Abstract
Objectives
Long non-coding RNAs (lncRNAs) play a crucial part in cancer progression. However, in epithelial ovarian carcinoma (EOC), the role of SNHG22 needs to be further explained.
Methods
Quantitative real-time PCR was used to detect the expression of SNHG22. EOC cells were stably transfected with lentivirus approach and cell proliferation, glycolysis and cell apoptosis, as well as tumorigenesis in animal were performed to assess the effects of SNHG22 in EOC. Chromatin immunoprecipitation (ChIP) and luciferase reporter assay were conducted to confirm the relationship between SP1 and SNHG22.
Results
Higher expressed SNHG22 was associated with a poor prognosis in EOC tissues. SNHG22 facilitated glycolysis and proliferation. Mechanistically, LDHA deficiency and glycolysis inhibitor (2-DG, 3-BG) partly rescued proliferation. SP1 mediated SNHG22 expression at the transcriptional level and the SNHG22 promoter region (−900~ −600) was necessary for SP1 binding. Hypoxia and HIF-1α also upregulated SNHG22 expression.
Conclusion
SNHG22 is an independent prognostic biomarker for EOC. SNHG22 promotes EOC progression and is a prospective therapeutic target.
Keywords: hypoxia, SNHG22, SP1, epithelial ovarian carcinoma, glycolysis
Introduction
In recent years, the incidence of ovarian cancer has declined.1,2 As of 2018, ovarian cancer is the seventh most common female cancer in the world, with approximately 240,000 new cases.3 It is frustrating that a great many cases are diagnosed with advanced stages, which greatly hinders treatment success, mainly because of lacking effective early detecting methods.4,5 Therefore, there is an urgent clinical need to explore new therapeutic options for EOC patients.
LncRNAs are a type of RNA, defined as being transcripts with lengths exceeding 200 nucleotides that are not translated into protein, distinguished from microRNAs, siRNAs, piRNAs, and snoRNAs. Recently, lncRNAs have been confirmed in a great number of cellular processes with tissue- and disease-specific expression. For example, CDKN2BAS predicts poor prognosis and promotes metastasis in hepatocellular carcinoma.4 RMRP facilitates proliferation, migration and invasion in bladder cancer.5 Although many high expressed lncRNAs have been verified, their biological function is not clearly demonstrated in epithelial ovarian carcinoma (EOC).
Small nucleolar RNA host gene 22 (SNHG22) is located in the 18q21.1 region of human genome, it is a novel lncRNA which was discovered as highly expressed and as a tumor-promoter in epithelial ovarian carcinoma cells through the miR-2467/ Gal-1 axis.6 The regulatory mechanism and function of SNHG22 in the phenotype of glycolysis have not been explained. Our findings showed that SNHG22 facilitates cell glycolysis and proliferation and could be a potential therapeutic target.
Materials and Methods
Clinical Samples
Patients of EOC were enrolled from the Second People’s Hospital of Baise and Cancer Hospital of China Medical University between March 2010 and November 2015. No patients were received chemotherapy or radiotherapy before surgery. The including criteria were as follow: patients were with definite pathological diagnosis. Serum samples were collected from EOC patients and healthy control. The tumor and paired non-tumor tissues were also collected after lesion excision with 30 mins and stored in liquid nitrogen, then transferred to −80°C refrigerator. The characteristics of cases were thoroughly noted. All patients involved in this study (Tables 1 and S4) gave their informed consent. Institutional review board (IRB) approval of our hospital was obtained for this study.
Table 1.
Clinicopathologic Characteristics of Patients in EOC
| Characteristics | Number of Cases (%) |
|---|---|
| Age (years) | |
| ≤60 | 53 (44.2) |
| >60 | 67 (55.8) |
| Tumor size (cm) | |
| ≤5 | 84 (70.0) |
| > 5 | 36 (30.0) |
| Tumor invasion depth | |
| T1-2 | 28 (23.3) |
| T3-4 | 92 (76.7) |
| Lymph node metastasis | |
| N0 | 45 (37.5) |
| N1-2 | 75 (62.5) |
| Vessel invasion | |
| Yes | 69 (57.5) |
| No | 51 (42.5) |
| TNM stage | |
| I–II | 28 (23.3) |
| III–IV | 92 (76.7) |
| Differentiation | |
| High | 38 (31.6) |
| Moderate and low | 82 (68.3) |
Cell Lines Culture
The human ovarian cancer cell lines of ES-2, HO8910, OVCAR-3, A2780 and human normal ovarian epithelial (HNOE) were from American Type Culture Collection (ATCC). All of the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, Ca) and supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and penicillin/streptomycin (Invitrogen) with 5% concentration of CO2 at 37◦C.
Total RNA Isolation, qRT-PCR, Transfection and GSEA Analysis
The expression levels of RNA were calculated by the qRT-PCR system. Total RNA was extracted by TRIzol reagent (Invitrogen), and 1 μg of total RNA was reverse transcribed using the PrimeScriptP RT Reagent Kit (Perfect Real-Time; Takara). SNHG22 cDNA, shRNA interference vector lentiviruses were purchased from GeneChem Corporation (Shanghai, China). SP1 siRNAs were purchased from Oligobio Company (Beijing, China). Ovarian cancer cells were transfected with plasmids in the presence of Lipofectamine 3000 (Invitrogen). After 48 h of transfection, cells were gathered for further use in the following experiments. The gene expression quantity was calculated using the 2-ΔΔCt method. The detail is in Tables S1, S2. Gene set enrichment analysis (GSEA) was performed with Broad’s GSEA algorithm.
Cell Proliferation Assay
Cell viability assay was carried out to analyze cell proliferation. Cell viability was estimated using CCK8 (CK04, DOJINDO, Beijing, China), based on the manufacturer’s instruction. Cells were seeded in 96‐well culture plates. After incubation for the indicated time, CCK8 reagent (10 μL) was added to each well. Cell viability was measured with a microplate reader for the absorbance at a wavelength of 450 nm.
Apoptosis Analysis
For apoptosis analysis, transfected cells were collected by trypsin 48 h after transfection and then stained with Annexin V-FITC and PI using Cell apoptosis Analysis Kit (Beyotime Biotechnology, China). Cell apoptosis was analyzed by flow cytometry (FACScan; BD Biosciences) using CellQuest software (BD Biosciences).
Glucose Uptake and Lactate Secretion Detection
The levels of glucose and lactate were calculated with a Glucose Colorimetric Assay Kit (BioVision, CA) and a Lactate Assay Kit (BioVision, CA) in line with the instructions of the manufacturer.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed with the protocol of SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) (#9003). Cells are fixed with formaldehyde and lysed, and chromatin is fragmented by partial digestion with Micrococcal Nuclease to obtain chromatin fragments of 1 to 5 nucleosomes. Chromatin immunoprecipitations are performed using ChIP-validated antibodies and ChIP-Grade Protein G Magnetic Beads. After reversal of protein-DNA cross-links, the DNA was purified using DNA purification spin columns and was quantified by real-time qPCR analysis.
Dual-Luciferase Reporter Assay
The SNHG22 promoter after PCR amplification was inserted into the downstream of the firefly luciferase gene in pGL3-Basic vector (Promega, USA) to construct pGL3-SNHG22 promoter reporter vector. Cells were co-transfected with a reporter construct (pGL3-basic plasmid or pGL3-SNHG22 plasmid) and shRNA or negative control. Dual-Luciferase Reporter Assay System (Promega) was used to measure the luciferase activity 48 h after transfection. The relative luciferase activity was contrasted with the Renilla luciferase and firefly luciferase activity.
Animal Study
This study had been approved by the Committee on the Ethics of Animal Experiments of the Second People’s Hospital of Baise. The mice were randomly assigned into five groups with six mice in each group of vector and SNHG22. ES-2 cells were transfected with SNHG22 1×107 indicated cells were subcutaneously injected into 4-week-old nude mice. After 35 days, the 8 mice were sacrificed by excessive carbon dioxide asphyxiation euthanasia. The tumor size was examined by calculating the length (L) and width (W) with calipers every 5 days, and the tumor volume was analyzed utilizing the method: (L × W2)/2. The animal study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the NIH.
Statistical Analysis
All the data were shown as the mean ± standard deviation, at least three independent experiments. Data were compared using the chi-square (χ2 test), Student’s t-test or one-way analysis of variance (ANOVA). ROC curve was applied to evaluate the predictive ability of ovarian cancer diagnosis. Univariate and multivariate analyses were performed to ascertain independent factors for the diagnosis of ovarian cancer. Kaplan–Meier method was applied to measure the overall survival (OS). SPSS 22.0 (SPSS Inc., Chicago, IL, USA) was used to conduct statistical analyses, and differences were ensured when P-value was <0.05.
Results
SNHG22 is Enhanced in EOC
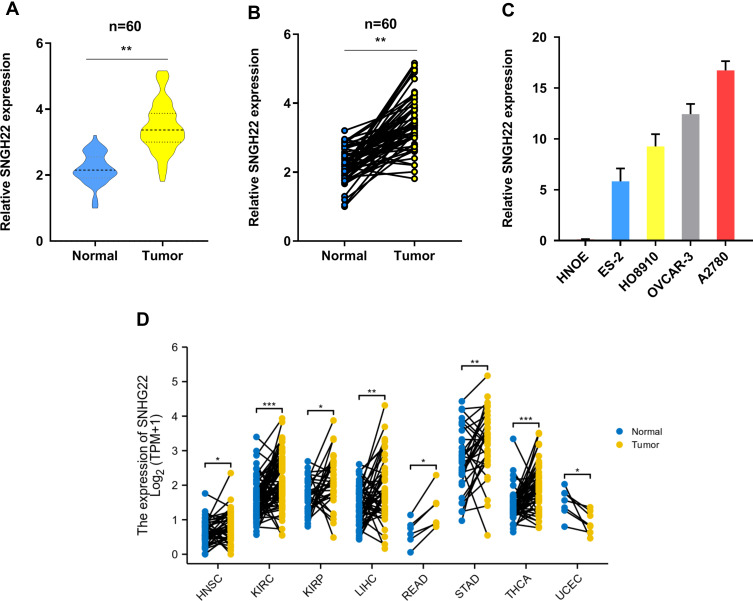
To explore whether SNHG22 was a long non-coding RNA, we examined the open-reading frames applying the ORF finder. No ORF was identified in SNHG22. Five other different online metrics also found that SNHG22 could not code protein (Table S3). We examined SNHG22 expression in 60 paired and non-tumor samples of EOC by qRT-PCR, the characteristics of EOC patients were showed in Table 1. The expression of SNHG22 was upregulated in EOC tissues compared with normal tissues (Figure 1A and B), and the cell lines of ES-2, HO8910, OVCAR-3, A2780 and HNOE were explored for SNHG22 expression. Significantly, the SNHG22 expression was increasing in cancer cell lines compared with HNOE cells (Figure 1C). Moreover, we explored the TCGA database and found SNHG22 expression was upregulated in many malignant tumors (Figure 1D). These results indicated that SNHG22 is overexpressed in EOC.
Figure 1.
SNHG22 is upregulated in ovarian cancer. (A, B) the qRT-PCR analysis was used to evaluate SNHG22 expression in 60 paired tumor and paired adjacent non-tumor tissues. (C) The expression profiles of SNHG22 in ES-2, HO-8910, OVCAR-3, A2780 and human normal ovarian epithelial (HNOE) were detected with qRT-PCR. (D) The expression of SNHG22 in HNSC, KIRC, KIRP, LIHC, READ, STAD, THCA and UCEC. *P < 0.05, **P < 0.01, ***P < 0.001.
Ectopic Expressed SNHG22 Correlates with Dismal Prognosis
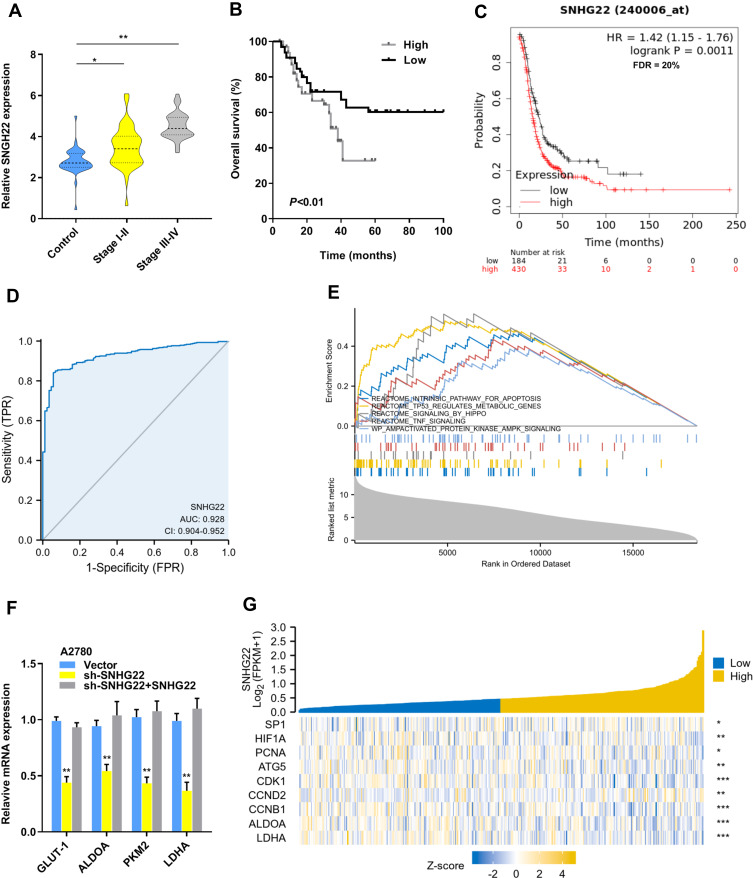
To examine the relationship between SNHG22 expression and clinicopathologic characteristics, SNHG22 expression was divided into low-expression and high-expression groups through the basis of median expression. And the SNHG22 expression was linked with vessel invasion (P < 0.05) and TNM stage (P < 0.01), while age (P = 0.68), gender (P = 0.56), tumor size (P = 0.06), tumor invasion depth (P = 0.23), or lymph node metastasis (p=0.08) were not (Table 2). And stage III–IV (advanced stage) had a higher SNHG22 level than stage I–II (early stage) (Figure 2A). Kaplan–Meier survival curves showed that patients with higher SNHG22 expression had a shorter survival time (Figure 2B) and predicted a poor prognosis of progress-free survival (PFS) in the KM-plotter (Figure 2C). The univariate analysis showed that overexpressing SNHG22 led to a dramatic risk of death (P < 0.01) (Table 3), and SNHG22 expression was an independent prognostic factor in multivariate analysis (P < 0.01).
Table 2.
Correlation Between SNHG22 Expression and Clinicopathologic Characteristics of EOC Patients
| Variables | SNHG22 Expression | P-value | ||
|---|---|---|---|---|
| Total (n=120) | High Expression | Low Expression | ||
| Age (years) | ||||
| ≤60 | 53 | 27 | 26 | 0.94 |
| >60 | 67 | 32 | 35 | |
| Tumor size (cm) | ||||
| ≤5 | 84 | 47 | 37 | 0.06 |
| > 5 | 36 | 16 | 24 | |
| Tumor invasion depth | ||||
| T1-2 | 28 | 12 | 16 | 0.23 |
| T3-4 | 92 | 53 | 39 | |
| Lymph node metastasis | ||||
| N0 | 45 | 25 | 20 | 0.08 |
| N1-2 | 75 | 36 | 39 | |
| Vessel invasion | ||||
| Yes | 69 | 49 | 20 | 0.03 |
| No | 51 | 20 | 31 | |
| TNM stage | ||||
| I–II | 28 | 6 | 22 | <0.01 |
| III–IV | 92 | 51 | 41 | |
| Differentiation | ||||
| High | 38 | 20 | 18 | 0.80 |
| Moderate and low | 82 | 42 | 40 | |
| Smoking status | ||||
| Ever | 46 | 24 | 22 | 0.92 |
| Never | 74 | 36 | 38 | |
| Drinking status | ||||
| Ever | 58 | 28 | 30 | 0.96 |
| Never | 62 | 30 | 32 | |
Figure 2.
Overexpressed SNHG22 correlates with poor prognosis in ovarian cancer. (A) Comparison of the expression level of SNHG22 in control, stage I–II (early stage) and stage III–IV (advanced stage) of EOC patients. (B) SNHG22 expression and survival predicted poor prognosis of OS in a cohort of 60 paired cases. (C) SNHG22 expression and survival predicted poor prognosis of PFS in the KM-plotter (http://kmplot.com/analysis/). (D) ROC curve analysis of the diagnostic value of SNHG22. (E) Gene set enrichment analysis of TCGA OV dataset showed enrichment of five pathways gene panels in the SNHG22 high expression group. (F) Analysis of the glycolic gene expression in SNHG22 knockdown or SNHG22 re-expression in A2780 cells. (G) SNHG22 co-expression heat map, TCGA (https://portal.gdc.cancer.gov/) OV, level 3 HTSeq-FPKM. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 3.
Univariate and Multivariate Analyses of Clinicopathologic Characteristics for Correlations with Overall Survival
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| SNHG22 expression | 1.91 (1.32–2.75) | <0.01 | 2.54 (1.18–4.23) | <0.01 |
| Vessel invasion | 1.92 (1.27–2.89) | <0.01 | 1.65 (1.12–2.54) | <0.01 |
| TNM stage | 1.64 (1.21–2.45) | <0.01 | 1.32(0.95–1.79) | 0.64 |
Subsequently, the ROC curve was used to assess the diagnostic value of SNHG22, the normal adjacent counterparts were used as a control, the area under the ROC curve (AUC) was 0.928 (95% CI: 0.904–0.952), with the sensitivity of 84.1% and specificity 94.3%, respectively (Figure 2D). Therefore, we speculated that forced SNHG22 expression predicted the unsatisfactory prognosis in EOC.
SNHG22 Promotes Glycolysis and Proliferation in EOC
To gain insight into the biological function of SNHG22 in EOC, the SNHG22 was overexpressed and silenced in ES-2 or A2780 cells by transforming overexpressed plasmids and shRNA targeting SNHG22 into them (Figure S1a). SNHG22 depletion might regulate glycolysis through the Gene set enrichment analysis (GSEA) (Figure 2E). The genes (GLUT-1, ALDOA, PKM2 and LDHA) related to glycolysis were silenced by SNHG22 knockdown and SNHG22 re-expression could recover these genes expression (Figure 2F). Moreover, SNHG22 was correlated with glycolytic related genes (ALDOA, LDH), proliferation-related genes (PCNA), cell cycle-related genes (CDK1, CCND2, CCNB1), and apoptosis-related genes (ATG5) from the TCGA-OV database (Figure 2G), suggesting that SNHG22 could regulate these phenotypes.
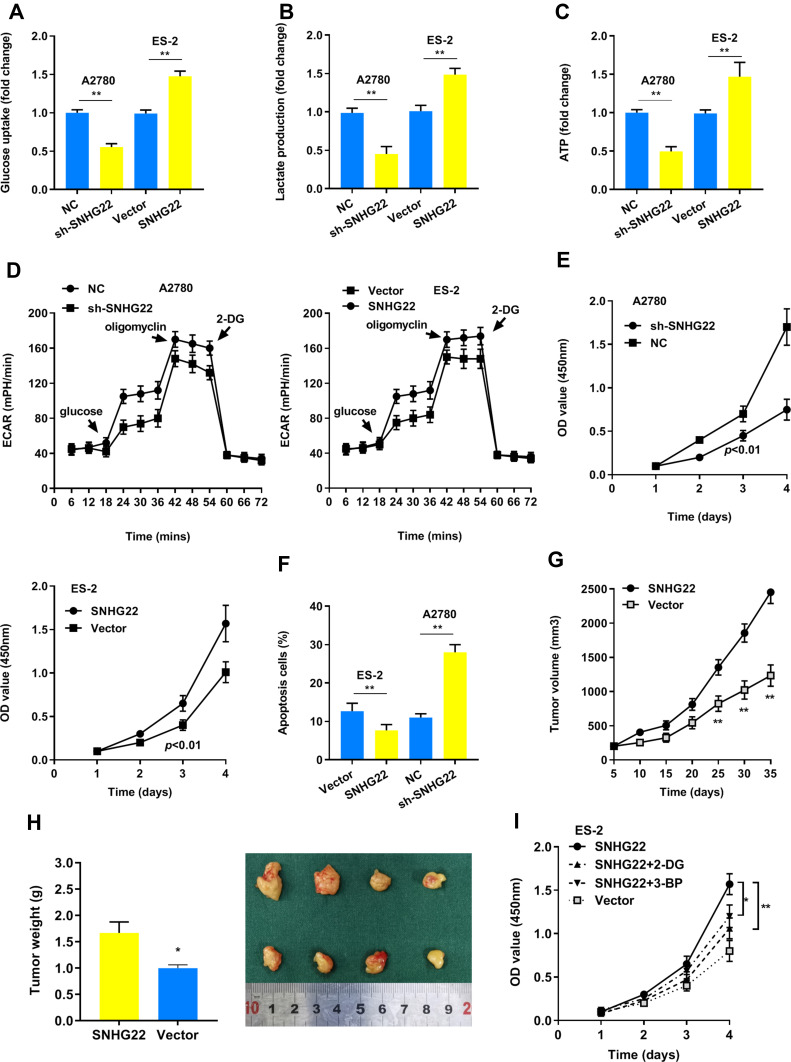
Our results found that SNHG22 depletion repressed glucose uptake, lactate production, ATP level and ECAR level, whereas the opposite outcome was observed after enforced expression of PTTG3P (Figure 3A-D). Thus, SNHG22 could regulate glycolysis. We also indicated that silenced SNHG22 suppressed cell proliferation and facilitated apoptosis (Figure 3E and F).
Figure 3.
SNHG22 promotes glycolysis and proliferation. (A) Glucose uptake analysis, (B) lactate production analysis, and (C) ATP analysis explored the glucose uptake, lactate production, and ATP accumulation. (D) Extracellular acidification rate (ECAR) analysis tested the glycolytic capacity in ovarian cancer cells transfected with sh-SNHG22 or overexpressed SNHG22. (E) CCK8 assay detected the proliferation of A2780 and ES-2 cells transfected with sh-SNHG22 or overexpressed SNHG22. (F) Flow cytometry assays revealed that SNHG22 affected cell apoptosis (G) Tumor volume and (H) weight were measured in vivo when injected with overexpressed SNHG22 in A2780 cells. (I) CCK8 assay detected the proliferation of ES-2 cells transfected with overexpressed SNHG22 and treated with 2.5mM 2-DG or 100 μM 3-BP. *P < 0.05, **P < 0.01.
In the animal study, we confirmed that SNHG22 overexpression facilitated tumor growth (Figure 3G and H). Next, we hypothesized that glycolysis might play a vital role in cell proliferation. As shown in Figure 3I, the glycolic inhibitors 2-DG or 3-BP could partly recover SNHG22 induced proliferation. Taken together, our findings recommended that SNHG22 facilitated cancer cell growth, and blocking glycolysis could partially rescue this effect.
SP1 Binds to the Promoter of SNHG22 and Promotes Its Transcription
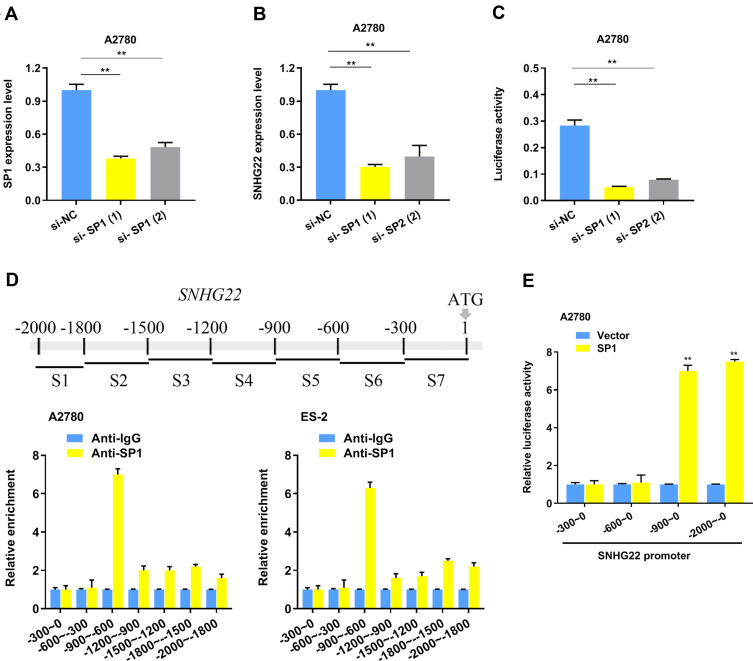
To explore the SNHG22 expression profiles in A2780 cells, we discovered that no genomic amplification was found (Figure S1b), and DNA methyltransferase inhibition induced no increase of SNHG22 expression (Figure S1c). Then we hypothesized that transcription factors played a role in regulating the expression of SNHG22. The JASPAR and UCSC website analysis showed that SNHG22 might be a direct target of SP1. First, we silenced the SP1 expression in A2780 cells using siRNA (Figure 4A) and found that the SNHG22 expression was decreased (Figure 4B). Second, the relative luciferase activity was significantly inhibited in A2780 cells transfected with si-SP1 (Figure 4C). According to the Jaspar analysis (Table S5), we divided the SNHG22 promoter to 7 regions (−300~ 0, −600~ −300, −900~ −600, −1200~ −900, −1500~ −1200, 1800~ −1500, −2000~ −1800), and ChIP analysis indicated SP1 markedly enriched in the promoter region of −900~ −600 (Figure 4D).
Figure 4.
SP1 regulates SNHG22 expression by serving as a transcription activator. (A) The SP1 interference efficiency using 2 different siRNAs and negative control detected by qRT-PCR. (B) qRT-PCR analysis was used to explore SNHG22 expression after SP1 knockdown in A2780 cells. (C) The luciferase activity was measured by transfecting SNHG22 promoter in A2780 cells. (D) SP1 mainly enriched on the promoter (− 900 bp~ − 600 bp) of SNHG22 in A2780 and ES-2 cells. (E) The luciferase activity was measured by transfecting different SNHG22 promoter region in A2780 cells. *P < 0.05, **P < 0.01.
Subsequently, we speculated that SNHG22 promoter region (−900~ −600) might be necessary for SP1 binding, then we discovered that SNHG22 without −900~ −600 promoter could not be regulated by SP1 (Figure 4E).
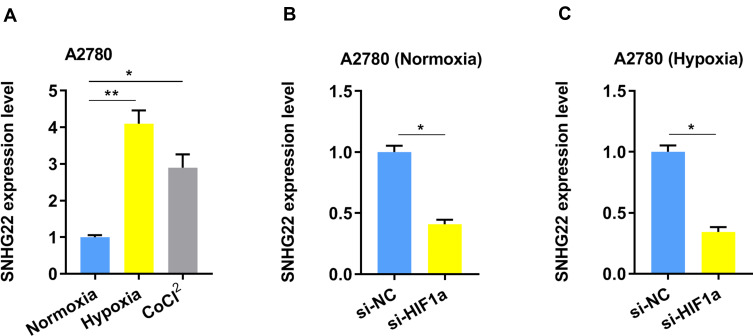
Additionally, tumor hypoxia refers to the lack of oxygen in tumor cells. The blood supply cannot meet the rapid growth of the tumor, causing the oxygen concentration in some areas of the tumor to be significantly lower than that of healthy tissue. We used hypoxia or CoCl2 (hypoxia chemical inducer) to treat the A2780 cells and found the SNHG22 expression was elevated (Figure 5A). While, ablation of HIF-1α dramatically ameliorated SNHG22 expression in both normoxia and hypoxia conditions (Figure 5B and C).
Figure 5.
The SNHG22 expression is regulated by hypoxia. (A) The expression of SNHG22 in A2780 cells was measured after culturing under normoxia, hypoxia (1% O2) or CoCl2 (100 μM) for 24 h by qRT-PCR. (B) The expression of SNHG22 was evaluated by qRT-PCR in A2780 cells after knockdown of HIF-1α under normoxia condition. (C) The expression of SNHG22 was evaluated by qRT-PCR in A2780 cells after knockdown of HIF-1α under hypoxia condition. *P < 0.05, **P < 0.01.
SNHG22 Plays a Crucial Role in EOC Immunology
The glucose uptake and glycolysis were higher in cancer cells, caused a lower level of the content of glucose and the production of IFN-r by CD8+T cells in the tumor. The tumor immune environment broadly participates in different malignant tumors, including EOC. Recently, the treatment of EOC with immune checkpoint inhibitors (ICI) has provided us a potential clinical treatment. Our findings suggested that low expression of the SNHG22 related to Th2 and neutrophils cells infiltration in the microenvironment of EOC, based on the TCGA database (Figure S2).
Discussion
The custom annotation pipeline of microarray data was analyzed in 217 early-stage Binet A CLL patients and 26 different subpopulations of normal B-cells and we found that SNHG22 may play a role in this process.7 Next, Koduru et al8 proposed that the level of SNHG22 could be served as a biomarker during the malignant transformation of hepatocellular carcinoma. Another, Fang et al9 documented that SNHG22 regulated triple-negative breast cancer via the miR-324-3p/SUDS3 axis and provided a promising targeted therapy. Recently, SNHG22 acts as a ceRNA to regulate Gal-1 expression by competitively binding to miR-2467 in EOC.6 But the upstream factor of SNHG22 and whether SNHG22 could modulate glycolysis is unclear in EOC.
Our study found that SNHG22 has a high expression in a total of 60 paired tumor and non-tumor tissues, and four EOC cell lines. The EOC patients were discriminated from the healthy controls based on the expression of SNHG22, with an AUC of 0.824 (95% CI: 0.747–0.900). Then we indicated that higher expression of the SNHG22 is correlated with clinicopathological features and led to shorter survival time. Further, SNHG22 expression is suggested to be an independent prognostic indicator of survival through multivariate analysis. Hence, our results proposed that SNHG22 is a promising diagnostic marker for patients with EOC. The up-regulation of glycolysis and acid resistance gives cancer cells a strong growth advantage, which can promote unrestricted proliferation and invasion.10–12 Otto Warburg first described this phenomenon in the 1920s, which is known as the Warburg effect.13 The Warburg hypothesis suggests that cancer is prominently caused by mitochondrial metabolism disorder. And emerging evidence indicates that targeting lactate metabolism is a promising approach for cancer therapeutics.12 In our approach, we demonstrated that SNHG22 regulates several genes linked with metabolic pathways and SNHG22 could facilitate glycolysis according to the elevated level of ECAR and lactic acid. Using a calorie-restricted ketogenic diet that shifts energy metabolism from glucose to a ketone-based fuel source may provide a means to starve cancer cells and suppress tumor progression.14,15 However, the detailed molecular mechanisms of SNHG22 in glycolysis of EOC remain to be explained.
Transcription factors can promote (as activators) or block (as repressors) the recruitment of RNA polymerase to promote or inhibit the rate of transcription of genetic information from DNA to messenger RNA. SP1 is a famous transcription factor. Our study demonstrated that SP1 could elevate SNHG22 expression by binding to its promoter, standing in line with other studies. For instance, SP1 can activate the transcription by binding to the GCMA promoter region in gastric cancer.16 Another study showed that SP1 can bind to the promoter of genes such as LMCD1-AS1 and SPRY4-IT1, and upregulate the transcription of them.17,18 In our study, we verified that SP1 induced SNHG22 overexpression in EOC. The SNHG22 region of nt −500~ 1 is indispensable for SP1 regulating SNHG22. We tested the expression of SNHG22 under hypoxia, for SNHG22 modulate glycolysis, hypoxia or the transcription factor hypoxia-inducible factor 1 (HIF-1) could greatly affect SNHG22 expression.
In conclusion, our findings documented that SNHG22 participates in EOC progression and can be a potential therapeutic target for EOC patients.
Acknowledgments
Thank you for li deng to fulfill some part of the statistical analysis. Ning Guan and Haiying Zheng are the co-first authors for this study.
Funding Statement
Natural Science Foundation of Liaoning Province, Project No. 2019-ZD-0588. Ethical approval No. 2019S1111.
Abbreviations
SNHG22, Small nucleolar RNA host gene 22; EOC, Epithelial ovarian carcinoma; ROC, Receiver operating curve; OS, Overall survival; lncRNA, Long non-coding RNA; GSEA, Gene set enrichment analysis; HNOE, Human normal ovarian epithelial; ChIP, Chromatin immunoprecipitation; ORF, Open reading frame; HIF-1, Hypoxia-inducible factor 1.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Statement of Ethics
Patients have given their written informed consent in our study. And ethics committee of the Second People’s Hospital of Baise and Liaoning cancer hospital approved the study protocol.
Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Walker JL, Powell CB, Chen L, et al. Society of Gynecologic Oncology recommendations for the prevention of ovarian cancer. Cancer. 2015;121(13):2108–2120. [DOI] [PubMed] [Google Scholar]
- 2.Webb PM, Green AC, Jordan SJ. Trends in hormone use and ovarian cancer incidence in US white and Australian women: implications for the future. Cancer CausesControl. 2017;28(5):365. [DOI] [PubMed] [Google Scholar]
- 3.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. [DOI] [PubMed] [Google Scholar]
- 4.Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. [DOI] [PubMed] [Google Scholar]
- 5.Bhosale P, Peungjesada S, Wei W, et al. Clinical utility of positron emission tomography/computed tomography in the evaluation of suspected recurrent ovarian cancer in the setting of normal CA-125 levels. Int J Gynecol Cancer. 2010;20:6. [DOI] [PubMed] [Google Scholar]
- 6.Zhang PF, Wu J, Luo JH, et al. SNHG22 overexpression indicates poor prognosis and induces chemotherapy resistance via the miR-2467/Gal-1 signaling pathway in epithelial ovarian carcinoma. Aging. 2019;11(19):8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronchetti D, Manzoni M, Agnelli L, et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016;6(9):e468–e468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koduru SV, Leberfinger AN, Kawasawa YI, et al. Non-coding RNAs in various stages of liver disease leading to hepatocellular carcinoma: differential expression of miRNAs, piRNAs, lncRNAs, circRNAs, and sno/mt-RNAs. Sci Rep. 2018;8(1):1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang X, Zhang J, Li C, et al. Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int. 2020;20(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. [DOI] [PubMed] [Google Scholar]
- 11.Ou J, Miao H, Ma Y, et al. Loss of abhd5 promotes colorectal tumor development and progression by inducing aerobic glycolysis and epithelial-mesenchymal transition. Cell Rep. 2014;9(5):1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123(9):3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz L, Seyfried T, Alfarouk K O, et al. Out of Warburg effect: An effective cancer treatment targeting the tumor specific metabolism and dysregulated pH. Seminars in cancer biology. Academic Press. 2017;43:134–138. [DOI] [PubMed] [Google Scholar]
- 15.Maroon J, Bost J, Amos A, et al. Restricted calorie ketogenic diet for the treatment of glioblastoma multiforme. J Child Neurol. 2013;28(8):1002–1008. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y, Ma R, Sun Y, et al. SP1-activated long noncoding RNA lncRNA GCMA functions as a competing endogenous RNA to promote tumor metastasis by sponging miR-124 and miR-34a in gastric cancer. Oncogene. 2020;39(25):4854–4868. [DOI] [PubMed] [Google Scholar]
- 17.He J, Li D, Zhou J, et al. SP1-mediated upregulation of lncRNA LMCD1-AS1 functions a ceRNA for miR-106b-5p to facilitate osteosarcoma progression. Biochem Biophys Res Commun. 2020;526(3):670–677. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Yao Y, Jiang X, et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]