Abstract
BACKGROUND:
Hemoglobin C, D Punjab, E or S trait can interfere with hemoglobin A1c (HbA1c) results. We assessed whether they affect results obtained with 15 current assay methods.
METHODS:
Hemoglobin AA (HbAA), HbAC, HbAD Punjab, HbAE and HbAS samples were analyzed on 2 enzymatic, 4 ion-exchange HPLC and 9 immunoassay methods. Trinity Premier Hb9210 boronate affinity HPLC was the comparative method. An overall test of coincidence of least-squared linear regression lines was performed to determine if HbA1c results were statistically significantly different from those of HbAA samples. Clinically significant interference was defined as >6% difference from HbAA at 6 or 9% HbA1c compared to Premier Hb9210 using Deming regression.
RESULTS:
All methods showed statistically significant effects for one or more variants. Clinically significant effects were observed for the Tosoh G11 variant mode (HbAD), Roche b 101 (HbAC and HbAE) and Siemens DCA Vantage (HbAE and HbAS). All other methods (Beckman Coulter B93009 and B00389 on DxC700AU, and Unicel DxC, Ortho Clinical Vitros 5.1, Roche cobas c 513, Siemens Dimension RxL and Vista, and Enzymatic on Advia and Atellica, Tosoh G8 5.24 and 5.28, and GX) showed no clinically significant differences.
CONCLUSIONS:
A few methods showed interference from one or more variants. Laboratories need to be aware of potential HbA1c assay interferences.
Keywords: Hemoglobin A1c, Hemoglobin Variants, Diabetes, HPLC, Capillary Electrophoresis, Immunoassay, Enzymatic
1. Introduction:
The global prevalence of diabetes mellitus has been increasing rapidly. In 2019, approximately 463 million adults were living with diabetes; by 2045, this number is projected to rise to 700 million (1). In the US, 34.2 million people or 10.5% of the population have diabetes; more than 7 million of these are undiagnosed (2). Hemoglobin A1c (HbA1c) is an important indicator of mean glycemia in patients with diabetes, and it has been shown to be strongly predictive of diabetes complications (3-4). Treatment goals for HbA1c have been established, and more recently the test has been recommended for use in diagnosing diabetes (5-6). Therefore, accurate and precise measurement of HbA1c is extremely important.
There has been considerable improvement in the quality of HbA1c testing over the past several years. Laboratories are encouraged to choose methods that are National Glycohemoglobin Standardization Program (NGSP) certified and to review method performance in accuracy-based proficiency testing. The gradual tightening of NGSP certification criteria as well as College of American Pathologists’ proficiency testing limits have helped to maintain continued improvement in HbA1c method performance (7). At this time there are several methods that have shown excellent performance over time. However, there are some analytical interferences with some HbA1c methods, specifically those from Hb variants, that require continuing further study and may influence a laboratory’s decision when selecting a method. These interferences could be a deciding factor when choosing a HbA1c method. The most common hemoglobin variants worldwide are HbS, HbE, HbC and HbD traits (8). Previous studies have shown method-specific analytic interference with HbA1c results from these heterozygous hemoglobin variants (9-11).
2. Materials and Methods:
2.1. Samples
Whole blood samples from individuals homozygous for HbA (n=48) and heterozygous for HbC (n=49), HbD Punjab (n=39), HbE (n=43), or HbS (n=48) trait and with and without diabetes representing a range of 4-12% HbA1c were collected in EDTA tubes, divided into aliquots, and stored at −70°C. Aliquots of each sample were shipped on dry ice to 8 designated laboratories and manufacturers for analysis. Due to logistical limitations, not all samples could be analyzed by all methods. Samples were obtained with IRB approval from Quest Diagnostics and the approval of the ethics review committee at DynaLIFE Medical Labs in Edmonton, Canada.
2.2. Assay Methods
Assay methods evaluated included the G8 variant mode running software version 5.24 (pending FDA approval in the US) and 5.28 (current version outside US), GX ver. 1.24 and G11 variant mode ver. 3.06 (Tosoh Bioscience), the Vitros MicroTip HbA1c Assay on Vitros 5,1 FS (Ortho Clinical Diagnostics), cobas HbA1c Test on cobas b 101 (Roche Diagnostics International Ltd.), Tina-quant HbA1c Gen.3 on cobas c 513 (Roche Diagnostics), Dimension RxL, Dimension Vista, Enzymatic Hemoglobin A1c on Advia and Atellica (Siemens Healthcare Diagnostic Inc.), DCA Vantage (Siemens Healthineers), Unicel DxC (Beckman Coulter), B93009 HbA1c Advanced and B00389 HbA1c on DxC700AU (Beckman Coulter AU systems, Co.).
The 4 Tosoh methods are ion-exchange HPLC. The Siemens Advia and Atellica methods are enzymatic. All other methods are immunoassay methods. Analyses were performed by the respective manufacturers with the exception of the Roche cobas c 513 which was analyzed by DynaLIFE Medical Labs and the Tosoh G8 5.24 analyzed at the Diabetes Diagnostic Laboratory, University of Missouri. The Trinity Premier Hb9210 boronate affinity HPLC (Trinity Biotech) at the University of Missouri-Columbia was used as the comparative method after being validated against the previous Trinity ultra2 method which has previously been shown to be unaffected by the hemoglobin variants tested (12-13). Hemoglobin variants were initially identified by ion-exchange HPLC and/or electrophoresis at the institutions where the samples were collected, and also presumptively identified using the Sebia Capillarys 2 Flex Piercing Hemoglobin(e) method (Sebia) at the University of Missouri-Columbia. Presumptive identification was based upon expected elution or migration time and variant peak proportion.
2.3. Data Analyses
For each method an overall test of coincidence of least-squared linear regression lines was used to determine if results for each variant were statistically significantly different (P<0.05) from those of HbAA samples. Deming regression was used to determine if the bias for each variant ves HbAA was clinically significant at 6% (42 mmol/mol) or 9% (75 mmol/mol) HbA1c; clinical significance was defined as a difference exceeding ±6% (14). In IFCC units (mmol/mol), clinical significance was defined as a difference exceeding 9.3% at 42 mmol/mol or ±7.9% at 75 mmol/mol; these limits are equivalent to ±6% at 6 and 9% HbA1c in National Glycohemoglobin Standardization Program (NGSP)/Diabetes Control and Complications Trial (DCCT) units (15). Data analyses were performed using SAS 9.4 and Graphpad Prism 5.0. A clinically significant difference of ±6% is tighter than the limits used in our previous study (11) because current accuracy-based CAP limits are now at ±6%. This is wider than current NGSP limits (±5%) but still reasonable for assessment of Hb variant interference.
2.4. Manufacturer Claims
For most of the methods evaluated the manufacturers claim no significant interference from any of the variants tested. However, the manufacturers’ acceptance level for interference is not always clear.
3. Results
3.1. Validation of Trinity Premier for HbA1c Measurement in Samples with Variants
Samples with HbAA (n=18), HbAC (n=26), HbAD (n=18), HbAE (n=21) and HbAS (n=29) representing a range of 4-12% HbA1c were analyzed on both the Trinity ultra2 and Trinity Premier Hb9210. There were no statistically significant differences in the relationships between each variant and HbAA samples (p>0.05 for all, Fig. 1). Deming regression showed differences of <0.15% HbA1c at 6% and 9% HbA1c for all variants vs. HbAA (Table 1).
Fig. 1. Relationships between the Premier Hb9210 and ultra2 for HbAA, HbAC, HbAD, HbAE and HbAS.
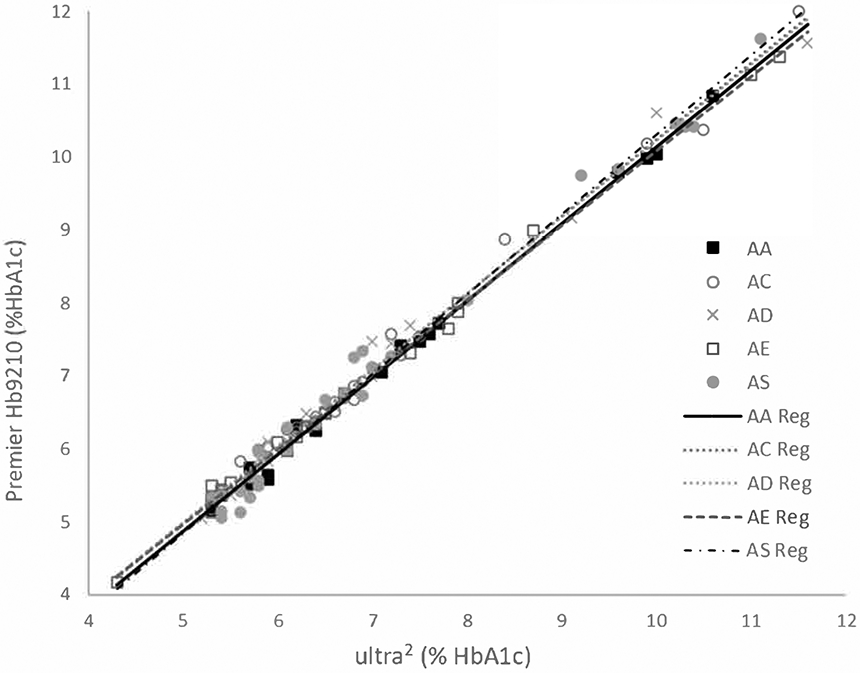
Linear regression for HbAA and each of the hemoglobin variants evaluated.
Table 1.
Deming Regression Results for Premier Hb9210 vs. ultra2.
| Premier %HbA1c | Bias vs. AA (%HbA1c) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ultra2 %HbA1c |
Premier AA %HbA1c |
AC | AD | AE | AS | AC | AD | AE | AS |
| 6 | 5.92 | 6.02 | 6.04 | 5.99 | 5.93 | 0.10 | 0.12 | 0.07 | 0.01 |
| 9 | 9.08 | 9.19 | 9.19 | 9.07 | 9.22 | 0.11 | 0.11 | −0.01 | 0.14 |
3.2. Statistical Significance
Fig. 2 shows boxplots of the differences versus Premier Hb9210 for HbAA and each variant for each method and indicates both statistically and clinically significant differences versus HbAA. While all methods showed statistically significant differences for one or more variants, only a few showed any clinically significant differences.
Fig. 2. Box-plots summarizing the absolute differences (%HbA1c and mmol/mol HbA1c) between each test method and the comparison method for HbAA, HbAC, HbAD, HbAE and HbAS.
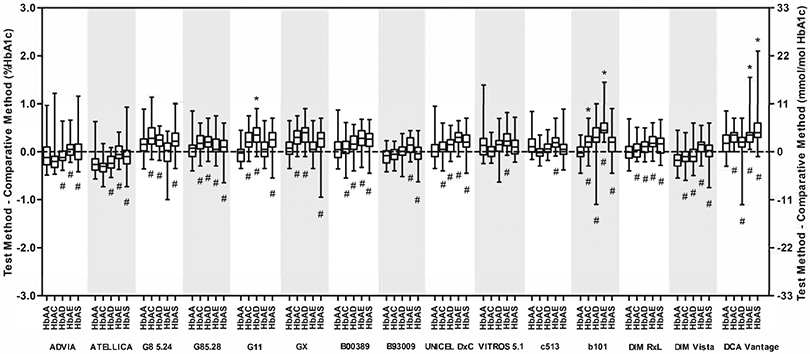
The horizontal line in each box is the median difference between the test and comparison methods. The limits of each box correspond to the 25th and 75th percentiles of the differences. The highest and lowest horizontal bars represent the most extreme individual differences between the test and comparison methods. Differences from HbAA that are statistically significant are indicated (#) below each bar where appropriate; clinically significant differences are indicated (*) above each bar where appropriate.
3.3. Clinical Significance
Table 2 shows the actual biases for each variant versus HbAA at 6 and 9% HbA1c (supplementary table 3 shows the biases in IFCC units at 42 and 75 mmol/mol) and along with Fig. 2 indicates which differences were clinically significant. The methods showing clinically significant differences were the b 101 (HbAC and HbAE), the DCA Vantage (HbAE and HbAS) and the G11 (HbAD). For all other methods, there were no clinically significant differences. For the DCA Vantage, the difference for HbAE was no longer clinically significant after the exclusion of the single HbAE data point >10% HbA1c.
Table 2:
Mean differences between test and comparative methods.a
| Method | HbAA | HbC trait | HbD trait | HbE trait | HbS trait | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | n | 6% HbA1c |
9% HbA1c |
n | 6% HbA1c |
9% HbA1c |
n | 6% HbA1c |
9% HbA1c |
n | 6% HbA1c |
9% HbA1c |
|
|
|
|
|
|
|
|||||||||
| Enzymatic | |||||||||||||
| Advia | 48 | 46 | 0.10 | 0.02 | 37 | 0.14 | 0.09 | 41 | 0.35 | 0.40 | 47 | 0.32 | 0.49 |
| Atellica | 48 | 44 | 0.00 | −0.20 | 37 | 0.10 | −0.10 | 40 | 0.32 | 0.33 | 47 | 0.28 | 0.42 |
| Ion-Exchange HPLC | |||||||||||||
| G8 5.24 | 48 | 49 | 0.26 | 0.29 | 38 | 0.15 | 0.02 | 42 | −0.06 | 0.09 | 48 | 0.21 | 0.38 |
| G8 5.28 | 48 | 44 | 0.23 | 0.29 | 38 | 0.23 | 0.02 | 42 | 0.11 | −0.06 | 44 | 0.15 | 0.16 |
| G11 | 48 | 44 | 0.29 | 0.40 | 38 | 0.41b | 0.32 | 43 | 0.10 | 0.00 | 44 | 0.30 | 0.46 |
| GX | 48 | 44 | 0.28 | 0.34 | 38 | 0.32 | 0.12 | 43 | 0.04 | −0.06 | 44 | 0.21 | 0.38 |
| Immunoassay | |||||||||||||
| B00389 | 48 | 42 | 0.08 | −0.26 | 35 | 0.19 | −0.21 | 38 | 0.29 | 0.13 | 44 | 0.27 | 0.16 |
| B93009 | 48 | 41 | 0.07 | −0.24 | 35 | 0.18 | −0.23 | 39 | 0.29 | 0.13 | 41 | 0.27 | 0.17 |
| Unicel DxC | 48 | 46 | 0.09 | −0.25 | 39 | 0.19 | 0.00 | 43 | 0.34 | 0.19 | 48 | 0.26 | 0.24 |
| Vitros 5,1 FS | 44 | 37 | 0.03 | 0.18 | 34 | 0.07 | 0.02 | 29 | 0.27 | 0.48 | 35 | 0.10 | 0.22 |
| c 513 | 48 | 46 | −0.07 | −0.22 | 39 | 0.02 | −0.09 | 43 | 0.19 | 0.29 | 46 | 0.05 | 0.18 |
| b 101 | 48 | 45 | 0.28 | 0.57b | 39 | 0.34 | 0.41 | 43 | 0.61b | 1.24b | 46 | 0.21 | 0.40 |
| Dimension RxL | 48 | 46 | 0.10 | 0.06 | 39 | 0.13 | 0.10 | 43 | 0.26 | 0.33 | 48 | 0.23 | 0.41 |
| Dimension Vista | 48 | 45 | 0.08 | −0.25 | 37 | 0.15 | −0.07 | 41 | 0.31 | 0.14 | 48 | 0.24 | 0.23 |
| DCA Vantage | 46 | 45 | 0.29 | 0.41 | 39 | 0.10 | −0.35 | 43 | 0.33 | 0.62b | 48 | 0.50b | 1.10b |
Deming regression analysis was performed using Premier Hb9210 as the comparative method. The biases (%HbA1c) for each method at clinical decision cutoffs of 6% and 9% HbA1c were calculated for each variant. To correct for inter-method calibration differences, the mean biases between each test method and the comparative method for homozygous HbA samples was subtracted from those calculated for the variant samples.
Clinically significant differences (>0.36% or >0.54% HbA1c at 6% or 9% HbA1c, respectively).
4. Discussion
While most of the methods evaluated did not show clinically significant interference with HbA1c results in the presence of the tested variants, the Roche b 101 POC method showed a clinically significant differences for HbAC and HbAE. Although the bias for HbAC was only marginally clinically significant at 9% HbA1c, the biases for HbAE were >10% at both 6 and 9% HbA1c, respectively. This finding for HbAE is consistent with other recent studies (16-17). The b101 product insert states that there is no significant interference from any of the variants tested, including HbAE. The Siemens DCA Vantage POC method showed a difference of >8% and >12% at 6 and 9% HbA1c, respectively for HbAS. This method was investigated in 2000 (18) showing differences of 2.8% and 5.8% at 6 and 9% HbA1c, respectively. However, in 2019, the CAP GH5 survey included a sample with HbAS and the DCA Vantage showed a 5.7% difference (0.32% HbA1c at 5.66% HbA1c) (19); this was higher than expected and consistent with the current results. We do not know what changes may have been made to the assay since 2000 that would account for the current HbAS interference.
The Tosoh G11 showed a difference of 6.8% at 6% HbA1c for HbAD. Unfortunately, the G11 separates HbS, HbC, and HbD in the same variant peak window, thus it cannot effectively differentiate between them. This means that although the G11 did not show clinically significant differences for the other variants, HbAC and HbAS, the laboratory would not be able to determine which of these three common variants is present without further investigation. Interestingly, the G11 did not show differences >6% at HbA1c levels of 6 and 9% for HbAD in our previous interference study (10), but it showed clinically significant interference from HbAC (the criterion for the previous study was ±7% at 6 and 9% HbA1c), which we did not observe in the current study. The G8 method showed clinically significant interference from all four variants in that previous study, but the newer software versions (5.24 and 5.28) addressed that issue and we did not see clinically significant interferences for the G8 or for the GX method in the current study. As mentioned in the previous study, we have noted that variant interferences can change over time as reagent and column lots, as well as software versions, change (10).
A limitation of the current study is the small numbers of variant samples at the upper end of the HbA1c range. However, further analysis using Passing Bablok regression, as well as exclusion of data points above 10% HbA1c, did not alter our conclusions regarding clinically significant interferences with the exception of HbAE on the DCA Vantage.
Although we focused on the most common variants, there are many other variants that can affect HbA1c results. Many of these cause method-specific interferences; however, homozygous or double heterozygous variants that are associated with reduced erythrocyte lifespan (e.g., HbSS, HbCC, HbSC) cause falsely lower results regardless of assay methodology. Rare variants that cause altered glycation rates, such as Hb Raleigh, also affect HbA1c results (20). Also, other conditions which can cause decreased erythrocyte lifespan (e.g. hemolytic anemia, blood loss) or increased erythrocyte lifespan (e.g. polycythemia, postsplenectomy) will tend to cause falsely lowered or elevated results, respectively (21-22).
It is important that healthcare providers consider potential variant interferences when interpreting HbA1c results. Laboratories can play an important role by considering the potential of hemoglobin variants in their patient populations when selecting assay methods, and by making clinicians aware of the potential of interferences with their assay methods.
Supplementary Material
Highlights.
Hemoglobin variants can affect hemoglobin A1c results
Fifteen HbA1c methods were tested for interference from Hb C, D, E and S traits
Three methods showed clinically significant interference from one or more variants
Laboratories need to be aware of potential interference from hemoglobin variants
Acknowledgements
We thank Siemens, Tosoh Corp., Beckman Coulter, Ortho Clinical Diagnostics, Roche International/PHC Corporation, and DynaLIFE Medical Labs for performing sample analyses. We thank DynaLIFE Medical Labs and Quest Diagnostics for providing variant samples for this study. We also would like to thank Greg Petroski at the University of Missouri (Columbia) for assisting with statistical analyses. This study was supported by the NIH/NIDDK.
Abbreviations
- CE
Capillary Electrophoresis
- NGSP
National Glycohemoglobin Standardization Program
- DCCT
Diabetes Control and Complications Trial
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].International Diabetes Federation, Diabetes Atlas 9th Edition, 2019, https://www.diabetesatlas.org/en/sections/worldwide-toll-of-diabetes.html, (accessed 16 December 2020).
- [2].Centers for Disease Control and Prevention, Diabetes Report Card, 2019. http://www.cdc.gov/diabetes/library/reports/congress.html, (accessed 17 December 2020).
- [3].DCCT Research Group, The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus, N. Engl. J. Med 329 (1993) 977–986. 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- [4].UK Prospective Diabetes Study UKPDS Group, Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33), Lancet 352 (1998) 837–853. 10.1016/S0140-6736(98)07019-6. [DOI] [PubMed] [Google Scholar]
- [5].American Diabetes Association, Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes, Diabetes Care 44 (Suppl. 1) (2021) S15–S33. 10.2337/dc21-S002. [DOI] [PubMed] [Google Scholar]
- [6].World Health Organization, Use of Glycated Haemoglobin (HbA1c) in the Diagnosis of Diabetes Mellitus, Abbreviated Report of a WHO Consultation, World Health Organization, Geneva, 2011. https://www.who.int/diabetes/publications/report-hba1c_2011.pdf?ua=1. [PubMed] [Google Scholar]
- [7].Little RR, Rohlfing C, Sacks DB, The National Glycohemoglobin Standardization Program: Over 20 Years of Improving Hemoglobin A1c Measurement, Clin. Chem 65 (2019) 839–848. 10.1373/clinchem.2018.296962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cataldo F, Immigration and changes in the epidemiology of hemoglobin disorders in Italy : an emerging public health burden, Ital J Pediatr 38, 32 (2012) 10.1186/1824-7288-38-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin CN, Emery TJ, Little RR, Hanson SE, Rohlfing CL, Jaisson S, Gillery P, Roberts WL, Effects of hemoglobin C, D, E, and S traits on measurements of HbA1c by six methods, Clin. Chim. Acta 413 (2012) 819–821. 10.1016/j.cca.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mongia SK, Little RR, Rohlfing CL, et al. , Effects of hemoglobin C and S traits on the results of 14 commercial glycated hemoglobin assays, Am. J. Clin. Pathol 130 (2008) 136–140. 10.1309/1YU0D34VJKNUCGT1. [DOI] [PubMed] [Google Scholar]
- [11].Rohlfing C, Hanson S, Weykamp C, Siebelder C, Higgins T, Molinaro R, Yip PM, Little RR, Effects of hemoglobin C, D, E and S traits on measurements of hemoglobin A1c by twelve methods, Clin. Chim. Acta 455 (2016) 80–83. 10.1016/j.cca.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Little RR, Vesper H, Rohlfing CL, Ospina M, Safar-Pour S, Roberts WL, Validation by a mass spectrometric reference method of use of boronate affinity chromatography to measure glycohemoglobin in the presence of hemoglobin S and C traits, Clin. Chem 51 (2005) 264–265. 10.1373/clinchem.2004.043141. [DOI] [PubMed] [Google Scholar]
- [13].Rohlfing C, Connolly S, Hanson S, Higgins T, Little RR, Validation of the Use of Trinity Biotech ultra2 as a Comparative Method for Hemoglobin A1c Measurements in the Presence of HbE and HbD-Punjab Traits, Clin Chem. 63 (2017) 608–610. 10.1373/clinchem.2016.266221. [DOI] [PubMed] [Google Scholar]
- [14].College of American Pathologists, Hemoglobin A1c (5 Challenge) GH5-B 2020, CAP, Northfield, IL, 2020. [Google Scholar]
- [15].Weykamp CW, Mosca A, Gillery P, Panteghini M, The analytical goals for hemoglobin A(1c) measurement in IFCC units and National Glycohemoglobin Standardization Program units are different, Clin. Chem 57 (2011) 1204–1206. 10.1373/clinchem.2011.162719. [DOI] [PubMed] [Google Scholar]
- [16].Lenters-Westra E, English E. Are hemoglobin A1c point-of-care analyzers fit for purpose? The story continues. Clin. Chem. Lab. Med 59 (2021):765–774. 10.1515/cclm-2020-1308 [DOI] [PubMed] [Google Scholar]
- [17].Yong S, Liu H, Lum CLT, et al. Impact of heterozygous hemoglobin E on six commercial methods for hemoglobin A1c measurement. PeerJ Analytical Chemistry 3:e9 (2021). DOI 10.7717/peerj-achem.9 [DOI] [Google Scholar]
- [18].Frank EL, Moulton L, Little RR, Wiedmeyer HM, Rohlfing C, Roberts WL,. Effects of hemoglobin C and S traits on seven glycated hemoglobin methods, Clin. Chem 46 (2000):864–867. 10.1093/clinchem/46.6.864. [DOI] [PubMed] [Google Scholar]
- [19].NGSP, College of American Pathologists (CAP) Survey Data. http://ngsp.org/CAP/CAP19a.pdf, (accessed 17 December 2020). [Google Scholar]
- [20].Little RR, Roberts WL, A review of variant hemoglobins interfering with hemoglobin A1c measurement, J. Diabetes Sci. Technol 3 (2009):446–451. doi: 10.1177/193229680900300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sacks DB, Arnold M, Bakris GL, et al. , Guidelines and Recommendations for Laboratory Analysis in the Diagnosis and Management of Diabetes Mellitus, Clin. Chem 57 (2011): e1–e47. 10.1373/clinchem.2010.161596. [DOI] [PubMed] [Google Scholar]
- [22].Takeuchi M, Kawakami Koji, Association between Hemoglobin and Hemoglobin A1c: A Data-Driven Analysis of Health Checkup Data in Japan, J. Clin. Med 7 (2018) 539. 10.3390/jcm7120539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


