Abstract
The molecular mechanisms of aging are unsolved fundamental biological questions. Caenorhabditis elegans is an ideal model organism for investigating aging. PUF-8, a PUF (Pumilio and FBF) protein in C. elegans, is crucial for germline development through binding with the 3′ untranslated regions (3′ UTR) in the target mRNAs. Recently, PUF-8 was reported to alter mitochondrial dynamics and mitophagy by regulating MFF-1, a mitochondrial fission factor, and subsequently regulated longevity. Here, we determined the crystal structure of the PUF domain of PUF-8 with an RNA substrate. Mutagenesis experiments were performed to alter PUF-8 recognition of its target mRNAs. Those mutations reduced the fertility and extended the lifespan of C. elegans. Deep sequencing of total mRNAs from wild-type and puf-8 mutant worms as well as in vivo RNA Crosslinking and Immunoprecipitation (CLIP) experiments identified six PUF-8 regulated genes, which contain at least one PUF-binding element (PBE) at the 3′ UTR. One of the six genes, pqm-1, is crucial for lipid storage and aging process. Knockdown of pqm-1 could revert the lifespan extension of puf-8 mutant animals. We conclude that PUF-8 regulate the lifespan of C. elegans may not only via MFF but also via modulating pqm-1-related pathways.
INTRODUCTION
The molecular and cellular mechanisms of aging and lifespan are a complex biological process which involve numerous factors and regulators. Due to the short and reproducible lifespan (∼18 days at 20°C), Caenorhabditis elegans is an ideal model organism to investigate the aging process. The entire genome of C. elegans has been sequenced, and most of the signaling pathways are evolutionarily conserved. The aging process is widely studied in C. elegans. One well-characterized pathway is the insulin/insulin-like growth factor 1 (IGF-1)-like signaling (IIS) pathway (1,2). IIS pathway comprises many genes, including DAF-2, an insulin/IGF-1 receptor homolog. Signals from insulin/IGF-1 phosphorylate and suppress the activity of DAF-16, a forkhead transcription factor (FOXO) (2,3). The daf-2 mutation leads to the de-phosphorylation and nuclear accumulation of DAF-16, which in turn regulates numerous target genes at transcriptional level and regulate various processes, including lifespan (4). Reduced IIS extends the C. elegans lifespan via a series of up-regulated (class I) and down-regulated genes (class II) simultaneously (4). DAF-16 is responsible for the activation of class I genes through the DAF-16-binding element (DBE) (5). Another transcription factor PQM-1 is responsible for the activation of class II genes through the DAF-16-associated element (DAE) (5). DAF‐16 and PQM‐1 work together in regulating the longevity, although their functions are antagonistic (6). The signaling pathway of DAF‐16 has been well characterized in the past decade, yet the mechanism and regulation of PQM-1 remains to be elucidated.
The PUF (Pumilio and FBF) family are posttranscriptional regulators that modulate various processes throughout the eukaryotic kingdom (7–9). PUF proteins regulate mRNA stability and translation by directly binds to the 3′ UTR of target mRNAs via the PUM homology domain (PUM-HD), also named as PUF domain (7,10). The classical PUM-HD consists of eight tandem PUF repeats (namely REP-1,2…,8 hereafter) (11). Each REP contains approximately 36 amino acids which fold into three α-helixes (12). Generally, each REP recognizes one nucleotide. The specificity of RNA base recognition is determined by three key residues within a five-residue RNA recognition motif (PUF motif) at the second helix of the REP (13,14). Triumvirate residues at positions 1, 2 and 5 of the PUF motif directly interact with one specific RNA nucleotide base. The first and fifth residues bind the RNA nucleotide base via hydrogen bonds or hydrophobic interactions; the second residue forms a stacking interaction with the adjacent nucleotide base, and the residues at positions 3 and 4 are hydrophobic and positioned at the back of the RNA binding surface (15). The ‘code’ for RNA recognition by each PUF repeat has been deduced from the structure of the PUF proteins. The amino acids of PUF repeats can therefore be engineered to develop RNA binding proteins that recognize desired RNA sequences (14).
PUF-8 is one of the PUF proteins which is the homologous of Homo sapiens PUM2 (HsPUM2). PUF-8 contains 535 amino acid residues and can be divided into two parts: an N-terminal low-complexity (LC) region (1–171 aa) and a C-terminal PUF domain (172–535 aa) (16). The LC region is essential for the interaction of PUF-8 with other proteins (17). The PUF domain contains eight PUF repeats, REP-1 to -8. The putative PUF-8 target mRNAs contain PUF-binding elements (PBEs, 5′-UGUANAUA-3′ or 5′-UGUAHAUA-3′; N refers to A/U/C/G, and H refers to A/U/C) at their 3′ UTR (16,18,19). RNA binds to PUF-8 in an antiparallel form, the 5′ terminal of RNA binds to the C-terminal of the PUF domain (13). PUF-8 and HsPUM2 share high similarity in both protein sequences and their substrate PBE sequence (5′-UGUANAUA-3′) (18). The crystal structure of PUF-8 together with PBE-5U has been determined recently (16).
PUF-8 is highly expressed in the gonad and enriched in germline stem cells (GSCs) (20), which is crucial for the development of germline stem and progenitor cells (21,22) by promoting germline stem cell mitosis (23), regulating the sperm-oocyte transition process (24,25), maintaining GSCs from repressing myogenesis (22), promoting the meiotic progression of spermatocytes (26), and regulating the development and morphology of the endoplasmic reticulum (27). Recently, D’Amico et al. (28) found that the mouse RNA binding protein Pumilio2 (PUM2) is induced upon aging and acts as a negative regulator of lifespan. PUM2 inhibits the translation of the mRNA encoding Mitochondrial Fission Factor (MFF), leading to aberrant mitochondrial dynamics, and reduced mitophagy and mitochondrial function in mice. This mechanism is conserved in C. elegans. Knockdown of puf-8 altered the mitochondrial dynamics and mitophagy in old nematodes and negatively regulate the longevity of C. elegans as well.
In C. elegans, lifespan is influenced by the proliferation of germline stem cells. Removing of the germline precursor cells Z2 and Z3 abolished the development of germline and extended the lifespan by 60% (29). These cells, together with downstream transcriptional regulators, influence aging in adults. We speculate that, in addition to inhibiting mff-1 mRNA translation, PUF-8 may function via other pathways to regulate the lifespan of C. elegans.
In this work, we determined the complex structure of PUF-8 with an RNA substrate and revealed non-classical structural features in REP-4 and REP-8. Then, we performed a series of mutagenesis experiments that altered PUF-8 recognition of its target mRNA. We performed RNA-seq experiments of wild-type and PUF-8 mutant worms. The mRNA of six genes (pqm-1, ctl-2, blmp-1, hlh-30, pha-4 and vhp-1) which contain at least one PBE at their 3′ UTR were identified. We confirmed the direct interaction of PUF-8 with these mRNAs in vivo by RNA-Crosslinking and Immunoprecipitation (CLIP) assay. pqm-1 is one of the six target genes. PUF-8 directly binds to the 3′ UTR of pqm-1 mRNA. The depletion of PUF-8 or disruption of its target recognition ability extended the lifespan of C. elegans. The knockdown of pqm-1 by RNAi reverted the lifespan extension effect in puf-8 mutant animals. Therefore, we conclude that PUF-8 not only regulates the development of germline stem and progenitor cells, but also controls the lifespan through multiple pathways, such as regulating pqm-1 and mff-1.
MATERIALS AND METHODS
Plasmid construction
DNA sequences of C. elegans PUF-8172–535 and PUF-8172–522 were amplified from the optimized puf-8 sequence in pUC57::PUF-81–535 (General Biology, Anhui, China) and cloned into a modified pET28a vector (Novagen) that contains an N-terminal 6× His tag, namely, p28a vector. Mutations of PUF-8 (Supplementary Table S1) were generated by the MutanBEST Kit (Takara) using p28a-PUF-8172–535 and p28a-PUF-8172–522 as the templates and verified by sanger sequencing (General Biology, Anhui, China).
Protein expression and purification
All the wild-type and mutant proteins of PUF-8172–535 and PUF-8172–522 were expressed in Escherichia coli BL21-GOLD (DE3) cells (Novagen). Cells were grown in Luria-Bertani (LB) medium at 37°C to an OD600 of 0.7–1.0 and induced with 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 22 h at 16°C. The cells were pelleted and resuspended in lysis buffer (25 mM Tris–HCl, pH 8.5, 500 mM NaCl, 20 mM imidazole, and 5% (v/v) glycerol) and lysed by sonication. The supernatant were purified by His tag purification resin (Roche), followed by size exclusion chromatography on a HiLoad Superdex 200 16/60 column (GE Healthcare) in buffer A (25 mM Tris–HCl, pH 8.5, 250 mM NaCl).
Native RNA: PBE-5A (5′-UGUAAAUA-3′), PBE-5U (5′-UGUAUAUA-3′), PBE-5C (5′-UGUACAUA-3′), PBE-5G (5′-UGUAGAUA-3′), PBE-1G (5′-GGUAAAUA-3′), PBE-2U (5′-UUUAUAUA-3′), RNActrl (5′-AUAAAUGU-3′), and the 5′-Fam-tagged RNAs (PBE-5A, PBE-5U, PBE-5C, PBE-5G, PBE-1G and Ctrl) were purchased from Takara Bio Inc. (Dalian, China). RNA oligomers were dissolved in diethyl pyrocarbonate (DEPC)-treated water to a concentration of 2 mM as the stock solution.
Crystallization and data collection
All the crystals of the PUF-8/RNA complexes and their mutants were grown by the hanging-drop vapor diffusion method at 10°C. The PUF-8 proteins were mixed with RNA at a molar ratio of 1:1 to a final concentration of 10 mg/ml protein. All of the protein/RNA complexes were crystallized in 1 M LiCl, 0.1 M citric acid, pH 5.0 and 15% PEG 6000.
The X-ray diffraction data were collected at beamline BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). The X-ray diffraction data are shown in Supplementary Table S2.
Structure determination and refinement
The diffraction data sets were indexed and scaled with HKL2000 (30). The initial model of the PUF-8/PBE-5U complex was determined by Phaser MR using the crystal structure model of human PUM2 (PDB ID: 3Q0Q, 43% sequence identity with PUF-8172–522). The structure of PUF-8/PBE-5U was modified and refined by a combination of the CCP4i suite (31,32), PHENIX (33,34) and Coot (35). The other PUF-8/RNA complexes were determined by molecular replacement using the crystal structure of PUF-8/PBE-5U as the initial model and modified and refined using the same method used for the PUF-8/PBE-5U complex structure. Refinement statistics are shown in Supplementary Table S1. Structural figures were generated using PyMOL (www.pymol.org).
Florescence polarization assay
The florescence polarization (FP) assays were performed by a SpectraMax M5 Microplate Reader System (Molecular Devices). The RNA fragments were synthesized with 5′-FAM (carboxy-fluorescein) by Takara Bio Inc. The RNA was dissolved in buffer A (25 mM Tris–HCl, pH 8.5, 250 mM NaCl). The reaction sample was 200 μl in total and was placed in a black 96-well plate. Each of the samples consisted of 40 nM RNA and logarithmically increasing concentrations of proteins from 5 nM to 2.56 μM. Every binding reaction plate was equilibrated for 5 min at 20°C before measurement. The fluorescence polarization P (preferred to use mP) was fit to the equation below:
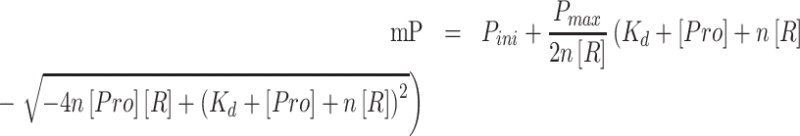 |
where Pini is the initial fluorescence polarization of the sample with RNA, Pmax is the maximum fluorescence polarization, [Pro] is the concentration of protein, [R] is the concentration of RNA, n referred to the binding stoichiometry (protein : RNA binding ratio, n fixed to 1 in our experiment as the 1:1 binding model) and Kd is the dissociation constant. The data and figures were analyzed and visualized by Origin 2019 (OriginLab).
Circular dichroism (CD)
Far-UV CD spectra were detected by J-1700 Circular Dichroism Spectrophotometer (JASCO). All measurements were carried out at 20 °C in the buffer containing 10 mM Tris–HCl, pH 7.4, and 100 mM NaCl, and the spectra were recorded at wavelengths ranging from 190 to 260 nm. All samples were recorded in triplicate and the final curve was the average of three scans.
C. elegans strains
Bristol strain N2 was used as the wild-type strain. Strains were maintained at 20°C. The strains used in this study are shown in Supplementary Table S6. To generate the plasmid containing the homologous arm sequence, we first amplified the endogenous sequences of PUF-8 from the N2 worms that included the 1500 bp upstream and downstream homologous arm sequences. The amplicons were then cloned into the optimized pCFJ151 vector (containing only the Ori and Amp+ sites, namely, pCFJOA hereafter) using the ClonExpress MultiS One Step Cloning Kit (Vazyme C113-02, Nanjing). The point mutations at Rep-4 and Rep-8 on pCFJOA-PUF-8 were constructed with the MutanBEST Kit (TAKARA, R401) to generate the plasmids pCFJOA-PUF-8 (MUT-4A), pCFJOA-PUF-8 (MUT-4N) and pCFJOA-PUF-8 (MUT-8G).
Similarly, we constructed the plasmids pCFJOA-PUF-8::gfp::3xflag and -PUF-8(Mut-8G)::gfp::3xflag. The gfp::3xflag sequence was inserted at the C-terminus of PUF-8 via a linker sequence (5′-GGAGGTGGAGGTGGAGCT-3′, which was inserted between PUF-8 and GFP::3 × FLAG).
CRISPR/Cas9 plasmid cloning
We constructed mutant worms by CRISPR/Cas9 gene editing technology. Small guide RNAs targeting the puf-8 genes were designed by CRISPOR (36). The primers that were used for the sgRNAs were designed and cloned into the Pu6 plasmid to obtain the following plasmids: sg-1∼6 (Supplementary Table S8), which were subsequently used for the construction of the MUT-4A, 4N, 8G (sg-1∼3) and GFP-3xFLAG tag (sg-4∼6) fusion mutant worms.
mRNA isolation, deep sequencing and data analysis
Animals were synchronized and collected at the young adult stage. Worms were sonicated with a Bioruptor-100 in sonication buffer (20 mM Tris–HCl, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40 and 1% RNase inhibitor (TAKARA)). The eluate was incubated with TRIzol reagent (Invitrogen), and RNA was precipitated with isopropanol. Then, the samples were subjected to library construction (Novogene, Beijing, China). The mRNA was then deep sequenced by Novogene.
Raw data were processed through in-house Perl scripts by Novogene. The reference genome was built by Hisat2 v2.0.5, and the number of reads per gene was counted by FeatureCounts v1.5.0-p3. The differential expression analysis was performed by the DESeq2 R package (1.16.1). The resulting P-values were adjusted using Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed (Supplementary data 1).
A gene list related to adult lifespan was downloaded from WormBase (GO: 0008340). The numbers of PBEs in the 5′ UTR, CDS, and 3′ UTR of the genes were counted by using custom Python scripts (Supplementary data 2).
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected. P <0.05 was considered statistically significant (Supplementary data 3 and 4).
Western blot
Primary antibodies for western blots were anti-FLAG monoclonal antibody clone M2 (Sigma, F3165) at 1:1000, and anti-β-actin monoclonal antibody produced in mouse (clone DM1A, Sigma) used at 1:1000. Secondary antibodies used for western blots were ImmunPure Peroxidase Conjugated Goat anti-Mouse IgG (H + L) (PIERCE, 31430) used at 1:2000.
RNA-crosslinking and immunoprecipitation (CLIP)
The RNA-crosslinking and immunoprecipitation were performed as previously described (28,37,38). All the steps were carried out at 4°C. Briefly, approximately 10 000 synchronized young adult worms were collected and rinsed to a new unseeded 10 cm NGM plates (pre-cooled at 4°C). Worms were then irradiated at 254 nm with a power of 400 mJ/cm2 in a UV crosslinker for 2 min for three times (SCIENTZ 03-II, Scientz Biotechnology). The worms were then rinsed from the plates with cold CLIP Buffer A (20 mM Tris (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2, 10% Glycerol, 0.5% NP-40 and 0.1% SDS) and pelleted at 200 RCF for 1 min. The supernatant was discarded and the worms were resuspended with CLIP Buffer B (CLIP Buffer A, supplemented with 0.05% BSA, 0.2 μg/ml yeast tRNA (Thermo Fisher), protease inhibitor (Roche) and Ribolock RNase Inhibitor (Thermo Fisher)). Worms were lysed by sonication (5 s on/15 s off, 20 cycle). Lysate were centrifuged at 14 000 g for 20 min, and pre-cleared by Protein A beads (BBI life sciences). Five percent supernatant were collected as the input. The rest of the supernatant were incubated with Anti-FLAG M2 magnetic beads (sigma) overnight. Beads were washed five times with CLIP buffer B and one time with buffer A. Beads were treated with DNase I for 20 min at 37°C and followed by Proteinase K (50 μg, Protein Biotechnologies) treatment for 30 min at 37°C. The RNAs were isolated from the beads using TRIzol reagent (Invitrogen), and precipitated with isopropanol. The input RNA was dissolved in 50 μl DEPC-treated water, and 12 μl were used for reverse transcription. The CLIP-RNA were dissolved in 12 μl DEPC-treated water, and used for reverse transcription. Four biological repeats were performed and averaged.
Quantitative real-time PCR (qRT-PCR)
RNAs were extracted as above, and reversed transcribed to cDNA by the GoScript Reverse Transcription System (Promega, A5001) following the manufacturer's instructions. The qRT-PCR reaction was performed by MyiQ2 (BIO-RAD) and analyzed by the iQ5 Optical System Software V2.1 (BIO-RAD) and SYBR Green chemistry (Vazyme). All the PCR results were presented relative to the mean of input data, negative control pie-1 mRNA (39) and mean of N2 worms (Supplementary data 6 and 7). Primers for qRT-PCR analyses are shown in Supplementary Table S9. The data and figures were analyzed and visualized by Origin 2019 (OriginLab).
Brood size assay
Brood size assays were performed at 20°C. Adult animals were selected and transferred onto an NGM plate to generate the first progeny. The L3–L4 staged worms were transferred to a new NGM plate and then transferred every day until no eggs were found on the plate. The hatched worms were counted and summarized. Each experiment was repeated at least 20 times.
RNAi
RNAi experiments were conducted as previously described (40).
Lifespan assay
Lifespan experiments were performed at 20°C. Animals were first grown on NGM plates until the worms reached the L4 stage. The animals were transferred to a new NGM plate, and L1 stage progeny were transferred to another plate, which was counted as day 0 in the lifespan assay. At the beginning, animals were transferred every 2 days until they stopped breeding (normally at days 5–7). Then, the animals were transferred to new plates every day, and the number of survivors was counted.
Animals that had an ‘exploded vulva’ phenotype and crawled off the plate were checked and excluded. Over 100 worms were used for each strain. The RNAi-treated lifespan experiments were performed using the same procedure, except the E. coli strain was changed from OP50 to HT115. The lifespan data and statistical analysis were evaluated using OASIS2 survival analysis software (41), the analysis data are shown in Supplementary data 5.
DAPI staining of gonads
Dissection and fixation of gonads of young adult animals for staining with the DNA-binding dye, 4,6-diamidino-2-phenylindole (DAPI) were performed as previously described (23).
Statistical analysis
Experimental data are expressed as mean ± standard deviation (s.d.) or standard error of mean (s.e.). Statistical significance was made by two-sided unpaired Student's t-test on two experimental conditions with P < 0.05 considered statistically significant. The level of statistical significances was denoted as follow: * P < 0.05; ** P < 0.01; *** P < 0.001. No statistical methods were used to predetermine sample size. For data presented without statistics, experiments were repeated multiple times (N ≥ 3) to ensure reproducibility unless otherwise stated.
RESULTS
Complex structure of PUF-8 with PBE-5U RNA
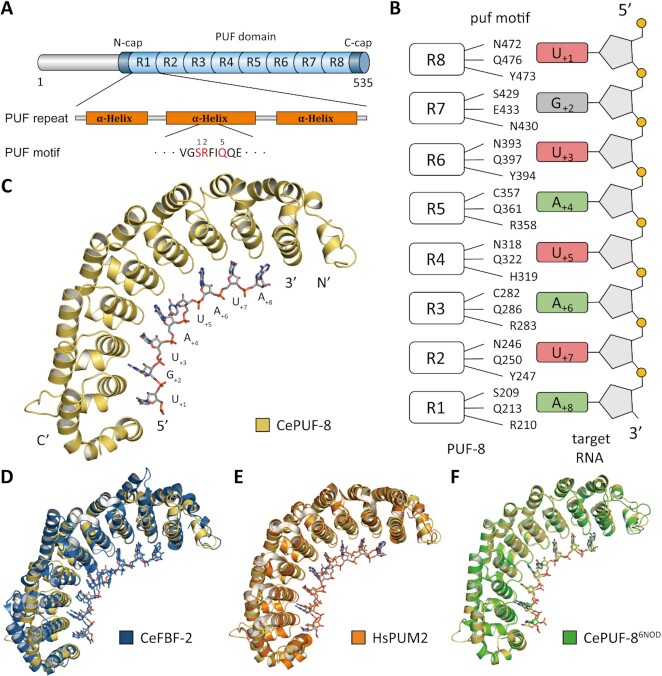
The PUF domain of PUF-8 has eight PUF repeats (REP-1 to -8). Each REP recognizes one RNA nucleotide in the target mRNA sequence, named the pum binding element (PBE) (Figure 1A) (16). Three residues on each REP, which are called PUF motifs, are crucial for direct binding to the RNA nucleotide base (Figure 1B). The PUF motif of each repeat is similar to those of the homology HsPUM1/2 and CeFBF1/2, and was predicted to bind PBE sequence: 5′- UGUANAUA -3′ (9,18,42). To investigate the structural bases and functions of PUF-8 in gene regulation. We determined the crystal structure of the PUF domain (residues 172–522) of PUF-8 in complex with PBE-5U (resolution 2.70 Å) (Figure 1C) (The X-ray crystal structure statistics are presented in Supplementary Table S2). The crystal structure of PUF-8 was previously determined by Hall's laboratory (PDB ID: 6NOD, namely PUF-86NOD hereafter) (Supplementary Figure S1A) (16). Superposition of our PUF-8-PBE-5U complex structure with those of the PUF-86NOD, HsPUM2 (PDB ID: 3K5Q) and CeFBF-2 (PDB ID: 3Q0Q) respectively, revealed that the overall structure of the PUF domain in complex with RNAs is similar (Figure 1D-F). The RMSDs (root-mean-square deviations) for Cα atoms between the PUF domains of PUF-8 with HsPUM2 and PUF-86NOD are about 1.57 and 0.633 Å, respectively (Supplementary Figure S1).
Figure 1.
Crystal structure of PUF-8. (A) The domain structure of PUF-8. The core residues that recognize RNA bases are numbered and marked in red. (B) Sequence alignment of REP-4 of PUF proteins. The recognition motif N318H319Q322 is conserved in paralogs. (C) Overall structure of PUM-HD in PUF-8 with PBE-5U (5′-UGUAUAUA-3′), the protein exhibits a crescent shape, and RNA binds to the inner concave region of the domain with its base group. (D) Alignment of the RNA complex structure of PUM-HD in PUF-8 (yellow) and CeFBF-2 (blue, PDB ID: 3K5Q). (E) Alignment of the RNA complex structure of PUM-HD in PUF-8 (yellow) and HsPUM2 (orange, PDB ID: 3Q0Q). (F) Alignment of the RNA complex structures of PUM-HD in PUF-8 (in this study, yellow) and PUF-8Hall (green, PDB ID: 6NOD).
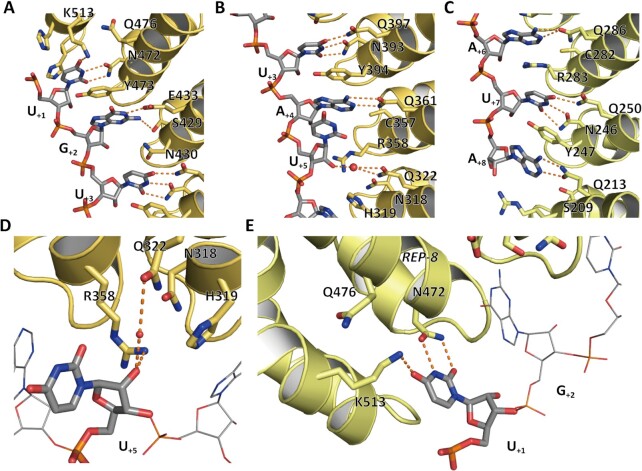
The RNA nucleotide bases are sequentially recognized by the motif of PUM repeat 8 to 1 (called REP-8 to -1 hereafter) and stacked into the inner surface of the protein. G+2, U+3, A+4, A+6, U+7 and A+8 are recognized by the three conserved residues in the motif of the corresponding repeat in a classical mode (18). The PUF motif of REP-2 and REP-6 are N1Y2Q5, which prefers to recognize uracil and binds to the U+7 and U+3, respectively. The side chains of residues Asn1 and Gln5 in each motif bind to a uracil via hydrogen bonds. In REP-1, -3 and -5, three residues in PUF motif, C(S)1R2Q5, interact with A+8, A+6 and A+4, respectively. G+2 is recognized S1N2E5 in REP-7. The hydroxide side chain groups of S429 and E433 in REP-1, -3, and -5 form hydrogen bonds with the guanine (Figure 2A–C). These repeats, except REP-4 and REP-8, were shown to bind to the above RNA in a common mode.
Figure 2.
The special mode of REP-4 and REP-8 in RNA recognition. The interface between PUF-8 and PBE-5U is shown. (A) RNA binds to PUM-HD with an antiparallel form. The U+1, G+2 and U+3 bases are recognized by the three conserved residue in REP-8, REP-7 and REP-6, respectively. (B, C) The U+4, A+6, U+7 and A+8 bases are recognized by similar motifs in REP-5, REP-3, REP-2 and REP-1, respectively. (D) The base group of U+5 is positioned in the solvent and atypically binds to Q322 in REP-4 with its 2′-OH group on ribose via a water molecule. (E) K513 at the C-terminal cap recognizes the base group of U+1 instead of Q476, which is the key residue in the recognition motif of REP-8.
The RNA binding modes of REP-4 and REP-8 are unique
According to the structure of the PUF motif and its target nucleotide base, the N1H2Q5 of the PUF motif in REP-4 is predicted to bind to a uracil (13,14). However, our complex crystal structure shows that the base group of U+5 is positioned in the solvent instead of forming hydrogen bonds with N318 and Q322. The Q322 in REP-4 interacts with the 2′-OH group on the ribose of U+5 via a water molecule (Figure 2D). This conformation is consistent with previously reported PUF-8 structure (Supplementary Figure S1E) (16).
The U+1 is predicted to be recognized by the three conserved residues (N472, Y473 and Q476) in REP-8. However, our structure showed that the side chain of Q476 in REP-8 faced away from the recognition pocket (Figure 2E). The base group of U+1 forms a hydrogen bond with the side chain of K513, which is located in the C-terminal cap of the PUF domain. The K513 of PUF-86NOD shows similar conformation, but the side chain of Q476 in PUF-86NOD is positioned to the binding surface and interacts both with K513 and U+1 (Supplementary Figure S1F).
Mutations in REP-4 and REP-8 changed the RNA recognition affinity but did not significantly change the sequence recognition preferences
To test whether the mutations in REP-4 and REP-8 alter their substrate RNA sequences preferences. We constructed five mutant PUF-8172–535 proteins: MUT-4A (N318C/H319R), MUT-4C (N318C/H319Y/Q322R), MUT-4G (N318S/H319N/Q322E), MUT-4N (N318A/H319A/Q322A) and MUT-8G (N472S/Y473N/Q476E) (Figure 3A and Supplementary Table S1). Consistent with the target recognition pattern of the PUF motif, MUT-4N was not able to recognize a specific base. MUT-4A, -4C and -4G changed its putative target RNA from U+5 to A+5, C+5 and G+5 respectively. MUT-8G changed its putative target RNA from U+1 to G+1 (14,43). To examine whether the mutations alter the secondary structure of PUF-8, we performed the Circular Dichroism (CD) spectra assay. Every mutant protein has similar secondary structure to that of wild-type PUF-8 (Supplementary Figure S3A).
Figure 3.
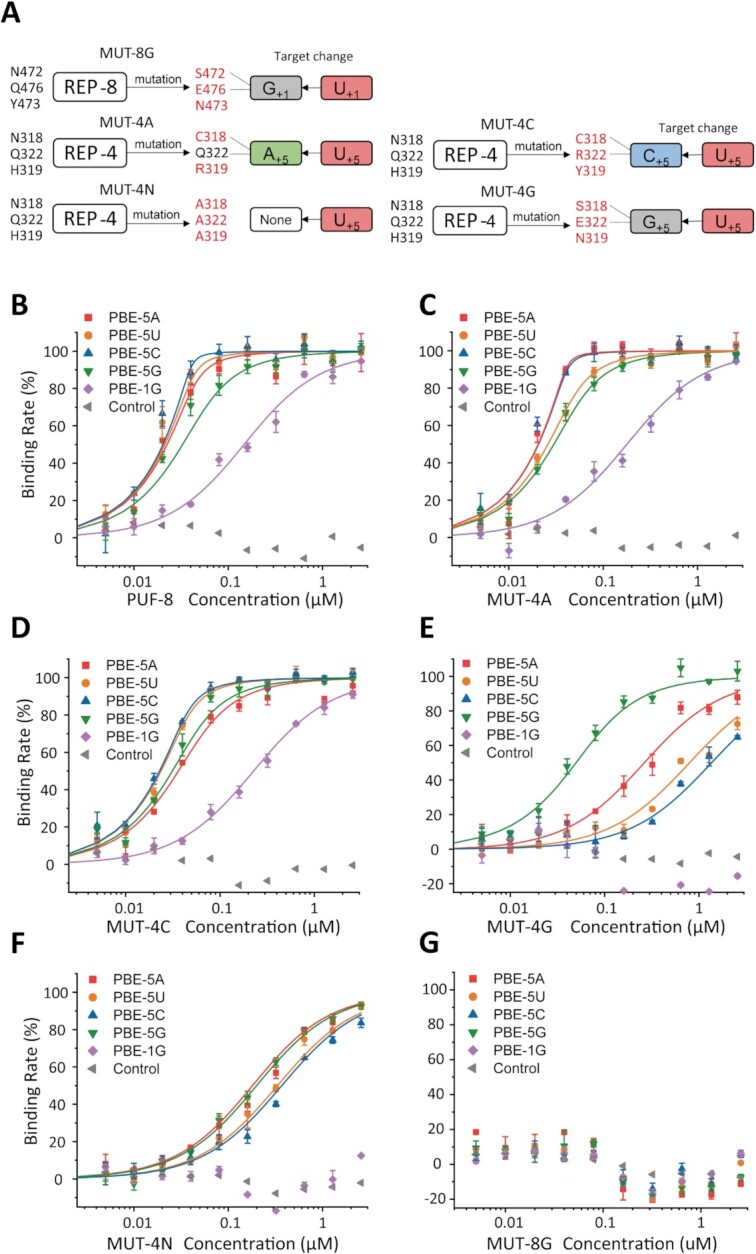
Binding affinity of PUF-8 mutants with distinct PBE sequence substrates. Fluorescence polarization assay of the indicated PUF-8 mutants with different PBE sequence substrates. Six RNAs were used, and the control RNA (gray, left triangle) sequence was the reverse sequence of the normal PBE (5′-AUACAUGU-3′). (A) Schematic of the mutations at REP-4 and REP-8. The mutated residues in REP-4 and REP-8 are marked in red, and the predicted binding nucleotides are shown on the right. (B) Wild-type PUF-8 reveals a high affinity in binding with PBE-5N and weak binding affinity with PBE-1G and control RNA. (C) MUT-4A shows a similar binding ability to that of PUF-8. (D) MUT-4C shows no preference to bind PBE-5N RNAs, but the binding affinity slightly decreased in all of the target RNAs. (E) MUT-4G prefers to bind PBE-5G rather than the other PBE-RNAs. (F) The binding affinity of MUT-4N with PBE-RNAs are significantly decreased. (G) MUT-8G has no detectable affinity for all RNAs, including PBE-1G (5′-GGUACAUA-3′), which is the putative target of MUT-8G.
Then, Six PBE-RNA were generated: PBE-5A (which means the fifth base group is an adenine) (5′-UGUAAAUA-3′), PBE-5U (5′-UGUAUAUA-3′), PBE-5C (5′-UGUACAUA-3′), PBE-5G (5′-UGUAGAUA-3′), PBE-1G (5′-GGUACAUA-3′) and the control sequence (5′-AUACAUGU-3′). PBE-5A, -5U, -5C and -5G are putative targets of wild-type PUF-8 and MUT-4A, -4C and -4G, respectively. PBE-1G is the putative target of MUT-8G.
Fluorescence polarization (FP) assays were performed to explore the binding affinity of PUF-8 mutants with various PBE sequences. The binding curves of each PUF-8 mutant protein with different PBE-RNAs are shown in Figure 3, the Kd value are listed in Table 1. The base group of + 5 nucleotide rotates and faces to the solvent in the crystal structure (Figure 2D), indicating that PUF-8 protein may bind PBE-5N (5′-U+1G+2U+3A+4N+5A+6U+7A+8–3′). Wild-type PUF-8 binds to the PBE-5A, -5U, -5C with a higher binding affinity (Kd ≈ 1 nM) compared to PBE-5G (Kd ≈ 12 nM) (Figure 3B and Table 1).
Table 1.
Fluorescence polarization assay of PUF-8 REP4 Mutants with RNAs
| Protein | RNA | RNA sequence | Kd (nM)* | Relative Krel** |
|---|---|---|---|---|
| wild type | PBE-5A | UGUAAAUA | 2.49 ± 1.50 | 1 |
| PBE-5U | UGUAUAUA | 1.27 ± 1.16 | 1 | |
| PBE-5C | UGUACAUA | 0.84 ± 1.08 | 1 | |
| PBE-5G | UGUAGAUA | 12.76 ± 5.63 | 1 | |
| PBE-1G | GGUACAUA | 128.31 ± 25.05 | 1 | |
| Control RNA | AUACAUGU | ND*** | – | |
| MUT-4A | PBE-5A | UGUAAAUA | 0.44 ± 0.39 | 0.18 |
| PBE-5U | UGUAUAUA | 5.85 ± 1.64 | 4.61 | |
| PBE-5C | UGUACAUA | 0.64 ± 0.20 | 0.76 | |
| PBE-5G | UGUAGAUA | 9.99 ± 3.11 | 0.78 | |
| PBE-1G | GGUACAUA | 158.51 ± 34.06 | 1.23 | |
| Control RNA | AUACAUGU | ND | – | |
| MUT-4C | PBE-5A | UGUAAAUA | 16.26 ± 4.64 | 6.53 |
| PBE-5U | UGUAUAUA | 3.72 ± 0.94 | 2.93 | |
| PBE-5C | UGUACAUA | 2.91 ± 0.36 | 3.46 | |
| PBE-5G | UGUAGAUA | 11.32 ± 3.43 | 0.89 | |
| PBE-1G | GGUACAUA | 207.40 ± 22.99 | 1.62 | |
| Control RNA | AUACAUGU | ND | – | |
| MUT-4G | PBE-5A | UGUAAAUA | 238.69 ± 48.03 | 95.86 |
| PBE-5U | UGUAUAUA | 774.60 ± 276.44 | 609.92 | |
| PBE-5C | UGUACAUA | 1349.1 ± 506.58 | 1606.07 | |
| PBE-5G | UGUAGAUA | 29.77 ± 9.14 | 2.33 | |
| PBE-1G | GGUACAUA | ND | – | |
| Control RNA | AUACAUGU | ND | – | |
| MUT-4N | PBE-5A | UGUAAAUA | 160.90 ± 21.98 | 64.62 |
| PBE-5U | UGUAUAUA | 312.53 ± 58.21 | 246.09 | |
| PBE-5C | UGUACAUA | 349.40 ± 83.24 | 415.95 | |
| PBE-5G | UGUAGAUA | 179.85 ± 22.81 | 14.09 | |
| PBE-1G | GGUACAUA | ND | – | |
| Control RNA | AUACAUGU | ND | – | |
| MUT-8G | PBE-5A | UGUAAAUA | ND | – |
| PBE-5U | UGUAUAUA | ND | – | |
| PBE-5C | UGUACAUA | ND | – | |
| PBE-5G | UGUAGAUA | ND | – | |
| PBE-1G | GGUACAUA | ND | – | |
| Control RNA | AUACAUGU | ND | – |
*Mean ± s.d., n = 3.
**Krel are calculated for each protein and normalized to that of wild type protein.
*** ND represents not detected.
MUT-4A shows similar binding affinity with PBE-5C, -5G and -1G as those of wild-type PUF-8. Yet MUT-4A has a stronger binding affinity with PBE-5A (Kd = 0.44 nM, Krel = 0.18) but lower binding affinity with PBE-5U (Kd = 5.85 nM, Krel = 4.61) (Figure 3C and Table 1). MUT-4C has reduced PBE binding affinity compared to that of the wild-type protein (Figure 3D). MUT-4G strongly reduced the PBE-RNA binding affinity (Krel > 95), except for its putative target PBE-5G (Kd = 29.77 nM, Krel = 2.33) (Figure 3E). MUT-4N lost its RNA binding sequence preference and exhibited a similar Kd and Krel to all of the PBE-RNA sequences (Figure 3F).
According to the target recognition rule of the PUF proteins, MUT-8G was predicted to bind PBE-1G with a higher affinity than other PBE-RNAs. However, FP experiment failed to detect the binding of MUT-8G to any of the PBE-RNAs (Figure 3G). Suggesting that the N472S/Y473N/Q476E mutation may have strongly reduced the RNA binding ability of the PUF domain.
Together, we concluded that mutation of the conserved residues at REP-4 altered the RNA recognition module, and changed the preferred target RNA sequences. However, mutation of REP-8 strongly reduced the RNA binding ability to all PBE-RNAs.
MUT-4A changed the RNA recognition mode of REP-4
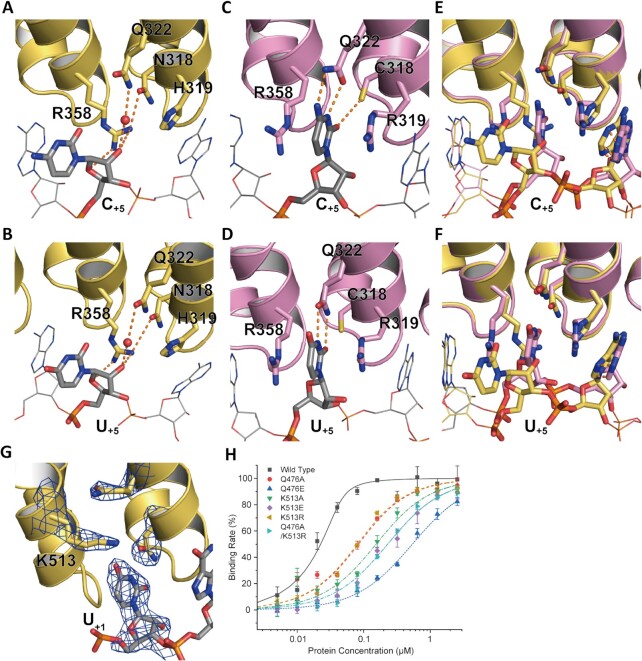
Although REP-4 exhibited a special RNA binding mode (Figure 2), the FP assay of MUT-4A revealed similar binding affinity to PBE-RNAs, comparing to those of wild-type protein (Figure 3 and Table 1). To explore how the residues in the PUF motif of REP-4 contribute to the RNA recognition, we solved the complex structures of PUF-8 and MUT-4A with PBE-5A, -5U, -5C and -5G, respectively (Crystal statistics: Supplementary Table S2).
There is a conformational change of the fifth RNA nucleotide base group at the portion of REP-4 between in the MUT-4A and wild-type structures. In the wild-type PUF-8, the 5th pyrimidine base faces outward toward the concave surface of the protein, and R358 (R2 in the motif of REP-5) is inserted into this surface. R358 forms two hydrogen bonds with the ribose 2′-OH group and 4′-OH group of the 5th nucleotide and forms another hydrogen bond with N318 on the other side (Figure 4A, B and Supplementary Figure S2). In addition, Q322 forms a hydrogen bond with the 2′-OH group of U+5 via a water molecule. In the MUT-4A/PBE complex, the C1R2Q5 motif of REP-4 formed a traditional pocket. The pyrimidine base of the 5th nucleotide is precisely stacked into this pocket (Figure 4C, D). The differences in binding with PBE-5C/PBE-5U between wild-type REP-4 and MUT-4A are also observed (Figure 4E, F). R319 in MUT-4A provides a narrow space, which restrains the position of A+4. For purine bases, the guanine groups and adenine groups exhibited similar binding modes with the wild-type REP-4 and MUT-4A proteins, but an orientation change occurred at the base groups between the wild-type and MUT-4A proteins (Supplementary Figure S2). The complex conformation of REP-4 with PBE-5A RNA is similar to that of HsPUM2 (PDB: 3Q0Q) (Supplementary Figure S2I, J).
Figure 4.
Interaction of the RNA base with PUF-8 and MUT-4A. The inter position of REP-4 in PUF-8 (yellow) and MUT-4A (pink, N318C/H319R double mutant) shows a different binding mode with C+5 or U+5. The base groups of C+5 and U+5 are away from the recognition pocket, as the base-omission formation, where a water molecule occupies and links U+5 and Q322 with hydrogen bonds (A and B). However, in the crystal structure of MUT-4A with RNA PBE-5U and PBE-5C, the base groups of C+5 (C) and U+5 (D) are stacked into the binding pocket. (E and F) View of the alignment between PUF-8 and MUT-4A with the C+5 and U+5 bases. (G) The 2Fo – Fc electron density map contoured at the 1.2σ level for the U+1 RNA and key residues is shown as a blue grid line. K513 recognizes the base group of U+1. (H) FP affinity results of K513 and Q476 mutant PUF-8 reveal that K513 contributes to the binding of U+1 with REP-8.
Therefore, MUT-4A has a similar binding affinity with PBE RNAs compared to that of wild type protein, but exhibits a different binding mode to the fifth RNA base.
Residues Q476 and K513 recognize U+1 synergistically with residues in REP-8
In the PUF-8/PBE complex structure, K513 interacts with U+1 (Figures 2E and 4G), suggesting that K513 may also influence RNA recognition of REP-8. We found that K557 (homolog of K513 in PUF-8) in CeFBF-2 has the same conformation as K513 in PUF-8 (Supplementary Figure S1G). However, residue K1163 in HsPUM2 is located at the top of Q1126 (homolog of Q476 in PUF-8) and keeps Q1126 facing the basic group of U+1 by a hydrogen bond (Supplementary Figures S1H, S2K and S2L). These structural features indicate that K513 may function in RNA recognition.
We mutated K513 and Q476 in PUF-8 and measured the binding affinity to PBE-RNAs by the FP assay (Figure 4H, mutant information shown in Supplementary Table S1). K513A mutant binds to the PBE-5A RNA with an affinity 53.61-fold less than that of the wild-type protein. The binding affinity of Q476A mutant is 25.50-fold less than that of wild-type protein. The Q476A; K513A double mutant binds to PBE-5A with a weaker affinity than that of each single mutant (Table 2). Q476E mutant strongly reduced the RNA binding ability (Krel, 208.55).
Table 2.
Fluorescence polarization assay of K513/Q476 mutation of PUF-8 with RNA
| Protein | RNA | Kd (nM)* | Relative Krel** |
|---|---|---|---|
| Wild type | PBE-5A | 2.49 ± 1.50 | 1 |
| MUT-#9 (Q476A) | PBE-5A | 63.5 ± 11.07 | 25.50 |
| MUT-#10 (Q476E) | PBE-5A | 519.29 ± 63.10 | 208.55 |
| MUT-#11 (K513A) | PBE-5A | 133.48 ± 29.76 | 53.61 |
| MUT-#12 (K513E) | PBE-5A | 208.63 ± 29.07 | 83.79 |
| MUT-#13 (K513R) | PBE-5A | 60.04 ± 13.21 | 24.11 |
| MUT-#14 (Q476A/K513A) | PBE-5A | 209.66 ± 27.4 | 84.20 |
*Mean ± s.d., n = 3.
**Krel are calculated for each protein and normalized to that of wild type protein.
Therefore, we concluded that both K513 and Q476 in REP-8 are essential in recognizing substrate RNAs.
PUF-8 mutants reduced the fertility and increased the lifespan of C. elegans
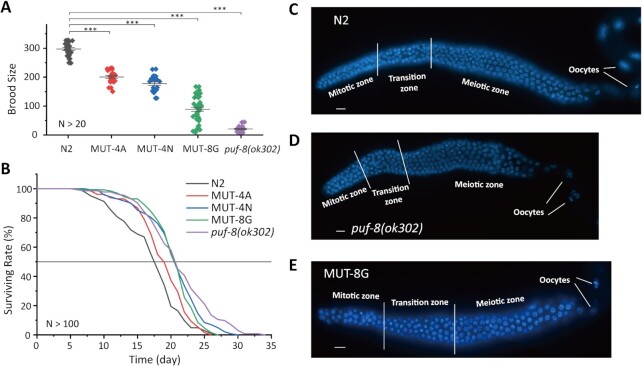
To investigate whether PUF-8 mutants affect the biological functions of PUF-8, we generated several PUF-8 mutant worms, including MUT-4A, MUT-4N and MUT-8G, by oligonucleotide-guided CRISPR/Cas9 gene editing technology (Supplementary Table S3). The puf-8(ok302) allele, which deleted the sequence from the middle of REP-2 to the end of the PUF domain, was considered as a null allele (20,22).
We first examined the brood size and lifespan of these PUF-8 mutant worms. The brood sizes decreased in the mutant worms (Figure 5A and Table 3). In addition, all these PUF-8 mutant animals have an extended lifespan compared to that of wild-type animals. The lifespan of MUT-4N, MUT-8G and puf-8(ok302) animals increased ∼20% compared to that of wild-type worms, and the lifespan of MUT-4A animals increased ∼11% (Figure 5B and Table 3), suggesting that PUF-8 may contribute to lifespan regulation.
Figure 5.
Mutant PUF-8 reduces worm brood size and extends lifespan. (A) Brood size of indicated animals at 20°C. N > 20; mean ± s.e.; ***P < 0.0001. (B) Survival curves of the indicated animals. The gray line refers to the 50% survival rate of worms (N > 100). N2 versus MUT-4A, P = 0.02; N2 versus MUT-4N, P < 0.0001; N2 versus MUT-8G, P < 0.0001; N2 versus puf-8(ok302), P < 0.0001. (C–E) Dissected young adult gonads of the indicated genotype (C, N2; D, puf-8(ok302); E, MUT-8G) stained with DAPI. See more in Supplementary Figure S4. Scale bar: 10 μm.
Table 3.
Fertility and lifespan assay fed with OP50
| Strains | Description | Brood size* | Fold change | Lifespan** (tmean, day) | Fold change |
|---|---|---|---|---|---|
| N2 | Wild type | 296.0 ± 24.51 | 1 | 17.22 ± 0.45 | 1 |
| MUT-4A | N318C/H319R | 198.3 ± 24.96 | 0.66 | 18.98 ± 0.34 | 1.10 |
| MUT-4N | N318A/H319A/Q322A | 175.5 ± 28.84 | 0.59 | 20.55 ± 0.37 | 1.19 |
| MUT-8G | N472S/Y473N/Q476E | 48.57 ± 34.68 | 0.16 | 20.53 ± 0.37 | 1.19 |
| JH1521 | ok302 | 17.8 ± 9.85 | 0.06 | 21.05 ± 0.43 | 1.22 |
*N > 20 animals, mean ± s.d.
**N > 100 animals, mean ± s.d.
We conducted DAPI staining for the gonad of wild-type N2 and mutant animals (Figure 5C–E). The number of germ cells were counted (Figure 5C–E and supplementary S4A–D). The result shows that the number of germ cells of puf-8 mutant worms were similar in compared with that of wild-type N2 worms at young adult stage (cultured at 20°C) (Supplementary Figure S4E).
Identification of PUF-8 target genes
To investigate how PUF-8 regulates the aging process of C. elegans, we isolated total RNAs from young adult wild-type and mutant animals, and performed mRNA-seq analysis.
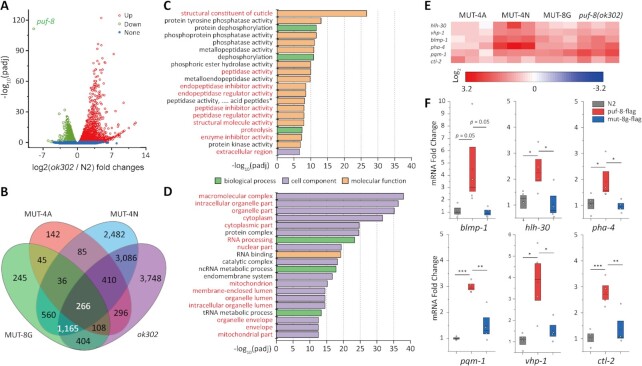
Compared to that of wild-type animals, 4638 genes were up regulated and 4855 genes were down regulated in the puf-8(ok302) null mutant (Figure 6A and Supplementary Table S4). GO enrichment analysis of the up and down regulated genes showed that molecular function (MF) related genes are mainly up regulated (Figure 6C), whereas most down regulated genes are clustered in cell component (CC) (Figure 6D).
Figure 6.
Identification of PUF-8 target genes. (A) Volcano view of the differential mRNA expression in wild-type and puf-8(ok302) animals. (B) Venn diagram of differentially expressed genes in the indicated animals. The white-colored numbers refer to group Gr266, which included genes collectively regulated by all the mutant. Group Gr1165 contains genes that were collectively regulated by all the worms except MUT-4A. (C, D) GO enrichment analysis of the up regulated (C) and down regulated (D) genes for puf-8(ok302) vs N2, top 20 pathways are shown (the overlapped pathways of MUT-4A vs N2 or MUT-8G vs N2 are marked in red, see also in Supplementary Figure S6). (E) Six genes that contain PBE sequence elements in the 3′ UTR are identified and are up regulated in mutant worms. ctl-2 is included in the Gr266 group. Other genes are included in the Gr1165 group. (F) RNA-CLIP assay followed by qRT-PCR reveals that PUF-8 can bind to the lifespan-regulating genes in vivo. mean ± s.e.; *P < 0.05; **P < 0.01; ***P < 0.001.
We then screened for the genes that contain at least one PBE at the 5′ UTR, 3′ UTR or CDS in the up regulated and down regulated genes using 5′-UGUANAUA-3′ as the target PBE sequence. We found that more than 500 genes contained the PBE sequence in both groups of the genes, and nearly 60% of the genes contained one PBE at their 3′ UTR (Supplementary Figure S5A), which is consistent with previous research showing that PUF-8 preferentially binds to the 3′ UTR of mRNAs (22,44).
We compared the mRNA expression levels in MUT-4A, MUT-4N, MUT-8G and puf-8(ok302) mutants. The number of genes that were changed in puf-8(ok302) and MUT-4N were similar (9501 and 8090 genes, respectively), and more than half of the genes (4927) were similarly regulated. MUT-4A and MUT-8G influenced 1388 and 2829 mRNAs, respectively (Supplementary Table S4). GO enrichment analysis showed that MUT-4A, MUT-8G and puf-8(ok302) share similar MF pathways in the up regulated genes and CC-related pathways in the down regulated genes (Supplementary Figure S6). MUT-4N reveals distinct GO enrichment pathways in which the biological process (BP) pathways and CC pathways were more enriched in the up regulated genes (Supplementary Figure S6). Venn diagram analysis showed that 266 genes (namely, Gr266 hereafter) were regulated in all the mutants, and 1165 genes (Gr1165 hereafter) were regulated in all mutants except MUT-4A worms (Figure 6B). We compared the combination of Gr266 and Gr1165 dataset with iCLIP data of C. elegans FBF-1 and FBF-2 (Supplementary Figure S7A), 99 genes and 58 genes were overlapped in FBF-1 versus PUF-8 and FBF-2 versus PUF-8, respectively. The overlapped genes of FBF-1 and PUF-8 are involved in organelle function (terms: organelle lumen and membrane-enclosed lumen), germline development (terms: meiotic cell cycle, P granule and germ plasm), and mRNA metabolism (terms: translation and regulation of translation) (Supplementary Figure S7B). The overlapped genes of FBF-2 and PUF-8 are enriched in mRNA metabolism (term: translation, RNA processing and regulation of translation) (Supplementary Figure S7C). The comparison of PUM2 (iCLIP) (45) vs PUF-8 (Gr266 and Gr1165) showed 93 overlapped genes, which are involved in transcription regulation (Supplementary Figure S7D, E).
To investigate how PUF-8 controls the lifespan of C. elegans, a list of 252 genes was collected from WormBase using the GO term determination of adult lifespan (GO: 0008340). Among these 252 genes, 65 contained putative PBE sequences (5′-UGUMNAWA-3′; M refers to A/C, W refers to A/T, and N refers to A, U, C and G) in the 3′ UTRs of the mRNAs. Six genes are significantly up regulated in PUF-8 mutant worms among these 65 genes (Figure 6E and Supplementary Figure S5B). Five genes, pqm-1, pha-4, blmp-1, vhp-1 and hlh-30, are collectively up regulated in Gr1165, while ctl-2 is up regulated in Gr266. The six up regulated genes are also confirmed by qRT-PCR in the puf-8 mutants (Supplementary Figure S8).
To test whether these genes interact with PUF-8 in vivo, we constructed puf-8::gfp::3xflag and MUT-8G::gfp::3xflag worms. The protein levels of these two Flag tagged protein are similar (Supplementary Figure S9A) and the puf-8::gfp::3xflag and MUT-8G::gfp::3xflag revealed similar expression pattern in the germline (Supplementary Figure S9B). Therefore, we performed the CLIP assay followed by qRT-PCR. pie-1 mRNA was used as a negative control (39). We found that the six mRNAs were all enriched in PUF-8::GFP::3xFLAG but not effectively in the MUT-8G::GFP::3xFLAG mutant worms (Figure 6F and Supplementary Figure S9C). These data suggest that PUF-8 can interact with these mRNAs in vivo.
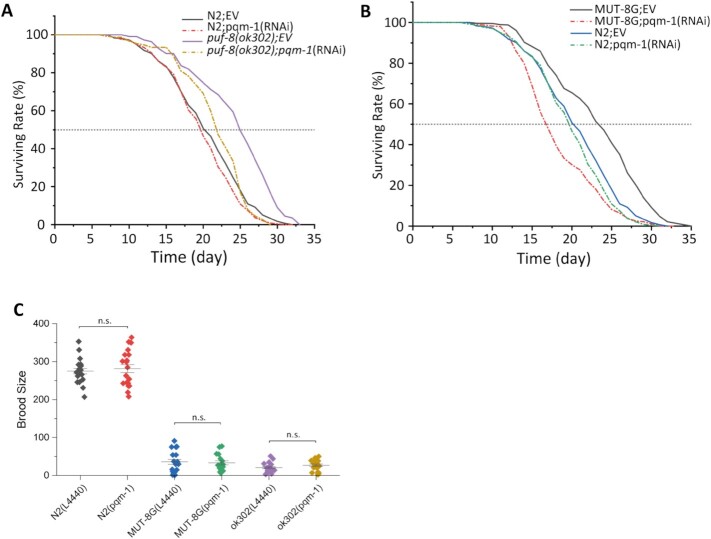
Knockdown of PQM-1 by RNAi suppressed the lifespan extension of puf-8(ok302)
To further confirm that PUF-8 target genes contributing to the lifespan extension in puf-8(ok302) worms, we fed N2 and puf-8(ok302) worms with dsRNA targeting these genes (Supplementary Figure S10A).
RNAi knockdown of pqm-1 significantly reduced the lifespan of puf-8(ok302) worms but not of N2 worms. The average lifespan of puf-8(ok302) was reduced about 10% (24.01 days (EV) to 21.68 days (pqm-1)) (Figure 7A and Supplementary Table S5). Meanwhile, the average lifespan of MUT-8G worms after knock down of pqm-1 was significantly reduced from 23.35 to 18.62 days (around 20% reduction) (Figure 7B and Supplementary Table S5). The brood size of different worms shows no significant difference between EV and the pqm-1 knockdown worms (Figure 7 and Supplementary Table S7).
Figure 7.
PUF-8 regulates lifespan by targeting pqm-1. (A) Knockdown of pqm-1 reduced the lifespan extension of puf-8(ok302) mutant. See also supplementary data 5. N2 (EV) versus N2 (pqm-1), P < 0.05; N2 (EV) versus puf-8(ok302) (EV), P < 0.0001; N2 (EV) vs puf-8(ok302) (pqm-1), P = 0.36; puf-8(ok302) (EV) vs puf-8(ok302) (pqm-1), P < 0.0001. (B) Lifespan curves of MUT-8G and N2 worms fed with HT115 empty vector (EV, L4440) and pqm-1, versus N > 250. MUT-8G (EV) versus N2 (EV), P < 0.0001; MUT-8G (pqm-1) versus N2 (EV), P < 0.0001; MUT-8G (EV) versus MUT-8G (pqm-1), P < 0.0001. (C) The brood size of N2, MUT-8G and puf-8(ok302) worms. mean ± s.e.; n.s., not significant.
Interestingly, the MUT-8G worms reduced fertile rate when fed with HT115 bacterium instead of OP50 (both in empty vector or pqm-1 RNAi vector) (Supplementary Figure S11A). Moreover, MUT-8G revealed high ‘exploded vulva’ phenotype when fed HT115 comparing to fed OP50 (Supplementary Figure S11B, C). This phenotype led to a large amount of abnormal dead MUT-8G worms during day 9 to 12.
The spermatogenesis initiation of puf-8(ok302) was delayed (17). To exclude the influence of development delay on the lifespan extension analysis, we also examined the lifespan of these worms from L4 stage (Supplementary Table S5). We tested the development time of N2, puf-8(ok302) and MUT-8G animal fed with pqm-1 RNAi. The development time delayed in puf-8(ok302) and MUT-8G compared with N2. Knock down of pqm-1 in N2, puf-8(ok302) and MUT-8G worms have no effect on the development time (Supplementary Figure S11D).
Knockdown of ctl-2 partially reverted the lifespan extension of puf-8(ok302) (Supplementary Figure S10C). There were no significant lifespan differences after depletion of pha-4 in either N2 or puf-8(ok302) worms (Supplementary Figure S10D). The depletion of hlh-30, vhp-1 and blmp-1 by RNAi induced animal lethality (Supplementary Figure S10B).
Together, these data suggested that PUF-8 may control lifespan by regulating pqm-1.
DISCUSSION
The complex structure of PUF-8 with an RNA substrate revealed unique structural features in REP-4 and REP-8
The structure of PUF proteins and the complex structure of PUF proteins with target RNAs have been extensively studied over the past decade (11,42,46,47). The RNA binding module, the PUM repeat, recognizes bases through its specific residue motif (46,48,49). Therefore, PUF proteins can be designed to recognize a given RNA sequence and used as a tool to manipulate the stability and translation efficiency of target RNAs (50–52).
In this study, we determined the crystal structures of PUF-8 in complex with a number of RNA substrates and found unique structural features in REP-4 and REP-8. We generated PUF-8 mutants in REP-4 and REP-8 to alter the affinity and sequence specificity of the target RNAs. These mutants reduced the fertility and increased the lifespan of C. elegans.
The MUT-8G mutation (N472S/Y473N/Q476E) significantly reduced its RNA binding ability rather than altered the sequence specificity of target RNAs. In addition, K513 of PUF-8 in the cap region, together with REP-8, is essential for U+1 base recognition. It is speculated that other residues in the cap region may also facilitate the recognition between REP-8 and RNA bases.
REP-4 plays important role in the recognition of RNA bases. From the FP assay, although the wild-type REP-4 (N1H2Q5), MUT-4A (C1R2Q5) and MUT-4C (C1Y2R5) proteins could bind all four types of PBE-RNAs, the conformation of the + 5 base was different in MUT-4A. MUT-4G (S1N2E5) preferentially binds to PBE-5G but not to PBE-5A, -5C, or -5U. The result suggested that the motif of S1N2E5 has a stronger selectivity for guanine than for other bases. Consistently, Bhat, V.D. et al. found that a REP-4 mutation in FBF-2 could change the length of the target RNA and regulates distinct biological processes (16).
The function of the RNA binding ability of PUF-8
PUF-8 protein plays fundamental roles in the germline development (20–22). Depletion of puf-8 in ok302 strain revealed lower brood size, deficiency in spermatogenesis, and delayed larva development (17). The puf-8(ok302) allele contained a 1249 bp deletion that removed all the 3′UTR and nearly all the PUM-HD. The germline development was regulated by numerous RNA binding factors, for example, puf-8, mex-3, glp-1, fbf-1/2 and tcer-1(17,21,23,25).
The function of PUF domain remains to be elucidated. We found that PUF domain mutation can influence the brood size and lifespan of animals as well as puf-8(ok302) deletion allele, yet the number of germ cells are similar. Previous report revealed that the brood size reduction of puf-8 mutant was temperature-dependent and was due to defective sperm (20). Interestingly, only MUT-8G revealed a higher ‘exploded vulva’ phenotype when fed with HT115 comparing to feed with OP50 (Supplementary Figure S10), yet the reason is unclear.
The mRNA-seq data showed that plenty of genes have been changed in different PUF-8 mutant worms, suggesting that the RNA binding ability of PUF-8 is crucial for the function of PUF-8. In addition, there are also a number of genes that are differently regulated between MUT-4N, MUT-8G and puf-8(ok302). We speculated that the differences in the miss-regulated genes may due to the protein expression level and the function of N-terminal domain of PUF-8, which was reported in mediating protein-protein interaction and the accumulation of PUF-8 in P granule (17,23).
PUF-8 regulates the lifespan of C. elegans through multi-pathways
PUF-8 was previously reported as a negative regulator that controls the lifespan of C. elegans by regulating the mRNA stability of MFF-1 and leads to aberrant mitochondrial dynamics and mitophagy (28).
In addition to inhibiting mff-1 mRNA stability, we found that PUF-8 may also control other pathways to negatively regulate the lifespan. Six genes, pqm-1, pha-4, blmp-1, vhp-1, hlh-30 and ctl-2, that were identified to bind PUF-8. These genes have been reported previously to regulate aging processes in C. elegans (5,53–56). Depletion or mutation of PUF-8 up-regulated the mRNA levels of these genes.
HLH-30 is a TFEB transcription factor that regulates the aging process by controlling lipid metabolism and regulating the ‘sperm competition’ process (56). HLH-30 regulates autophagy processes and longevity cross-talking with DAF-16/FOXO under harmful conditions (57,58). CTL-2 is a peroxisomal catalase, and ctl-2 mutation shortens the lifespan of long-lived mutant worms (53). VHP-1 and PHA-4 control lifespan by regulating the JNK signaling pathway (54) and TOR signaling pathway (55), respectively.
In this study, we found that pqm-1 is essential for the lifespan extension of puf-8 mutant worms. PQM-1 is a transcription factor and regulates class II aging-related genes via the DAE (5,56). DAF‐16 and PQM‐1 work together in regulating the longevity, yet their functions are antagonistic (6). The signaling pathway of DAF‐16 has been well elucidated in the past decade, but the signaling pathway of PQM-1 remains to be elucidated. Here, we show that RNAi knockdown of pqm-1 significantly reduces the lifespan extension of puf-8(ok302) worms and MUT-8G worms (Figure 7), indicating that PUF-8 may control the lifespan of C. elegans by regulating pqm-1.
PQM-1 is a fat metabolic regulator (59,60), and positively regulates fat transport to develop oocytes through vitellogenins (59). As lipids provide energy, fat storage and mobilization from intestinal cells to germline cells have an intimate relationship with oogenesis and early embryogenesis. Previously, Wang et al. reported that fat metabolism linked to the development of germline stem cell to longevity (61). We speculates that PUF-8 may modulate lipid metabolism, and the development of germline stem cells via pqm-1 to regulate the lifespan.
The germline development is highly correlated with lifespan of C. elegans (61,62). PUF-8 plays important roles both in germline development (21–25) and lifespan (28). Our study showed that PUF-8 can not only regulate mff-1 but also regulate multiple target genes, including pqm-1, to modulate the lifespan of C. elegans. In addition, knockdown of hlh-30 by RNAi, caused remarkable shortening of lifespan, both in N2 and puf-8 mutants, suggesting the importance of hlh-30 in the lifespan regulation. Further investigations are required to examine the precisely mechanisms by which PUF-8 identifies and regulates specific target genes to control aging and lifespan processes.
DATA AVAILABILITY
The atomic coordinates and structure factors (PDB IDs: 7CGF, 7CGG, 7CGI, 7CGH, 7CGJ, 7CGK, 7CGM and 7CGL) have been deposited in the Protein Data Bank (http://www.pdb.org/). The high throughput RNA-sequencing data have been deposited in the GEO repository (accession code: GSE165061).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the staff of Beamlines BL17U, BL18U and BL19U at SSRF for assisting in the data collection. We thank Drs. Fudong Li and Yiyang Jiang for helping with structure determination.
Contributor Information
Zheng Xu, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
Jie Zhao, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
Minjie Hong, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
Chenming Zeng, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
Shouhong Guang, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
Yunyu Shi, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230027, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB39010300, XDB39010600]; Ministry of Science and Technology of China [2019YFA0508403, 2016YFA0500700]; Chinese National Natural Science Foundation [32090040, 31870760, 91940303, 31870812, 32070619, 31871300, 31900434].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bluher M., Kahn B.B., Kahn C.R.. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003; 299:572–574. [DOI] [PubMed] [Google Scholar]
- 2.Lin K., Hsin H., Libina N., Kenyon C.. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 2001; 28:139–145. [DOI] [PubMed] [Google Scholar]
- 3.Tullet J.M.DAF-16 target identification in C. elegans: past, present and future. Biogerontology. 2015; 16:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay A., Oh S.W., Tissenbaum H.A.. Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 2006; 41:928–934. [DOI] [PubMed] [Google Scholar]
- 5.Tepper R.G., Ashraf J., Kaletsky R., Kleemann G., Murphy C.T., Bussemaker H.J.. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013; 154:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tepper R.G., Murphy C.T., Bussemaker H.J.. DAF-16 and PQM-1: partners in longevity. Aging (Albany NY). 2014; 6:5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickens M., Bernstein D.S., Kimble J., Parker R.. A PUF family portrait: 3′ UTR regulation as a way of life. Trends in Genetics. 2002; 18:150–157. [DOI] [PubMed] [Google Scholar]
- 8.Miller M.A., Olivas W.M.. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip. Rev. RNA. 2011; 2:471–492. [DOI] [PubMed] [Google Scholar]
- 9.Lu G., Dolgner S.J., Hall T.M.. Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 2009; 19:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quenault T., Lithgow T., Traven A.. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011; 21:104–112. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Zamore P.D., Hall T.M.. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001; 7:855–865. [DOI] [PubMed] [Google Scholar]
- 12.Spassov D.S., Jurecic R.. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function?. IUBMB Life. 2003; 55:359–366. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., McLachlan J., Zamore P.D., Hall T.M.. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 14.Cheong C.G., Hall T.M.. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl Acad. Sci. U.S.A. 2006; 103:13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall T.M.De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016; 36:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat V.D., McCann K.L., Wang Y., Fonseca D.R., Shukla T., Alexander J.C., Qiu C., Wickens M., Lo T.W., Tanaka Hall T.M.et al.. Engineering a conserved RNA regulatory protein repurposes its biological function in vivo. Elife. 2019; 8:e43788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pushpa K., Kumar G.A., Subramaniam K.. PUF-8 and TCER-1 are essential for normal levels of multiple mRNAs in the C. elegans germline. Development. 2013; 140:1312–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opperman L., Hook B., DeFino M., Bernstein D.S., Wickens M.. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005; 12:945–951. [DOI] [PubMed] [Google Scholar]
- 19.Martinez J.C., Randolph L.K., Iascone D.M., Pernice H.F., Polleux F., Hengst U.. Pum2 shapes the transcriptome in developing axons through retention of target mRNAs in the cell body. Neuron. 2019; 104:931–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramaniam K., Seydoux G.. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Current Biology. 2003; 13:134–139. [DOI] [PubMed] [Google Scholar]
- 21.Racher H., Hansen D. PUF-8, a Pumilio homolog, inhibits the proliferative fate in the Caenorhabditis elegans germline. G3 (Bethesda). 2012; 2:1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mainpal R., Priti A., Subramaniam K.. PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 2011; 360:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariz M., Mainpal R., Subramaniam K.. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 2009; 326:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datla U.S., Scovill N.C., Brokamp A.J., Kim E., Asch A.S., Lee M.H.. Role of PUF-8/PUF protein in stem cell control, sperm-oocyte decision and cell fate reprogramming. J. Cell. Physiol. 2014; 229:1306–1311. [DOI] [PubMed] [Google Scholar]
- 25.Bachorik J.L., Kimble J.. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl Acad. Sci. U.S.A. 2005; 102:10893–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priti A., Subramaniam K.. PUF-8 functions redundantly with GLD-1 to promote the meiotic progression of spermatocytes in Caenorhabditis elegans. G3 (Bethesda). 2015; 5:1675–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maheshwari R., Pushpa K., Subramaniam K.. A role for post-transcriptional control of endoplasmic reticulum dynamics and function in C. elegans germline stem cell maintenance. Development. 2016; 143:3097–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Amico D., Mottis A., Potenza F., Sorrentino V., Li H., Romani M., Lemos V., Schoonjans K., Zamboni N., Knott G.et al.. The RNA-Binding protein PUM2 Impairs mitochondrial dynamics and mitophagy during aging. Mol. Cell. 2019; 73:775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenyon C.A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2010; 1204:156–162. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwart P.H., Afonine P.V., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., McKee E., Moriarty N.W., Read R.J., Sacchettini J.C.et al.. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008; 426:419–435. [DOI] [PubMed] [Google Scholar]
- 34.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Concordet J.P., Haeussler M.. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018; 46:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad A., Porter D.F., Kroll-Conner P.L., Mohanty I., Ryan A.R., Crittenden S.L., Wickens M., Kimble J.. The PUF binding landscape in metazoan germ cells. RNA. 2016; 22:1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huppertz I., Attig J., D’Ambrogio A., Easton L.E., Sibley C.R., Sugimoto Y., Tajnik M., Konig J., Ule J.. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014; 65:274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaid S., Ariz M., Chaturbedi A., Kumar G.A., Subramaniam K.. PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development. 2013; 140:1645–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu F., Feng X., Chen X., Weng C., Yan Q., Xu T., Hong M., Guang S.. A cytoplasmic argonaute protein promotes the inheritance of RNAi. Cell Rep. 2018; 23:2482–2494. [DOI] [PubMed] [Google Scholar]
- 41.Han S.K., Lee D., Lee H., Kim D., Son H.G., Yang J.S., Lee S.V., Kim S. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016; 7:56147–56152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu C., Bhat V.D., Rajeev S., Zhang C., Lasley A.E., Wine R.N., Campbell Z.T., Hall T.M.T.. A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity. Elife. 2019; 8:e48968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamala K.P., Martin-Alarcon D.A., Boyden E.S.. Programmable RNA-binding protein composed of repeats of a single modular unit. Proc. Natl Acad. Sci. U.S.A. 2016; 113:E2579–E2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Voronina E.. Diverse roles of PUF proteins in germline stem and progenitor cell development in C. elegans. Front. Cell Dev. Biol. 2020; 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M.et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta Y.K., Nair D.T., Wharton R.P., Aggarwal A.K.. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008; 16:549–557. [DOI] [PubMed] [Google Scholar]
- 47.Kaymak E., Wee L.M., Ryder S.P.. Structure and function of nematode RNA-binding proteins. Curr. Opin. Struct. Biol. 2010; 20:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Opperman L., Wickens M., Hall T.M.. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20186–20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu G., Hall T.M.. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011; 19:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilsner J., Linnik O., Christensen N.M., Bell K., Roberts I.M., Lacomme C., Oparka K.J.. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 2009; 57:758–770. [DOI] [PubMed] [Google Scholar]
- 51.Dong S., Wang Y., Cassidy-Amstutz C., Lu G., Bigler R., Jezyk M.R., Li C., Hall T.M., Wang Z.. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 2011; 286:26732–26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhury R., Tsai Y.S., Dominguez D., Wang Y., Wang Z.. Engineering RNA endonucleases with customized sequence specificities. Nat. Commun. 2012; 3:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petriv O.I., Rachubinski R.A.. Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. J. Biol. Chem. 2004; 279:19996–20001. [DOI] [PubMed] [Google Scholar]
- 54.Mizuno T., Hisamoto N., Terada T., Kondo T., Adachi M., Nishida E., Kim D.H., Ausubel F.M., Matsumoto K.. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004; 23:2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandit A., Jain V., Kumar N., Mukhopadhyay A.. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging (Albany NY). 2014; 6:835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi C., Booth L.N., Murphy C.T.. Insulin-like peptides and the mTOR-TFEB pathway protect Caenorhabditis elegans hermaphrodites from mating-induced death. Elife. 2019; 8:e46413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin X.X., Sen I., Janssens G.E., Zhou X., Fonslow B.R., Edgar D., Stroustrup N., Swoboda P., Yates J.R. 3rd, Ruvkun G.et al.. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat. Commun. 2018; 9:4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerisch B., Tharyan R.G., Mak J., Denzel S.I., Popkes-van Oepen T., Henn N., Antebi A.. HLH-30/TFEB is a master regulator of reproductive quiescence. Dev. Cell. 2020; 53:316–329. [DOI] [PubMed] [Google Scholar]
- 59.Dowen R.H., Breen P.C., Tullius T., Conery A.L., Ruvkun G.. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 2016; 30:1515–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Brien D., Jones L.M., Good S., Miles J., Vijayabaskar M.S., Aston R., Smith C.E., Westhead D.R., van Oosten-Hawle P.. A PQM-1-mediated response triggers transcellular chaperone signaling and regulates organismal proteostasis. Cell Rep. 2018; 23:3905–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang M.C., O’Rourke E.J., Ruvkun G.. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008; 322:957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heimbucher T., Hog J., Gupta P., Murphy C.T.. PQM-1 controls hypoxic survival via regulation of lipid metabolism. Nat. Commun. 2020; 11:4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors (PDB IDs: 7CGF, 7CGG, 7CGI, 7CGH, 7CGJ, 7CGK, 7CGM and 7CGL) have been deposited in the Protein Data Bank (http://www.pdb.org/). The high throughput RNA-sequencing data have been deposited in the GEO repository (accession code: GSE165061).