Abstract
Background
Ischemic stroke is a disease with high rate of death and disability worldwide. CircRNAs, as a novel type of non-coding RNAs, lacking 5’ caps and 3’ poly-A tails, has been associated with ischemic stroke. This study aimed to investigate key circRNAs related to ischemic stroke.
Methods
RNA sequencing was performed obtain the circRNA expression profiles from peripheral whole blood of three ischemic stroke patients and three healthy individuals. Through bioinformatic analysis, differentially expressed circRNAs (DEcircRNAs) were identified, and GO and pathway analyses for the host genes of DEcircRNAs were conducted. The expression levels of selected circRNAs were analyzed with qRT-PCR. To further explore the functions of key circRNAs, a DEcircRNA-miRNA interaction network was constructed.
Results
A total of 736 DEcircRNAs were detected in ischemic stroke. Functional annotation of host genes of DEcircRNAs revealed several significantly enriched pathways, including Fc epsilon RI signaling pathway, B cell receptor signaling pathway, and T cell receptor signaling pathway. The qRT-PCR results were largely in keeping with our RNA-seq data. The ROC curve analyses indicated that hsa_circ_0000745, hsa_circ_0001459, hsa_circ_0003694 and hsa_circ_0007706 with relatively high diagnostic value. A circRNA-miRNA network, including 1544 circRNA-miRNA pairs, 456 circRNAs and 4 miRNAs, was obtained.
Conclusions
The results of our study may help to elucidate the specific mechanism underlying ischemic stroke.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12883-021-02397-0.
Keywords: Ischemic stroke, CircRNA, High-throughput sequencing, Host gene
Background
Stroke is a leading cause of long-term disability and life-threatening disease [1]. Ischemic stroke, which accounts for approximately 80 % of total strokes, results in reduced cerebral blood flow due to cerebral artery occlusion, leading to rapid loss of brain function [2]. However, at present, there is no effective treatment available for cerebral ischemic diseases; this deficiency is attributable to many factors, particularly the rapid development of brain injury following ischemia, molecular changes following acute ischemic stroke, complex interplays among signaling pathways, and the narrow therapeutic window for specific targets [3]. Hence, it is urgently important to elucidate the pathogenesis and underlying mechanisms of cerebral ischemic injury and develop effective treatment strategies for this disease.
Lacking 5’ caps and 3’ poly-A tails, circular RNAs (circRNAs) are resistant to RNaseR treatment (which degrades essentially all linear forms of RNAs), which makes circRNAs more stable than linear RNAs [4]. In addition, circRNAs are evolutionarily conserved among diverse species [4]. These features enable circRNAs to perform a variety of physiological functions, including mediating alternative splicing of mRNAs, regulating the transcription of parental genes and binding to miRNAs as competing endogenous RNAs [5]. Three models of circRNA regulating the expression of parental gene: (1) The intron-only ciRNA binds to elongating RNA Pol II and promotes host transcription in a cis-acting manner; (2) EIciRNA binds to U1 snRNP through specific RNA-RNA interaction between U1 snRNA and EIciRNA, and then the EIciRNA-U1 snRNP complexes might interact with RNA Pol II transcription complex to promote host gene expression; (3) CircRNA shares some miRNA binding sites with 3’-UTR of the transcript from their parental gene, and then CircRNA acts as miRNA sponge and increases the translations of the transcript from its parental gene [6].
CircRNAs have been reported to confer functions in multiple pathogenic processes, including cancers and stroke [7]. Bazan et al. reported that a high ratio of serum circR-284 to miR-221 is observed in acute ischemic stroke patients, indicating the potential diagnostic value of circRNAs in cerebrovascular ischemia [8]. Bai et al. indicated that significantly decreased circDLGAP4 was present in both acute ischemic stroke patients and a mouse stroke model [9]. Three circRANs (circFUNDC1, circPDS5B, and circCDC14A) were reported to be positively correlated with infarct volume, suggesting the 3 circRNAs may be envisioned as potential biomarkers for acute ischemic stroke diagnosis [10]. Li et al. suggested that hsa_circ_0000607 may play a crucial role in the pathogenesis and progression of acute ischemic stroke by regulating the miR-337-3p/Bcl2 axis [11]. Significantly decreased CircSCMH1 in the plasma of patients with acute ischemic stroke is of great significance in predicting stroke outcomes [12].
In the current study, we employed high-throughput RNA sequencing (RNA-seq) to investigate the circRNA expression profiles of ischemic stroke patients. Bioinformatic analysis was applied to identify differentially expressed circRNAs (DEcircRNAs) and DEcircRNA-miRNA interaction networks. In addition, the expression levels of selected circRNAs were validated with quantitative real-time polymerase chain reaction (qRT-PCR). By doing this, the results of our study may help to elucidate the pathogenesis and underlying mechanisms of ischemic stroke.
Methods
Patients and samples
The cohort subjected to RNA-Seq consisted 3 ischemic stroke patients and 3 healthy individuals. The etiology of stroke was classified according to Trial of Org 10,172 in Acute Stroke Treatment (TOAST) criteria [13]. Three patients with large-artery atherosclerosis were admitted at 2, 4, and 7 days after the onset of stroke, respectively. The inclusion criteria were ischemic stroke patients with initial onset aged 18–75 years who were diagnosed according to Magnetic Resonance Imaging (MRI) or computed tomography (CT) scan of the brain and clinical diagnostic criteria. Patients with other neurological diseases, including transient ischemic attack, cardiogenic cerebral embolism, hemorrhagic infarction, traumatic cerebrovascular disease, and occult cerebral vascular malformation, were excluded from the study. Healthy individuals were recruited from people who underwent a routine medical examination at the hospital. Individuals with a history of stroke, surgery, heart surgery, head trauma, or neurological disease were excluded. Table 1 describes the characteristics of these individuals. The subjects were matched for age and gender. All samples were collected after obtaining written informed consent. This study was approved by the ethics committee of the First Affiliated Hospital of Shantou University Medical College and performed in accordance with the principles of the Declaration of Helsinki. Peripheral whole blood (2.5 ml) drawn from each patient and each control subject was used for RNA extraction.
Table 1.
Patient characteristics
| Case (n = 3) | Control (n = 3) | p-value | |
|---|---|---|---|
| Age (mean (SD)) | 61.33 (8.39) | 63.67 (4.51) | 0.693 |
| Gender = Male (%) | 2 (66.7) | 2 (66.7) | 1 |
| Hypertension = Yes (%) | 2 (66.7) | 0 (0.0) | 0.386 |
| Hyperlipidemia = Yes (%) | 2 (66.7) | 0 (0.0) | 0.386 |
| Diabetes = Yes (%) | 1 (33.3) | 0 (0.0) | 1 |
| Triglycerides (mean (SD)) | 2.09 (0.73) | 1.21 (0.25) | 0.122 |
| HDL (mean (SD)) | 1.09 (0.14) | 1.35 (0.31) | 0.259 |
| LDL (mean (SD)) | 3.62 (0.86) | 3.63 (0.60) | 0.979 |
LDL low-density lipoprotein; HDL high-density lipoprotein
RNA sequencing and identification of DEcircRNAs
Using TRIzol reagent, total RNA was extracted from samples. RNA integrity and concentration were evaluated with an Agilent 2100 Bioanalyzer. Total RNA samples used in subsequent experiments fulfilled the following requirements: RNA integrity number (RIN) > 7.0 and 28 S/18S ≥ 1. In brief, total RNA was subjected to ribosomal RNA (rRNA) removal using the Ribo-Zero. To remove linear RNAs, total RNA was digested with RNase R. A total amount of 3 µg RNA was used for library preparation. Libraries for sequencing were constructed according to the manufacturer’s protocol. The quality of the libraries was determined using an Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System. Libraries measuring 100–200 bp were selected. RNA sequencing was performed based on HiSeq 10X-150PE and 10 GB RNA-seq data per sample was generated. Low-quality data, adapter sequences and sequences with N base rate of raw reads > 1 % were filtered using SOAPnukev1.5.2 (parameters: -l 15 -q 0.2 -n 0.01 –i) [14]. Then, the remaining clean reads with high quality were subsequently aligned to the human reference genome (hg19) using BWA [15]. CIRI (v2.0.5) [16] uses BWA and find_circ (v1.2) [17] uses Bowtie2 (http://bowtie-bio.sourceforge.net/index.shtml) with default parameters to detect circRNAs, respectively. In this study, circRNA expression was calculated according to the junction reads count at both ends of the circRNA, and the final junction reads count takes the average value of the two software results. The junction reads per billion mapped reads were applied to normalize all samples. DEGseq, an R package to identify differentially expressed genes or isoforms for RNA-seq data from different samples, takes uniquely mapped reads from RNA-seq data for the two samples with a gene annotation as input [18]. With DEGseq v 1.43.0, DEcircRNAs were identified. Compared with controls, the DEcircRNAs in ischemic stroke were defined with p-value < 0.05 & |log2 FoldChange| > 2. Hierarchical clustering analysis of DEcircRNAs was performed with R package “pheatmap”. Then, enrichment analysis for host genes of DEcircRNAs was performed by GeneCodis3 with R package ggplot2.
In vitro validation
Following the manufacturer’s protocol, total RNA was isolated from blood samples of 15 ischemic stroke patients and 15 healthy controls with the TRIzol reagent. RNA integrity and concentration were evaluated by NanoVue Plus. By using FastQuant cDNA (Tiangen, Beijing, China), we generated cDNA from 1 µg extracted RNA. The qRT-PCR analyses were performed in an ABI 7300 Real-time PCR Detection System with SuperReal PreMix Plus (Invitrogen, USA). The qRT-PCR thermal cycling parameters were as follows: an initial denaturation step of 15 min at 95℃, followed by 40 cycles of 10 s at 95℃ and 30 s at 55℃, 32 s at 72℃, and 15 s at 95℃, 60 s at 60℃, 15 s at 95℃. Relative gene expression was calculated with the 2−ΔΔCT method. Statistical significance was assessed by t-test. GAPDH was utilized as an internal control. The characteristics of these individuals were presented in Table S1. We designed specific divergent primers spanning the back-splice junction sites of circRNAs. The PCR primers were displayed in Table 2.
Table 2.
The primers used in qRT-PCR experiments
| circRNA | Pimers |
|---|---|
| GAPDH | Forward: 5’ TCGACAGTCAGCCGCATCTTCTTT 3’ |
| Reverse: 5’ ACCAAATCCGTTGACTCCGACCTT 3’ | |
| hsa_circ_0001459 | Forward: 5’ ACAGCCCAATACTCATCACCAG 3’ |
| Reverse: 5’ TCTTCACTCTTGAGCACTGAATCT 3’ | |
| hsa_circ_0007706 | Forward: 5’ TGAAGAAGAAGAAGTCACCAAAGGA 3’ |
| Reverse: 5’ GCAAACTGTGGGAAAGCCAT 3’ | |
| hsa_circ_0000745 | Forward: 5’ ATGTTGAAAGTAGCCCGAGCAG 3’ |
| Reverse: 5’ TGGGAGTGTTGGAAGAAGTTGG 3’ | |
| hsa_circ_0003694 | Forward: 5’ AGCAGGAATTGAGCCACAGAG 3’ |
| Reverse: 5’ AGAAGCTGTAAAGGCCTGTTGT 3’ | |
| hsa_circ_0037852 | Forward: 5’ TTCTCAGACCACATCCGCTG 3’ |
| Reverse: 5’ AGTCGCTGTTATCAGCTATTCTCT 3’ |
miRNA prediction of validated circRNAs
Pathways with p_adj < 0.05 were defined as significantly enriched pathways. To further investigate the functions of circRNAs, the target miRNAs of circRNAs were predicted based on the RNAhybrid database (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) with -sc > 150 and –en < 7. The circRNA-miRNA interaction network was visualized with Cytoscape (http://www.cytoscape.org).
Results
Identification of DEcircRNAs
A total of 22,434 circRNAs were detected in this study. Compared with normal controls, 736 DEcircRNAs (307 up-regulated and 429 down-regulated DEcircRNAs) were detected in ischemic stroke with p-value < 0.05 and |log2 FoldChange| > 2 (Fig. 1). These DEcircRNAs were widely distributed across almost all human chromosomes, including the sex chromosomes (Figure S1). Among these circRNAs, hsa_circ_0001459 and hsa_circ_0037852 were the most up-regulated and down-regulated, respectively (Table 3). The raw data have been deposited in the Gene Expression Omnibus database (GSE178764, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178764).
Fig. 1.
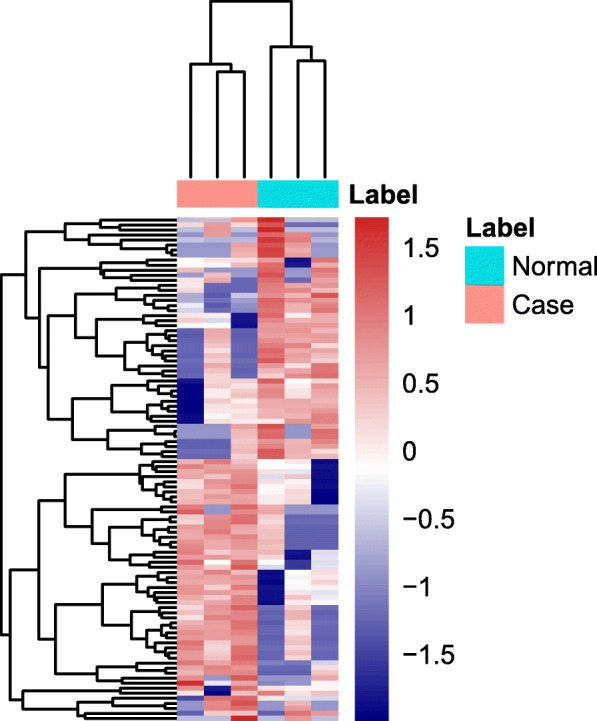
The heatmap of top 100 up-regulated and down-regulated differentially expressed circRNAs in ischemic stroke patients. Rows and columns represented DEcircRNAs and samples, respectively. The color scale represented the expression levels. Red and blue color indicated up- and down-regulation, respectively
Table 3.
Top 10 up- and down-regulated DEcircRNAs between patients with ischemic stroke and normal controls
| GeneID | circBaseID | log2FC | p-value | Regulation | Host gene | N1 | N2 | N3 | P1 | P2 | P3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr4:178,274,461|178,274,882 | hsa_circ_0001459 | 3.037662 | 0 | up | NEIL3 | 0 | 0 | 253 | 1296 | 395 | 725 |
| chr17:53,478,829|53,481,229 | hsa_circ_0002015 | 2.576615 | 0 | up | MMD | 1059 | 108 | 92 | 1819 | 2522 | 4393 |
| chr12:58,340,777|58,347,472 | hsa_circ_0000412 | 2.298732 | 0 | up | ATP23 | 648 | 0 | 391 | 1371 | 2061 | 2513 |
| chr21:37,711,076|37,717,005 | hsa_circ_0001189 | 2.129201 | 0 | up | MORC3 | 130 | 496 | 460 | 1620 | 1052 | 2853 |
| chr9:6,880,011|6,893,232 | hsa_circ_0001839 | 2.1205 | 0 | up | KDM4C | 259 | 237 | 506 | 50 | 2390 | 2627 |
| chr17:20,107,645|20,109,225 | hsa_circ_0000745 | 2.025537 | 0 | up | SPECC1 | 583 | 1444 | 529 | 8198 | 1096 | 2808 |
| chr3:169,840,378|169,847,340 | hsa_circ_0067900 | 3.239944 | 3.03E-277 | up | PHC3 | 151 | 0 | 0 | 1096 | 110 | 453 |
| chr18:196,636|199,316 | hsa_circ_0007706 | 3.034998 | 9.43E-260 | up | USP14 | 130 | 43 | 0 | 648 | 548 | 453 |
| chr1:205,238,077|205,239,012 | hsa_circ_0003344 | 2.098331 | 4.26E-231 | up | TMCC2 | 43 | 172 | 230 | 1047 | 241 | 928 |
| chr1:29,362,337|29,379,824 | hsa_circ_0011173 | 3.928555 | 3.58E-223 | up | EPB41 | 65 | 0 | 0 | 50 | 263 | 838 |
| chr16:11,214,471|11,217,803 | hsa_circ_0037852 | -5.09647 | 0 | down | CLEC16A | 281 | 431 | 759 | 50 | 0 | 0 |
| chr18:45,391,429|45,396,935 | hsa_circ_0003694 | -3.13386 | 0 | down | SMAD2 | 562 | 625 | 1402 | 299 | 44 | 0 |
| chr7:104,925,455|104,937,980 | hsa_circ_0003656 | -2.84711 | 0 | down | SRPK2 | 540 | 970 | 965 | 199 | 88 | 113 |
| chr15:50,330,964|50,339,661 | hsa_circ_0035197 | -2.41562 | 0 | down | ATP8B4 | 1664 | 2478 | 2896 | 797 | 307 | 430 |
| chr20:60,737,807|60,738,678 | hsa_circ_0003231 | -3.34158 | 1.31E-276 | down | SS18L1 | 216 | 388 | 782 | 0 | 0 | 159 |
| chr10:11,312,628|11,330,515 | hsa_circ_0017680 | -2.63557 | 8.20E-247 | down | CELF2 | 1102 | 453 | 0 | 0 | 132 | 159 |
| chr9:126,519,981|126,531,842 | hsa_circ_0088474 | -4.5753 | 1.71E-220 | down | DENND1A | 130 | 151 | 621 | 0 | 44 | 0 |
| chr6:83,667,030|83,754,378 | hsa_circ_0008236 | -3.23452 | 9.18E-219 | down | UBE3D | 346 | 43 | 736 | 50 | 44 | 45 |
| chr19:58,904,342|58,904,854 | hsa_circ_0005598 | -2.15791 | 9.64E-214 | down | RPS5 | 605 | 474 | 621 | 0 | 307 | 136 |
| chr17:20,910,208|20,914,622 | hsa_circ_0042458 | -3.45041 | 2.27E-212 | down | USP22 | 238 | 129 | 667 | 0 | 110 | 0 |
DEcircRNAs differentially expressed circRNAs; FC fold change, N1-3 normal controls; P1-3 patients with ischemic stroke
Functional annotation
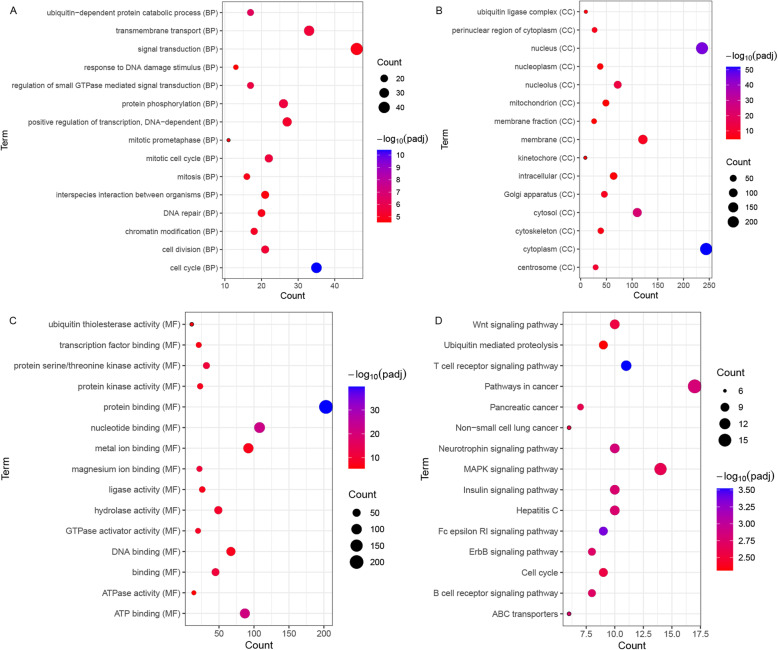
In total, 607 host genes of DEcircRNAs were identified. GO analysis indicated several significantly enriched terms, such as, cell cycle (p = 2.65E-14), ubiquitin-dependent protein catabolic process (p = 7.03E-10), cytoplasm (p = 3.74E-55) and protein binding (p = 3.08E-43) (Fig. 2 A-C). KEGG pathway enrichment analysis indicated that several pathways were significantly enriched, including T cell receptor signaling pathway (p = 1.92E-06), Fc epsilon RI signaling pathway (p = 5.72E-06), B cell receptor signaling pathway (p = 3.72E-05) and Pathways in cancer (p = 3.93E-05) (Fig. 2D).
Fig. 2.
Significantly enriched GO terms and KEGG pathways of host genes of DEcircRNAs in ischemic stroke patients. (A) BP, biological process; (B) CC, cellular component; (C) MF, molecular function; (D) KEGG pathways. The x-axis shows counts of DEGs enriched in GO terms or KEGG pathways and the y-axis shows GO terms or KEGG pathways. The color scale represented -lg p_adj
In vitro validation
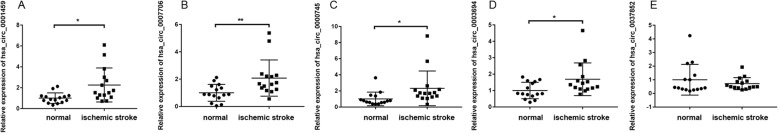
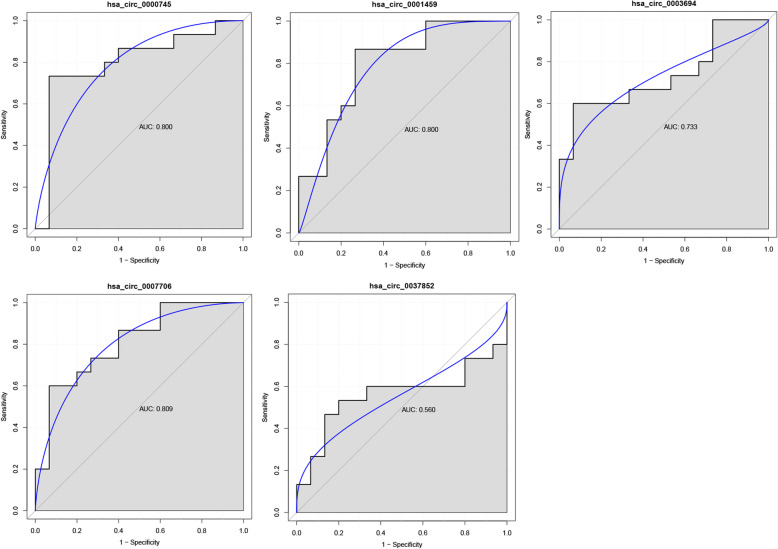
Five circRNAs, specifically three up-regulated (hsa_circ_0001459, hsa_circ_0007706 and hsa_circ_0000745) and two down-regulated DEcircRNAs (hsa_circ_0003694 and hsa_circ_0037852), were selected randomly from the top 10 dysregulated circRNAs (two were the most dysregulated, and three were randomly chosen) for qRT-PCR analysis. Three of these circRNAs at the level of significance and a trend in the same direction was observed for another. In other words, except for hsa_circ_0003694, the expression of the others in the qRT-PCR results generally exhibited the same pattern as that in our RNA-seq results (Fig. 3). Then, by using pROC package in R language, we performed the ROC analysis to assess the diagnostic value of DEcircRNAs. The area under the curve (AUC) was further calculated. And the ROC curve analyses indicated that the AUC of hsa_circ_0000745 (0.800), hsa_circ_0001459 (0.800), hsa_circ_0003694 (0.733) and hsa_circ_0007706 (0.809) was more than 0.7, and with relatively high diagnostic value (Fig. 4).
Fig. 3.
The qRT-PCR results of the DEcircRNAs in patients with ischemic stroke. (A) hsa_circ_0001459, (B) hsa_circ_0007706, (C) hsa_circ_0000745, (D) hsa_circ_0003694, (E) hsa_circ_0037852. * represented p < 0.05, ** represented p < 0.01
Fig. 4.
The ROC curves of DEcircRNAs in ischemic stroke. (A) hsa_circ_0000745, (B) hsa_circ_0001459, (C) hsa_circ_0003694, (D) hsa_circ_0007706, (E) hsa_circ_0037852
DEcircRNA-miRNA interaction network
A total of 1544 circRNA-miRNA pairs, involving 456 circRNAs and 4 miRNAs were obtained (Table S2). The subnetwork of hsa_circ_0001459, hsa_circ_0007706, hsa_circ_0000745, hsa_circ_0037852 and hsa_circ_0003694 were displayed in Fig. 5. In addition, NEIL3 was the host gene of hsa_circ_0001459, USP14 was the host gene of hsa_circ_0007706, SPECC1 was the host gene of hsa_circ_0000745, CLEC16A was the host gene of hsa_circ_0037852, and SMAD2 was the host gene of hsa_circ_0003694, respectively.
Fig. 5.
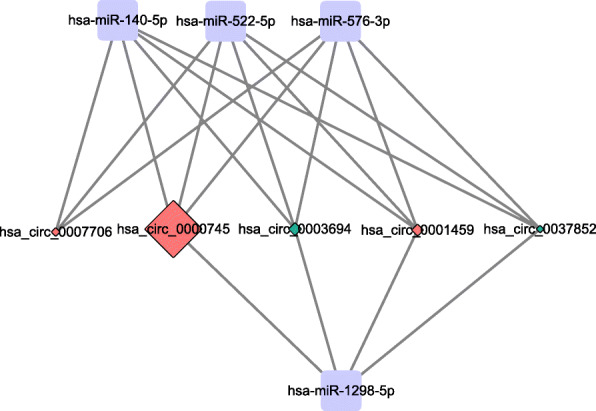
Subnetwork of hsa_circ_0001459, hsa_circ_0007706, hsa_circ_0000745, hsa_circ_0003694, and hsa_circ_0037852. The rhombic nodes and rectangle nodes indicate DEcircRNAs and miRNAs, respectively. Red and green color represent up-regulation and down-regulation, respectively. Nodes with black border were DEcircRNA derived from top 10 up-regulated and down-regulated DEcircRNA in ischemic stroke patients. The node size represents the expression level of circRNA. DEcircRNA, differentially expressed circRNA
Discussion
CircRNAs were once regarded as by-products of aberrant splicing, evolutionary junk or transcriptional noise with little functional potential [19]. Recently, these RNAs have attracted increasing attention, not only because of their conserved expression among species but also due to their functions. Ostolaza et al. suggested that hsa_circRNA_102488 may serve as an interesting candidate marker which showed a statistically significant change in expression between stroke etiology subtypes [20]. Lu et al. found that circBBS2 and circPHKA2 were differentially expressed in the blood of acute ischemic stroke patients and demonstrated that blood circRNAs may serve as potential biomarkers for acute ischemic stroke diagnosis [21]. In this study, a total of 736 DEcircRNAs were detected in ischemic stroke patients by high-throughput sequencing.
The hsa_circ_0001459, a circRNA derived from NEIL3, was identified to be up-regulated in both the RNA-seq results and qRT-PCR results in this analysis. Oxidative stress is known as one of the molecular mechanisms underlying cerebral ischemic stroke [22]. Chronic exposure to higher concentrations of reactive oxygen species (ROS) may cause damage to the DNA structures of brain cells, which results in an increased risk of stroke [23]. Nei like DNA glycosylase 3 (NEIL3) is a DNA glycosylase, that plays a vital role in repairing the DNA structure damaged by ROS [24]. Knockout of NEIL3 induces severe neuropathy, and NEIL3 knockout mice exhibited poor outcomes after ischemic stroke [25]. If cultured neural stem/progenitor cells lack NEIL3, the repair of oxidative DNA base lesions is significantly impaired [25]. Jalland et al. indicated that NEIL3 also promotes neurogenesis and reduces oxidative DNA damage to protect against prion disease [26]. The studies mentioned above suggested a close link between NEIL3 and ischemic stroke. Hence, we speculated that hsa_circ_0001459 may exert its effect by regulating its host gene NEIL3 in ischemic stroke.
Hsa_circ_0007706 was another significantly down-regulated circRNA in this analysis. Stroke is associated with over-production of misfolded and aggregating proteins [27]. Ubiquitin specific peptidase 14 (USP14), a deubiquitinated protein, negatively regulates proteasome activity and inhibits the degradation of ubiquitin-protein conjugates [28]. In vitro, IU1 inhibits USP14 activity to enhance proteasome activity, thereby reducing cell death induced by oxidative stress [29, 30]. IU1 is correlated with decreased protein aggregates and is thought to be a therapeutic target in the stroke model [27]. It has been suggested that miR-124 conferred a neuroprotective function by directly targeting USP14 after cerebral ischemia [31]. Taken together, these findings indicate that hsa_circ_0007706, derived from USP14, may be implicated in ischemic stroke.
It is worth noting that hsa_circ_0037852 is the most significantly down-regulated circRNA. C-type lectin domain containing 16 A (CLEC16A) has been shown to be highly expressed in immune cells, including B lymphocytes, dendritic cells and natural killer cells [32]. Fujimaki et al. indicated that CLEC16A may be a susceptibility locus for myocardial infarction in Japanese individuals without chronic kidney disease [33]. Yoshida et al. showed that the polymorphism of CLEC16A (rs9925481) was closely related to myocardial infarction in individuals without hypertension [34]. Similarly, hsa_circ_0037852, derived from CLEC16A, may also be involved in ischemic stroke.
Hsa_circ_0003694 was the second significantly down-regulated circRNA in ischemic stroke. In response to stroke, astrocytes convert to a reactive phenotype (known as reactive astrogliosis) [35, 36]. Reactive astrogliosis and glial scar formation are among the primary causes of the difficulty in achieving functional recovery after ischemic stroke, as their presence inhibits the regeneration of neurons [37, 38]. Zhang et al. suggested that RGMa suppresses neurological functional recovery and promotes reactive astrogliosis and glial scar formation, which was associated with SMAD family member 2 (SMAD2) [39]. Lu et al. indicated that GDF11 may promote the neurogenesis and angiogenesis after stroke, which was also related to SMAD2 [40]. SMAD2 is the host gene of hsa_circ_0003694, which was down-regulated in the RNA-seq results and up-regulated in the qRT-PCR validation. Hence, the precise role played by hsa_circ_0003694 in ischemic stroke has not been determined.
Hsa_circ_0000745 has been linked with various types of cancer. It was suggested that hsa_circ_0000745 was down-regulated in gastric cancer and considered as a diagnostic marker for gastric cancer [41]. Jiao et al. demonstrated that hsa_circ_0000745 could promote cervical cancer and was a candidate target for the treatment of cervical cancer in the clinic [42]. In this study, hsa_circ_0000745 exhibited increased expression in ischemic stroke, which was the first report to link hsa_circ_0000745 with ischemic stroke. However, further research on the correlation between hsa_circ_0000745 and ischemic stroke is required to confirm these findings.
The DEcircRNA-miRNA interaction network demonstrated that there was a shared miR-140-5p target in hsa_circ_0001459, hsa_circ_0007706, hsa_circ_0000745, hsa_circ_0037852 and hsa_circ_0003694. It has been suggested that angiogenesis is implicated in neurological functional recovery [43]. An in vitro study indicated that miR-140-5p inhibits angiogenesis after cerebral ischemia [44]. Sørensen et al. observed up-regulated miR-140-5p in the stroke group and indicated that it may be related to ischemic stroke [45]. It is widely recognized that circRNAs can regulate gene expression by serving as ceRNAs by sponging miRNAs. Therefore, these findings further highlighted the important roles of the DEcircRNAs mentioned above in the stroke.
In this study, GO and KEGG analyses predicted and analyzed the potential circRNA function and biological pathways. The stroke-related biological processes included the immune process (such as, T cell receptor signaling pathway, B cell receptor signaling pathway and Fc epsilon RI signaling pathway) and signal transduction pathways (such as, MAPK signaling pathway and Wnt signaling pathway). It is known that immunity is integral parts of the pathogenic processes provoked by ischemia and reperfusion [46]. Consistently, previous studies have been demonstrated that MAPK signaling pathway and Wnt signaling pathway were involved in ischemic stroke [47–49]. Our study is consistent with what is known about stroke-related pathology.
Conclusions
In conclusion, to the best of our knowledge, these five DEcircRNAs were reported for the first time that may be associated with ischemic stroke by high-throughput sequencing in this study. In addition, these five DEcircRNAs are all intron-type, suggesting they regulate their host genes through the first mechanism described above. Inevitably, the current study has some limitations. First, the sample size for RNA-seq and qRT-PCR validation was small. Second, there are differences in the distribution of hypertension and hyperlipemia between these ischemic stroke groups and healthy control groups. Third, no significant differences in the expression levels of hsa_circ_0037852 in qRT-PCR were observed between ischemic stroke groups and healthy control groups and the opposite results between RNA-seq and qRT-PCR of circ-0003694 were observed. This discrepancy probably arose because of the relatively small sample size, technical bias, the differential distribution of hypertension, hyperlipemia between these two groups, and the heterogeneity among samples. More samples that strictly meet the requirements need to be involved in our research for further verification and functional experiments of the significance of circRNAs in ischemic stroke. In addition, the ratio of the circRNA to linear expression, the effect of polymorphism on circRNA, and the specific mechanism of circRNA regulation of its host genes would be included in our future work plan.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- AUC
Area under the curve
- circRNA
Circular RNA
- CLEC16A
C-type lectin domain containing 16 A
- CT
Computed tomography
- DEcircRNA
Differentially expressed circRNA
- MRI
Magnetic Resonance Imaging
- NEIL3
Nei like DNA glycosylase 3
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RIN
RNA integrity number
- ROS
Reactive oxygen species
- SMAD2
SMAD family member 2
- TOAST
Trial of Org 10,172 in Acute Stroke Treatment
- USP14
Ubiquitin specific peptidase 14
Authors’ contributions
WH contributed to the study conception and design. Material preparation and experiments were performed by DW, JC, XC and SW. Data collection, analysis and interpretation were performed by ZC, YH and ZS. The first draft of the manuscript was written by DW, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the "Dengfeng Project" for the construction of high-level hospitals in Guangdong Province-the First Affiliated Hospital of Shantou University Medical College Supporting Funding, Jiacheng Li Foundation crossresearch project in 2020 The continuous medical service management for stroke patients based on miRNA chip research, combined with Internet APP platform and artificial intelligence technology and medical, nursing and pharmaceutical “linkage” mode (2020LKSFG09C), Trial Implementation Measures for supporting Project Scientific Research Special Plan of the First Affiliated Hospital of Shantou University Medical College (Revised) (2019-70) and Publicity of The 2021 Guangdong Science and Technology Special Fund ("major special project + Task list ") project of Shantou Science and Technology Bureau (2021010206). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the corresponding author on reasonable request. The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus database (GSE178764, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178764).
Declarations
Ethics approval and consent to participate
All samples were collected after obtaining written informed consent. This study was approved by the ethics committee of the First Affiliated Hospital of Shantou University Medical College and performed in accordance with the principles of the Declaration of Helsinki.
Consent for publication
Written informed consent for publication of clinical details was obtained from all of the patients.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hu X, De Silva TM, Chen J, Faraci FM. Cerebral Vascular Disease and Neurovascular Injury in Ischemic Stroke. Circulation research. 2017;120(3):449–71. doi: 10.1161/CIRCRESAHA.116.308427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beal CC. Gender and stroke symptoms: a review of the current literature. The Journal of neuroscience nursing: journal of the American Association of Neuroscience Nurses. 2010;42(2):80–7. doi: 10.1097/JNN.0b013e3181ce5c70. [DOI] [PubMed] [Google Scholar]
- 3.Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, et al. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiology of disease. 2010;38(1):17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature biotechnology. 2014;32(5):453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL. The biogenesis and emerging roles of circular RNAs. Nature reviews Molecular cell biology. 2016;17(4):205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 6.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer letters. 2015;365(2):141–8. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Wu N, Liu J, Wu Z, Dong D. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagnostic pathology. 2015;10:131. doi: 10.1186/s13000-015-0310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ, Jr., Woods TC. Carotid Plaque Rupture Is Accompanied by an Increase in the Ratio of Serum circR-284 to miR-221 Levels. Circulation Cardiovascular genetics. 2017;10(4):e001720. doi: 10.1161/CIRCGENETICS.117.001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2018;38(1):32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuo L, Zhang L, Zu J, Wang Z, Han B, Chen B, et al. Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke. 2020;51(1):319–23. doi: 10.1161/STROKEAHA.119.027348. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Chen L, Xu C, Qu X, Qin Z, Gao J, et al. Expression profile and bioinformatics analysis of circular RNAs in acute ischemic stroke in a South Chinese Han population. Scientific reports. 2020;10(1):10138. doi: 10.1038/s41598-020-66990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation. 2020;142(6):556–74. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 13.Adams HP, Jr., Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35–41. doi: 10.1161/01.STR.24.1.35. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome biology. 2015;16(1):4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics (Oxford, England) 2010;26(1):136–8. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 19.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacology & therapeutics. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Ostolaza A, Blanco-Luquin I, Urdánoz-Casado A, Rubio I, Labarga A, Zandio B, et al. Circular RNA expression profile in blood according to ischemic stroke etiology. Cell & bioscience. 2020;10:34. doi: 10.1186/s13578-020-00394-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu D, Ho ES, Mai H, Zang J, Liu Y, Li Y, et al. Identification of Blood Circular RNAs as Potential Biomarkers for Acute Ischemic Stroke. Frontiers in neuroscience. 2020;14:81. doi: 10.3389/fnins.2020.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Stetler RA, Leak RK, Shi Y, Li Y, Yu W, et al. Oxidative stress and DNA damage after cerebral ischemia: Potential therapeutic targets to repair the genome and improve stroke recovery. Neuropharmacology. 2018;134(Pt B):208–17. doi: 10.1016/j.neuropharm.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He W, Huang P, Liu D, Zhong L, Yu R, Li J. Polymorphism of the XRCC1 Gene Is Associated with Susceptibility and Short-Term Recovery of Ischemic Stroke. International journal of environmental research and public health. 2016;13(10):1016. doi: 10.3390/ijerph13101016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He W, Wang Q, Gu L, Zhong L, Liu D. NOX4 rs11018628 polymorphism associates with a decreased risk and better short-term recovery of ischemic stroke. Experimental and therapeutic medicine. 2018;16(6):5258–64. doi: 10.3892/etm.2018.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, et al. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(46):18802–7. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jalland CM, Scheffler K, Benestad SL, Moldal T, Ersdal C, Gunnes G, et al. Neil3 induced neurogenesis protects against prion disease during the clinical phase. Scientific reports. 2016;6:37844. doi: 10.1038/srep37844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min JW, Lu L, Freeling JL, Martin DS, Wang H. USP14 inhibitor attenuates cerebral ischemia/reperfusion-induced neuronal injury in mice. Journal of neurochemistry. 2017;140(5):826–33. doi: 10.1111/jnc.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nature genetics. 2002;32(3):420–5. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 29.Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes & development. 2016;30(15):1718–30. doi: 10.1101/gad.285122.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, et al. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta neuropathologica. 2013;126(2):251–65. doi: 10.1007/s00401-013-1142-5. [DOI] [PubMed] [Google Scholar]
- 32.(IMSGC) IMSGC The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes and immunity. 2009;10(1):11–4. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimaki T, Kato K, Yokoi K, Yoshida T, Oguri M, Watanabe S, et al. Genetic risk for myocardial infarction in Japanese individuals with or without chronic kidney disease. International journal of molecular medicine. 2010;25(5):743–9. doi: 10.3892/ijmm_00000400. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, Kato K, Yokoi K, Oguri M, Watanabe S, Metoki N, et al. Association of genetic variants with myocardial infarction in individuals with or without hypertension or diabetes mellitus. International journal of molecular medicine. 2009;24(5):701–9. doi: 10.3892/ijmm_00000282. [DOI] [PubMed] [Google Scholar]
- 35.Cregg JM, DePaul MA, Filous AR, Lang BT, Tran A, Silver J. Functional regeneration beyond the glial scar. Experimental neurology. 2014;253:197–207. doi: 10.1016/j.expneurol.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, Chopp M. Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Progress in neurobiology. 2016;144:103–20. doi: 10.1016/j.pneurobio.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pekny M, Pekna M. Astrocyte reactivity and reactive astrogliosis: costs and benefits. Physiological reviews. 2014;94(4):1077–98. doi: 10.1152/physrev.00041.2013. [DOI] [PubMed] [Google Scholar]
- 38.Rossi D. Astrocyte physiopathology: At the crossroads of intercellular networking, inflammation and cell death. Progress in neurobiology. 2015;130:86–120. doi: 10.1016/j.pneurobio.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, Wu Y, Xie F, Zhong Y, Wang Y, Xu M, et al. RGMa mediates reactive astrogliosis and glial scar formation through TGFbeta1/Smad2/3 signaling after stroke. Cell death and differentiation. 2018;25(8):1503–16. doi: 10.1038/s41418-018-0058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu L, Bai X, Cao Y, Luo H, Yang X, Kang L, et al. Growth Differentiation Factor 11 Promotes Neurovascular Recovery After Stroke in Mice. Frontiers in cellular neuroscience. 2018;12:205. doi: 10.3389/fncel.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World journal of gastroenterology. 2017;23(34):6330–8. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao J, Zhang T, Jiao X, Huang T, Zhao L, Ma D, et al. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. Journal of cellular physiology. 2020;235(2):1287–95. doi: 10.1002/jcp.29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck H, Plate KH. Angiogenesis after cerebral ischemia. Acta neuropathologica. 2009;117(5):481–96. doi: 10.1007/s00401-009-0483-6. [DOI] [PubMed] [Google Scholar]
- 44.Sun J, Tao S, Liu L, Guo D, Xia Z, Huang M. miR1405p regulates angiogenesis following ischemic stroke by targeting VEGFA. Molecular medicine reports. 2016;13(5):4499–505. doi: 10.3892/mmr.2016.5066. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Translational stroke research. 2014;5(6):711–8. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 46.Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nature medicine. 2011;17(7):796–808. doi: 10.1038/nm.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fann DY, Lim YA, Cheng YL, Lok KZ, Chunduri P, Baik SH, et al. Evidence that NF-κB and MAPK Signaling Promotes NLRP Inflammasome Activation in Neurons Following Ischemic Stroke. Molecular neurobiology. 2018;55(2):1082–96. doi: 10.1007/s12035-017-0394-9. [DOI] [PubMed] [Google Scholar]
- 48.Jean LeBlanc N, Menet R, Picard K, Parent G, Tremblay M, ElAli A. Canonical Wnt Pathway Maintains Blood-Brain Barrier Integrity upon Ischemic Stroke and Its Activation Ameliorates Tissue Plasminogen Activator Therapy. Molecular neurobiology. 2019;56(9):6521–38. doi: 10.1007/s12035-019-1539-9. [DOI] [PubMed] [Google Scholar]
- 49.Song S, Huang H, Guan X, Fiesler V, Bhuiyan MIH, Liu R, et al. Activation of endothelial Wnt/β-catenin signaling by protective astrocytes repairs BBB damage in ischemic stroke. Progress in neurobiology. 2021;199:101963. doi: 10.1016/j.pneurobio.2020.101963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the corresponding author on reasonable request. The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus database (GSE178764, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178764).