Abstract
The CCR4-NOT complex (1 mDa in size), consisting of the proteins CCR4, CAF1, and NOT1 to NOT5, regulates gene expression both positively and negatively and is distinct from other large transcriptional complexes in Saccharomyces cerevisiae such as SNF/SWI, TFIID, SAGA, and RNA polymerase II holoenzyme. The physical and genetic interactions between the components of the CCR4-NOT complex were investigated in order to gain insight into how this complex affects the expression of diverse genes and processes. The CAF1 protein was found to be absolutely required for CCR4 association with the NOT proteins, and CCR4 and CAF1, in turn, physically interacted with NOT1 through its central amino acid region from positions 667 to 1152. The NOT3, NOT4, and NOT5 proteins had no significant effect on the association of CCR4, CAF1, and NOT1 with each other. In contrast, the NOT2, NOT4, and NOT5 interacted with the C-terminal region (residues 1490 to 2108) of NOT1 in which NOT2 and NOT5 physically associated in the absence of CAF1, NOT3, and NOT4. These and other data indicate that the physical ordering of these proteins in the complex is CCR4-CAF1-NOT1-(NOT2, NOT5), with NOT4 and NOT3 more peripheral to NOT2 and NOT5. The physical separation of CCR4 and CAF1 from other components of the CCR4-NOT complex correlated with genetic analysis indicating partially separate functions for these two groups of proteins. ccr4 or caf1 deletion suppressed the increased 3-aminotriazole resistance phenotype conferred by not mutations, resulted in opposite effects on gene expression as compared to several not mutations, and resulted in a number of synthetic phenotypes in combination with not mutations. These results define the CCR4-NOT complex as consisting of at least two physically and functionally separated groups of proteins.
The CCR4-NOT complex from Saccharomyces cerevisiae displays both positive and negative roles in the regulation of diverse genes and processes (6, 9, 20, 25). This complex, distinct from other large transcriptionally important complexes such as SNF/SWI, SAGA, SRB-containing polymerase II holoenzyme, and TFIID (10, 13, 20), consists of two forms, a 1.9 × 106-Da (1.9-mDa) and 1-mDa complex (20). The smaller complex consists of CCR4, CAF1 (POP2) (24), the five NOT proteins, and several unidentified proteins (20, 22, 23). Defects in components of this complex reduce expression of ADH2 and other nonfermentative genes, affect the expression of genes involved in cell wall integrity, and suppress spt10-induced expression at the ADH2 locus (9, 11, 13, 20, 22). Furthermore, mutations in CCR4 or CAF1 affect cell cycle progression in late mitosis (22). The NOT genes, in turn, were originally identified as repressing HIS3 expression from a noncanonical TATA (TATA-less) element (5, 6), as well as affecting a number of other genes and processes (1, 8, 16). The recent demonstration that not and caf1 mutations can suppress a defect in SRB4, a key component of the RNA polymerase II holoenzyme required for the transcription of most genes in yeast (19), further indicates a very general repressor role for the CCR4-NOT complex. It has been proposed that the NOT proteins inhibit transcriptional initiation by affecting TATA binding protein access to TATA-less sequences (4), a model in agreement with the fact that NOT1 has been found to associate with TATA binding protein (TBP) (19).
Of the proteins of the CCR4-NOT complex, only NOT1 is an essential protein (5). The C-terminal residues 1319 to 2108 of NOT1 are sufficient, however, for cells to remain viable (26). Pairwise combinations of not mutations do not in general lead to synthetic lethality (except for not4 with not5), suggesting that they form a complex displaying overlapping functions (6, 23). However, CCR4 and CAF1 appear in certain contexts to be distinct from the other NOT proteins. Mutations in the five NOT genes result in increased resistance to 3-aminotriazole (3-AT) in a partially defective GCN4 background (6). This phenotype is not associated with CCR4 or CAF1 defects (20). Moreover, not mutations tend to increase HO-lacZ and FKS1-lacZ expression, whereas a ccr4 or caf1 deletion reduces expression or has little effect on these promoters (20). The CCR4 and CAF1 proteins also appear to be strongly associated; partial disruption of CAF1 inhibits the association of CCR4 with the NOT1 and NOT2 proteins (20). Therefore, while not alleles have several phenotypes in common with ccr4 and caf1 defects (20), notably caffeine, temperature, and magnesium sensitivities, effects on ADH2 and CYC1 gene expression, and suppression of spt10-enhanced expression, CCR4 and CAF1 proteins may functionally and physically represent a separate group of proteins within the CCR4-NOT complex.
We have analyzed the association of CCR4, CAF1, and the NOT proteins and related these associations to the phenotypes of the constituents of this complex. The central segment of NOT1 (residues 667 to 1152) binds CCR4 and CAF1, whereas the C-terminus of NOT1 (1490 to 2108) associates with NOT2, -4, and -5. We provide evidence that the arrangement of the proteins in the complex is CCR4-CAF1-NOT1-(NOT2, NOT5), with NOT3 and NOT4 peripheral to NOT2 and NOT5. Further, the physical separation of CCR4 and CAF1 from the other NOT proteins correlates, in general, with phenotypes associated with defects in CCR4 and CAF1 compared to the other NOT mutations. The CCR4-NOT complex appears, therefore, to be composed of at least two physically separate groups of proteins that can function differently depending on the promoter context.
MATERIALS AND METHODS
Yeast strains, growth conditions, and enzyme assays.
Yeast strains (Table 1) were grown at 30°C on YEP medium (2% yeast extract, 1% Bacto Peptone) or selective medium (7) supplemented with 5% glucose or with 2% galactose and 2% raffinose unless otherwise indicated. β-Galactosidase assays and alcohol dehydrogenase (ADH) assays were carried out as described previously (12). Assay values represent the averages of at least three independent assays. The yeast transformation protocol was as described previously (7, 17).
TABLE 1.
Yeast strains used
| Strain | Genotype |
|---|---|
| A790 | MATα aro7 his3 leu2 ura3 |
| A792 | MATα aro7 his3 leu2 ura3 pop2-Δ3::LEU2 |
| KY803 | MATa leu2-PET56 trp1-Δ1 ura3-52 gal2 gcn4-Δ1 |
| KY803-1 | Isogenic to KY803 except ccr4::URA3 |
| KY803-c1 | Isogenic to KY803 except caf1::LEU2 |
| MY8 | Isogenic to KY803 except not1-2 |
| MY8-1d | Isogenic to MY8 except ccr4::URA3 |
| MY8-c1c | Isogenic to MY8 except caf1::LEU2 |
| MY1737 | Isogenic to KY803 except his3::TRP1 not1::LEU2 pRS426-NOT1(396–2108) |
| MY1738 | Isogenic to KY803 except MATα not1::LEU2 pRS426-NOT1(1319–2108) |
| MY16 | Isogenic to KY803 except not2-1 |
| MY16-1c | Isogenic to MY16 except ccr4::URA3 |
| MY508 | Isogenic to KY803 except not3::URA3 |
| MY508-c1b | Isogenic to MY508 except caf1::LEU2 |
| MY25 | Isogenic to KY803 except not3-2 |
| MY25-1b | Isogenic to MY25 except ccr4::URA3 |
| MY537 | Isogenic to KY803 except not4::URA3 |
| MY537-1 | Isogenic to MY537 not4::ura3 |
| MY537-1-1b | Isogenic to MY537-1 ccr4::URA3 |
| MY1735 | Isogenic to KY803 except not5::URA3 |
| EGY188 | MATa ura3 his3 trp1 LexA-LEU2 |
| EGY188-1 | Isogenic to EGY188 except ccr4::URA3 |
| EGY188-c1 | Isogenic to EGY188 except caf1::URA3 |
| EGY188-1-1 | Isogenic to EGY188 except ccr4::ura3 |
| EGY188-c1-1 | Isogenic to EGY188 except caf1::ura3 |
| 787-6b | MATα adh1-11 ADR1-5C-TRP1 ura3 his3 leu2 trp1 |
| 1076-2c-1 | MATα ura3 his3 leu2 trp1 ccr4::ura3::TRP1 |
| 1278-5d | MATa ura3 his3 leu2 trp1 caf1::LEU2 |
| 1393-4a | MATα his3/his2 leu2 ura3 not2::TRP1 |
| 1402-1a | MATα ura3 his3 leu2 trp1 not4::URA3 |
| 1402-4a | MATα ura3 his3 leu2 trp1 not4::URA3 |
| 1422-21 | MATα adh1-11 ura3 his3 leu2 trp1 not1::LEU2 pRS426-NOT1(1490–2108) |
| 1469-2-1c | MATa ura3 his3 leu2 trp1 ccr4::ura3::TRP1 |
| 1462-3c | MATa ura3 his3 leu2 trp1 not5::URA3 |
DNA constructions.
The LexA-NOT1 plasmids containing various length of NOT1 were constructed as follows. For expression of LexA-NOT1(667–1152), pLexA-NOT1 was cut with BamHI and XhoI and the segment carrying codons 667 to 1152 was ligated with pLexA202-4 (7) cut with the same two enzymes. For expression of LexA-NOT1(1480–2108), pET28a-NOT1 (XbaI-SalI) was cut with BamHI and SalI and the segment carrying codons 1480 to 2108 was inserted into the BamHI and SalI sites of pLexA-202-2. For expression of LexA-NOT1(1–667), pLexA-NOT1 was cut with BamHI and SalI, the ends were filled in with the large subunit of Escherichia coli DNA polymerase (Klenow), and the plasmid was religated. For expression of LexA-NOT1(1–1152), pLexA-NOT1 was cut with EcoRI and XhoI and the fragment encoding residues 1 to 1152 of NOT1 was ligated with pLexA202-2 cut with EcoRI and SalI. For expression of LexA-NOT1(667–2108), pLexA-NOT1 was cut with BamHI and SalI and the piece encoding residues 667 to 2108 was inserted into the BamHI and SalI sites of pLexA202-2. Expression of all fusion proteins was confirmed by Western blot analysis.
Antibodies and immunoprecipitation.
For Western analysis, the antibodies were directed against glutathione S-transferase (GST)–CAF1, GST-NOT2, His6-NOT5, GST-NOT1(1480–2108), and GST-DHH1(267–506) fusion proteins. Western analysis was conducted as described previously (13). Immunoprecipitations were carried out as previously described (12). The CAF1 antibody was partially purified as described elsewhere (14).
Gel filtration chromatography.
The procedure for gel filtration chromatography using a Superose 6 10/30 column was performed as described in detail elsewhere (20) except that the running buffer consisted of 50 mM Tris, 150 mM potassium acetate, and 0.02% Tween 20 only. The flow rate was 0.2 ml/min, and 0.5 ml was collected per fraction. Molecular weights for each fraction were calculated based on the elution volumes of blue dextran (7.5 ml), thyroglobin (12 ml), and bovine serum albumin (16 ml).
RESULTS
CAF1 is required for CCR4 association with the NOT proteins.
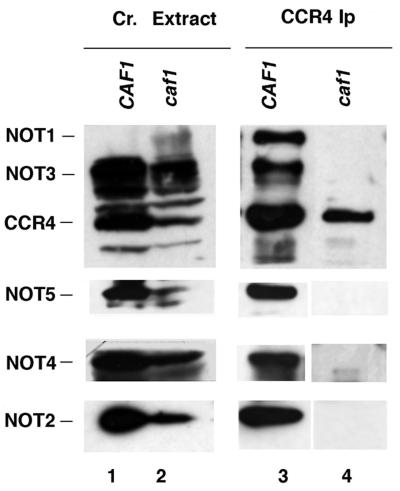
We had shown previously that deleting CAF1 removed CCR4 completely from the 1-mDa CCR4-NOT complex and reduced significantly but did not eliminate CCR4 association in the 1.9-mDa complex (20). We have also shown that CCR4 is dependent on CAF1 in its association with NOT1 and NOT2 (20). Using a complete deletion of CAF1, we further investigated the dependency on CAF1 for CCR4 for interaction with the other NOT proteins. In a wild-type strain, immunoprecipitation of CCR4 with anti-CCR4 antibody brought down NOT1 through NOT5 (Fig. 1, lane 3). In contrast, in a caf1 deletion strain, none of the NOT proteins coimmunoprecipitated with CCR4 (lane 4), although all NOT proteins were present in the crude extract (lane 2). CAF1 protein is therefore required for CCR4 to associate with all NOT proteins.
FIG. 1.
CCR4 requires CAF1 to immunoprecipitate NOT proteins. Immunoprecipitations with CCR4 antibody were conducted in caf1 (A792, pop2) and wild-type (A790) strains. Lanes 1 and 2, protein extracts from strains A790 and A792, respectively; lanes 3 and 4, immunoprecipitated (Ip) proteins analyzed by Western analysis using antibodies directed against NOT1 through NOT5 and CCR4. The NOT1 antibody used in these experiments could not detect NOT1 protein in crude (cr.) extracts (lanes 1 and 2), but other results indicate that NOT1 is present in both CAF1- and caf1-containing strains (20).
Physical interactions of CCR4-CAF1-NOT1 are independent of the NOT3, NOT4, and NOT5 proteins.
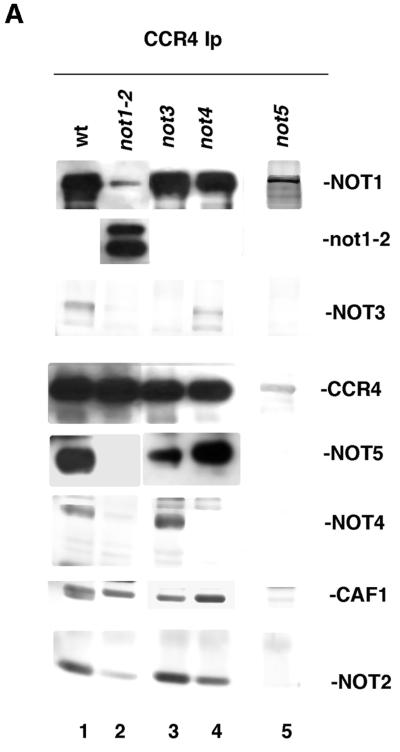
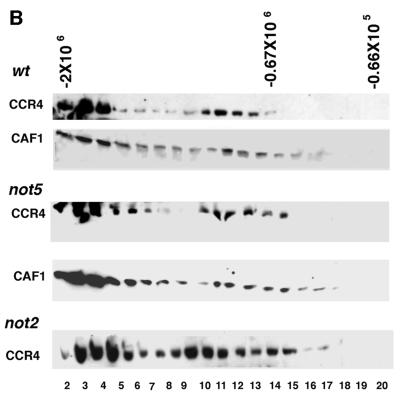
Since the above results suggest that the arrangement of proteins is CCR4-CAF1-NOTs, we subsequently examined what factors were required for CCR4, CAF1, and NOT1 to associate. Immunoprecipitation of CCR4 showed that NOT1 and CAF1 can be coimmunoprecipitated in not3-, not4-, or not5-deleted backgrounds (Fig. 2A, lanes 3 to 5). (For lane 5, the CAF1 protein was clearly visible in the original Western results.) The same results were obtained when anti-CAF1 antibody was used for immunoprecipitation (data not shown). The observation that the CCR4-CAF1 interaction was not dependent on NOT5 was further confirmed by gel filtration analysis (Fig. 2B, middle panel). In a not5 strain, CCR4 and CAF1 cofractionated in both 1.9-mDa (fractions 3 and 4) and 0.8-mDa (fractions 11 to 13) complexes which have been previously described for CCR4 and CAF1 (20) (Fig. 2B, top panel, in which CCR4 migrates in fractions 3 and 4 and fractions 10 to 12). It should be noted that the smaller complex in the not5 strain runs at a slightly smaller size (0.8 mDa) than the wild-type strain (0.9 mDa), probably due to loss of the NOT5 and other potential proteins. Also, the 0.9-mDa CCR4-NOT complex observed for the wild-type strain runs at slightly smaller size than in the strains used in our previous study (20).
FIG. 2.
CCR4 associates with CAF1 in the absence of NOT3, NOT4, NOT5, and the N-terminal 396 residues of NOT1. (A) Immunoprecipitations (Ip) were conducted with anti-CCR4 antibody. Western analysis was conducted with antibody directed against NOT1, NOT3, CCR4, NOT4, CAF1, NOT2, or NOT5 as indicated. An enhanced chemiluminescence-based system was used for NOT1, CCR4, and NOT5 Western blots for lanes 1 to 4, whereas an alkaline phosphatase-based system was used for the remainder of the results. Strains: wild type (wt), KY803; not1-2, MY8; not3, MY508; not4, MY537; not5, MY1735. (B) Yeast extracts from KY803 (wild type [wt]), 1393-4a (not2), and MY1735 (not5) were analyzed by gel filtration chromatography using a Superose 6 10/30 column. The protein extracts were precleared by centrifugation at 100,000 × g for 1 min, and 200 μl of sample was loaded onto the column. The flow rate was 0.2 ml/min, and a 0.5-ml volume was collected in each fraction; 100 μl from each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using CCR4 and CAF1 antibodies. Molecular weight markers for the gel filtration experiment were blue dextran (2 × 106 Da), thyroglobulin (0.67 × 106 Da), and bovine serum albumin (6.6 × 104 Da). (C) Immunoprecipitations were conducted in strain MY1737 [not1 pNOT1(396–2108)] and wild-type backgrounds with CAF1 antibody. Lanes 1 and 2 contain 1/10 of the crude extract (Cr. Ex.) protein input used for the immunoprecipitations (Ip) in lanes 3 and 4. Western analysis was conducted with anti-CCR4 and anti-NOT antibodies as indicated. NOT4 and NOT2 proteins in the crude extracts in lanes 1 and 2 were visible in the original Western blots and were in equal abundance for the two strains. IgG, immunoglobulin G.
We also conducted gel filtration analysis in not3 and not4 backgrounds. In a not3 deletion strain, CCR4 and CAF1 still migrated in large and medium complexes but the proteins were clearly more spread out, suggesting that their stability in the complexes was being compromised (data not shown). A not4 deletion had no apparent effect on CCR4 or CAF1 migration in the 1.9- and 0.8-mDa complexes (data not shown). CCR4 and CAF1 can therefore still associate in the CCR4-NOT complex in the absence of not3, not4, or not5, although these deletions may cause subtle effects on the structure and integrity of the complex. It should also be mentioned that in contrast to CCR4 and CAF1, the association of several other components of the CCR4-NOT complex could not be determined by gel filtration chromatography. NOT2 protein was hardly detectable after Superose 6 chromatography, NOT3 tended to migrate at its monomeric size, and NOT5 and NOT1 did not tend to migrate in well-defined peaks as was observed for CCR4 and CAF1 (data not shown).
The above results indicate that CCR4 and CAF1 can associate with NOT1 in the absence of NOT3, NOT4, or NOT5. In a not2 strain, however, the association of CCR4 with CAF1 and the NOT proteins could not be ascertained by immunoprecipitation due to the inability to immunoprecipitate sufficient levels of CCR4 and CAF1 proteins. This may be partially the result of the very low amount of CAF1 and several of the NOT proteins present in the extracts (data not shown; see also Fig. 5), but it may also result from overall instability of the complex and susceptibility to proteolytic degradation in a not2 background. Our gel filtration analysis indicated that CCR4 still migrated in 1.9- and 0.9-mD complexes in a not2 background (bottom panel of Fig. 2B), but the presence of CAF1 in the 0.9-mDa complex could not be determined and only a very small amount of CAF1 was visible in the 1.9-mDa complex (data not shown). In a not2-1 strain background, CCR4 and CAF1 migrated in both 1.9- and 0.9-mDa complexes (data not shown). These results suggest that NOT2 affects the overall integrity of the complex but may not be required for CCR4 association in the CCR4-NOT complex.
FIG. 5.
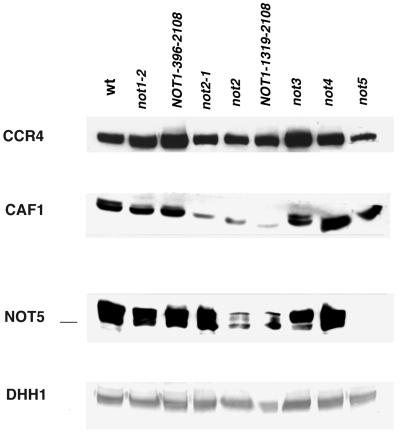
CCR4, CAF1, and NOT5 protein levels in not mutant backgrounds. All strains were grown to mid-log phase in YEP medium containing 5% glucose. Cells were harvested and lysed, and 40 μg of total protein was loaded in each lane. The RNA helicase homolog DHH1 was used as an internal control to demonstrate equivalent loading on the sodium dodecyl sulfate-polyacrylamide gel. Strains used: KY803 (wild type [wt]); MY8 (not1-2); MY1737 [not1 pNOT1(396–2108)]; MY1738 [not1 pNOT1(1319–2108)]; MY16 (not2-1); 1393-4a (not2); MY508 (not3); MY537 (not4); MY1735 (not5). Western blot analysis was conducted with antibodies directed against CCR4, CAF1, NOT5, and DHH1 as indicated.
It is also apparent in Fig. 2A that when CCR4 is immunoprecipitated, NOT4 is not required for NOT2, -3, and -5 to associate with CCR4, CAF1, and NOT1, and similarly, NOT3 is not required for NOT2, -4, -5, to associate with CCR4, CAF1, and NOT1. In a not5 background, however, because of the decrease in abundance of NOT3, NOT4, and NOT2 protein levels in the crude extracts used for the immunoprecipitation (data not shown), we could not ascertain if these three NOT proteins associated with CCR4, CAF1, and NOT1 following the immunoprecipitation. These results confirm that CCR4 and CAF1 are tightly if not directly linked, that NOT3 to NOT5 are not required for CCR4 and CAF1 association or with their association with NOT1, and that neither NOT3 nor NOT4 is required for NOT2 and NOT5 association with CCR4, CAF1, and NOT1.
The C terminus of NOT1 is not required for CCR4 and CAF1 association.
We used several complementary approaches to identify the region of NOT1 which interacted with CCR4 and CAF1. Since a not1 deletion is lethal, we initially used two truncated versions of NOT1 to assess the NOT1 requirement for CCR4 and CAF1 association. First, we analyzed the not1-2 allele. The not1-2 allele results in a NOT1 protein that is about 120 kDa in size (Fig. 2A, lane 2) and has been reported to be the result of a stop codon located in the region between residues 396 and 1318 of NOT1 (26). About 10% of the not1-2 protein is full length, which is apparently sufficient for the yeast to survive. The truncated not1-2 protein still coimmunoprecipitated with either CCR4 or CAF1 (Fig. 2A, lane 2, and data not shown). However, NOT5 and NOT4 no longer immunoprecipitated with CCR4 in a not1-2 strain (Fig. 2A, lane 2), and the amount of NOT2 was significantly reduced in the immunoprecipitation. The reduced amount of NOT2 that coimmunoprecipitated could be derived from NOT2 binding to the full-length NOT1 protein (Fig. 2A, lane 2). NOT5, NOT2, and NOT4 appear, therefore, to interact with the C-terminal region of NOT1, a result confirmed by other results described below.
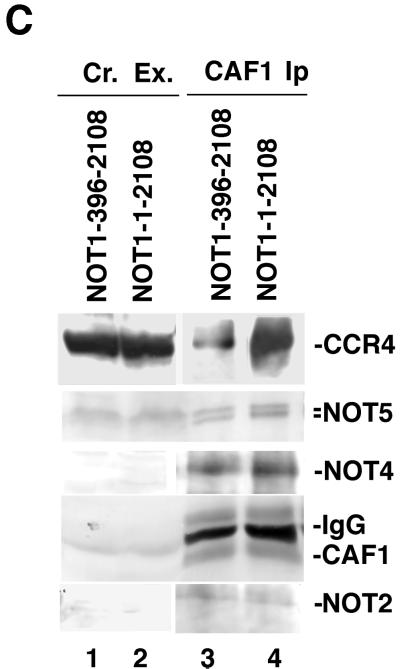
Second, we examined whether the CCR4-CAF1 interaction required the N-terminal 395 codons of NOT1. Immunoprecipitating CAF1 coimmunoprecipitated a significant amount of CCR4 in a strain carrying NOT1(396–2108) (Fig. 2C, lane 3). The central region of NOT1 (residues 396 to about 1100) appears sufficient, therefore, for CCR4 interaction with CAF1, although the N-terminal 395 residues of NOT1 appear to aid the stable association of CCR4 with CAF1 (Fig. 2C; compare lane 3 with lane 4).
An internal segment (residues 667 to 1152) of NOT1 is sufficient for binding CCR4 and CAF1.
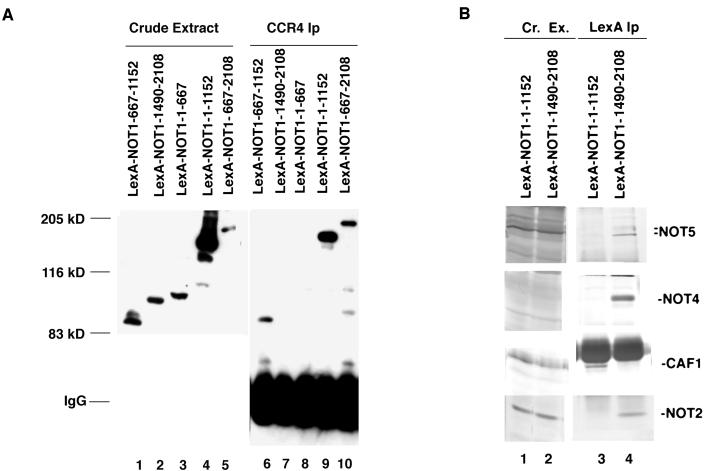
To examine more thoroughly the region of NOT1 that interacted with CCR4 and CAF1, we expressed in yeast several LexA-NOT1 fusions and determined their ability to be coimmunoprecipitated with CCR4 and CAF1. Three LexA-NOT1 fusions, LexA-NOT1(667–1152), LexA-NOT1(667–2108), and LexA-NOT1(1–1152), were coimmunoprecipitated with CCR4 (Fig. 3A, lanes 6, 9, and 10) or CAF1 antibody (data not shown). In contrast, the LexA-NOT1(1–667) and LexA-NOT1(1490–2108) could not be coimmunoprecipitated with CCR4 or CAF1 (Fig. 3A, lanes 7 and 8, and data not shown). Residues 667 to 1152 of NOT1 are, therefore, sufficient for binding CCR4 and CAF1, a conclusion that agrees with the ability of CCR4 and/or CAF1 to immunoprecipitate both the truncated not1-2 protein and NOT1(396–2108).
FIG. 3.
Localization of the NOT1 protein region that is sufficient for binding CCR4. (A) LexA-NOT1(667–1152) is sufficient for binding to CCR4 and CAF1. LexA-NOT1 fusions as indicated were expressed in strain EGY188, and immunoprecipitations were conducted with anti-CCR4 antibody. Western analysis was conducted with anti-LexA antibody. The crude protein extracts in lanes 1 to 5 contain 1/10 of the amount of extract used for the immunoprecipitations (Ip) displayed in lanes 6 to 10, respectively. IgG, immunoglobulin G. (B) The C terminus of NOT1 binds NOT2, NOT4, and NOT5. Strain EGY188 containing either LexA-NOT1(1–1152) or LexA-NOT1(1490–2108) was immunoprecipitated with LexA antibody. Western analysis using the antibodies as indicated was conducted as detailed in Fig. 1. Cr. Ex., crude extract.
The N-terminal 1318 residues of NOT1 are required for association of CCR4 with CAF1 in the 0.9-mDa complex.
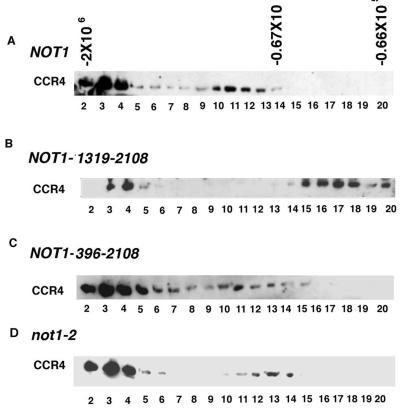
We further analyzed CCR4-CAF1-NOT1 interactions by Superose 6 gel filtration. Removing the N-terminal half of NOT1 (residues 1 to 1318) caused the dissociation of CCR4 from the 0.9-mDa complex peak fractions 10 to 12 (compare Fig. 4B with Fig. 4A), suggesting that residues 1 to 1318 of NOT1 are required for the physical integrity of CCR4 in this complex. Very little CAF1 protein could be detected following Superose 6 chromatography (data not shown). The total CAF1 protein level was reduced in the strain carrying NOT1(1318–2108) (Fig. 5), but that reduction alone cannot explain the extremely low level of total CAF1 in all Superose 6 fractions. The CAF1 protein may be particularly sensitive to degradation without the presence of the N terminus of NOT1. Deleting the N-terminal 395 codons of NOT1 had no effect on CCR4 migration in either the 1.9- or 0.9-mDa complex (Fig. 4C). The gel filtration profile of CCR4 in the not1-2 strain showed that CCR4 migrated at 1.9 mDa and about 700 kDa (fractions 12 to 14). The shift of the 0.9-mDa peak may be the result of both the truncated not1-2 protein being about 70 kDa smaller than NOT1 and the loss of the NOT5 and NOT4 proteins (Fig. 2A). CAF1 was also found to migrate in 1.9- and 0.9-mDa peaks in the not1-2 strain and in 1.9- and 0.9-mDa complexes in the NOT1(396–2108) strain (data not shown). These gel filtration results agree with the above immunoprecipitation results and indicate that in order for the CCR4-CAF1-NOT1 proteins to associate in the 0.9-mDa complex, NOT1 must contain residues 396 to about 1100.
FIG. 4.
Gel filtration analysis of CCR4 in NOT1 mutant backgrounds. (A) Strain KY803 (wild type); (B) strain MY1738 [not1 pNOT1(1319–2108)]; (C) strain MY1737 [not1 pNOT1(396–2108)]; (D) strain MY8 (not1-2). Gel filtration chromatography was conducted as described for Fig. 2B. Anti-CCR4 antibody was used to detect the CCR4 protein.
The C-terminal region from residues 1490 to 2108 of NOT1 interacts with NOT2, NOT4, and NOT5.
Because the C-terminal part of NOT1 was required for NOT5, NOT4, and NOT2 to associate with CCR4, CAF1, and NOT1 (Fig. 2A, lane 2), we initially used two-hybrid analysis to examine interactions of LexA-NOT1(1490–2108) with the NOT components of the CCR4-NOT complex (20). B42-NOT2, B42-NOT4, and B42-NOT5 all interacted with LexA-NOT1(1490–2108) (410, 520, and 77 U of β-galactosidase/mg, respectively). As expected from the immunoprecipitation analysis, neither B42-CAF1 nor B42-CCR4 interacted with the C-terminal region of NOT1 (5.5 and 3.0 U of β-galactosidase/mg, respectively; values for B42-NOT1, B42-NOT3, and B42 were 5.8, 4.5, and 3.2 U of β-galactosidase/mg, respectively). To further analyze the interaction of the C terminus of NOT1 with other components of the CCR4-NOT complex, we immunoprecipitated LexA-NOT1(1–1152) and LexA-NOT1(1490–2108) with LexA antibody. As displayed in Fig. 3B, lane 4, the C-terminal portion of NOT1(1490–2108) was able to immunoprecipitate NOT2, NOT5, and NOT4. In contrast, LexA-NOT1(1–1152) did not immunoprecipitate these proteins and instead immunoprecipitated CAF1 (Fig. 3B, lane 3) and a small amount of NOT3 (not shown). While CCR4 did not immunoprecipitate with either LexA fusion, in Fig. 3A it clearly interacted with residues 667 to 1152 to NOT1. The above immunoprecipitation, gel filtration, and two-hybrid analysis indicate, therefore, that the NOT1 protein contains two separable domains, 667 to 1152 for binding CAF1 and CCR4 and 1490 to 2108 for interacting with NOT2, NOT4, and NOT5.
The effects of deleting the different regions of NOT1 on ADH2 expression were subsequently analyzed. NOT1(1318–2108) resulted in a two- to threefold decrease in ADH II activity compared to NOT1(396–2108) or full-length NOT1 (Table 2). LexA-NOT1(1490–2108) resulted in a similar low level of ADH2 expression (Table 2). Coexpressing in yeast LexA-NOT1(1–1152) along with NOT1(1318–2108) allowed a twofold increase in ADH II activity. In contrast, coexpressing LexA-NOT1(1–667) was insufficient for recovering the ability of NOT1(1318–2108) to fully activate ADH2. The 667–1152 region of NOT1 that binds CAF1 and CCR4 appears, therefore, to be necessary for recovering in trans NOT1(1318–2108) function and confirms the existence of two distinct functional regions of NOT1.
TABLE 2.
Effects of not1 truncations on ADH2 expressiona
| NOT1 plasmid | LexA plasmid | ADH II activity (mU/mg; mean ± SEM)b |
|---|---|---|
| NOT1(396–2108) | None | 2,900 ± 450 |
| NOT1(1319–2108) | None | 910 ± 150 |
| LexA-NOT1(1–2108) | 2,300 ± 150 | |
| LexA-NOT1(1–1152) | 1,800 ± 120 | |
| LexA-NOT1(1–667) | 830 ± 100 | |
| None | LexA-NOT1(1–2108) | 2,200 ± 320 |
| LexA-NOT1(1490–2108) | 1,000 ± 230 |
ADH II assays were conducted after growth of strain 1422–21 on YEP medium containing 3% ethanol. ADH II activities in a strain carrying an integrated full-length NOT1 gene is generally 2,500 to 3,000 mU/mg. LexA plasmids contain NOT1 sequences fused to LexA-202 (7). NOT1 plasmids were pRS426 derivatives.
Average of at least three separate transformants.
NOT2 and NOT5 associate closely and in the absence of CAF1, NOT3, or NOT4.
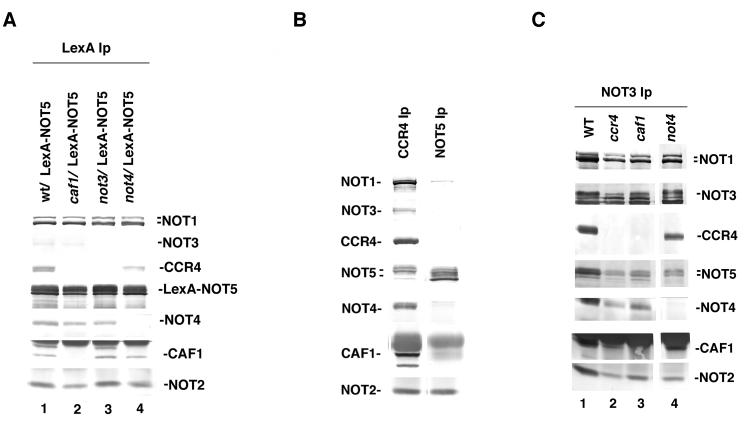
NOT5-NOT2 interactions were also analyzed following the observation that when LexA-NOT5 is immunoprecipitated with anti-LexA antibody, all components of the CCR4-NOT complex can be coimmunoprecipitated (Fig. 6A, lane 1). Immunoprecipitation of LexA alone does not coimmunoprecipitate any of these proteins (reference 20 and data not shown), indicating that it is the NOT5 moiety which is interacting with these proteins. As shown in Fig. 6A, lanes 2 to 4, NOT2 was capable of immunoprecipitating with LexA-NOT5 in the absence of CAF1, NOT3, or NOT4. As expected no CCR4 was capable of coimmunoprecipitating with LexA-NOT5 in the absence of CAF1 (Fig. 6A, lane 2). It should be noted that in the original results (Fig. 6A, lane 3) some CCR4 protein was immunoprecipitated with LexA-NOT5 in a not3 strain. These results are in agreement with the physical separation of CAF1 and CCR4 from the NOT2 and NOT5 proteins and the dependency on CAF1 for CCR4 association with these other factors. Moreover, NOT3 and NOT4 had no effect on the ability of LexA-NOT5 to coimmunoprecipitate CAF1, NOT2, or NOT1, although NOT3 may play a role in stabilizing CCR4 interactions with the complex. We subsequently used anti-NOT5 antibody to analyze more completely the NOT5-NOT2 association. Anti-NOT5 antibody immunoprecipitated only NOT5, NOT2, and a small amount of NOT1 (Fig. 6B, lane 2). While the NOT5 antibody may interfere with the association of NOT5 with the rest of the CCR4-NOT protein components, its ability to immunoprecipitate NOT2 confirms a close physical association of NOT2 and NOT5.
FIG. 6.
NOT5 coimmunoprecipitates with NOT2 in the absence of CAF1, NOT3, and NOT4. (A) LexA-NOT5 immunoprecipitates NOT2 in the absence of CAF1, NOT3, and NOT4. Immunoprecipitations (Ip) were conducted with anti-LexA antibody, and Western analysis used antibodies as indicated; LexA-NOT5 is full-length NOT5 fused to LexA(1–202). Lanes: 1, KY803 (wild type [wt]); 2, KY803-c1 (caf1); 3, MY508 (not3); 4, MY537 (not4). In the original Western blots, CCR4 was immunoprecipitated in lane 3. (B) NOT5 antibody immunoprecipitates NOT2. Antibody against CCR4 (lane 1) or NOT5 (lane 2) was used for immunoprecipitations from strain KY803. (C) NOT3 immunoprecipitates NOT1, NOT2, and NOT5 in the absence of CCR4, CAF1, and NOT4. Anti-NOT3 antibody was used to conduct the immunoprecipitations. WT (wild type), strain KY803; ccr4, KY803-1; caf1, KY803-c1; not4, MY537. Western analysis was conducted as detailed above.
NOT3 associates with the CCR4-NOT complex independently of CCR4, CAF1, or NOT4.
Immunoprecipitating NOT3 was also found to coimmunoprecipitate the whole CCR4-NOT complex (Fig. 6C, lane 1). We therefore investigated the effects of various deletions in CCR4-NOT components on NOT3 immunoprecipitation of the complex. Deleting CCR4 had no effect on NOT3 associations (Fig. 6C, lane 2), whereas caf1, as expected, resulted in only CCR4 not being able to associate in the complex (lane 3). Deleting NOT4 also had no effect on NOT3 interaction with the other components of the complex (Fig. 6C, lane 4). In not1-2, not2, or not5 strains, NOT3 could not be immunoprecipitated, suggesting that NOT2, NOT5, and the C-terminal region of NOT1 are required for stable NOT3 association with the rest of the complex or existence in an immunoprecipitable form.
CAF1 and CCR4 can act phenotypically opposite NOT2, NOT4, and NOT5.
Previously we had shown that a caf1 or ccr4 deletion resulted in no or very little increased 3-AT resistance (20) whereas mutation or deletion of the NOT genes is known to cause increased 3-AT resistance, indicative of increased HIS3 gene expression (6, 23). The not4 and not5 deletions also resulted in increased ADR1-5C activation of ADH2 under glucose growth conditions (ADH II activities of 370 ± 24 and 220 ± 10 mU/mg, respectively, versus 94 ± 7.6 mU/mg for ADR1-5C), whereas a caf1 deletion had no effect on ADR1-5C activation of ADH2 and a ccr4 deletion reduced twofold the ability of ADR1-5C to activate (ADH II activities of 95 ± 7.4 and 45 ± 1.4 mU/mg, respectively). Similar differences between the effects of caf1 and ccr4 effects on gene expression and the effects of the not alleles were reported previously (20). The most salient of these is the reduction in HO-lacZ and FKS1-lacZ expression caused by ccr4 and caf1 alleles and the two- to threefold increases in HO-lacZ or FKS1-lacZ expression caused by not1-2, not2, and not4 defects (20). These phenotypic effects support the existence of the separate location of these groups of proteins within the CCR4-NOT complex.
Since deleting NOT1 is lethal, we examined the effects of deleting components from the two separate groups of proteins in the CCR4-NOT complex. Suitable crosses were made between either ccr4 or caf1 deletions and not deletions, and the viability of different deletion combinations was analyzed by tetrad analysis. As shown in Table 3, not2 and not5 were lethal in combination with either ccr4 or caf1. Lethality was confirmed in all cases by ascertaining whether a plasmid-borne copy of one of the deleted genes could rescue the lethality. In addition, in all cases where the lethality was rescued by the plasmid-borne gene, the plasmid could not be lost from the cell, confirming the lethality of the double deletion. These synthetic lethalities are consistent with the importance of NOT2 and NOT5 to the integrity and function of the CCR4-NOT complex and to a role that is in addition to and/or separate from CCR4 and CAF1. Whereas deleting not3 did not result in synthetic phenotypes with either caf1 or ccr4, the not3-2 mutation clearly resulted in exacerbated growth phenotypes with ccr4 (Table 3). not1-2 also displayed synthetic growth defects with a ccr4 or caf1 deletion (Table 3), consistent with the observation that in a not1-2 strain, NOT2, NOT4, and NOT5 associate in the complex less well due to the increased levels of C-terminally truncated NOT1 protein (Fig. 2A, lane 2).
TABLE 3.
Synthetic lethalities between deletions in ccr4 and caf1 and mutations in the not genesa
| Construct | Growth
|
||
|---|---|---|---|
| wt | ccr4 | caf1 | |
| wt | + | + | + |
| not1-2 | |||
| 30°C | + | + | + |
| 34°C | + | − | − |
| pNOT1(1490–2108) | |||
| not1 | + | Lethal | ND |
| not2 | + | Lethal | Lethal |
| not2-1 | |||
| 30°C | + | + | ND |
| 34°C | + | − | ND |
| not3 | + | + | + |
| not3-2 | |||
| 30°C | + | + | ND |
| 37°C | + | − | ND |
| not4 | + | + | + |
| not5 | + | Lethal | Lethal |
Growth was determined at 30°C on YEP medium supplemented with 2% glucose unless otherwise indicated. +, growth; −, no growth; lethal, the gene pair resulted in cell death; ND, not done. Isogenic strains used: wild type (wt), KY803; not1-2, MY8; not1-2 ccr4, MY8-1d; not1-2 caf1, MY8-c1c; not3, MY508; not3 caf1, MY508-c1b; not2-1, MY16; not2-1 ccr4, MY16-1c; not3, MY508; not3 caf1, MY508-c1b; not3-2, MY25; not3-2 ccr4, MY25-1b; not4, MY537-1; not4 ccr4, MY537-1-1b. not3 ccr4 segregants were obtained from 1076-2c-1 × MY508 crosses, and not4 caf1 segregants were obtained from 1278-5d × 1402-1a and 1278-5d × 1402-4a crosses. pNOT1(1480–2108) not1 ccr4 lethality was confirmed by analysis of segregants of diploid 1471 [1422-21/pNOT1(1480–2108)/pLexA-NOT1 × 1469-2-1c], ccr4 not2 lethality was confirmed from strain 1441 (188-1/pLexA-NOT2 × 1393-4a), ccr4 not5 lethality was confirmed from strain 1469 (1076-2c-1/pLexANOT5 × 1462-3a), caf1 not2 lethality was confirmed from strain 1442 (188-c1/pLexA-CAF1 × 1393-4a), and caf1 not5 lethality was confirmed from strain 1469 (1076-2c-1/pLexA-NOT5 × 1462-3c).
The hallmark of the not alleles is their increased resistance to 3-AT, yet the ccr4 and caf1 alleles do not display this phenotype (20). Because of the possible antagonistic behavior of CCR4 and CAF1 in relation to the other NOT proteins, we tested the effect of a ccr4 or caf1 deletion on the ability of not alleles to confer increased 3-AT resistance. We observed that a caf1 or ccr4 deletion suppressed the increased 3-AT resistance of not1-2, not3-2, and not4 alleles (Table 4), confirming that CCR4 and CAF1 can act in an opposite manner to the NOT proteins in certain promoter contexts.
TABLE 4.
caf1 and ccr4 disruptions suppress not-induced 3-AT phenotypesa
| Relevant genotype | Growth on 3-AT (mM)
|
||
|---|---|---|---|
| 0 | 5 | 10 | |
| Wild type | + | − | − |
| caf1 | + | − | − |
| ccr4 | + | − | − |
| not1-2 | + | + | + |
| not1-2 caf1 | + | w | − |
| not1-2 ccr4 | + | − | − |
| not3-2 | + | + | + |
| not3-2 ccr4 | + | − | − |
| not4 | + | + | + |
| not4 caf1 | + | − | − |
| not4 ccr4 | + | − | − |
All strains are isogenic to KY803 except for the indicated allele. Growth was monitored on minimal medium lacking histidine and supplemented with 3-AT as indicated.
DISCUSSION
CCR4-CAF1 interact with the N-terminal 1152 residues of NOT1, whereas NOT2, NOT4, and NOT5 interact with the C-terminal 1490–2108 region of NOT1.
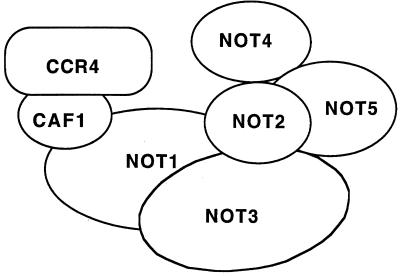
It was shown previously that disrupting the CAF1 gene blocked the ability of CCR4 to associate with NOT1 (20). In this report, we showed that CAF1 is required for CCR4 to associate with all of the NOT proteins. NOT3, NOT4, and NOT5 were, in turn, found not to be required for CCR4-CAF1-NOT1 association or for CCR4 and CAF1 association in the 1-mDa complex. Relatedly, immunoprecipitating NOT3 or LexA-NOT5 did not coimmunoprecipitate CCR4 when CAF1 was deleted. These results clearly indicate that CCR4 binds through CAF1 to associate with NOT1 and the other components of the complex (Fig. 7 summarizes the interactions in the CCR4-NOT complex).
FIG. 7.
Model for protein contacts in the CCR4-NOT complex. Based on the results presented herein, CAF1 is presumed to bind to residues 667 to 1152 of NOT1, CCR4 binds to CAF1, and NOT2 and NOT5 interact with the C-terminal residues 1490 to 2108 of NOT1 in no particular order. NOT4 is placed on the periphery of NOT2 and NOT5, and it is presumed that NOT3 makes contacts with both NOT2, NOT5, or NOT4 and the N terminus of NOT1.
The region of NOT1 with which CCR4 and CAF1 physically associate was found to be distinct from that bound by the NOT2, NOT4, and NOT5 proteins. Several lines of evidence indicate that the region of NOT1 binding CAF1 is localized to residues 667 to 1152. First, CCR4 or CAF1 could immunoprecipitate an internal segment of NOT1 (residues 667 to 1152). Second, when the immunoprecipitation was conducted in the reverse direction, LexA-NOT1(1–1152) immunoprecipitated CAF1 but LexA-NOT1(1490–2108) did not. Third, in a not1-2 strain wherein the major NOT1 species contains only the N-terminal 1,000 residues or so of NOT1, both CCR4 and CAF1 immunoprecipitated with the not1-2 protein. Fourth, removing the N-terminal 395 residues of NOT1 did not abrogate the ability of CAF1 to immunoprecipitate CCR4, although the N-terminal segment of NOT1 clearly played some role in stabilizing the CCR4-CAF1 interactions. Finally, when the N-terminal 1,318 residues of NOT1 were removed, neither CCR4 nor CAF1 was able to associate in the 1-mDa complex.
In contrast to the above results, the region of NOT1 that associates with NOT2, NOT4, and NOT5 was localized to the C-terminal portion of NOT1 (residues 1490 to 2108). The NOT2, NOT4, and NOT5 were found to display two-hybrid interactions with LexA-NOT1(1490–2108), and immunoprecipitating LexA-NOT1(1490–2108) coimmunoprecipitated NOT2, NOT4, and NOT5. As a comparison, immunoprecipitating LexA-NOT1(1–1152) failed to coimmunoprecipitate these proteins. Moreover, in the not1-2 strain, NOT5 and NOT4 did not immunoprecipitate with CCR4 or CAF1 and only a limited amount of NOT2 coimmunoprecipitated (possibly due to association with the residual full-length NOT1 still present in the cell). Finally, NOT2, NOT4, and NOT5 were coimmunoprecipitated in the absence of CAF1 when either NOT3 or LexA-NOT5 was immunoprecipitated. These results indicate that it is the C-terminal portion of NOT1 protein (residues 1490 to 2108) that binds NOT2, NOT4, and NOT5. These data establish a clear physical separation of CCR4 and CAF1 from NOT2, NOT4, and NOT5 through their binding to separate regions of NOT1. Other limited interactions between these two groups of proteins can not be excluded, however.
Our data further show that NOT2 and NOT5 are closely linked. First, deletion of either of these components tends to result in decreased abundance of several other components in the complex. NOT2 and NOT5 appear to have general effects on the integrity or stability of the CCR4-NOT complex. Second, both proteins interacted in the two-hybrid system with the C-terminal segment of NOT1 and immunoprecipitated with the same segment of NOT1. Third, NOT3, NOT4, CAF1, or CCR4 defects did not affect the ability of LexA-NOT5 to immunoprecipitate NOT2. Fourth, NOT5 antibody coimmunoprecipitated NOT2, some NOT1, and no other CCR4-NOT component. These data implicate a close physical association between NOT2 and NOT5 that is important to the stability of the CCR4-NOT complex.
While NOT3 and NOT4 appear to be peripheral to NOT2 and NOT5, the location of the NOT3 protein could not be readily determined. For one thing, in all mutant strains that we analyzed by gel filtration analysis, NOT3 migrated at or near its monomeric size. NOT3 appears to be less stably associated with the CCR4-NOT complex. Immunoprecipitating CCR4 and CAF1 showed that NOT3 failed to coimmunoprecipitate in a not1-2 strain, suggesting NOT3 associated with the C terminus of NOT1. However, when LexA-NOT1(1–1152) was immunoprecipitated, a small and reproducible amount of NOT3 was coimmunoprecipitated. No NOT3 was observed to coimmunoprecipitate with LexA-NOT1(1490–2108). Also, NOT3 did not require NOT4 for association with NOT1, CCR4, or CAF1. Since neither a not3 nor a not4 deletion affected the ability of LexA-NOT5 to immunoprecipitate NOT1, CCR4, CAF1, or NOT2, it appears that NOT3 and NOT4 are peripheral to NOT2 and NOT5. However, a not3 deletion did reduce the ability of LexA-NOT5 to immunoprecipitate CCR4. In Fig. 7, we therefore assign NOT3 a place that includes contacts to the N terminus of NOT1 and binding to the outskirts of NOT2 and NOT5.
In our model for the physical arrangement of the components of the CCR4-NOT complex based on the above data (Fig. 7), CCR4 associates at one end of the complex through binding CAF1, which in turn binds the central portion of NOT1 (residues 667 to 1152). NOT2 and NOT5 bind the C-terminal region of NOT1 (residues 1490 to 2108). NOT4 is on the outside of NOT2 and NOT5. NOT3 may display multiple contacts both to the N terminus of NOT1 and to NOT2 and NOT5.
CCR4 and CAF1 differ phenotypically from NOT2 to NOT5.
The above biochemical data define CCR4 and CAF1 as a separate group of proteins within the CCR4-NOT complex that contacts the NOT1 protein in a distinct location from the NOT2, NOT4, and NOT5 proteins. Based on these identified separate locations within the complex, we would expect these subgroups of proteins to exhibit specific differences in both function and the proteins with which they interact. Phenotypically, CCR4 and CAF1 can display a number of functions that are different from those displayed by the other NOT proteins. This distinction correlates with the above-defined physical interactions. First, the NOT1 protein [NOT1(1490–2108)] which lacks the binding site for CAF1 and CCR4 is defective in ADH2 derepression. Adding back to yeast in trans the N-terminal segment of NOT1 (residues 1 to 1152) can rescue this defect in ADH2 expression, implicating the binding of CAF1 and CCR4 to this region as important for full ADH2 expression. Second, the not4 and not5 disruptions augment ADR1-5C-induced ADH2 expression under glucose growth conditions, whereas caf1 has no effect and ccr4 causes a reduction in expression. Third, at other promoters such as HO-lacZ and FKS1-lacZ, the not1, not2, and not4 defects (not5 was not tested) result in increased expression whereas caf1 and ccr4 result in defects in expression (20). Fourth, the not alleles were identified in a genetic screen for increased 3-AT resistance. In contrast, little or no effect on 3-AT resistance is observed for caf1 or ccr4 defects (20). Fifth, whereas not alleles cause increased 3-AT resistance, ccr4 and caf1 defects can suppress these effects.
In addition, the ccr4 and caf1 defects when combined with not alleles resulted in synthetic phenotypes. ccr4 or caf1 were lethal with either not2 or not5 deletions, whereas ccr4 caf1 and not2 not5 double knockouts are viable. Although other explanations are possible with respect to synthetic growth defects, the synthetic defect of ccr4 and caf1 with that of not2 and not5 is consistent with the idea that these two groups of proteins display separate functions that are important for the overall integrity and activity of the CCR4-NOT complex. It is also possible that NOT2 and NOT5 display functions redundant with those of CCR4 and CAF1, although we feel that this is unlikely because of their dissimilarity in protein sequence and their actual physical separation and distinctiveness within the CCR4-NOT complex. Similarly, ccr4 not1-2 and caf1 not1-2 knockouts also displayed synthetic phenotypes. not1-2 results from a stop codon (26) that causes 90% of the NOT1 protein to be about 1,100 amino acids long, which confirms that loss of the C terminus of the not1-2 protein in combination with ccr4 or caf1 defects results in a synthetic phenotype. The nonlethality of this combination is most likely due to the existence of some full-length NOT1 protein. These data also suggest that the lethality that results from deleting the not1 gene may be caused by the combined loss of essential parts of the CCR4-CAF1 components and the NOT2 to NOT5 components. NOT1 may, therefore, be an essential protein due to its structural role in forming and maintaining the CCR4-NOT complex. However, the C-terminal region of NOT1 (residues 1490 to 2108) encompassing the site of binding NOT2, -4, and -5 by itself can complement a not1 disruption, unlike the region of NOT1 binding CCR4 and CAF1 (residues 667 to 1152). The C terminus of NOT1 may be essential due to its ability to bind multiple NOT proteins, and/or the C terminus of NOT1 conveys another, as yet undetermined essential function.
It should also be noted that not4 or not3 deletions in combination with ccr4 or caf1 did not result in synthetic phenotypes. Therefore, although NOT2 and NOT5 appear important to the integrity of the complex, the lethality between not2 or not5 and that of ccr4 or caf1 is not simply due to loss of structural roles for NOT2 and NOT5 and their presumed importance for binding NOT3 and NOT4. Instead, NOT2 and NOT5 must play an important biochemical role independent of their mere physical presence in the CCR4-NOT complex. The observation that not3 and not4 deletions do not display synthetic growth defects with that of ccr4 and caf1 suggest either that these proteins as a group are not required for the function of any essential genes or that they actually function in the same pathway. It should be noted, though, that the not3-2 allele in combination with ccr4 resulted in a synthetic growth defect, suggesting that the not3-2 protein affects a particular interaction in a negative manner that is worse than the complete loss of the NOT3 protein.
Roles of the 1.9- and 0.9-mDa CCR4-NOT complexes.
CCR4, CAF1, NOT1, NOT3, and NOT5 have all been found to associate in 1.9- and 0.9-mDa complexes (reference 20 and results herein), and NOT2 has also been shown to migrate at 0.9-mDa (unpublished observation). The association of CCR4 in the 0.9-mDa complex requires CAF1 (20) and, as we have shown here, the N-terminal region of NOT1. A caf1 deletion reduced significantly the ability of CCR4 to associate in the 1.9-mDa complex (20) but did not eliminate it entirely. CCR4 must, therefore, be able to interact in the 1.9-mDa complex independently of CAF1 or its association with NOT1. The contacts and function for CCR4 in the 1.9-mDa complex may be separable from or additional to those it displays in the 0.9-mDa complex. Relatedly, CCR4 must express a function separate from its presence in the 0.9-mDa complex since a ccr4 deletion was lethal when combined with the strain expressing only pNOT1(1490–2108). It remains possible that CCR4 exists in multiple 1.9-mDa complexes (3).
Other proteins that have been found to associate with CCR4, CAF1, or NOT proteins are DHH1 (15), CAF4, CAF16 (21), DBF2 (22), and MOB1 (18). These proteins may all be candidates for components of the 1.9-mDa complex. DHH1, CAF16, and MOB1 (3a) do not immunoprecipitate with CCR4 and are not components of the 0.9-mDa complex. However, CAF16 (21), DBF2, and MOB1 (unpublished observation) have been found to migrate in a 1.9-mDa complex. The complete components, assembly, and functional significance of the two CCR4-NOT complexes remain to be clarified. Characterizing the 1.9-mDa complex, understanding how the CCR4-NOT complexes interact with other transcriptional factors, and defining the particular roles of CCR4 and CAF1 in contrast to NOT2, NOT5, NOT3, and NOT4 should lead to a better conception of how the CCR4-NOT proteins function in both activated and repressed transcription.
ACKNOWLEDGMENTS
This research was supported by NIH grant GM41215, NSF grant MCB95-13412, and HATCH project H291 to C.L.D. and Swiss National Science Foundation grant 31-49808.96 to M.A.C.
Footnotes
Scientific contribution no. 1999 from the New Hampshire Agricultural Experiment Station.
REFERENCES
- 1.Cade R M, Errede B. NOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3139–3149. doi: 10.1128/mcb.14.5.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Chang M, French-Corney D, Fan H, Klein H, Denis C L, Jaehning J A. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Chen, J. Personal communication.
- 4.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collart M A, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart M A, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3 and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 7.Cook W J, Mosley S P, Audino D C, Mullaney D L, Rovelli A, Stewart G, Denis C L. Mutations in the zinc-finger region of the yeast regulatory protein ADR1 affect both DNA binding and transcriptional activation. J Biol Chem. 1994;269:9374–9379. [PubMed] [Google Scholar]
- 8.de Barros Lopes M, Ho J Y, Reed S I. Mutations in cell division cycle genes CDC36 and CDC39 activated the Saccharomyces cerevisiae mating pheromone response pathway. Mol Cell Biol. 1990;10:2966–2972. doi: 10.1128/mcb.10.6.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis C L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denis C L, Draper M P, Liu H-Y, Malvar T, Vallari R C, Cook W J. The yeast CCR4 protein is neither regulated by nor associated with the SPT6 and SPT10 proteins and forms a functionally distinct complex from that of the SNF/SWI transcription factors. Genetics. 1994;138:1005–1013. doi: 10.1093/genetics/138.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denis C L, Malvar T. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics. 1990;124:283–291. doi: 10.1093/genetics/124.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draper M P, Liu H Y, Nelsbach A H, Mosley S P, Denis C L. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draper M P, Salvadore C, Denis C L. Identification of a mouse protein, whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulating complex. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagop Y. Immunoaffinity purification of antibodies against GST fusion proteins. Bio/Technology. 1998;24:198–202. doi: 10.2144/98242bm05. [DOI] [PubMed] [Google Scholar]
- 15.Hata H, Mitsui H, Liu H, Bai Y, Denis C L, Shimizu Y, Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irie K, Yamaguchi K, Kawase K, Matsumoto K. The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-pheromone-responsive genes. Mol Cell Biol. 1994;14:3150–3157. doi: 10.1128/mcb.14.5.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komarnistsky S I, Chiang Y C, Luca F C, Chen J, Toyn J H, Winey M, Johnston L H, Denis C L. DBF2 protein kinase binds to and acts through the cell-cycle-regulated MOB1 protein. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H-Y, Badarinarayana V, Audino D, Rappsilber J, Mann M, Denis C L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, H.-Y., Y.-C. Chiang, J. Pan, C. Salvadore, J. Chen, V. Badarinarayana, B. Anderson, and C. L. Denis. 1999. Unpublished data.
- 22.Liu H-Y, Toyn J H, Chiang Y C, Draper M P, Johnston L H, Denis C L. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5189–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberholzer U, Collart M A. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene. 1998;207:61–69. doi: 10.1016/s0378-1119(97)00605-7. [DOI] [PubMed] [Google Scholar]
- 24.Sakai A, Chibazakura T, Shimizu Y, Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu, C. Personal communication.