Abstract
Background
Lumpy skin disease (LSD) is an important viral disease causing significant economic losses in commercial livestock production. In mid‐2019, an outbreak of LSD has been reported in cattle population from different parts of Bangladesh including Chattogram division. A cross‐sectional surveillance study was undertaken from August 2019 to December 2019 to investigate the prevalence and associated risk factors of LSD in cattle in Chattogram district.
Methods
A total of 3,327 cattle from 19 commercial farms were examined for the LSD specific skin lesions and associated risk factors. A total of 120 skin biopsies were collected from the suspected animal for the confirmation of the disease using molecular detection and histopathological examination. Partial genome sequencing and phylogenetic analyses were performed on selected viral isolates.
Results
The overall clinical prevalence of LSD in the study population was 10% (95% confidence interval [CI]: 9.4%–11%) where the highest farm level outbreak frequency was 63.33% (95% CI: 45.51%–78.13%) and the lowest 4.22% (95% CI: 3.39%–5.25%). Crossbred and female cattle showed a significantly higher prevalence of the disease compared to their counterparts. Introduction of new animals in farms was found to be one of the most significant risk factors in the transmission of the disease. All suspected skin biopsies were positive for LSD virus (LSDV) infection with granulomatous and pyogranulomatous dermatitis was revealed on histopathology. Phylogenetic analysis based on the inverted terminal repeat region of the LSDV gene suggested that the locally circulating strain was closely related to the strains isolated from the Middle East and North African countries.
Conclusions
The data generated in this study would be beneficial to the field veterinarians and animal health decision makers in the country as well as it will aid in taking appropriate measures to prevent further relapse or outbreak of this disease in future.
Keywords: cattle, Chattogram of Bangladesh, lumpy skin disease, phylogenetic analyses, prevalence
Lumpy Skin Disease (LSD) is an important viral disease capable of incurring significant economic losses in commercial livestock production. This investigation summarises the clinical outbreaks of LSD in the commercial cattle population in Chattogram district of Bangladesh unrevealing the disease burden and associated risk factors.
![]()
1. INTRODUCTION
Lumpy skin disease (LSD) is a viral disease caused by LSD virus (LSDV) that belongs to the family Poxviridae and genus Capripoxvirus. The disease affects a wide range of domestic animals including cattle, buffalo, sheep and goats (Alkhamis & VanderWaal, 2016; El‐Nahas et al., 2011), and the main symptoms are fever and nodular lesions on the skin, mucous membrane of respiratory and digestive tracts (Coetzer & Tuppurainen, 2004). The World Organization for Animal Health (OIE) included the disease in notifiable transboundary disease list due to its substantial economic losses in terms of reduced productivity, poor hide quality, poor growth rate, infertility and even death (Anonymous, 2021; Tuppurainen et al., 2017; Tuppurainen & Oura, 2012). LSDV is believed to be transmitted mainly biting arthropods such as mosquitoes, flies and ticks (Magori‐Cohen et al., 2012). Higher incidence of this disease is observed in crossbred young animals with communal grazing and during the wet season when the arthropods vectors are abundant. Introduction of new animals is another important risk factor (Al Rammahi & Jassim, 2015; Alemayehu et al., 2013; Chihota et al., 2001; El‐khabaz, 2014; Kiplagat et al., 2020; Ochwo et al., 2019). Zambia is the first country where LSD was first identified in 1929 that was followed by many African and Middle Eastern countries (Kasem et al., 2018). Although many countries have experienced the outbreak of LSD, but which strain or variants will be the perfect match for vaccine production is remained as a debatable issue (Ayelet et al., 2013; Ben‐Gera et al., 2015). Some recent articles claimed the potentiality of vaccine candidates for LSD prevention (Klement et al., 2020; Wolff et al., 2020; Zhugunissov et al., 2020). In contrast, vaccine strains were also found during the disease outbreak in Russia which raised further doubts on the vaccine candidate and its efficacy (Kononov et al., 2019).
In Bangladesh, an outbreak of an unknown syndrome with nodular skin lesions was reported by local veterinary services authority in mid‐2019 in commercial and backyard cattle population in some locations (Anwara, Karnaphuli and Patiya) of Chattogram district (Anonymous, 2019). Same pattern of clinical onset was reported later in different districts of the country (Giasuddin et al., 2020; Khalil et al., 2021). The outbreak report was preliminary confirmed based on clinical signs and later using the reverse transcription polymerase chain reaction (RT‐PCR) test by the Department of Livestock Services (DLS), Bangladesh and notified the disease as LSD to OIE in August, 2019 (Anonymous, 2019). Therefore, a cross‐sectional surveillance study was undertaken on clinically suspected LSD cases throughout Chattogram district; the south‐eastern part of Bangladesh. The aim of the present study was to confirm the disease occurrence based on clinical, molecular and pathological identification and unveiled the plausible risk factors of LSDV infection in this region. We further analysed the sequence data of the circulating LSDV strains to identify the probable geographical origin of this strain.
2. MATERIALS AND METHODS
2.1. Study design
The study was conducted over a period of 5 months (August to December 2019) in Chattogram District of Bangladesh at the onset of a lumpy skin disease outbreak. A cross‐sectional study was designed to collect the samples, and individual animal was considered as the sampling unit. A standard questionnaire was used to collect demographic data such as breed, age, sex and other data (e.g., introduction of new animals, source of water supply in the farm, etc.). Selected animals were categorized as Holstein Friesian crossbred (Bos taurus X B. indicus) and zebu cattle (B. indicus). Age of the animals was categorized as calf: ≤1 year; heifer: >1 to ≤2.5 years for crossbred and >1 to ≤3.5 years for indigenous cattle; cow: >2.5 years for crossbred and >3.5 years for indigenous cattle and bull (≥1 year) (Alim et al., 2012). Selection of study areas and animals were based on the suspected cases reported by the local veterinarians and physical visit to the farms. A case was considered positive for LSD when an animal showed two or more of the following signs such as nodular characteristic lesions on the skin, fever, lameness, lymphadenopathy, edema and decreased production (e.g., reduction of milk yield) (Magori‐Cohen et al., 2012). A total of 19 commercial farms from Chattogram district (six farms from Pahartali area, three farms from Sitakunda and two farms from each of Chattogram Port, Double Mooring, Hathazari, Panchlaish and Chadgaon area) were selected (Figure 2). Farms comprising less than 15 cattle were excluded from the present survey (Sikder et al., 2012). Sample from affected animals was collected from the individual farms using a simple randomization technique. A farm was considered positive for ectoparasites (flies, ticks, lice) when at least one animal was infested by one of these parasites.
FIGURE 2.
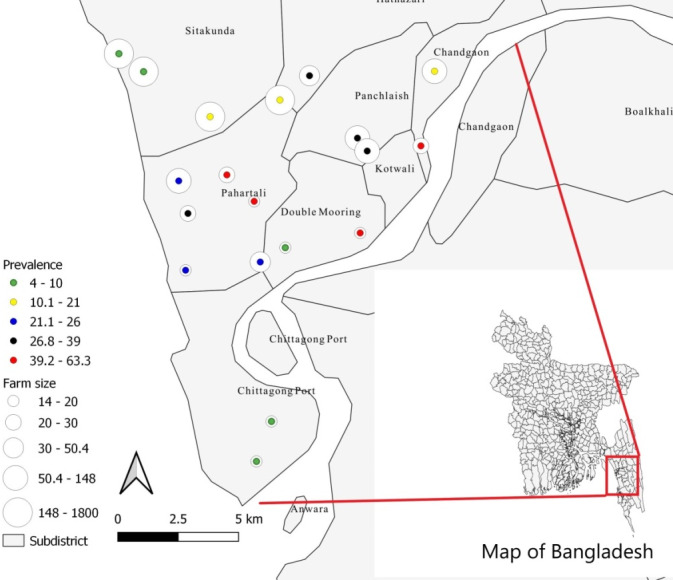
Map showing the location and farm size (circle) along with the number infected animals (farm level frequency, %) in each farm
2.2. Sample collection and preservation
A total of 19 farms having 3,327 animals were considered consisting of 669 calves, 281 heifers, 2,272 cows and 105 bulls. Data were collected by face to face interview of the animal attendants of the particular farm and physical examination of the cattle. Among the diseased or suspected cattle (Figure 1a,b), a total of 120 skin biopsies from nodular lesions were collected aseptically using punch biopsy techniques (Kasem et al., 2018). Briefly, the biopsy site was shaved by the sterile blades, and a small punch was taken deeply in the skin so that all layers along with the subcutaneous tissue were collected. Half of the skin biopsy specimen was kept in neutral buffered formalin (10%) for histological examination following conventional haematoxylin and eosin (H&E) staining (Fischer et al., 2008). The rest half of the skin biopsy samples were preserved in −20°C for molecular confirmation of the infection.
FIGURE 1.
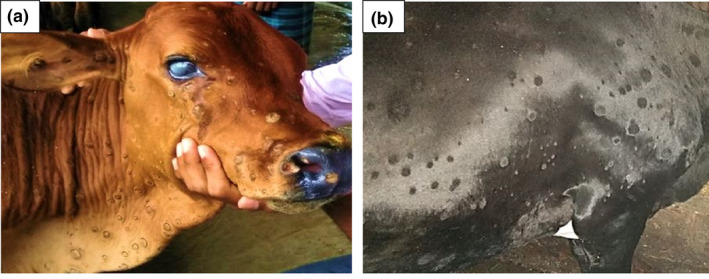
Nodular lesions of LSD affected calf (a) and cow (b)
2.3. DNA extraction and PCR presence of LSDV
Total genomic DNA was extracted from all suspected skin biopsies using commercially available kits following manufacturer's instruction with some modifications (DNeasy Blood & Tissue Kits®). PCR was then performed to confirm the presence of LSDV using a set of published primers (forward; GTGGAAGCCAATTAAGTAGA and reverse; GTAAGAGGGACATTAGTTCT) targeting the inverted repeat region (ITR) of the genome (Stram et al., 2008). In brief, PCR reactions were set up in 50‐μl final volumes containing 25‐μl master mix, 2.5‐μl forward primer, 2.5‐μl reverse primer, 5.0‐μl DNA template and 15‐μl nuclease free water. The PCR conditions were follows as an initial denaturation step of 95°C for 1 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 49°C for 30 s, extension at 72°C for 70 s and a final extension step at 72°C for 5 min. Then, 5‐μl of amplified amplicons were taken and stained using 0.05% ethidium bromide (Sigma‐Aldrich®) and a visualization of the band (1,237 bp) after agarose gel (1%) electrophoresis.
2.4. Nucleotide sequencing and phylogenetic analysis
Four randomly selected LSDV PCR amplicons were gel purified using Wizard® SV Gel and PCR Clean‐Up System (Promega) and sequenced by sanger dideoxy sequencing (Macrogen®). The sequence read data were then manually cleaned up using the chromatogram software (Geneious Prime version 2020) and deposited in GenBank. NCBI BLAST was performed for each of the sequence reads and the ITR from a diverse range related LSDV and other poxvirus genome sequences (Nt sequence identity 100%–70%) were retrieved (N = 63) and aligned with MAFFT v7.017 using G‐INS‐i (gap open penalty 1.53; offset value 0.123) alignment algorithm (Katoh et al., 2002). jModelTest program 2.1.3 favoured a general‐time‐reversible model with gamma distribution rate variation and a proportion of invariable sites (GTR + I + G4) for the phylogeny (Darriba et al., 2012). Maximum likelihood (ML) phylogenetic trees was reconstructed using the program PhyML v3.1 (Guindon & Gascuel, 2003), and FigTree v1.4 was used to generate the consensus tree (Smith et al., 2009). The proportion of bootstrap support (%) was showed in each branch while multiple taxa showing polytomy, and closely related isolates were collapsed for better visualization.
2.5. Statistical analysis
All data were inserted and coded in Microsoft office Excel 2016 spreadsheet, and both univariable and multivariable analyses were performed using generalized linear mixed models in STATA‐IC 13. Farm was included in the model as random effect. Backward elimination procedure was followed, and a p value ≤ 0.05 was considered significant in both univariable and multivariable analyses. Prevalence map along with location and size of the farms was created using QGIS 3.12.0.
3. RESULTS
3.1. Clinical prevalence of LSD
The overall clinical prevalence of LSD was 10% (95% CI: 9.4%–11%) in the study farms. The farm level highest frequency was 63.33% in one of the farms located in Chadgaon region, and the lowest was 4.22% in a farm at Sitakunda region of Chattogram District (Figure 2). The clinical prevalence ranges from 20% to 40% in five farms of Pathartali, two farms of Panchlaish, and one farm of Hathazari and Chandgaon area of Chattogram district. This clinical prevalence ranges 41%–63% in two farms of Chattogram port area and one farm of Pahartali, Double mooring and Chandgaon area. Three farms of Sitakunda and one farm of Pahartali and Double mooring area of the same district showed the clinical prevalence below 10% (Figure 2).
3.2. Risk factors associated with the occurrence of LSD
The clinical prevalence was observed the lowest in bulls (5%) (95% CI: 2%–10%). Univariable analysis showed that odds ratio (OR) of having the disease in calves, cows and heifers were 1.37 (CI: 0.53–3.55), 2.52 (CI: 1.02–6.26) and 3.51(CI: 1.35–9.14) times higher compared to bulls, respectively (Table 1). Females were in higher risk (OR = 2.26, CI: 1.28–4.0) than males. In terms of lactation, with increasing lactation number decrease in prevalence was observed; odds of having the disease in first lactation was 7 times higher compared to fourth lactation. The univariable analysis also showed that local cattle were less susceptible than the crossbred. Besides, introduction of new animals, sources of water supply and floor types (brick or cemented) in the farm act as potential risk factors of the disease (Table 1). In multivariable model, crossbred (p = .0080, OR = 3.58, CI: 1.40–9.17) and female (p ≤ .0001, OR = 3.96, CI: 2.16–7.27) cattle had a significantly higher risk of getting the disease compared to their counterparts (Table 2).
TABLE 1.
Risk factors associated with lumpy skin disease in cattle farms of Chattogram district of Bangladesh from the univariable logistic regression analysis
| Variables | Level | N (animals) | Positive N (%) | OR (95% CI) | p value |
|---|---|---|---|---|---|
| Breed | Cross | 3,220 | 340 (11) | 2.40 (0.97–5.95) | .05 |
| Local | 107 | 5 (5) | Ref | ||
| Animal category | Calf | 669 | 43 (6) | 1.37 (0.53–3.55) | <.0001 |
| Heifer | 281 | 42 (15) | 3.51 (1.35–9.14) | ||
| Cow | 2,272 | 255 (11) | 2.52 (1.02–6.26) | ||
| Bull | 105 | 5 (5) | Ref | ||
| Sex | Female | 3,071 | 332 (11) | 2.26 (1.28–4.00) | 0 |
| Male | 256 | 13 (5) | Ref | ||
| Lactationa | 1 | 107 | 98 (92) | 7.70 (6.04–9.37) | <.0001 |
| 2 | 267 | 105 (39) | 4.25 (2.77–5.73) | ||
| 3 | 1,780 | 50 (3) | 1.10 (0.37–2.57) | ||
| 4 | 118 | 2 (2) | Ref | ||
| Introduction of new animals | Yes | 62 | 13 (21) | 2.34 (1.25–4.36) | .007 |
| No | 3,265 | 332 (10) | Ref | ||
| Water source | Pond | 50 | 14 (28) | 3.46 (1.84–6.48) | <.0001 |
| Underground (tubewell) | 3,277 | 331 (10) | Ref | ||
| Floor | Brick | 72 | 13 (18) | 1.93 (1.05–3.57) | .03 |
| Cemented | 3,255 | 332 (10) | Ref | ||
| Overall | 3,327 | 345 (10) |
Abbreviations: CI, confidence interval; OR, odds ratio.
This OR was calculated only including lactating cows.
TABLE 2.
Risk factors associated with lumpy skin disease in cattle farms of Chattogram district of Bangladesh using a logistic regression analysis
| Variables | Level | Estimates | SEM | OR (95% CI) | p value |
|---|---|---|---|---|---|
| Intercept | −3.71 | ||||
| Breed | Cross | 1.277 | 0.605 | 3.58 (1.40–9.17) | .0080 |
| Local | 0 | Ref | |||
| Sex | Female | 1.377 | 0.479 | 3.96 (2.16–7.27) | <.0001 |
| Male | 0 | Ref | |||
| Random effect of farm | 1.003 | 0.186 |
Abbreviations: CI, confidence interval; OR, odds ratio; SEM, standard error of the mean.
3.3. Molecular identification of LSDV
All of the collected skin biopsies were PCR positive for LSDV. Among them, a total of 4 samples were sequenced randomly (GenBank accession: MT070969‐72). The ML tree reconstructed from the ITR region of closely related poxviruses revealed that most LSD_CVASU isolates belong to a strongly supported (100% bootstrap value) clade dominated by LSDV strains. LSDV isolated from different regions of the world (Africa and Middle East) over three decades (1997–2019) timeframe. Isolate LSD_CVASU_M1 and M3 (MT070969 and MT070971) together formed a sister branch to the LSDV isolate from Egypt (EU350218) with a moderate bootstrap support (55%), while for isolate M4 (MT070972), the phylogenetic resolution was not clear and demonstrated some relatedness (58% bootstrap support) with sheep pox reference sequence (CAPIS1ITR) (Figure 3). On the other hand, Isolate M2 on the other hand showed stronger bootstrap support (80%) towards recent isolates of LSDV from Egypt (KF588352, KR052866 and KF58835). The phylogenetic reconstruction thus reaffirm that the viral isolates from the nodular skin biopsies were LSDV genotypes most closely related to those from Egypt (Figure 3).
FIGURE 3.
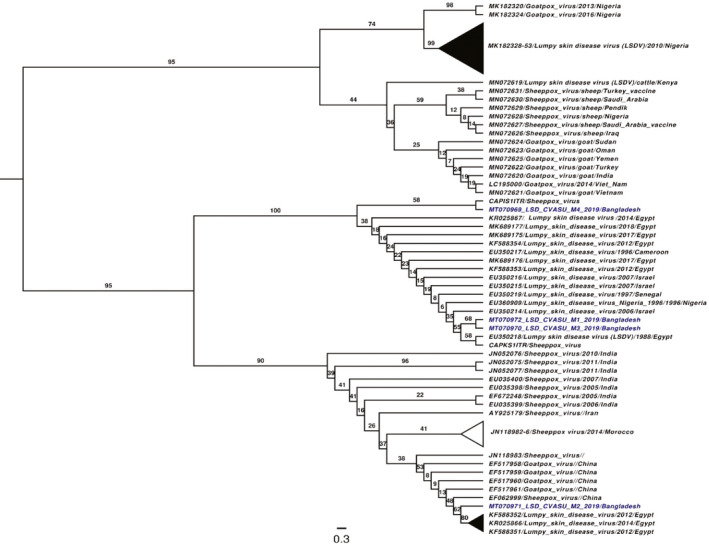
Maximum likelihood (ML) tree rooted at midpoint with proportionally arranged branches based on the ITR region of poxvirus genome demonstrating phylogenetic relatedness of LSDV isolates form Chattogram, Bangladesh (blue taxa). Clades suggesting polytomies were collapsed, and shown in cartoon, the bootstrap statistics (percentage) were shown as branch support numbers
3.4. Histological features of the skin biopsies
All cases involved granulomatous and pyogranulomatous dermatitis with multifocal to diffuse deep dermal necrosis and panniculitis (Figure 4). The superficial and deep dermis was infiltrated with variable numbers of lymphocytes, plasma cells, macrophages and relatively fewer neutrophils. However, multifocal small dermal abscesses also observed in the samples. Acanthosis and orthokertatotic hyperkeratosis were common features of the nodules with instances of epidermal ulceration. Hair follicles of the skin biopsies were partially destroyed and replaced by necrotic epithelium, mixed cellular infiltrates and keratinaceous debris from ruptured follicles (furunculosis) (Figure 4). Pannicular infarction and subcutaneous vasculitis were present in the samples.
FIGURE 4.
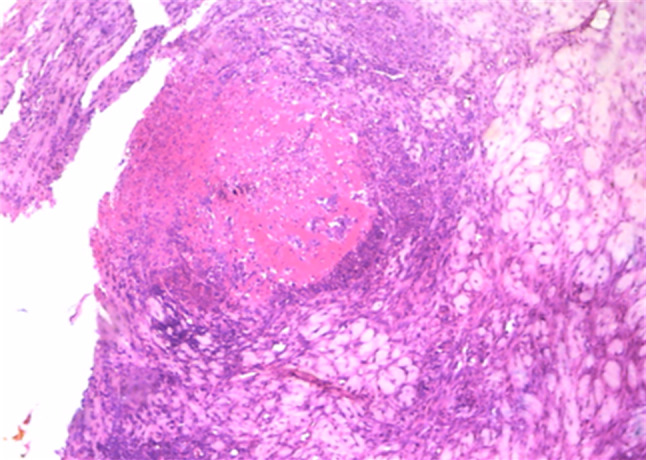
Histological feature of LSD affected nodular lesions. Figure demonstrated deep dermis and subcutis; focal granulomatous lesion comprised of necrotic debris and encircling mononuclear cell infiltration (magnification of image 100×)
4. DISCUSSION
Bangladesh was free of LSD before mid of 2019, and the very first LSDV infection was reported in Anwara, Karnaphuli and Patiya Upazila (subdistrict), Chattogram to OIE in August 2019 (Anonymous, 2019). The present investigation summarises the clinical outbreaks of LSD in the commercial cattle population in Chattogram District unrevealing the disease burden and associated risk factors. The epidemiological data were supported by histopathological features of the clinically characteristic nodular skin lesions as well as PCR‐based molecular identification and phylogenetic analyses.
The overall clinical prevalence of LSD in Chattogram District was 10% similar to some previous studied in Saudi Arabia, Ethiopia and Turkey who reported 6%–12% prevalence in their cattle population (Abera et al., 2015; Al‐Salihi & Hassan, 2015; Kasem et al., 2018; Şevik & Doğan, 2017). Body et al., (2012) observed a much higher prevalence (27.9%) in cattle of Oman which was higher than the overall prevalence of the current study. The higher or lower prevalence of disease might have been influenced by many factors such as geography, farm management and biosecurity, seasons, availability of arthropods vectors, importation of animal from infected areas, disposal of the dead animals. Although we didn't observe any mortality in the study population, some of the previous studies reported 0.99%–2.12% of mortality (Gari et al., 2010; Kasem et al., 2018). Comparatively shorter duration of the actual study period and culling of diseased animals might be a reason for the paucity of mortality. However, clinical form of LSD is generally associated with economic loss in terms of production and treatment expenditure (Babiuk et al., 2008a; 2008b).
Risk factors analyses suggests that crossbred cattle were more susceptible to LSD than indigenous cattle which was consistent with the findings of previous studies (Al Rammahi & Jassim, 2015; Kiplagat et al., 2020; Klement et al., 2018). Higher susceptibility of crossbred cattle might be due to lower disease resistance capability in comparison to indigenous breeds (Tageldin et al., 2014). Further, the higher number of crossbred animals (96.79%) was sampled over local (3.21%) cattle might explain the variation of the results. Heifers were affected largely with LSD in comparison to bulls, calves and cows. In previous studies, a higher morbidity was recorded in younger cattle (<2 years) in Saudi Arabia (Kasem et al., 2018) and calves (0.5–1 year) in Ethiopia (Molla et al., 2018). This might be due to management system of the farms where heifer was kept in poor hygienic conditions in comparison to other animals (calf, cow or bull). Females were more prone to LSD compared to males which was consistent with previous research (Ayelet et al., 2014; Magori‐Cohen et al., 2012; Salib & Osman, 2011). Higher frequency of LSD in female cattle could be due to their exposure to many stress conditions, e.g., pregnancy, parturition and sometimes less amount of feed supplied compared to their actual requipment (Kasem et al., 2018). We observed an inverse relationship with lactation number in the occurrence of LSD in cattle although we were unable to identify the possible reasons. Farm‐specific risk factors such as introduction of new animals to the farm demonstrated a significantly higher risk to be infected with the virus or its transmission which was supported by previous findings (Gari et al., 2010; Macpherson, 1994; Munyeme et al., 2008).
The histopathological features of the suspected nodular skin biopsies demonstrated granulomatous and pyogranulomatous dermatitis with vasculitis and pannicular involvement (Figure 4) which are merely non‐specific lesions. However, similar histological features of suspected skin lesions were documented in many prior studies with confirmed LSD cases (Abdallah et al., 2018; Body et al., 2012; El‐khabaz, 2014; Stram et al., 2008). Unsurprisingly, we could not identify any intracytoplasmic inclusion bodies or so‐called ‘sheep pox’ cells (SPCs) or ‘cellules claveleuses’ of Borrel in any of the skin biopsies (El‐Neweshy et al., 2013). Although SPCs or intracytoplasmic inclusions often considered confirmatory histopathological findings for LSD (Abdallah et al., 2018; Body et al., 2012), they are rarely found in natural LSD cases (El‐Neweshy et al., 2013; House et al., 1990) and often associated only with acute phase infections. In the present study, all tissue sections were stained with H&E, and the histological features leaned towards subacute to chronic stage infections. Future studies should incorporate immunohistochemistry of the tissue section using anti‐LSDV monoclonal antibodies to reveal replicating virus particles in macrophages and epithelial cells of dermis.
The PCR‐based molecular test targeting ITR region of the LSDV has successfully confirmed all suspected cases of LSD in this study, and the local genotype circulating in Chattogram district was deposited in GenBank as well (Gene bank accession no. MT070969‐MT070972). The ML tree reconstructed from the ITR region of all related poxviruses showed that the LSDV strains circulating in Bangladesh are closely related to that in Middle East and North Africa as three out of four sequences had closest phylogenetic relationship with isolates from Egypt. However, the ITR region of the genome used for amplification and sequencing of LSDV is a pseudogene, relatively conserved and homologous to many other poxvirus genomes (Gershoni & Black, 1989). Therefore, the phylogenetic reconstruction had lack of discriminatory resolution, as presented by relatively low bootstrap supports in many branches and positioning LSDV genotypes with sheep pox and goat poxvirus strains in some clades (Figure 3). This limitation might have imposed a negative implication defining the possible source of the outbreak based on evolutionary relatedness of geographically distant strains. Further studies should incorporate sequencing at least three different core gene groups along with concatenation or partitioning approach for alignment and subsequent phylogenetic analyses to reconstruct a comprehensive evolutionary tree with better discriminatory power and resolution.
In Bangladesh, there is no previous outbreak of LSD in any of the susceptible species including cattle. Many factors might have been involved with the current outbreak and transmission across the country. There are both legal and illegal cattle trading occurs every year from neighbouring countries, namely, India and Myanmar. Further, throughout the year, livestock mobility across the country is quite high which usually reaches a peak during Eid‐ul‐Adha (a holy festival of Muslim) as thousands of temporary wet markets are established to meet the demand (Khatun et al., 2016). It is also mentionable that this outbreak was reported just a month after the festival. It is plausible that unregulated and illegal import of live animals without prior health check or quarantine measures have embarked the clinical outbreak of LSD. Unrestricted in‐country movements of livestock even after the first reporting might have significantly aggravated the viral transmission (Tuppurainen et al., 2017). However, outbreak of this disease occurred in China and Odisha of India in August 2019 (Anonymous, 2019; Sudhakar et al., 2020), and this could be an unexplored link to this outbreak as cattle movements were speculated as a risk factor (Klausner et al., 2017). Within farm LSDV transmission is further related with the biosecurity measures and other management practices. We found a positive correlation between the communal water supply as well as the floor made of brick as observed by others (Babiuk et al., 2008a; 2008b; Tuppurainen & Oura, 2012). We have observed ectoparasites in almost all the farms which may play a role in the transmission of this virus as reported by previous research (Ince et al., 2016). Future research should be directed for identification of the specific vectors to overcome the limitation of this study. However, the present study may have some inherent limitations of cross‐sectional study despite designed carefully. Particularly, this study design might lead to selection bias, recall bias and temporal sequences between exposure and outcome cannot be evaluated. We recorded most of the clinical data by visual observation that might have minimized the recall bias and major number of variables would not be affected by the temporal sequence, such as sex, breed, type of animal, etc.
In summary, the current study investigated the outbreak of LSDV infection in commercial farms of Bangladesh unveiling the overall clinical prevalence and risk factors associated the disease. This study also suggests a plausible source of the outbreak based on limited genomic data and evolutionary assays. As there is no effective vaccine of this economically important disease, further research should be focused on the molecular characterization of the whole genome of the local strain of LSDV for developing a suitable vaccine candidate. The data generated in this study would be beneficial to the field veterinarians and animal health decision makers in Bangladesh, and also it will aid in taking appropriate measures to prevent further relapse or outbreak of this disease in future.
5. ETHICAL APPROVAL STATEMENT
Ethical approval was obtained from the institutional ethical approval committee of Chattogram Veterinary and Animal Sciences University (CVASU) [CVASU/Dir (R&E) EC/2019/126(13)].
CONFLICT OF INTEREST
There are no conflicts of interest between the authors.
AUTHOR CONTRIBUTION
Farazi Muhammad Yasir Hasib: Data curation; Investigation; Methodology; Project administration; Visualization; Writing‐original draft; Writing‐review & editing. Mohammad Sirazul Islam: Data curation; Investigation; Methodology; Project administration; Writing‐original draft. Tridip Das: Data curation; Formal analysis; Methodology. Eaftekhar Ahmed Rana: Formal analysis; Investigation; Methodology. Mohammad Helal Uddin: Data curation; Investigation; Methodology. Mohammad Bayzid: Data curation; Investigation; Methodology. Chandan Nath: Data curation; Investigation; Methodology. Mohammad Alamgir Hossain: Conceptualization; Funding acquisition; Resources; Validation; Visualization; Writing‐review & editing. Mohammad Masuduzzaman: Conceptualization; Funding acquisition; Resources; Visualization; Writing‐review & editing. Shubhagata Das: Conceptualization; Formal analysis; Methodology; Resources; Software; Validation; Writing‐original draft; Writing‐review & editing. Md Abdul Alim: Conceptualization; Data curation; Funding acquisition; Investigation; Methodology; Project administration; Supervision; Validation; Writing‐original draft; Writing‐review & editing.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.524.
ACKNOWLEDGEMENTS
Authors would like thanks to the University Grants Comission and Chattogram Veterinary and Animal Sciences University (CVASU), Bangladesh for funding to this project. We also would like to acknowledge Professor Dr. Sharmin Chowdhury, Department of Pathology and Parasitology, Faculty of Veterinary Medicine, CVASU for analyses of the data and review of the manuscript.
Hasib FMY, Islam MS, Das T, et al. Lumpy skin disease outbreak in cattle population of Chattogram, Bangladesh. Vet Med Sci. 2021;00:1616–1624. 10.1002/vms3.524
DATA AVAILABILITY STATEMENT
Data files associating this research are available online (https://doi.org/10.6084/m9.figshare.12619868.v1).
REFERENCES
- Abdallah, F. M., El Damaty, H. M., & Kotb, G. F. (2018). Sporadic cases of lumpy skin disease among cattle in Sharkia province, Egypt: Genetic characterization of lumpy skin disease virus isolates and pathological findings. Veterinary World, 11(8), 1150–1158. 10.14202/vetworld.2018.1150-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abera, Z., Degefu, H., Gari, G., & Kidane, M. (2015). Sero‐prevalence of lumpy skin disease in selected districts of West Wollega zone, Ethiopia. BMC Veterinary Research, 11(1), 1–9. 10.1186/s12917-015-0432-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rammahi, H. M., & Jassim, A. (2015). Epidemiological and diagnostic study of first lumpy skin disease outbreak in southern Baghdad district. Asian Academic Research Journal of Multidisciplinary, 1(30), 196–205. [Google Scholar]
- Alemayehu, G., Zewde, G., & Admassu, B. (2013). Risk assessments of lumpy skin diseases in Borena bull market chain and its implication for livelihoods and international trade. Tropical Animal Health and Production, 45(5), 1153–1159. 10.1007/s11250-012-0340-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alim, M. A., Das, S., Roy, K., Masuduzzaman, M., Sikder, S., Hassan, A. Z., & Hossain, M. A. (2012). Prevalence of hemoprotozoan diseases in cattle population of Chittagong division, Bangladesh. Pakistan Veterinary Journal, 32(2), 221–224. [Google Scholar]
- Alkhamis, M. A., & VanderWaal, K. (2016). Spatial and temporal epidemiology of lumpy skin disease in the Middle East, 2012–2015. Frontiers in Veterinary Science, 3, 19. 10.3389/fvets.2016.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Salihi, K. A., & Hassan, I. Q. (2015). Lumpy skin disease in Iraq: Study of the disease emergence. Transboundary and Emerging Diseases, 62(5), 457–462. 10.1111/tbed.12386 [DOI] [PubMed] [Google Scholar]
- Anonymous . (2019). Situation Report: Lumpy skin disease in Bangladesh. https://fscluster.org/sites/default/files/documents/sitrep_lsd_20191210.pdf [Google Scholar]
- Anonymous . (2021), OIE‐Listed diseases, infections and infestations in force in 2021. https://www.oie.int/en/animal‐health‐in‐the‐world/oie‐listed‐diseases‐2021/ [Google Scholar]
- Ayelet, G., Abate, Y., Sisay, T., Nigussie, H., Gelaye, E., Jemberie, S., & Asmare, K. (2013). Lumpy skin disease: Preliminary vaccine efficacy assessment and overview on outbreak impact in dairy cattle at Debre Zeit, central Ethiopia. Antiviral Research, 98(2), 261–265. 10.1016/j.antiviral.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Ayelet, G., Haftu, R., Jemberie, S., Belay, A., Gelaye, E., Sibhat, B., Skjerve, E., & Asmare, K. (2014). Lumpy skin disease in cattle in central Ethiopia: Outbreak investigation and isolation and molecular detection of the virus. Revue Scientifique Et Technique De l'OIE, 33(3), 877–887. [DOI] [PubMed] [Google Scholar]
- Babiuk, S., Bowden, T. R., Boyle, D. B., Wallace, D. B., & Kitching, R. P. (2008). Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transboundary and Emerging Diseases, 55(7), 263–272. 10.1111/j.1865-1682.2008.01043.x [DOI] [PubMed] [Google Scholar]
- Babiuk, S., Bowden, T. R., Parkyn, G., Dalman, B., Manning, L., Neufeld, J., Embury‐Hyatt, C., Copps, J., & Boyle, D. B. (2008). Quantification of lumpy skin disease virus following experimental infection in cattle. Transboundary and Emerging Diseases, 55(7), 299–307. 10.1111/j.1865-1682.2008.01024.x [DOI] [PubMed] [Google Scholar]
- Ben‐Gera, J., Klement, E., Khinich, E., Stram, Y., & Shpigel, N. Y. (2015). Comparison of the efficacy of Neethling lumpy skin disease virus and x10RM65 sheep‐pox live attenuated vaccines for the prevention of lumpy skin disease–The results of a randomized controlled field study. Vaccine, 33(38), 4837–4842. 10.1016/j.vaccine.2015.07.071 [DOI] [PubMed] [Google Scholar]
- Body, M., Singh, P. K., Hussain, H. M., Al‐rawahi, A., Al‐maawali, M., Al‐lamki, K., & Al‐habsy, S. (2012). Clinico‐histopathological findings and PCR based diagnosis of lumpy skin disease in the Sultanate of Oman. Pakistan Veterinary Journal, 32(2), 206–210. [Google Scholar]
- Chihota, C. M., Rennie, L. F., Kitching, R. P., & Mellor, P. S. (2001). Mechanical transmission of lumpy skin disease virus by Aedes . Epidemiology and Infection, 126, 317–321. 10.1017/S0950268801005179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzer, J. A. W., & Tuppurainen, E. (2004). Lumpy skin disease. Infectious Diseases of Livestock, 1268–1276. [Google Scholar]
- Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐khabaz, K. A. S. (2014). Rapid laboratory diagnosis of lumpy skin disease by using PCR technique. Assiut Veterinary Medicine Journal, 60(143), 37–41. [Google Scholar]
- El‐Nahas, E. M., El‐Habbaa, A., El‐Bagoury, G., & Radwan, M. E. (2011). Isolation and identification of lumpy skin disease virus from naturally infected buffaloes at Kaluobia, Egypt. Global Veterinaria, 7(3), 234–237. [Google Scholar]
- El‐Neweshy, M. S., El‐Shemey, T. M., & Youssef, S. A. (2013). Pathologic and immunohistochemical findings of natural lumpy skin disease in Egyptian cattle. Pakistan Veterinary Journal, 33(1), 60–64. [Google Scholar]
- Fischer, A. H., Jacobson, K. A., Rose, J., & Zeller, R. (2008). Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harbor Protocols, 2008(5), pp.pdb‐prot4986. 10.1101/pdb.prot4986 [DOI] [PubMed] [Google Scholar]
- Gari, G., Waret‐Szkuta, A., Grosbois, V., Jacquiet, P., & Roger, F. (2010). Risk factors associated with observed clinical lumpy skin disease in Ethiopia. Epidemiology and Infection, 138(11), 1657–1666. 10.1017/S0950268810000506 [DOI] [PubMed] [Google Scholar]
- Gershoni, P. D., & Black, D. N. (1989). A capripoxvirus pseudogene whose only intact homologs are in other poxvirus genomes. Virology, 172(1), 350–354. 10.1016/0042-6822(89)90138-4 [DOI] [PubMed] [Google Scholar]
- Giasuddin, M., Yousuf, M., Hasan, M., Rahman, M., Hassan, M., & Ali, M. (2020). Isolation and molecular identification of Lumpy Skin Disease (LSD) virus from infected cattle in Bangladesh. Bangladesh Journal of Livestock Research, 26(1–2), 15–20. 10.3329/bjlr.v26i1-2.49933 [DOI] [Google Scholar]
- Guindon, S., & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52(5), 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- House, J. A., Wilson, T. M., Nakashly, S. E., Karim, I. A., Ismail, I., Danaf, N. E., Moussa, A. M., & Ayoub, N. N. (1990). The isolation of lumpy skin disease virus and bovine herpesvirus‐ from cattle in Egypt. Journal of Veterinary Diagnostic Investigation, 2(2), 111–115. 10.1177/104063879000200205 [DOI] [PubMed] [Google Scholar]
- Ince, Ö. B., Çakir, S., & Dereli, M. A. (2016). Risk analysis of lumpy skin disease in Turkey. Indian Journal of Animal Research, 50(6), 1013–1017. 10.18805/ijar.9370 [DOI] [Google Scholar]
- Kasem, S., Saleh, M., Qasim, I., Hashim, O., Alkarar, A., Abu‐Obeida, A., Gaafer, A., Hussien, R., AL‐Sahaf, A., Al‐Doweriej, A., Bayoumi, F., Hodhood, A., & Abdelatif, M. (2018). Outbreak investigation and molecular diagnosis of lumpy skin disease among livestock in Saudi Arabia 2016. Transboundary and Emerging Diseases, 65(2), e494–e500. 10.1111/tbed.12769 [DOI] [PubMed] [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K., & Miyata, T. (2002). MAFFT: A novel method for rapid MAFFT: Multiple sequence alignment based on fast Fourier transforms. Nucleic Acids Research, 30(14), 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, M. I., Sarker, M. F. R., Hasib, F. M. Y., & Chowdhury, S. (2021). Outbreak investigation of lumpy skin disease in dairy farms at Barishal, Bangladesh. Turkish Journal of Agriculture ‐ Food Science and Technology, 9(1), 205–209. 10.24925/turjaf.v9i1.205-209.3827 [DOI] [Google Scholar]
- Khatun, R., Ahmed, S., Hasan, M. A., Islam, M. N., Uddin, A. S. M. A., & Mahmud, M. S. (2016). Baseline survey on cattle imports through different peripheral areas of Bangladesh. American Journal of Experimental Agriculture, 13(6): 1‐9. [Google Scholar]
- Kiplagat, S. K., Kitala, P. M., Onono, J. O., Beard, P. M., & Lyons, N. A. (2020). Risk factors for outbreaks of lumpy skin disease and the economic impact in cattle farms of Nakuru County, Kenya. Frontiers in Veterinary Science, 7, 259. 10.3389/fvets.2020.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner, Z., Fattal, E., & Klement, E. (2017). Using synoptic systems' typical wind trajectories for the analysis of potential atmospheric long‐distance dispersal of lumpy skin disease virus. Transboundary and Emerging Diseases, 64(2), 398–410. 10.1111/tbed.12378 [DOI] [PubMed] [Google Scholar]
- Klement, E. (2018). Economic impact of lumpy skin disease. Lumpy skin disease. Springer. 10.1007/978-3-319-92411-3_3 [DOI] [Google Scholar]
- Klement, E., Broglia, A., Antoniou, S.‐E., Tsiamadis, V., Plevraki, E., Petrović, T., Polaček, V., Debeljak, Z., Miteva, A., Alexandrov, T., Marojevic, D., Pite, L., Kondratenko, V., Atanasov, Z., Gubbins, S., Stegeman, A., & Abrahantes, J. C. (2020). Neethling vaccine proved highly effective in controlling lumpy skin disease epidemics in the Balkans. Preventive Veterinary Medicine, 181, 104595. 10.1016/j.prevetmed.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Kononov, A., Byadovskaya, O., Kononova, S., Yashin, R., Zinyakov, N., Mischenko, V., Perevozchikova, N., & Sprygin, A. (2019). Detection of vaccine‐like strains of lumpy skin disease virus in outbreaks in Russia in 2017. Archives of Virology, 164(6), 1575–1585. 10.1007/s00705-019-04229-6 [DOI] [PubMed] [Google Scholar]
- Macpherson, C. N. L. (1994). The effect of transhumance of the epidemiology of animal diseases, 1994. Kenya Veterinarian, 18(2), https://www.sciencedirect.com/science/article/pii/0167587795005390 [Google Scholar]
- Magori‐Cohen, R., Louzoun, Y., Herziger, Y., Oron, E., Arazi, A., Tuppurainen, E., Shpigel, N. Y., & Klement, E. (2012). Mathematical modelling and evaluation of the different routes of transmission of lumpy skin disease virus. Veterinary Research, 43(1), 10.1186/1297-9716-43-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla, W., Frankena, K., Gari, G., Kidane, M., Shegu, D., & de Jong, M. C. M. (2018). Seroprevalence and risk factors of lumpy skin disease in Ethiopia. Preventive Veterinary Medicine, 160, 99–104. 10.1016/j.prevetmed.2018.09.029 [DOI] [PubMed] [Google Scholar]
- Munyeme, M., Muma, J. B., Skjerve, E., Nambota, A. M., Phiri, I., Samui, K. L., Dorny, P., & Tryland, M. (2008). Risk factors associated with bovine tuberculosis in traditional cattle of the livestock/wildlife interface areas in the Kafue basin of Zambia. Preventive Veterinary Medicine, 85(3–4), 317–328. 10.1016/j.prevetmed.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Ochwo, S., VanderWaal, K., Munsey, A., Nkamwesiga, J., Ndekezi, C., Auma, E., & Mwiine, F. N. (2019). Seroprevalence and risk factors for lumpy skin disease virus seropositivity in cattle in Uganda. BMC Veterinary Research, 15(1), 236. 10.1186/s12917-019-1983-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salib, F. A., & Osman, A. H. (2011). Incidence of lumpy skin disease among Egyptian cattle in Giza Governorate, Egypt. Veterinary World, 4(4), 162–167. 10.5455/vetworld.2011.162-167 [DOI] [Google Scholar]
- Şevik, M., & Doğan, M. (2017). Epidemiological and molecular studies on lumpy skin disease outbreaks in turkey during 2014–2015. Transboundary and Emerging Diseases, 64(4), 1268–1279. 10.1111/tbed.12501 [DOI] [PubMed] [Google Scholar]
- Sikder, S., Rahman, A. K. M. A., Faruque, M. R., Alim, M. A., Das, S., Gupta, A. D., Das, B. C., Uddin, M. I., & Prodhan, M. A. M. (2012). Bovine brucellosis: An epidemiological study at Chittagong, Bangladesh. Pakistan Veterinary Journal, 32(4), 499–502. [Google Scholar]
- Smith, G. J. D., Vijaykrishna, D., Bahl, J., Lycett, S. J., Worobey, M., Pybus, O. G., Ma, S. K., Cheung, C. L., Raghwani, J., Bhatt, S., Peiris, J. S. M., Guan, Y. I., & Rambaut, A. (2009). Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature, 459(7250), 1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Stram, Y., Kuznetzova, L., Friedgut, O., Gelman, B., Yadin, H., & Rubinstein‐Guini, M. (2008). The use of lumpy skin disease virus genome termini for detection and phylogenetic analysis. Journal of Virological Methods, 151(2), 225–229. 10.1016/j.jviromet.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Sudhakar, S. B., Mishra, N., Kalaiyarasu, S., Jhade, S. K., Hemadri, D., Sood, R., Bal, G. C., Nayak, M. K., Pradhan, S. K., & Singh, V. P. (2020). Lumpy skin disease (LSD) outbreaks in cattle in Odisha state, India in August 2019: Epidemiological features and molecular studies. Transboundary and Emerging Diseases, 67(6), 2408–2422. 10.1111/tbed.13579 [DOI] [PubMed] [Google Scholar]
- Tageldin, M. H., Wallace, D. B., Gerdes, G. H., Putterill, J. F., Greyling, R. R., Phosiwa, M. N., Al Busaidy, R. M., & Al Ismaaily, S. I. (2014). Lumpy skin disease of cattle: An emerging problem in the Sultanate of Oman. Tropical Animal Health and Production, 46(1), 241–246. 10.1007/s11250-013-0483-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppurainen, E., Alexandrov, T., & Beltrán‐Alcrudo, D. (2017). Lumpy skin disease field manual – A manual for veterinarians. FAO Animal Production and Health Manual No. 20 (pp. 1–60). Rome: Food and Agriculture Organization of the United Nations (FAO). https://www.fao.org/3/i7330en/I7330EN.pdf [Google Scholar]
- Tuppurainen, E. S. M., & Oura, C. A. L. (2012). Review: Lumpy skin disease: An emerging threat to Europe, the Middle East and Asia. Transboundary and Emerging Diseases, 59(1), 40–48. 10.1111/j.1865-1682.2011.01242.x [DOI] [PubMed] [Google Scholar]
- Wolff, J., Moritz, T., Schlottau, K., Hoffmann, D., Beer, M., & Hoffmann, B. (2020). Development of a safe and highly efficient inactivated vaccine candidate against lumpy skin disease virus. Vaccines, 9(1), 4. 10.3390/vaccines9010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhugunissov, K., Bulatov, Y. E., Orynbayev, M., Kutumbetov, L., Abduraimov, Y. E., Shayakhmetov, Y. E., Taranov, D., Amanova, Z. H., Mambetaliyev, M., Absatova, Z. H., Azanbekova, M., Khairullin, B., Zakarya, K., & Tuppurainen, E. (2020). Goatpox virus (G20‐LKV) vaccine strain elicits a protective response in cattle against lumpy skin disease at challenge with lumpy skin disease virulent field strain in a comparative study. Veterinary Microbiology, 245, 108695. 10.1016/j.vetmic.2020.108695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data files associating this research are available online (https://doi.org/10.6084/m9.figshare.12619868.v1).


