antibodies. (C) Chromatin fractions, isolated 60 min after the addition of 2000 nuclei/μl in egg extracts were treated or not with λ phosphatase and subjected to western blotting with indicated antibodies. (D) Sperm nuclei were incubated in egg extracts in the absence or presence of the ATR/ATM inhibitor caffeine (5 mM) and the Chk1 inhibitor AZD7762 (0.5 μM), reactions were stopped at indicated times, chromatin was purified and subjected to western blotting with indicated antibodies. (E) Sperm nuclei were incubated in egg extracts in the absence or presence of the Cdc7 inhibitor PHA767491 (50 μM), reactions were stopped at 60 min, chromatin was purified and subjected to western blotting with indicated antibodies.
Chromatin Plk1 interactome during S phase
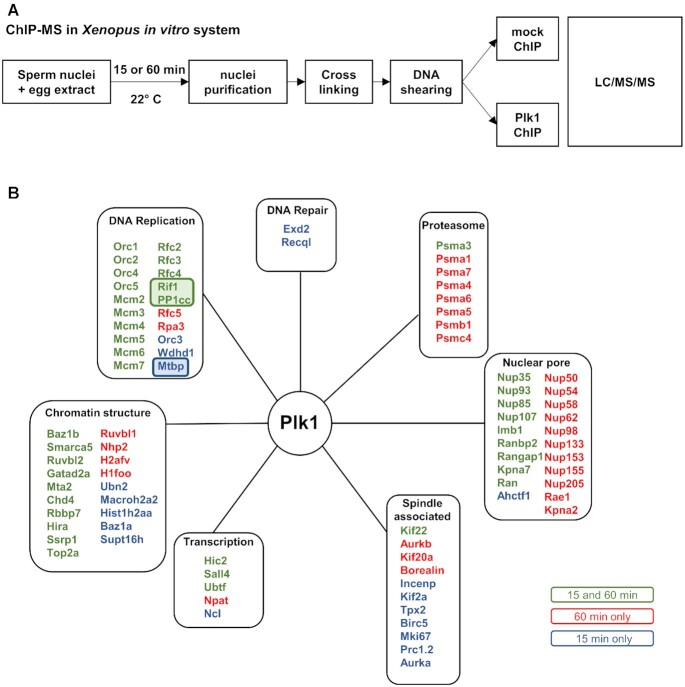
In mitosis, Plk1 has been shown to interact through its polo-box domain with several hundreds of proteins from different cellular compartments (58), but only few S phase interacting proteins have been identified so far. Our previous results show that Plk1 is recruited to chromatin at the time when the pre-RC assembly takes place (40), before DNA replication starts. In order to specifically identify Plk1 interactions with chromatin and nuclear proteins, we developed a Plk1-chromatin immunoprecipitation approach followed by a proteomic shotgun analysis (ChIP-MS) (Figure 7A). Nuclei were purified after pre-RC assembly before the start of DNA replication (15 min), then during DNA replication (60 min). Purified nuclei were crosslinked, solubilized and the DNA was fractionated by sonication. This was followed by immunoprecipitation using anti-Plk1 and control antibodies and LC/MS/MS analysis. More than 112 proteins were found enriched at least 3 fold in the Plk1 immunoprecipitate compared to the control (Supplementary Table S2). Among those proteins, several are known Plk1-interacting proteins such as topoisomerase 2, Orc2, MCM2, Aurora A or Kif2 (Figure 7B). In addition, we identified several new putative Plk1 interacting partners implicated in replication initiation or elongation such as Wdhd1/And-1, Rfc2-5, Rpa3 and interestingly MTBP, Rif1 and PP1. Chromatin remodeling factors were another novel class of proteins that we identified in the chromatin Plk1 network, especially the NuRD complex (Chd4, Mta2, HDAC, Rbbp7) and a FACT subunit SSRP1, all involved in the regulation of DNA replication in Xenopus (59,60). Rif1, for which we have already observed a significant increase on chromatin after Plk1 depletion, was the most interesting candidate since it is a major regulator of replication timing at the level of large replication domains (13,14). We therefore investigated in more detail the interaction between Rif1 and Plk1 during S phase in the Xenopus system.
Figure 7.
ChIP mass spectroscopy reveals potential Plk1 interactions with DNA replication proteins. (A) Experimental diagram of the ChIP MS approach from the Xenopus in vitro system after 15 min (pre-RC assembly) and 60 min (ongoing replication) incubation of sperm nuclei (4000 nuclei/μl) in egg extracts. (B) Identified nuclear proteins enriched more than 3-fold (spectral counts) in Plk1- versus mock-ChIP were classified into various functional groups according to GO-term location and functions retrieved from Uniprot database manually curated.
Plk1 interacts with Rif1 and phosphorylates Rif1 on several sites in vitro
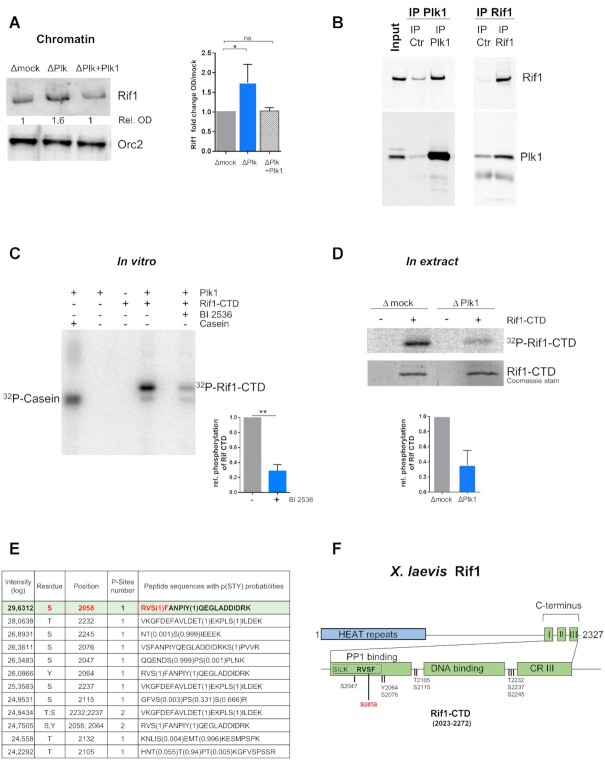
Immuno-detection analysis of chromatin fractions showed that Rif1 amounts were significantly increased (mean 1.7) after Plk1 depletion compared to mock (Figure 8A), confirming the Chromass result (Figure 4). This increase was abolished by the addition of recombinant Plk1. In order to confirm a direct interaction between Plk1 and Rif1, we performed co-immunoprecipitation experiments in cytoplasmic egg extracts with either Plk1 or Rif1 antibody coupled beads (Figure 8B). We found that Rif1 co-precipitated with Plk1 and conversely that Plk1 co-precipitated with Rif1. This reciprocal co-immunoprecipitation between Plk1 and Rif1 in cytoplasmic extracts was further confirmed in nucleoplasmic extract (Supplementary Figure S9). We were unable to obtain the full length 275 kDa Rif1 recombinant protein. To circumvent this potential limitation, we used the C-terminal domain of Rif1 (Rif1–CTD), known to bind PP1 via conserved SILK and RVSF consensus sites in metazoans. Because Rif1–CTD contains several Plk1 phosphorylation motifs and the gamma catalytic subunit of PP1 was found enriched in the proteomic Plk1-ChIP approach (Figure 7B), we tested if Plk1 could phosphorylate Rif1–CTD in vitro. Recombinant Rif1–CTD was incubated with recombinant active Xenopus Plk1 in the presence or absence of the Plk1 inhibitor, BI2536, and [γ32P]-ATP (Figure 8C and Supplementary Figure S10). We observed the phosphorylation of Rif1–CTD, which decreased in the presence of the Plk1 inhibitor. In order to demonstrate that endogenous Plk1 also phosphorylates Rif1–CTD in S phase egg extracts, we incubated Rif1–CTD in mock and Plk1 depleted egg extract in the presence of [γ32P]-ATP. Rif1–CTD was then purified using Ni-Sepharose beads and analyzed by SDS-PAGE (Figure 8D). As expected, we found that Rif1–CTD was phosphorylated in mock depleted egg extract, whereas its phosphorylation decreased in Plk1 depleted extracts. Rif1–CTD phosphorylated in vitro by recombinant Plk1 was analyzed by LC/MS/MS and ten highly confident phosphorylation sites were detected (Figure 8E and Supplementary Table S2). Amongst them, six residues are conserved between X. laevis and human Rif1 and four have been reported to be phosphorylated in vivo in human cells by mass spectrometry experiments on the curated PhosphoSitePlus website (http://www.phosphosite.org). The most abundant phosphorylated site in our analysis was Serine 2058, located in the PP1 binding motif RVSF (Figure 8F). This S2058 followed by a Phenylalanine forms a motif previously identified as a Plk phosphorylation site (p[S/T]F) (61). We conclude that Rif1 interacts with Plk1 and is phosphorylated on several sites by Plk1 within its C-terminal domain, particularly on a PP1 binding site, in vitro as well as in S phase Xenopus egg extracts in a Plk1 dependent manner.
Figure 8.
Plk1 interacts with Rif1 and phosphorylates the C-terminal region of Rif1. (A) Left: Chromatin was purified after 70 min incubation of sperm nuclei (2000 nuclei/μl) in extracts depleted of mock, Plk1 or Plk1 replenished with recombinant Plk1 (300 nM) (ΔP + Plk1) and analyzed by western blotting, right: mean (n = 17 and 8) fold change with SEM of Orc2 normalized Rif1 signal compared to mock depletions from four different experiments, Wilcoxon signed rank test, * indicates significant and ns non-significant differences. (B) Cytoplasmic extract, LSS (Input) was incubated with control or Plk1 antibody coupled to Sepharose beads (respectively IP Ctrl and IP Plk1) or with control or Rif1 antibody coupled to Sepharose beads (respectively IP Ctrl and IP Rif1). The input and immunoprecipitates were subjected to gel electrophoresis and western blot analysis using the indicated antibodies. (C) In vitro phosphorylation of recombinant Rif1–CTD (6 μM) or control substrate casein (6 μM) by recombinant Plk1 (60 nM) in the presence of [γ32P]-ATP with or without the Plk1 inhibitor BI2536 (100 nM); left, representative autoradiography of SDS-PAGE; right, mean with SD of relative phosphorylation of three independent experiments, t-test, P value 0.005. (D) Phosphorylation of Rif1–CTD in S phase egg extracts: Rif1–CTD was added to mock- or Plk1- depleted extract in the presence of [γ 32P]-ATP, purified using Ni-Sepharose beads and analyzed on SDS-PAGE. Top, autoradiography; middle, Coomassie stain; bottom, mean (n = 2) with SEM of relative phosphorylation of Rif1–CTD normalized to total Rif1–CTD pulled down (Coomassie). (E) Identified phosphopeptides and modified amino acids in Rif1–CTD phosphorylated in vitro by recombinant Plk1, as in (D), but in the presence of cold ATP, by LC/MS/MS analysis. (F) Diagrammatic representation of Xenopus Rif1 with Rif1–CTD composed of three C-terminal regions (CR) comprising PP1 binding motifs SILK and RVSF located in CRI, adapted from (74).
Acceleration of DNA replication after Rif1–CTD addition depends on PP1 interaction with Rif1-S2058 in the RVSF motif
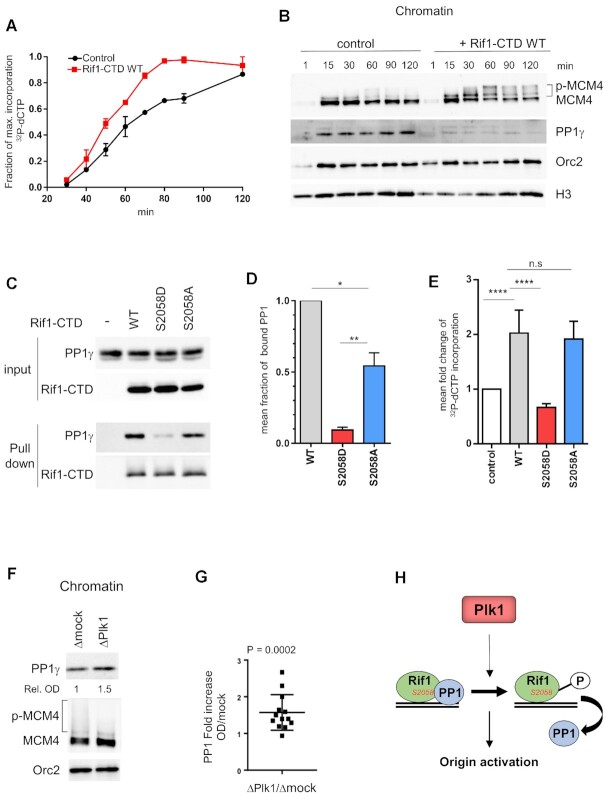
Next, we wondered whether the presence of Rif1–CTD would affect replication. For that purpose, we incubated sperm nuclei in egg extracts without or with Rif1–CTD (640 nM) in the presence of [α32P]-dCTP. DNA was purified at indicated times and analyzed by gel electrophoresis followed by the quantification of 32P incorporation into DNA (Figure 9A). We found that the addition of Rif1–CTD significantly increased DNA replication (Figure 9A and E). In parallel, chromatin fractions were analyzed by western blotting. Chromatin-bound PP1 was drastically lower in the presence of Rif1–CTD than in the control while the phosphorylation of MCM4 was strongly increased (Figure 9B). These results show that a six-fold excess of Rif1–CTD versus the endogenous Rif1 (62) in extracts interferes with PP1 binding to chromatin, maybe by titrating PP1 away, leading to a more active MCM complex, thereby increasing origin activation and accelerating DNA replication.
Figure 9.
Rif1–CTD accelerates DNA replication dependent on PP1 interaction with Rif1-S2058. (A) Rif1–CTD or buffer was added to sperm nuclei (2000 nuclei/μl) replicating in egg extracts in the presence of [α 32P]-dCTP, DNA was purified at indicated times and replication was analyzed on alkaline gel electrophoresis, the mean incorporation with SEM from two independent experiments was plotted. (B) Rif1–CTD or buffer was added to sperm nuclei replicating in egg extracts, chromatin was purified at indicated times, specified proteins were analyzed by western blotting. (C) Buffer (-), wild type (WT), 1 μM Rif1–CTD mutants respectively (S2058D) and (S2058A) were added to egg extracts, recovered by pull-down after 1h incubation using Ni-magnetic beads, then analyzed by western blotting using anti-PP1 (PP1γ) or anti-Rif1 (Rif1–CTD) antibodies. (D) PP1 bound to Rif1–CTD was quantified, normalized to Rif1, and expressed as mean fraction with SEM of PP1 bound to WT Rif1–CTD in three independent experiments, unpaired or one sample t-test. (E) As in (A) WT, S2058D and S2058A Rif1–CTD were incubated in replicating reactions and [α 32P]-dCTP incorporation was quantified after DNA gel electrophoresis then plotted as mean fold increase compared to control (buffer) with SEM, in 3 (for WT, S2058D) or 5 (for WT, S2058A) independent experiments with different time points within (n = 9, n = 15 respectively), Mann-Whitney test. (F) Rif1–CTD or buffer was added to sperm nuclei (2000 nuclei/μl) replicating in mock- (Δmock), or Plk1- (ΔPlk1) depleted extracts, chromatin was purified after 60 min and indicated proteins were analyzed by western blotting. (G) Fold increase of chromatin bound PP1 normalized to Orc2 OD on western blots in Plk1- versus mock- depleted extracts from five different replication reactions with different time points, scatter dot blot with mean (n = 13), Wilcoxon signed ranked test, P-value above blot. (H) Model of Plk1 regulating origin activation via phosphorylation of Rif1 which interrupts the Rif1-PP1 interaction.
In order to investigate if Rif1–CTD effectively binds PP1 in extracts and to test whether S2058 modifications within in the PP1 binding motif RVSF would affect this binding, we produced a non-phosphorylatable S2058A as well as a phosphomimetic S2058D mutant of Rif1–CTD. We retrieved PP1 bound to wild type Rif1–CTD from extract (Figure 9C), showing that the C-terminal domain of Rif1 contributes to PP1 binding (17,63). Both mutants showed decreased PP1 binding but to a quite different degree; whereas the phosphomimetic S2058D mutant strongly reduced PP1 binding (9%), the S2058A mutant was still able to bind substantial amounts of PP1 (54%) compared to the wild type Rif1–CTD (Figure 9 C and D).
We reasoned that if a functional PP1-Rif1–CTD interaction is responsible for the acceleration of DNA replication, then the S2058D mutant should fail to enhance DNA replication. As expected, we found that adding the phosphomimetic mutant S2058D did not increase DNA replication, on the other hand, the non-phosphorylatable mutant S2058A was still able to increase DNA replication to an extent similar as wild type Rif1–CTD (Figure 9E), in agreement with the observed differences in PP1 binding of these mutants (Figure 9D). Likewise, chromatin bound PP1 was higher in replication reactions after addition of the phosphomimetic S2058D mutant compared to the WT and the non-phosphorylatable S2058A mutant (Supplementary Figure S11). These different results demonstrate that the phosphorylation of S2058 inhibits the Rif1–CTD–PP1 interaction and can modulate DNA replication. If Plk1 directly regulates the Rif1–PP1 interaction by phosphorylation during DNA replication, then we would expect to see more PP1 to endogenous Rif1 on chromatin after Plk1 depletion. Indeed, we observed in Plk1 depleted replication reactions that the amount of chromatin bound PP1 significantly increased (1.6-fold) and that MCM4 phosphorylation decreased compared to the control (Figure 9F and G). Therefore, we conclude that the phosphorylation by Plk1 at S2058 in the RVSF motif of Rif1–CTD prevents its interaction with PP1, suggesting thereby a mechanism by which Plk1 could counteract the inhibitory action of Rif1-PP1 in the activation of replication origins (Figure 9H).
DISCUSSION
In this study, we have explored how Polo-like kinase 1 regulates the spatio-temporal replication program. Our findings indicate that Plk1 positively controls replication origin firing at the level of early, efficiently replicating genomic regions. First, DNA combing experiments showed that Plk1 depletion generates a reduction of the replication initiation frequency and an increase of eye-to-eye distances. Second, two different proteomic approaches together with Plk1 co-immunoprecipitation experiments identified the origin firing proteins Treslin/MTBP/TopBP1 as well as the replication timing regulator Rif1 as novel Plk1 interacting partners. Third, Plk1 phosphorylates Rif1 at its PP1 binding site and the binding of Rif1-PP1 on chromatin increased in a Plk1 dependent manner, suggesting a so far undescribed mechanism for regulation of origin firing by Plk1. Finally, our numerical simulations of replication parameters extracted from the DNA combing experiments, suggest that Plk1 promotes the synchrony of initiations within highly efficient domains and increases the overall initiation efficiency via modulating the availability of limiting replication factors.
Plk1 promotes replication origin activation in Xenopus
Our DNA combing study revealed that after Plk1 depletion replication fork density and initiation frequency decrease, whereas locally, distances between neighboring replication eyes increase. This strongly argues that Plk1 promotes origin activation. In addition, our data imply that the synchrony of firing of groups of early efficient origins firing is perturbed after Plk1 depletion, or in other words that Plk1 could interfere with the organization of the genome that is normally divided into early regions with efficient origins and later regions composed of less efficient origins. We also observed that eye lengths increased during mid and late S phase after Plk1 depletion. This could be explained either by the decrease of new initiations (small eyes) or by a higher fork speed to ensure complete replication in presence of a reduced number of activated origins (64). Alternatively, more origins could be initiated close to forks (65,66), rapidly merging and no longer appearing as single initiation events. Replication in the Xenopus in vitro system mimics very rapid embryonic S phase in early embryonic cell cycles before the onset of transcription at the mid-blastula transition (MBT). We recently found that the level of active, phosphorylated Plk1 is high before the MBT, but suddenly declines after the MBT when the DNA replication program slows down due to a decrease in fork density (40,67). The decrease of active Plk1 levels around the MBT could contribute to the decrease of origin activation at that stage of development.
Using Plk1-ChIP mass spectroscopy during S phase, we identified most of the members of the Orc and MCM complex, mentioned in earlier studies (31,37,39,68). In addition, we identified replication elongation proteins such as Rfc2-5, whose direct or indirect interaction with Plk1 could explain the possible effect on fork speed. The decrease in the initiation frequency after Plk1 depletion could be attributed to a more active replication checkpoint in the absence of Plk1 (40). However, the involvement of other parallel pathways could not be excluded. In fact, we identified in this study Rif1 and MTBP/Treslin/TopBP1 as new Plk1 interacting proteins, which are important regulators of both DNA replication and intra-S phase checkpoint.
Plk1–Rif1 interaction
Rif1 is a multi-functional protein known to regulate replication timing (13,14) and to individually inhibit the firing of late origins by targeting PP1 phosphatase to those origins, counteracting Cdc7/Dbf4 dependent MCM4 phosphorylation (15–17). A negative regulation of Rif1 by Plk1 could therefore account for the decrease in initiation frequency, the increase of eye-to-eye distances and the changes in synchrony of origin firing after Plk1 depletion. Three observations strongly support that Plk1 could regulate DNA replication, at least in part, via a direct interaction and a phosphorylation of Rif1: (i) Rif1 and Plk1 co-immunoprecipitated both in cytosolic egg extracts and in nuclear extracts. (ii) In vitro the C-terminal domain of Rif1 was phosphorylated by recombinant Plk1 and was also phosphorylated in a Plk1-dependent manner in extracts. (iii) Most of the in vitro phosphorylation in the Rif1 C-terminal domain by Plk1 occurred at S2058, in a highly conserved PP1 binding site RVSF which, when mutated, strongly decreased PP1 binding. In human cells, the homologous residue (S2205) was reported to be phosphorylated by Chk1 in a substrate screen (69), but also in response to ATR/Chk1 inhibition in a Cdk1 dependent manner (70), which makes the RVSF motif a key target site for Rif1/PP1 regulation. Mutation of Serine 2058 into the phosphomimetic Aspartate almost abolished PP1 binding to Rif1, consistent with findings in human cells (70), suggesting that the phosphorylation interrupts the Rif1-PP1 interaction. The non-phosphorylatable mutation still allows to bind substantial amounts PP1, although the interaction was decreased compared to the wild type. Similarly, in in vitro assays PP1 binds to a non-phosphorylated Rif1 peptide (71) and the S2205A mutation of a small human Rif1 peptide inhibited only slightly PP1 binding (63). In addition, the overexpression of Rif1–CTD in extracts led to an increase in DNA replication with a strong decrease of the binding of PP1 to chromatin and a MCM4 hyperphosphorylation. Those effects are similar to the effect of Rif1 immunodepletion in Xenopus (17) or to those of Rif1 knock-down in human cells (18). The addition of Rif1–CTD could titrate free PP1 and/or compete with endogenous Rif1 on chromatin. In strong support for this, we found that the addition of the phosphomimetic S2058D mutant of Rif1–CTD failed to increase DNA replication whereas the non-phosphorylatable S2058A mutant still supports an acceleration of DNA replication. This observation suggests that the phosphomimetic mutant cannot bind enough PP1, therefore cannot not titrate away PP1 and fails to enhance DNA replication. The non-phosphorylatable mutant would still be able to titrate enough PP1, because its interaction with PP1 remains efficient. After Plk1 depletion, PP1 is significantly increased on chromatin together with an hypophosphorylation of MCM4, concomitantly, we observed a small but significant increase of chromatin bound Rif1. We can hypothesize that lowering Rif1 Serine 2058 phosphorylation by depleting Plk1 would increase Rif1/PP1 interaction and stabilize this complex on chromatin thereby increasing MCM4 dephosphorylation.
Rif1 is also known to play a role as chromatin organizer (13,20), a role which was described as partially independent of its interaction with PP1 (72). Both the N- and C-terminal domain of Rif1 have been shown to mediate DNA binding (73,74). It cannot be excluded that the interaction between Rif1 and Plk1 interferes more indirectly with the replication timing by modifying the role of Rif1 as a chromatin organizer. On one hand, the direct phosphorylation of the Rif1 C-terminus by Plk1 could promote its removal from chromatin, necessary for the activation of late origins. On the other hand, Plk1 could also inhibit the checkpoint resulting in a decreased Cdk activity, regulating Rif1. Indeed, in Drosophila it has been shown that an increased Cdk1 activity induces Rif1 removal from chromatin during late S phase (75). However, in Xenopus Rif1 is not necessary for a response to replication stress (41) and seems to regulate S phase progression in parallel with ATR/Chk1 in a human cell line (17), independently of its role in the checkpoint. Therefore, the most likely analysis of our results is a direct regulation of Rif1 by Plk1 by negatively interfering with the Rif1–PP1 interaction.
Plk1-Treslin/MTBP/TopBP1 interaction
Our results show that Plk1 interacts with pre-IC proteins that may favor origin firing. Plk1 interacts with MTBP on chromatin and co-immunoprecipitates with MTBP/Treslin and TopBP1 in cytosolic egg extract. We found that MTBP is most likely not phosphorylated by Plk1 itself, since it remains phosphorylated after Plk1 depletion, but MTBP is phosphorylated both in a DDK and replication checkpoint dependent manner, as it has been evidenced for Treslin (23) and TopBP1 (69). The phosphorylation by a priming kinase may create a docking site for Plk1, via its polo-box domain, to bind to Treslin/MTBP/TopBP1 for further regulation, a mechanism already described for ATR which phosphorylates the Chk1 mediator, Claspin, allowing to bind Plk1 (35). Reciprocal co-immunoprecipitation has shown that Plk1 and TopBP1 interact in cytosolic extracts from asynchronous human culture cells and that this interaction increases in mitosis (36). A potential limitation of our study is that we do not establish which of the three pre-IC proteins could directly interact with Plk1 in S phase Xenopus extracts to help deciphering the biochemical nature and functionality of this complex. After Plk1 depletion, we observed an increase of Treslin/MTBP/TopBP1 on chromatin, especially at late stages of S phase, which could be the consequence of a slower S phase progression. Following this rational, an increase of Treslin/MTBP/TopBP1 should produce an increase of fork density in late S Phase of the Plk1 depleted sample compared to mock; we however measured the opposite, a decrease in fork density (Figure 1D and 2D). This suggests, but does not exclude that the increase of Treslin/MTBP/TopBP1 on chromatin is not an effect of a slower replication progression. In yeast, Sld3/Sld7/Dbp11 are rate limiting and dissociate from origins during CMG formation (9,76). The MTBP orthologue Sld7 is necessary to stabilize Sld3 (Treslin orthologue) and reduces its affinity for Cdc45 (53). In the absence of Sld7, the dissociation of GINS from the origin and S phase progression are delayed. Treslin and TopBP1 are also limiting factors during early Xenopus development (10). Our hypothesis is that the interaction between Plk1 and the Treslin/MTBP/TopBP1 proteins is necessary for the timely release of these limiting factors from the activated origins in order to recycle them to other yet un-activated pre-RCs, but further experiments are required to evidence this and to better characterize the mechanism involved.
A numerical model to explain how Plk1 regulates the spatio-temporal replication program
In order to enrich our different experimental observations, we made use of a newly developed numerical model (29). In this model, the probabilities of origin firing in the fraction θ of the genome (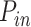 ) and close to replication forks (
) and close to replication forks (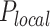 ) are independent of Chk1 action. Conversely, the background probability of origin firing in the remaining 1 – θ fraction of the genome is strongly regulated by Chk1 action. The fraction θ of the genome, with high probability of origin firing, increases as DNA replication progresses.
) are independent of Chk1 action. Conversely, the background probability of origin firing in the remaining 1 – θ fraction of the genome is strongly regulated by Chk1 action. The fraction θ of the genome, with high probability of origin firing, increases as DNA replication progresses.
After fitting this model to the DNA combing data obtained in the absence of Plk1, two parameters significantly decreased: (i) the fraction θ of the genome with high probability of origin firing, in other words, it reduces the total extent of early replicating regions and hence, changes the partitioning between regions of high and low firing probability. (ii) J, the import/activation rate of available limiting factors. The decrease of J suggests a direct implication of Plk1 in the availability of limiting replication factors such as MTBP, Treslin or TopBP1, which we also identified as Plk1 interacting partners.
Since MTBP has been shown to be enriched in regions which replicate preferentially early (77), the interaction between MTBP and Plk1 could favor the origin firing in early regions. In addition, Rif1 has been shown to control the replication timing domain structure through regulating higher order chromatin structure. It is therefore tempting to propose that the crosstalk between Rif1 and Plk1 could take part in the regulation of this replication parameter θ. Furthermore, the fact that local origin firing probabilities are independent of the presence or absence of Plk1 and Chk1 (29) rules out the possible regulation of Chk1 through an inhibitory action of Plk1 close to replication fork, as it has been proposed in case of replication stress (31,32,78). Therefore, Plk1 does not repress the Chk1 inhibition of origin firing, especially close to active replication forks, but seems to limit the inhibitory action of Chk1 on origin firing in a more global manner as indicated by the θ and J parameters. This minimal model with only seven parameters well reproduced our experimental data. However, we cannot exclude that more complex numerical models including additional or alternative assumptions may also fit our data.
The deregulation of S phase and its spatio-temporal DNA replication program often results in genomic instability and cancer development. Plk1 is frequently overexpressed during the growth of aggressive cancers (79), but its role has been mainly restrained to the deregulation of mitosis. We previously found that Plk1 overexpression accelerated DNA replication (40). Here, we provide novel molecular insights into how Plk1 controls the spatio-temporal DNA replication program in vertebrates during unperturbed S phase via the negative regulation of the Rif1-PP1 interaction. Further investigations remain necessary to better understand how Plk1 links three-dimensional chromatin architecture to replication origin firing.
DATA AVAILABILITY
The mass spectrometry proteomics data sets have been deposited to the ProteomeXchange Consortium via the PRIDE (80) https://www.ebi.ac.uk/pride: PXD022440 (Identification of Plk1 interacting proteins by XChIP-MS); PXD022572 (Chromass/ Protein recruitment on chromatin in the presence or absence of Plk1); PXD022410 (phosphorylation of Rif1 by Plk1).
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to A. Kumagai and B. Dunphy for the gift of C-terminal Xenopus Rif1 and the anti-XMTBP and anti-XTopBP1 antibodies. We thank Bruno Miroux for critical reading of the manuscript and Virginie Chiodelli for help with extract preparation and testing. Anti-Xenopus PP1γ antibody, developed by JL. Maller was obtained from the European Xenopus Resource Centre, curated with funding from the Wellcome Trust/BBSRC and maintained by the University of Portsmouth, School of Biological Sciences. This work has benefited from the facilities and expertise of the I2BC proteomic platform (Proteomic-Gif, SICaPS) supported by IBiSA, Ile de France Region, Plan Cancer, CNRS and Paris-Sud University.
Notes
Present address: Diletta Ciardo, Institut de Biologie de l’Ecole Normale Supérieure, Ecole Normale Supérieure, CNRS, INSERM, PSL Research University, Paris, France.
Contributor Information
Diletta Ciardo, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Olivier Haccard, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Hemalatha Narassimprakash, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
David Cornu, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Ida Chiara Guerrera, Proteomics platform Necker, Université de Paris - Structure Fédérative de Recherche Necker, INSERM US24/CNRS UMS3633, Paris 75015, France.
Arach Goldar, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Kathrin Marheineke, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondation de la Recherche Medicale (FRM) [DEI20151234404 to A.G.); Institut National du Cancer (INCa) [PLBIO16-302, A.G.]; Commissariat à l’Energie Atomique (CEA to A.G.); Centre Nationale de Recherche Scientifique (CNRS to K.M., O.H., D.C.]; IdEX Program of Paris-Saclay University for the interdisciplinary PhD fellowship (IDI) of DC (to K.M.). Funding for publication charge: Institut National du Cancer (INCa) [PLBIO16-302].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ganier O., Prorok P., Akerman I., Méchali M.. Metazoan DNA replication origins. Curr. Opin. Cell Biol. 2019; 58:134–141. [DOI] [PubMed] [Google Scholar]
- 2.Rhind N., Gilbert D.M.. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013; 5:a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G.. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010; 140:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansam C.L., Cruz N.M., Danielian P.S., Amsterdam A., Lau M.L., Hopkins N., Lees J.A.. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010; 24:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boos D., Sanchez-Pulido L., Rappas M., Pearl L.H., Oliver A.W., Ponting C.P., Diffley J.F.X.. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 2011; 21:1152–1157. [DOI] [PubMed] [Google Scholar]
- 6.Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G.. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 2011; 193:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boos D., Yekezare M., Diffley J.F.X.. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science. 2013; 340:981–984. [DOI] [PubMed] [Google Scholar]
- 8.Kumagai A., Dunphy W.G.. MTBP, the partner of Treslin, contains a novel DNA-binding domain that is essential for proper initiation of DNA replication. Mol. Biol. Cell. 2017; 28:2998–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantiero D., Mackenzie A., Donaldson A., Zegerman P.. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011; 30:4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collart C., Allen G.E., Bradshaw C.R., Smith J.C., Zegerman P.. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013; 341:893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H.. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012; 26:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peace J.M., Ter-Zakarian A., Aparicio O.M.. Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS ONE. 2014; 9:e98501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornacchia D., Dileep V., Quivy J.-P., Foti R., Tili F., Santarella-Mellwig R., Antony C., Almouzni G., Gilbert D.M., Buonomo S.B.. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012; 31:3678–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki S., Ishii A., Kanoh Y., Oda M., Nishito Y., Masai H.. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012; 31:3667–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiraga S. -i, Alvino G.M., Chang F., Lian H. -y., Sridhar A., Kubota T., Brewer B.J., Weinreich M., Raghuraman M.K., Donaldson A.D.. Rif1 controls DNA replication by directing protein phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014; 28:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davé A., Cooley C., Garg M., Bianchi A.. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014; 7:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alver R.C., Chadha G.S., Gillespie P.J., Blow J.J.. Reversal of DDK-mediated MCM phosphorylation by Rif1-PP1 regulates replication initiation and replisome stability independently of ATR/Chk1. Cell Rep. 2017; 18:2508–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraga S., Ly T., Garzón J., Hořejší Z., Ohkubo Y., Endo A., Obuse C., Boulton S.J., Lamond A.I., Donaldson A.D.. Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 2017; 18:403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainey M.D., Bennett D., O’Dea R., Zanchetta M.E., Voisin M., Seoighe C., Santocanale C.. ATR restrains DNA synthesis and mitotic catastrophe in response to CDC7 inhibition. Cell Rep. 2020; 32:108096. [DOI] [PubMed] [Google Scholar]
- 20.Foti R., Gnan S., Cornacchia D., Dileep V., Bulut-Karslioglu A., Diehl S., Buness A., Klein F.A., Huber W., Johnstone E.et al.. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol. Cell. 2016; 61:260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marheineke K., Hyrien O.. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 2004; 279:28071–28081. [DOI] [PubMed] [Google Scholar]
- 22.Shechter D., Costanzo V., Gautier J.. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004; 6:648–655. [DOI] [PubMed] [Google Scholar]
- 23.Guo C., Kumagai A., Schlacher K., Shevchenko A., Shevchenko A., Dunphy W.G.. Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol. Cell. 2015; 57:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platel M., Goldar A., Wiggins J.M., Barbosa P., Libeau P., Priam P., Narassimprakash H., Grodzenski X., Marheineke K.. Tight Chk1 levels control replication cluster activation in Xenopus. PLOS ONE. 2015; 10:e0129090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya-Mendoza A., Petermann E., Gillespie D.A.F., Caldecott K.W., Jackson D.A.. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007; 26:2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michelena J., Gatti M., Teloni F., Imhof R., Altmeyer M.. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J. Cell Biol. 2019; 218:2865–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blow J.J., Gillespie P.J., Francis D., Jackson D.A.. Replication origins in Xenopus egg extract Are 5-15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol. 2001; 152:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marheineke K., Hyrien O.. Aphidicolin triggers a block to replication origin firing in Xenopus egg extracts. J. Biol. Chem. 2001; 276:17092–17100. [DOI] [PubMed] [Google Scholar]
- 29.Ciardo D., Haccard O., Narassimprakash H., Arbona J.-M., Hyrien O., Audit B., Marheineke K., Goldar A.. Organization of DNA replication origin firing in Xenopus egg extracts: the role of intra-S checkpoint. Genes. 2021; 12:1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhind N., Yang S.C.-H, Bechhoefer J.. Reconciling stochastic origin firing with defined replication timing. Chromosome Res. 2010; 18:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trenz K., Errico A., Costanzo V.. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008; 27:876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yekezare M., Gómez-González B., Diffley J.F.X.. Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally. J. Cell Sci. 2013; 126:1297–1306. [DOI] [PubMed] [Google Scholar]
- 33.Ciardo D., Goldar A., Marheineke K.. On the interplay of the DNA replication program and the intra-S phase checkpoint pathway. Genes. 2019; 10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elia A.E., Cantley L.C., Yaffe M.B.. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003; 299:1228–1231. [DOI] [PubMed] [Google Scholar]
- 35.Yoo H.Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G.. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004; 117:575–588. [DOI] [PubMed] [Google Scholar]
- 36.Moudry P., Watanabe K., Wolanin K.M., Bartkova J., Wassing I.E., Watanabe S., Strauss R., Troelsgaard Pedersen R., Oestergaard V.H., Lisby M.et al.. TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity. J. Cell Biol. 2016; 212:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsvetkov L., Stern D.F.. Interaction of chromatin-associated Plk1 and Mcm7. J. Biol. Chem. 2005; 280:11943–11947. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z.-Q., Liu X.. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song B., Liu X.S., Davis K., Liu X.. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol. Cell. Biol. 2011; 31:4844–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciardo D., Haccard O., Narassimprakash H., Chiodelli V., Goldar A., Marheineke K.. Polo-like kinase 1 (Plk1) is a positive regulator of DNA replication in the Xenopus in vitro system. Cell Cycle. 2020; 19:1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S., Yoo H.Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G.. Role for Rif1 in the checkpoint response to damaged DNA in Xenopus egg extracts. Cell Cycle. 2012; 11:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blow J.J., Laskey R.A.. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986; 47:577–587. [DOI] [PubMed] [Google Scholar]
- 43.Marheineke K., Goldar A., Krude T., Hyrien O.. Use of DNA combing to study DNA replication in Xenopus and human cell-free systems. Methods Mol. Biol. Clifton NJ. 2009; 521:575–603. [DOI] [PubMed] [Google Scholar]
- 44.Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J.D., Ellenberger T.E., Schärer O.D., Walter J.C.. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008; 134:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Räschle M., Smeenk G., Hansen R.K., Temu T., Oka Y., Hein M.Y., Nagaraj N., Long D.T., Walter J.C., Hofmann K.et al.. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015; 348:1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karimi K., Fortriede J.D., Lotay V.S., Burns K.A., Wang D.Z., Fisher M.E., Pells T.J., James-Zorn C., Wang Y., Ponferrada V.G.et al.. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018; 46:D861–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 48.The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 50.Edwards M.C.MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in xenopus egg extracts. J. Biol. Chem. 2002; 277:33049–33057. [DOI] [PubMed] [Google Scholar]
- 51.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R.C., Edmondson R.D., Calzada A., Labib K.. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 2009; 28:2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruck I., Kaplan D.L.. Origin single-stranded DNA releases Sld3 protein from the Mcm2–7 complex, allowing the GINS tetramer to bind the Mcm2–7 complex. J. Biol. Chem. 2011; 286:18602–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka T., Umemori T., Endo S., Muramatsu S., Kanemaki M., Kamimura Y., Obuse C., Araki H.. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast: Sld3-Sld7 complex in DNA replication. EMBO J. 2011; 30:2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamimura Y., Tak Y.S., Sugino A., Araki H.. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001; 20:2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreira P., Höfer V., Kronshage N., Marko A., Reusswig K.-U., Tetik B., Dießel C., Köhler K., Tschernoster N., Altmüller J.et al.. MTBP phosphorylation controls DNA replication origin firing. Sci. Rep. 2021; 11:4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattarocci S., Shyian M., Lemmens L., Damay P., Altintas D.M., Shi T., Bartholomew C.R., Thomä N.H., Hardy C.F.J., Shore D.. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep. 2014; 7:62–69. [DOI] [PubMed] [Google Scholar]
- 57.Poh W.T., Chadha G.S., Gillespie P.J., Kaldis P., Blow J.J.. Xenopus Cdc7 executes its essential function early in S phase and is counteracted by checkpoint-regulated protein phosphatase 1. Open Biol. 2014; 4:130138–130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowery D.M., Clauser K.R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S.-E., Gammeltoft S., Carr S.A., Yaffe M.B.. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007; 26:2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christov C.P., Gardiner T.J., Szüts D., Krude T.. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol. Cell. Biol. 2006; 26:6993–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Falbo L., Raspelli E., Romeo F., Fiorani S., Pezzimenti F., Casagrande F., Costa I., Parazzoli D., Costanzo V.. SSRP1-mediated histone H1 eviction promotes replication origin assembly and accelerated development. Nat. Commun. 2020; 11:1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., Gerber S.A.. Quantitative phosphoproteomics identifies substrates and functional modules of aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011; 4:rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peshkin L., Wühr M., Pearl E., Haas W., Freeman R.M., Gerhart J.C., Klein A.M., Horb M., Gygi S.P., Kirschner M.W.. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell. 2015; 35:383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sukackaite R., Cornacchia D., Jensen M.R., Mas P.J., Blackledge M., Enervald E., Duan G., Auchynnikava T., Köhn M., Hart D.J.et al.. Mouse Rif1 is a regulatory subunit of protein phosphatase 1 (PP1). Sci. Rep. 2017; 7:2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti C., Seiler J., Pommier Y.. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007; 6:2760–2767. [DOI] [PubMed] [Google Scholar]
- 65.Guilbaud G., Rappailles A., Baker A., Chen C.-L., Arneodo A., Goldar A., d’Aubenton-Carafa Y., Thermes C., Audit B., Hyrien O.. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput. Biol. 2011; 7:e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Löb D., Lengert N., Chagin V.O., Reinhart M., Casas-Delucchi C.S., Cardoso M.C., Drossel B.. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun. 2016; 7:11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Platel M., Narassimprakash H., Ciardo D., Haccard O., Marheineke K.. Genome wide decrease of DNA replication eye density at the midblastula transition of Xenopus laevis. Cell Cycle. 2019; 18:1458–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuermer A., Hoehn K., Faul T., Auth T., Brand N., Kneissl M., Pütter V., Grummt F.. Mouse pre-replicative complex proteins colocalise and interact with the centrosome. Eur. J. Cell Biol. 2007; 86:37–50. [DOI] [PubMed] [Google Scholar]
- 69.Blasius M., Forment J.V., Thakkar N., Wagner S.A., Choudhary C., Jackson S.P.. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011; 12:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moiseeva T.N., Yin Y., Calderon M.J., Qian C., Schamus-Haynes S., Sugitani N., Osmanbeyoglu H.U., Rothenberg E., Watkins S.C., Bakkenist C.J.. An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:13374–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nasa I., Rusin S.F., Kettenbach A.N., Moorhead G.B.. Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal. 2018; 11:eaai8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gnan S., Flyamer I.M., Klein K.N., Castelli E., Rapp A., Maiser A., Chen N., Weber P., Enervald E., Cardoso M.C.et al.. Nuclear organisation and replication timing are coupled through RIF1–PP1 interaction. Nat. Commun. 2021; 12:2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu D., Muniandy P., Leo E., Yin J., Thangavel S., Shen X., Ii M., Agama K., Guo R., Fox D.et al.. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010; 29:3140–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sukackaite R., Jensen M.R., Mas P.J., Blackledge M., Buonomo S.B., Hart D.J.. Structural and biophysical characterization of murine Rif1 C terminus reveals high specificity for DNA cruciform structures. J. Biol. Chem. 2014; 289:13903–13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seller C.A., O’Farrell P.H. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLOS Biol. 2018; 16:e2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanemaki M., Labib K.. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006; 25:1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumagai A., Dunphy W.G.. Binding of the Treslin-MTBP complex to specific regions of the human genome promotes the initiation of DNA replication. Cell Rep. 2020; 32:108178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwab R.A., Blackford A.N., Niedzwiedz W.. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010; 29:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strebhardt K.Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat. Rev. Drug Discov. 2010; 9:643–660. [DOI] [PubMed] [Google Scholar]
- 80.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data sets have been deposited to the ProteomeXchange Consortium via the PRIDE (80) https://www.ebi.ac.uk/pride: PXD022440 (Identification of Plk1 interacting proteins by XChIP-MS); PXD022572 (Chromass/ Protein recruitment on chromatin in the presence or absence of Plk1); PXD022410 (phosphorylation of Rif1 by Plk1).