Abstract
The incidence and mortality of cancer are increasing each year. At present, the sensitivity and specificity of the blood biomarkers that were used in clinical practice are low, which make the detection rate of cancer decrease. With advances in bioinformatics and technology, some non-coding RNA as biomarkers can be easily detected through some traditional and new technologies. Circular RNAs (circRNAs) are non-coding RNAs, that is, they do not encode proteins, and have important regulatory functions. CircRNAs can remain stable in bodily fluids, such as in saliva, blood, urine, and especially plasma. The difference in the expression of plasma circRNAs between cancer patients and normal people may suggest that plasma circRNAs may play an important role in the occurrence and development of cancer. In this review, we summarized the clinical effect of plasma circRNAs in several high-incidence cancers. CircRNAs may be effective biomarkers for tumour diagnosis, treatment selection and prognosis evaluation.
Keywords: circular RNA, plasma, cancer, biomarkers
Introduction
Cancer is the leading cause of death worldwide and places a great burden on families and society. Currently, the diagnostic approach to cancer primarily includes imaging examination and laboratory tests. Among these approaches, blood sample testing is a relatively simple and less invasive way to detect biomarkers. For example, carcinoembryonic antigen (CEA), cancer antigen 19–9 (CA 19–9) and alpha-fetoprotein (AFP) in blood are common clinical diagnostic indicators, but they lack specificity.1,2 It is of great importance to search for effective and sensitive markers for the early diagnosis of cancer. An increasing number of studies have indicated that circRNAs are stably expressed in tissues, cells and plasma and may be potential biomarkers.3
Circular RNAs (circRNAs) are noncoding RNAs that form covalently closed rings by reverse shearing and widely participate in cell growth and development and regulate gene expression.4 Earlier studies suggested that circRNAs are formed by seemingly rare errors in RNA splicing.5,6 However, with the emergence of high-throughput sequencing technology, later studies confirmed that the circular structure of circRNAs is formed through specialized pre-mRNA splicing.7 In contrast to the structure of linear RNA, the closed circular structure of circRNAs also indicates a lack of polarity at their 5’ caps and 3’ polyadenylated tails.8 This circRNA structure is resistant to exonucleases and is not degraded by RNase R, and therefore, it is more stable than linear RNA.9 The circular structure of circRNAs enables them to be stable and highly preserved in plasma. And the half-life of circRNAs in plasma was greater than 48 hours.10 Winston Koh et al detected circRNAs in the plasma of pregnant women in different pregnancies, and their expression showed an increasing trend, which was positively correlated with classic indicators such as human chorionic gonadotropin (HCG). The concept of circRNAs as useful biomarkers was proposed.11 Zhang et al found that the high level of plasma hsa_circ_0007534 was closely related to tumor differentiation and TNM stage of colorectal cancer and indicated a poor prognosis.12
In recent years, an increasing number of circRNAs have been found to be involved in the occurrence and development of malignant tumours.13 Therefore, circRNAs may be ideal tumour markers for the clinical diagnosis and treatment of tumours.
Biogenesis of circRNAs
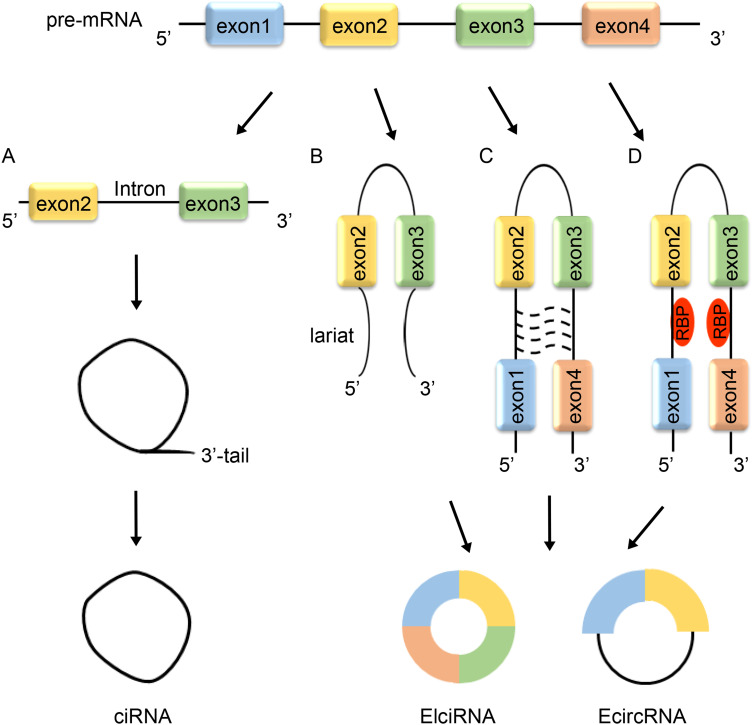
According to their composition, circRNAs can be divided into three categories: Circular intronic RNAs (ciRNAs), which contain only introns; Exon-intronic circRNAs (ElciRNAs), which have both exons and introns and can interact with U1 snRNP to regulate gene transcription; and Exonic circRNAs (EcircRNAs), comprised solely of exons.14,15
With the advances in technology, Jeck et al summarized three models of circRNA structure: lariat-driven circularization (model 1), intron-pairing-driven circularization (model 2) and RNA-binding protein (RBP)-mediated circularization (model 3).16 In model 1, researchers found that the lariat structure was formed by exon skipping as the RNA was folded. The lariat species undergoes rapid splicing to remove intron sequences and generate circRNAs. Thus, the model 1 process was named exon-skipping. In summary, EcircRNAs are generated when the 3’ splice of the exon is connected to the 5’ splice of another exon.17 In model 2, complementary intronic sequences at both ends of a pre-mRNA form pair. In model 3, RBPs promote the formation of circRNAs under specific conditions. For example, the double-chain RNA-editing enzyme ADAR antagonizes circRNA formation by editing intron sequences.18 The knockdown of ADAR promotes the formation of circRNAs.19 Quaking (QKI) is an alternative splicing factor that may induce RNA splicing and affect the abundance of many circRNA species20 (Figure 1).
Figure 1.
Biogenesis of circRNAs. (A) The pre-mRNA is folded to form an intron lariat, and then the 3’ tail is removed to form ciRNAs. (B) In lariat-driven circularization, RNA lariat is formed by skipping several exons during pre-mRNA splicing. (C) In intron-pairing-driven circularization, ElciRNA or EcircRNA was produced by complementary pairing of intron sequences. (D) In RBP-mediated circularization, RBP connects the upstream and downstream of introns to form the ElciRNAs or EcircRNAs.
Abbreviations: ciRNAs, circular intronic RNAs; ElciRNAs, exon-intronic circRNAs; EcircRNAs, exonic circRNAs; RBP, RNA-binding protein.
Function of circRNAs
The function of circRNAs is the basis of further research. According to the existing evidence, we summarize several circRNA actions: 1). sponging of miRNAs, 2) gene transcriptional regulation, 3) translation, and 4) protein-binding regulation.21 These biological functions are the bases of extensive research on circRNAs and are expected to be leveraged for clinical applications.
circRNA as miRNA Sponges
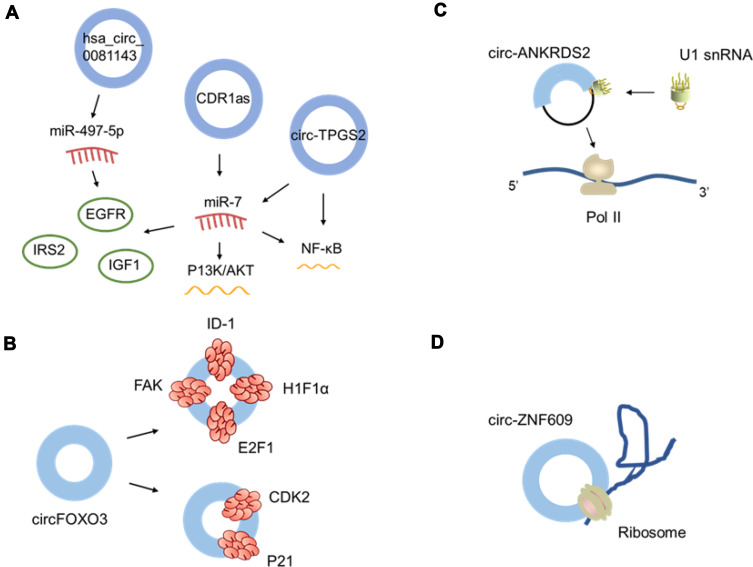
CircRNAs serve as competitive endogenous RNAs (ceRNAs) and can act as miRNA sponges. CircRNAs have multiple miRNA response elements (MREs) and competitively bind miRNAs to regulate the expression of downstream target genes.22 Hansen et al demonstrated that cerebellar degeneration-related protein 1 antisense transcript (CDR1as) was stably expressed in the brain. CDR1as acted as a miR-7 sponge containing more than 70 binding sites and significantly inhibited the expression of miR-7.23 With further research, CDR1as/miR-7 axis regulated the expression of target genes such as epidermal growth factor receptor (EGFR), insulin receptor substrate 2 (IRS2) and insulin-like growth factor 1 (IGF1),24,25 and participated in the occurrence and development of diseases by multiple signal pathway such as epithelial-mesenchymal transition (EMT), P13K/AKT and NF-kappaB.26,27 It was reported circ-TPGS2 acted as a sponge of miR-7 to activate the NF-kappaB pathway.28 Tang et at demonstrated that hsa_circ_0081143 acted as miR-497-5p sponge to regulate the expression of EGFR and EMT29(Figure 2A).
Figure 2.
Functions of circRNAs. (A) circRNA as miRNA sponges. (B) Protein-binding regulation. (C) Gene transcriptional regulation. (D) Translation.
Protein-Binding Regulation
CircRNAs could act as sponges not only for miRNAs but also for proteins. CircRNA has specific binding sites to interact with RBP. This mechanism of action is bidirectional, which can not only affect the expression of protein and regulate its gene transcription, but also have certain influence on the synthesis and degradation of circRNA.22,30 CircFOXO3 has a high affinity for cell cycle-related proteins such as cyclin-dependent kinase 2 (CDK2) and p21. A ternary complex blocks the cell cycle in G1 phase and prevents it from entering S phase.31 In addition, other studies have found that circFOXO3 played a certain role in the damage of cardiomyocytes. CircFOXO3 promoted cellular senescence by interacting with related proteins32 (Figure 2B).
Gene Transcriptional Regulation
Some circRNAs are located in the nucleus and play an important role in regulating gene transcription. ElciRNAs bind with U1 snRNP to form the ElciRNA-U1 snRNP complex and regulate the expression of host genes in the presence of polymerase II.33 Zhang et al found that circ-ANKRDS2 regulated parental gene transcription by interacting with RNA polymerase II. Knockdown of circ-ANKRDS2 decreased the expression of the parent gene. These results suggest that circ-ANRDS2 may be a positive regulator34 (Figure 2C).
Translation
Although most circRNAs cannot be translated by ribosomes, studies have reported that a few endogenous circRNAs can be translated into peptides or proteins.21,35 For example, circ-ZNF609 consists of an open reading frame with a long sequence and is translated into a protein in a splicing-dependent manner. Circ-ZNF609 plays a significant role in promoting the proliferation of muscles36 (Figure 2D).
Methods for Plasma circRNAs
Growing evidence suggests that circRNAs can be used as biomarkers in the clinical diagnosis of human diseases.21,37 However, the mechanism of circRNA function in plasma is still unclear. Maass et al found 57 differentially expressed circRNAs in plasma, and they speculated that circRNAs may be secreted into plasma.38 Koh et al used RNA-seq and microarray assays to detect circRNAs in the plasma of pregnant women at different stages of pregnancy. They proposed that these plasma circRNAs may be effective in assessing the health of the mother and foetus during pregnancy.39
In addition, because of their stability in plasma, circRNAs can ce detected effectively via liquid biopsy sampling followed by reverse transcriptase-digital droplet polymerase chain reaction (RT-ddPCR).40,41 Liquid biopsies have been an effective non-invasive measure of tumour detection in bodily fluids in recent years. Liquid biopsy samples are primarily used to detect cancer in its early stages through measures of several tumour components, such as nucleic acids, DNA, RNA and/or proteins.42 Compared with traditional solid biopsy samples, liquid biopsy samples can be used to screen the whole tumour, not merely a specific part. In addition, repeated sampling tests can be performed to dynamically monitor changes in a tumour.42,43 RT-ddPCR is a third-generation PCR assay that is used to detect the quantification of nucleic acids in the target sample through the water-oil fusion technique.44 A sample is divided into thousands of droplets, and fluorescence quantification is performed on each droplet. RT-ddPCR can be used for accurate detection of DNA/RNA, such as plasma circRNAs, at lower concentrations in bodily fluids than can be achieved through traditional PCR.45 Li et al confirmed that RT-ddPCR was more accurate and reliable in detecting plasma circRNAs. Among them, low expression of hsa_circ_0001017 and hsa_circ_0061276 were the most likely diagnostic markers for gastric cancer.46 The researchers used RT-ddPCR to test 272 plasma samples, including 231 lung cancer patients and 41 healthy subjects. They found that the expression of hsa_circ_0000190 in lung cancer was high and related to poor prognosis.47
In this review, we summarize the mechanisms of plasma circRNAs in various cancers that have been reported thus far. The receiver operating characteristic (ROC) curve is primarily used to evaluate the diagnostic value of medical biomarkers, and the area under the curve (AUC) is widely used as an evaluation indicator.
Plasma circRNAs in Cancer
Lung Cancer
Lung cancer is the most common cancer with the highest mortality in China and is the main cause of cancer-related deaths. Non-small cell lung cancer (NSCLC) accounts for 80% of these cases, and the main pathological type is lung adenocarcinoma (LUAD). To more effectively diagnose and treat lung cancer, the factors in plasma that have been discovered and applied in clinical practice are epidermal growth factor receptor (EGFR), Kirsten rat sarcoma viral oncogene homologue (KRAS), ROS proto-oncogene 1, and receptor tyrosine kinase (ROS1).48,49 EGFR is widely expressed in lung cancer, and EGFR mutants are specific targets for the treatment of lung cancer.50 The methods used to detect EGFR, KRAS and ROS1 are complex, and the positive detection rate for lung cancer is low.
Compared to these classical biomarkers, circRNAs are stable in plasma and can be detected with high specificity. CircRNAs can act as ceRNAs by sponging miRNAs and form a circRNA-miRNA-mRNA network. There have been many reports indicating that miRNAs, such as miR-21, miR-7 and miR-145, are involved in EGFR mutation, and thus affect the invasion and metastasis of lung cancer.51–53 Liu et al found that hsa_circ_0086414 was overexpressed in the plasma of lung cancer patients. The researchers were surprised to find that the expression of plasma hsa_circ_0086414 was related to an EGFR mutation. They further hypothesized that plasma hsa_circ_0086414 might induce EGFR mutations by binding to downstream miRNAs.54 Activation of EGFR significantly promoted the expression of miR-7 in plasma and tissues of lung cancer patients.52 Lin et al verified the mechanism of plasma circRNAs. They also found that plasma hsa_circ_0102537 can promote the activation of tumour signalling pathways such as the PI3K-Akt signalling pathway, thus affecting the invasion and migration of lung cancer cells.55
Among non-small cell lung cancer, lung adenocarcinoma has the highest incidence and is characterized by high metastasis rate and high recurrence rate.56 CircRNAs also played an important role in tumor metastasis. High levels of hsa_circ_0001715 were detected in patients with lung adenocarcinoma with lymph node metastasis, distant metastasis, or stage III–IV, respectively. The diagnostic sensitivity of hsa_circ_0001715 for lung adenocarcinoma with metastasis was 0.871.57
Plasma circRNAs can be detected at higher rates during development of lung cancer and play significant roles in tumorigenesis and development, and therefore, they show great diagnostic value.
Liver Cancer
To detect and diagnose liver cancer early, common blood laboratory tests measure the levels of alpha-fetoprotein (AFP), lens culinaris-agglutinin-reactive fraction of AFP (AFP-L3), and protein induced by vitamin K absence or antagonist-II (PIVKA-II).58 AFP is generally considered the most effective indicator of liver cancer. However, some studies have found that AFP was significantly increased in 60% of liver cancer patients, but the remaining 40% of liver cancer patients had levels within the normal range.59 Therefore, the diagnostic value of classic markers such as AFP, AFP-L3 and PIVKA-II combined with plasma circRNAs has been a focus of recent research. Zhang et al found that the level of plasma hsa_circ_0001445 can be measured for effectively distinguishing patients with hepatitis, cirrhosis, and liver cancer. The specificity of plasma hsa_circ_0001445 was 94.2% in the diagnosis of hepatocellular carcinoma (HCC). Moreover, according to stepwise logistic regression models, the accuracy of the combination of plasma hsa_circ_0001445 and AFP in the diagnosis of HCC was significantly higher than that of plasma hsa_circ_0001445 or AFP alone (AUC=0.970).60 Hsa_circ_0001445 can also serve as a miR-17-3p or miR-181b-5p sponge to facilitate TIMP3 expression and inhibit the growth and metastasis of HCC cells.61 In addition to acting as miRNA sponges, compared to miRNAs, circRNAs remain stable in human bodily fluids. Therefore, it is reasonable to believe that circRNAs are better choices for detecting plasma markers.
Hepatitis B virus infection (HBV) is the leading cause of liver cancer and accounts for 56% of liver cancer cases worldwide.62 In particular, there is a high incidence of hepatitis B virus infection in China. Zhu et al measured the upregulated expression of plasma hsa_circ_0027089 in 64 HBV-related patients with HCC and 40 patients with HBV-related liver cirrhosis and compared the results to those of 72 normal control patients. Although the sample size was small, plasma hsa_circ_0027089 can be considered a potential biomarker in HBV-related HCC.63 It has also been found that some plasma circRNAs are significantly upregulated in HBV-related HCC patients, but their expression is downregulated after HCC resection. In summary, changes in plasma circRNA expression can be used to evaluate the therapeutic effect of liver cancer treatments.64
Gastric Cancer
Different expression levels of circRNAs may affect the invasion and metastasis of gastric cancer. Rao et al detected a series of differentially expressed circRNAs in the plasma exosomes of patients with gastric cancer. They also screened 1060 plasma exosomal circRNAs involved in many cancer-related signalling pathways, such as PTEN/P13K/AKT, Wnt and thyroid hormone signalling, which are closely associated with the cancer metastasis.65 Ye et al verified that the expression of hsa_circ_0010882 was significantly upregulated in the plasma of patients with gastric cancer and was positively correlated with TNM stage and tumour differentiation. Further experiments showed that plasma hsa_circ_0010882 promoted the proliferation and metastasis of gastric cancer cells by interacting with the PI3K/AKT pathway.66 Hsa_circ_0078607 was stably overexpressed in the plasma of patients with gastric cancer. The researchers constructed a hsa_circ_0078607-miR-188-3p-RAP1B network on the basis of a bioinformatics analysis. The network analysis indicated that the ERK1/2/AKT pathway was involved in the metastasis of gastric cancer.67 Circ-Rangap1 was found to be highly expressed in the plasma of patients with gastric cancer. Its high expression was positively correlated with TNM stage and advanced lymph node metastasis. Circ-Rangap1 may be an effective biomarker to evaluate the prognosis of gastric cancer.68 Low expression of hsa_circ_0000745 in the plasma of patients with gastric cancer could also be used as a diagnostic marker and was closely related to tumor differentiation. The level of hsa_circ_0000745 combined CEA levels to improve diagnostic accuracy.69 Plasma circPSMC3 as a tumor suppressor in gastric cancer was positively relative to TNM stage and lymphatic metastasis. It was very sensitive in distinguishing between gastric cancer patients and normal people, with an AUC of 0.933.70 The expression of circ-KIAA1244 was decreased in the plasma of gastric cancer patients and was negatively correlated with tumor invasion and migration. The Kaplan-Meier overall survival curve results showed that gastric cancer patients with low expression of circ-KIAA1244 had a longer survival time. Therefore, circ-KIAA1244 was a key indicator to evaluate the prognosis of gastric cancer.71 Different expression levels of plasma circRNAs predict the survival period of cancer patients and can be used as an effective prognostic indicator.
Colorectal Cancer
Currently, clinical biomarkers in blood used for the diagnosis of colorectal cancer are mainly CEA, CA19-9 and some novel biomarkers, such as EGFR.72–74 CEA and CA19-9 are not specific markers for colorectal cancer since they are also elevated in gastric, breast, pancreatic and other cancers.75–77 However, CEA and CA19-9 show an increased sensitivity for advanced colorectal cancer but are not as effective for use in detecting early colorectal cancer.78 Therefore, the need to find new markers for the diagnosis of colorectal cancer is urgent. Li et al chose three upregulated plasma circRNAs (hsa_circ_0001900, hsa_circ_0001178 and hsa_circ_0005927) to design a circular RNA panel. The circular RNA panel showed great advantages in distinguishing patients with colorectal cancer from healthy people and showed higher diagnostic accuracy than CEA (AUC=0.698).12 Lin et al used the same method to examine the clinical significance of plasma circRNAs in colorectal cancer. Moreover, they concluded that a circular RNA panel combined with the biomarkers CEA and CA19-9 increased the detection rate of early colorectal cancer (AUC=0.855).79 Ye et al measured the changes in several highly expressed circRNAs (hsa_circ_0082182, hsa_circ_0000370 and hsa_circ_0035445) in the plasma of colorectal cancer patients before and after surgery. Their results showed that the expression of circRNAs in plasma decreased significantly after surgery and suggested that these circRNAs may be involved in the malignant progression of colorectal cancer.80 These results show that plasma circRNAs are of great value in diagnosing and evaluating the progression of colon cancer.
Breast Cancer
Breast cancer is one of the most common cancers in women. It is essential to find new biomarkers for the early detection and diagnosis of breast cancer. Yin et al used microarray technology to detect 41 abnormally expressed circRNAs in the plasma of breast cancer patients. Among these circRNAs, high expression of plasma hsa_circ_0001785 showed higher diagnostic accuracy than CEA or CA15-3, with an AUC of 0.784. In addition, the level of plasma hsa_circ_0001785 declined gradually after surgery. Plasma hsa_circ_0001785 may have potential diagnostic and therapeutic value.81 According to a univariate Cox analysis results, the expression of plasma hsa_circ_0008673 may significantly affect the overall survival (OS) and disease‐specific survival (DSS) of breast cancer patients.82 Li et al found that the expression of hsa_circ_0104824 was downregulated in the plasma of 84 breast cancer patients. The diagnostic sensitivity of this circRNA was 71.1%.83
Esophageal Squamous Cell Carcinoma
In China, a diverse diet contributes to the increasing annual incidence of esophageal cancer. Esophageal squamous cell carcinoma (ESCC) is the predominant pathological diagnosis in this population. Wang et al suggested that upregulation of plasma circ-TTC17 contributes to poor patient prognosis. The survival time for ESCC patients with high levels of plasma circ-TTC17 is significantly shorter than that of those with low levels.84 In addition, the expression of circ-DLG1 (hsa_circ_0007203) was found to be higher in both the tissues and plasma of ESCC patients compared to that of samples obtained from healthy individuals and promoted cancer cell proliferation and metastasis.85 Huang et al demonstrated that the circ-0004771-miR-339-5p-CDC25A axis facilitated the malignant progression of ESCC. Upregulation of plasma hsa_circ_0004771 was related to poor patient prognosis.86 The five-year survival rate of esophageal cancer is less than 13%, indicating a poor prognosis. Upregulation of circGSK3β significantly promoted the invasion and migration of ESCC cells. Plasma circGSK3β was highly sensitive to the diagnosis of early ESCC, with an AUC of 0.8012. What’s more, For ESCC patients who relapsed within 10 months after surgery, plasma circGSK3β expression was significantly increased. This reflected his importance in assessing the prognosis of ESCC.87
Pancreatic Cancer
Pancreatic cancer is one of the most aggressive tumours, and the survival of most patients is less than one year. Improving the early diagnosis rate of pancreatic cancer is the focus of current research. The combination of plasma circ-LDLRAD3 and CEA has shown higher accuracy and specificity in the diagnosis of pancreatic cancer. The AUC was 0.870. The expression of circ-LDLRAD3 was significantly increased in the plasma of pancreatic cancer patients with distant metastasis. Plasma circ-LDLRAD3 may be used as a biomarker to predict the progression of pancreatic cancer.88 Plasma hsa_circ_0001569 showed a high sensitivity for pancreatic cancer (AUC=0.716) and was closely associated with blood invasion and lymph node metastasis.89
Thyroid Cancer
Thyroid cancer is an indolent cancer compared to other malignant cancers. However, early detection remains the basis of successful thyroid cancer treatment. Hsa_circ_0124055 or hsa_circ_0101622, with high expression in the plasma of thyroid cancer patients, was found to modulate tumour proliferation, and plasma hsa_circ_0124055 (AUC=0.836) or hsa_circ_0101622 (AUC=0.805) may be used as potential markers for thyroid cancer treatment.90
Other Cancer
In addition to a few high incidence of cancers, plasma circRNAs have also been studied in other cancers. High expression of plasma circ-MTO1 indicated an increase in the malignancy of gallbladder carcinoma and a poor prognosis.91 Moreover, monitoring the levels of plasma hsa_circ_0066755 was helpful in distinguishing nasopharyngeal carcinoma from noncancerous conditions in patients, and its diagnostic accuracy was similar to that of imaging examination. Researchers have also shown that hsa_circ_0066755 acts as a miR-651 sponge to influence tumour progression.92 The roles of circRNAs in ten common cancers are summarized in Table 1.
Table 1.
The Expression of Plasma circRNA in Cancers
| Cancer | CircRNA | Expression | Significance | Function | AUC | Reference |
|---|---|---|---|---|---|---|
| Lung cancer | hsa_circ_0005962 | Up | Diagnosis | miR-1265 | 0.730 | [54] |
| hsa_circ_0086414 | Down | Diagnosis | miR-7 | 0.780 | [54] | |
| hsa_circ_0102537 | Down | Diagnosis | P13K/AKT | [55] | ||
| hsa_circ_0000190 | Up | Prognosis | miR-767-5p | 0.950 | [47] | |
| hsa_circ_0001715 | Up | Diagnosis | 0.871 | [57] | ||
| Hepatocellular carcinoma | hsa_circ_0001445 | Up | Diagnosis | miR-17-3p | 0.712 | [60] |
| hsa_circ_0027089 | Up | Diagnosis | miR-15b-3p | 0.765 | [63] | |
| circ-SMARCA5 | Down | Diagnosis | miR-17-3p | [100] | ||
| hsa_circ_0064428 | Down | Prognosis | miR-7977 | [101] | ||
| hsa_circ_0009582 | Up | Diagnosis | 0.688 | [64] | ||
| hsa_circ_0037120 | Up | Diagnosis | 0.742 | [64] | ||
| hsa_circ_0140117 | Up | Diagnosis | 0.762 | [64] | ||
| Gastric cancer | hsa_circ_0010882 | Up | Prognosis | PI3K/AKT | [66] | |
| hsa_circ_0001017 | Down | Diagnosis | 0.871 | [47] | ||
| hsa_circ_0061276 | Down | Diagnosis | 0.912 | [47] | ||
| hsa_circ_0078607 | Up | Diagnosis | miR-188-3p | [67] | ||
| hsa_circ_0000745 | Down | Diagnosis | miR-335-5p | 0.683 | [69] | |
| circ-Rangap1 | Up | Prognosis | miR-877-3p | 0.830 | [68] | |
| hsa_circ_0000181 | Up | Diagnosis | 0.756 | [99] | ||
| circ-PSMC3 | Down | Diagnosis | miR-296-5p | 0.933 | [70] | |
| circ-KIAA1244 | Down | Diagnosis | 0.748 | [71] | ||
| Colorectal cancer | hsa_circ_0001900 | Up | Diagnosis | 0.722 | [12] | |
| hsa_circ_0001178 | Up | Diagnosis | 0.718 | [12] | ||
| hsa_circ_0005927 | Up | Diagnosis | Wnt/β-catenin | 0.784 | [12] | |
| hsa_circ_0082182 | Up | Prognosis | 0.737 | [80] | ||
| hsa_circ_0000370 | Up | Prognosis | 0.815 | [80] | ||
| hsa_circ_0035445 | Up | Prognosis | 0.703 | [80] | ||
| hsa_circ_0006990 | Up | Diagnosis | miR-101 | 0.724 | [93] | |
| Breast cancer | hsa_circ_0001785 | Up | Diagnosis | miR-942 | 0.771 | [81] |
| hsa_circ_0008673 | Up | Prognosis | 0.833 | [82] | ||
| hsa_circ_0104824 | Down | Diagnosis | miR-1278 | 0.878 | [83] | |
| Esophageal cancer | circ-TTC17 | Up | Prognosis | miR-153 | 0.820 | [84] |
| hsa_circ_0007203 | Up | Diagnosis | miR-515 | 0.648 | [85] | |
| hsa_circ_0004771 | Up | Prognosis | miR-339-5p | 0.816 | [86] | |
| circGSK3β | Up | Diagnosis | 0.782 | [87] | ||
| hsa_circ_0066755 | Up | Prognosis | miR-651 | 0.853 | [92] | |
| circ-SLC7A5 | Up | Prognosis | 0.771 | [102] | ||
| Pancreatic cancer | circ-LDLRAD3 | Up | Diagnosis | miR-137-3p | 0.670 | [88] |
| hsa_circ_0001569 | Up | Diagnosis | miR-18a-5p | 0.716 | [89] | |
| Thyroid cancer | hsa_circ_0124055 | Up | Diagnosis | 0.836 | [90] | |
| hsa_circ_0101622 | Up | Diagnosis | 0.805 | [90] | ||
| Gallbladder cancer | circ-MTO1 | Up | Prognosis | miR-17 | 0.882 | [91] |
Summary and Prospects
In this review, we summarized the expression of many plasma circRNAs in different cancers and compared some clinical indicators. The combination of some plasma circRNAs with CEA and CA199 can improve the detection rate of early digestive tract tumours. The correlation between plasma circRNA levels and TNM stage, tumour size and degree of tumour cell differentiation was evaluated to assess tumour development and patient prognosis.
CircRNAs can competitively bind to miRNAs to relieve the inhibition of miRNAs on downstream target genes and promote the upregulation of target genes. CircRNAs act as miRNA sponges to mediate tumour progression. Li et al found that the level of hsa_circ_0006990 in the plasma of colorectal patients was upregulated. Hsa_circ_0006990 promoted carcinogenic characteristics and induced the proliferation, invasion and migration of tumour cells by sponging miR-101.93 MiRNAs that bind to proteins are released from cells into bodily fluids and remain stable.94 Many studies have suggested that plasma miRNAs can be used as effective diagnostic biomarkers for cancer, such as colorectal cancer, ovarian cancer and hepatocellular cancer.95–97 Plasma circRNAs and miRNAs are profoundly significant tumour markers, and their interactions are worthy of further study.
CircRNAs display tissue specificity and stability. CircRNAs can be stable in bodily fluids, such as saliva, blood, urine, and especially plasma.98 This property has led researchers to suggest that plasma circRNAs may be a promising biomarker for tumour diagnosis. The expression of plasma hsa_circ_0000181 in gastric cancer patients was significantly lower than that in healthy controls. Combined with clinical data, research results suggest that the level of plasma hsa_circ_0000181 is positively correlated with CEA level and the degree of tumour differentiation. The area under the ROC curve was 0.582, and the sensitivity was 99%. It was suggested that plasma hsa_circ_0000181 can be used as a biomarker for gastric cancer.99 The AUC of plasma circSMARCA5, which was used to diagnose HCC, was 0.847. Downregulation of plasma circSMARCA5 can be used for accurately distinguishing between healthy individuals, hepatitis patients, cirrhosis patients and HCC patients.100 Therefore, plasma circRNAs play important roles in the diagnosis and monitoring of cancer development.
Treatment is the most important focus of cancer patients. Immunotherapy is a novel and effective treatment for cancer patients. The mechanism by which immunotherapy affects the expression of circRNAs is unclear. Luo et al monitored changes in plasma hsa_circ_0000190 expression in several patients with advanced lung cancer receiving immunotherapy. Hsa_circ_0000190, which was originally highly expressed in patients with lung cancer, was reduced after treatment. Hsa_circ_0000190 may be a potential marker to assess the efficacy of immunotherapy treatment.47
As research progressed, most circRNAs were found to have carcinogenic or anticancer properties, which seriously affects the prognosis of cancer patients. Weng et al found that the higher the expression of tumour-infiltrating lymphocytes (TILs) is, the better the survival rate in HCC patients. However, plasma hsa_circ_0064428 expression was decreased in HCC patients with high TIL levels and predicted a poor patient prognosis.101 Plasma circ-SLC7A5 expression was found to be closely related to TNM staging of oesophageal cancer. Patients with high levels of plasma circ-SLC7A5 had shorter survival times than those with low levels of plasma circ-SLC7A5.102
According to current research, plasma circRNAs may be effective biomarkers for tumour diagnosis, treatment selection and prognosis evaluation. CircRNAs may be rising stars in the detection of the malignant progression of cancers. However, many challenges remain to be addressed. Multiple functions of circRNAs have been reported, but the pathophysiological processes of these functions in cancer need further investigation. In addition, due to the large number of circRNAs, screening noteworthy circRNAs is a lengthy process. The low concentration of circRNAs in plasma and the limited detection methods hinder their widespread clinical use. In conclusion, circRNAs have great potential for use as biomarkers of tumour development and are worthy of further exploration.
Conclusion
Numerous studies have shown that circRNAs can be effective markers of tumors. Different from previous studies in tumor tissues and cells, we summarized the different expression of circRNAs in plasma of tumor patients. The expression of circRNAs in plasma is stable and can be easily detected by novel techniques such as RT-ddPCR and liquid biopsy. Compared with traditional markers such as CEA, CA199, AFP and other indicators, plasma circRNA can be widely used in clinical diagnosis, treatment and prognostic evaluation with higher sensitivity and accuracy. However, more research is needed to explore the important role that circRNAs play in the plasma.
Acknowledgments
Qian Zhou and LinLing Ju contributed equally to this study. All authors have reviewed and agreed on the final version to be published. This research was funded by Nantong Science and Technology Bureau (MS12020023, MS22020014, MSZ20099, and JC2020011), Jiangsu Health and Health Committee (Z2020011), and Health Bureau of Nantong City (MA2020014, MA2020015).
Abbreviations
circRNAs, circular RNAs; CEA, carcinoembryonic antigen; CA 19-9, cancer antigen 19-9; AFP, alpha-fetoprotein; ciRNAs, circular intronic RNAs; ElciRNAs, exon-intronic circRNAs; EcircRNAs, exonic circRNAs; RBP, RNA-binding protein; ceRNAs, competitive endogenous RNAs; MREs, miRNA response elements; EGFR, epidermal growth factor receptor; IRS2, insulin receptor substrate 2; IGF1, insulin-like growth factor 1; HCG, human chorionic gonadotropin; RT-ddPCR, reverse transcriptase-digital droplet polymerase chain reaction; ROC, receiver operating characteristic; AUC, the area under the curve; HBV, Hepatitis B virus infection; HCC, hepatocellular carcinoma; AFP-L3, lens culinaris-agglutinin-reactive fraction of AFP; PIVKA-II, protein induced by vitamin K absence or antagonist-II; CEA, carcinoembryonic antigen; CA19-9, cancer antigen 19-9; OS, overall survival; DSS, disease‐specific survival.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and ferritin as diagnostic markers and factors of clinical parameters for colorectal cancer. Sci Rep. 2018;8(1):2732. doi: 10.1038/s41598-018-21048-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolscheid-Pommerich RC, Manekeller S, Walgenbach-Brünagel G, et al. Clinical Performance of CEA, CA19-9, CA15-3, CA125 and AFP in Gastrointestinal Cancer Using LOCI™-based Assays. Anticancer Res. 2017;37(1):353–359. doi: 10.21873/anticanres.11329 [DOI] [PubMed] [Google Scholar]
- 3.Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi: 10.7150/jca.30828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 6.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 7.Hou LD, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30(1):1–6. doi: 10.1177/0394632016686985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Yang L, Chen LL. The Biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 9.Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. 2018;9(9):1548–1559. doi: 10.7150/jca.24566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10(10):e0141214. doi: 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winston Koh W, Pan W, Gawad C, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A. 2014;111(20):7361–7366. doi: 10.1073/pnas.1405528111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Song Y, Wang J, Huang J. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer detection. Am J Transl Res. 2020;12(11):7395–7403. [PMC free article] [PubMed] [Google Scholar]
- 13.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 15.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 18.Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 19.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 21.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi: 10.7150/thno.42174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 24.Jiang L, Liu X, Chen Z, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432(1):199–205. doi: 10.1042/BJ20100859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Liu FB, Huang M, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18(6):580–586. doi: 10.1016/j.hbpd.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Li J, Zhou Y, et al. Down-regulated ciRS-7/up-regulated miR-7 axis aggravated cartilage degradation and autophagy defection by PI3K/AKT/mTOR activation mediated by IL-17A in osteoarthritis. Aging. 2020;12(20):20163–20183. doi: 10.18632/aging.103731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su C, Han Y, Zhang H, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22(6):3097–3107. doi: 10.1111/jcmm.13587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Feng X, Wang Y, Li Q, Li X. Dysregulation of tumour microenvironment driven by circ-TPGS2/miR-7/TRAF6/NF-κB axis facilitates breast cancer cell motility. Autoimmunity. 2021;1–10. doi: 10.1080/08916934.2021.1963958 [DOI] [PubMed] [Google Scholar]
- 29.Tang J, Zhu H, Lin J, Wang H. Knockdown of Circ_0081143 Mitigates Hypoxia-Induced Migration, Invasion, and EMT in Gastric Cancer Cells Through the miR-497-5p/EGFR Axis. Cancer Biother Radiopharm. 2021;36(4):333–346. doi: 10.1089/cbr.2019.3512 [DOI] [PubMed] [Google Scholar]
- 30.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. doi: 10.1002/jnr.24356 [DOI] [PubMed] [Google Scholar]
- 31.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 35.Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e27. doi: 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e29. doi: 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: a novel type of biomarker and genetic tools in cancer. Oncotarget. 2017;8(38):64551–64563. doi: 10.18632/oncotarget.18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maass P, Glažar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh W, Pan W, Gawad C, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A. 2014;111(20):7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med. 2018;96(1):85–96. doi: 10.1007/s00109-017-1600-y [DOI] [PubMed] [Google Scholar]
- 41.Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treatment. 2017;40(7–8):404–408. doi: 10.1159/000478018 [DOI] [PubMed] [Google Scholar]
- 42.Aghamir SMK, Heshmat R, Ebrahimi M, Khatami F. Liquid biopsy: the unique test for chasing the genetics of solid tumors. Epigenetics Insights. 2020;13:2516865720904052. doi: 10.1177/2516865720904052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. doi: 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- 44.Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abachin E, Convers S, Falque S, Esson R, Mallet L, Nougarede N. Comparison of reverse-transcriptase qPCR and droplet digital PCR for the quantification of dengue virus nucleic acid. Biologicals. 2018;52:49–54. doi: 10.1016/j.biologicals.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 46.Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med. 2018;96(1):85–96. [DOI] [PubMed] [Google Scholar]
- 47.Luo YH, Yang YP, Chien CS, et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers. 2020;12(7):584. doi: 10.3390/cancers12071740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villalobos P. Lung Cancer Biomarkers. Hematol Oncol Clin North Am. 2017;31(1):13–29. doi: 10.1016/j.hoc.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffy MJ, O’Byrne K. Tissue and blood biomarkers in lung cancer: a review. Adv Clin Chem. 2018;86:1–21. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Wang P, Zhang C, Ma Z. Epidermal growth factor receptor (EGFR): a rising star in the era of precision medicine of lung cancer. Oncotarget. 2017;8(30):50209–50220. doi: 10.18632/oncotarget.16854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiou YH, Liou SH, Wong RH, Chen CY, Lee H. Nickel may contribute to EGFR mutation and synergistically promotes tumor invasion in EGFR-mutated lung cancer via nickel-induced microRNA-21 expression. Toxicol Lett. 2015;237(1):46–54. doi: 10.1016/j.toxlet.2015.05.019 [DOI] [PubMed] [Google Scholar]
- 52.Chou YT, Lin HH, Lien YC, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–8831. doi: 10.1158/0008-5472.CAN-10-0638 [DOI] [PubMed] [Google Scholar]
- 53.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur j Cancer. 2009;45(12):2197–2206. doi: 10.1016/j.ejca.2009.04.039 [DOI] [PubMed] [Google Scholar]
- 54.Liu XX, Yang YE, Liu X, et al. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J Transl Med. 2019;17(1):50. doi: 10.1186/s12967-019-1800-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin S, Xiong W, Liu H, Pei L, Yi H, Guan Y. Profiling and integrated analysis of differentially expressed circular RNAS in plasma exosomes as novel biomarkers for advanced-stage lung adenocarcinoma. Onco Targets Ther. 2020;13:12965–12977. doi: 10.2147/OTT.S279710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith RA, Cokkinides V, Brawley OW. Cancer screening in the United States, 2009: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin. 2009;59(1):27–41. [DOI] [PubMed] [Google Scholar]
- 57.Lu GJ, Cui J, Qian Q, et al. Overexpression of hsa_circ_0001715 is a potential diagnostic and prognostic biomarker in lung adenocarcinoma. Onco Targets Ther. 2020;13:10775–10783. doi: 10.2147/OTT.S274932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park SJ, Jang JY, Jeong SW, et al. Usefulness of AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing hepatocellular carcinoma. Medicine. 2017;96(11):e5811. doi: 10.1097/MD.0000000000005811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worland T, Harrison B, Delmenico L, Dowling D. Hepatocellular Carcinoma Screening Utilising Serum Alpha-Fetoprotein Measurement and Abdominal Ultrasound Is More Effective than Ultrasound Alone in Patients with Non-viral Cirrhosis. J Gastrointest Cancer. 2018;49(4):476–480. doi: 10.1007/s12029-017-0006-y [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Zhou H, Jing W, et al. The Circular RNA hsa_circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and May Serve as a Diagnostic Biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 62.Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer. 2018;142(12):2471–2477. doi: 10.1002/ijc.31280 [DOI] [PubMed] [Google Scholar]
- 63.Zhu K, Zhan H, Peng Y, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41(3):296–302. doi: 10.1093/carcin/bgz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu C, Deng L, Zhuo H, et al. Circulating circRNA predicting the occurrence of hepatocellular carcinoma in patients with HBV infection. J Cell Mol Med. 2020;24(17):10216–10222. doi: 10.1111/jcmm.15635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rao M, Zhu Y, Qi L, Hu F, Gao P. Circular RNA profiling in plasma exosomes from patients with gastric cancer. Oncol Lett. 2020;20(3):2199–2208. doi: 10.3892/ol.2020.11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng YK, Pu K, Su HX, et al. Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(3):1142–1151. [DOI] [PubMed] [Google Scholar]
- 67.Bian W, Liu Z, Chu Y, Xing X. Silencing of circ_0078607 prevents development of gastric cancer and inactivates the ERK1/2/AKT pathway through the miR-188-3p/RAP1B axis. Anticancer Drugs. 2021. doi: 10.1097/CAD.0000000000001083 [DOI] [PubMed] [Google Scholar]
- 68.Lu J, Wang YH, Yoon C, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038 [DOI] [PubMed] [Google Scholar]
- 69.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18(1):25. doi: 10.1186/s12943-019-0958-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi: 10.1186/s12943-018-0888-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campos-da-paz M, Dórea JG, Galdino AS, Lacava ZGM. Carcinoembryonic Antigen (CEA) and Hepatic Metastasis in Colorectal Cancer: update on Biomarker for Clinical and Biotechnological Approaches. Recent Pat Biotechnol. 2018;12(4):269–279. doi: 10.2174/1872208312666180731104244 [DOI] [PubMed] [Google Scholar]
- 73.Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. World J Gastroenterol. 2016;22(5):1745–1755. doi: 10.3748/wjg.v22.i5.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36(3):1093–1102. [PubMed] [Google Scholar]
- 75.He B, Zhang HQ, Xiong SP, Lu S, Wan YY, Song RF. Changing patterns of Serum CEA and CA199 for Evaluating the Response to First-line Chemotherapy in Patients with Advanced Gastric Adenocarcinoma. Asian Pac J Cancer Prev. 2015;16(8):3111–3116. doi: 10.7314/APJCP.2015.16.8.3111 [DOI] [PubMed] [Google Scholar]
- 76.Zhao S, Mei Y, Wang Y, Zhu J, Zheng G, Ma R. Levels of CEA, CA153, CA199, CA724 and AFP in nipple discharge of breast cancer patients. Int J Clin Exp Med. 2015;8(11):20837–20844. [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Gao SG, Chen JM, et al. Serum CA242, CA199, CA125, CEA, and TSGF are biomarkers for the efficacy and prognosis of cryoablation in pancreatic cancer patients. Cell Biochem Biophys. 2015;71(3):1287–1291. doi: 10.1007/s12013-014-0345-2 [DOI] [PubMed] [Google Scholar]
- 78.Xie HL, Gong YZ, Kuang JA, Gao F, Tang SY, Gan JL. The prognostic value of the postoperative serum CEA levels/preoperative serum CEA levels ratio in colorectal cancer patients with high preoperative serum CEA levels. Cancer Manag Res. 2019;11:7499–7511. doi: 10.2147/CMAR.S213580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin J, Cai D, Li W, et al. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem. 2019;74:60–68. doi: 10.1016/j.clinbiochem.2019.10.012 [DOI] [PubMed] [Google Scholar]
- 80.Ye DX, Wang SS, Huang Y, Chi P. A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int. 2019;19:276. doi: 10.1186/s12935-019-0995-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi: 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 82.Hu Y, Song Q, Zhao J, et al. Identification of plasma hsa_circ_0008673 expression as a potential biomarker and tumor regulator of breast cancer. J Clin Lab Anal. 2020;34(9):e23393. doi: 10.1002/jcla.23393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Ma F, Wu L, et al. Identification of Hsa_circ_0104824 as a potential biomarkers for breast cancer. Technol Cancer Res Treat. 2020;19:1533033820960745. doi: 10.1177/1533033820960745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Q, Zhang Q, Sun H, et al. Circ-TTC17 promotes proliferation and migration of esophageal squamous cell carcinoma. Dig Dis Sci. 2019;64(3):751–758. doi: 10.1007/s10620-018-5382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rong J, Wang Q, Zhang Y, et al. Circ-DLG1 promotes the proliferation of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:6723–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang E, Fu J, Yu Q, et al. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics. 2020;12(7):587–603. doi: 10.2217/epi-2019-0404 [DOI] [PubMed] [Google Scholar]
- 87.Hu X, Wu D, He X, et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol Cancer. 2019;18(1):160. doi: 10.1186/s12943-019-1095-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. doi: 10.3748/wjg.v23.i47.8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen X, Chen Y, Li J, Huang H, Liu C, Zhou N. Identification of Circ_001569 as a Potential Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. Technol Cancer Res Treat. 2021;20:1533033820983302. doi: 10.1177/1533033820983302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun JW, Qiu S, Yang JY, Chen X, Li HX. Hsa_circ_0124055 and hsa_circ_0101622 regulate proliferation and apoptosis in thyroid cancer and serve as prognostic and diagnostic indicators. Eur Rev Med Pharmacol Sci. 2020;24(8):4348–4360. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Lin YK, Lu ZL, Li J. Circular RNA circ-MTO1 serves as a novel potential diagnostic and prognostic biomarker for gallbladder cancer. Eur Rev Med Pharmacol Sci. 2020;24(16):8359–8366. [DOI] [PubMed] [Google Scholar]
- 92.Wang J, Kong J, Nie Z, et al. Circular RNA Hsa_circ_0066755 as an Oncogene via sponging miR-651 and as a Promising Diagnostic Biomarker for Nasopharyngeal Carcinoma. Int J Med Sci. 2020;17(11):1499–1507. doi: 10.7150/ijms.47024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacotherapy. 2019;112:108611. doi: 10.1016/j.biopha.2019.108611 [DOI] [PubMed] [Google Scholar]
- 94.Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116(6):762–774. doi: 10.1038/bjc.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zanutto S, Ciniselli CM, Belfiore A, et al. Plasma miRNA-based signatures in CRC screening programs. Int J Cancer. 2020;146(4):1164–1173. doi: 10.1002/ijc.32573 [DOI] [PubMed] [Google Scholar]
- 96.Penyige A, Márton É, Soltész B, et al. Circulating miRNA Profiling in Plasma Samples of Ovarian Cancer Patients. Int J Mol Sci. 2019;20(18):87–95. doi: 10.3390/ijms20184533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bai X, Liu Z, Shao X, et al. The heterogeneity of plasma miRNA profiles in hepatocellular carcinoma patients and the exploration of diagnostic circulating miRNAs for hepatocellular carcinoma. PLoS One. 2019;14(2):e0211581. doi: 10.1371/journal.pone.0211581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang S, Zhang K, Tan S, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20(1):13. doi: 10.1186/s12943-020-01298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32(4):e22333. doi: 10.1002/jcla.22333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Zhou Y, Yang G, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37–44. doi: 10.1016/j.cca.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 101.Weng Q, Chen M, Li M, et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56(1):32–38. doi: 10.1136/jmedgenet-2018-105440 [DOI] [PubMed] [Google Scholar]
- 102.Wang Q, Liu H, Liu Z, et al. Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet. 2020;240:33–39. doi: 10.1016/j.cancergen.2019.11.001 [DOI] [PubMed] [Google Scholar]