Abstract
BACKGOUND:
B cells contribute to chronic transplant rejection by producing donor-specific antibodies and promoting T cell response, but how these processes are regulated at the transcriptional level remains unclear. Herein, we investigate the role of transcription factor interferon regulatory factor 4 (IRF4) in controlling B cell response during chronic transplant rejection.
METHODS:
We generated the Irf4gfp reporter mice to determine IRF4 expression in B cell lineage. We then used mice with B cell-specific IRF4 deletion to define the role of IRF4 in B cell response after NP-KLH immunization or allogeneic heart transplantation. In particular, graft survival and histology, as well as B and T cell responses, were evaluated after transplantation.
RESULTS:
IRF4 is dynamically expressed at different stages of B cell development and is absent in germinal center (GC) B cells. However, IRF4 ablation in the B cell lineage primarily eliminates GC B cells in both naïve and NP-KLH immunized mice. In the transplantation setting, IRF4 functions intrinsically in B cells and governs allogeneic B cell responses at multiple levels, including GC B cell generation, plasma cell differentiation, donor-specific antibody production, and support of T cell response. B cell-specific IRF4 deletion combined with transient CTLA4-Ig treatment abrogates acute and chronic cardiac allograft rejection in naïve recipient mice but not in donor skin-sensitized recipients.
CONCLUSIONS:
B cells require IRF4 to mediate chronic transplant rejection. IRF4 ablation in B cells abrogates allogeneic B cell responses and may also inhibit the ability of B cells to prime allogenic T cells. Targeting IRF4 in B cells represents a potential therapeutic strategy for eliminating chronic transplant rejection.
Introduction
Both B cells and T cells play important roles in the rejection of transplanted organs.1–3 Inhibition of T cell activation and expansion with immunosuppressive drugs has greatly improved transplant outcomes;1 however, T cell-based immunosuppression remains less effective to prevent chronic transplant rejection, a process that is mainly characterized by arteriosclerosis and interstitial fibrosis in allografts.4, 5 B cells have been shown to promote chronic rejection by producing donor-specific antibodies (DSA).6 Binding of DSA to donor endothelium activates complement and recruits phagocytic cells, and sequentially contributes to vascular injury and chronic graft loss.7–10 To prevent chronic rejection and graft loss over time, it is important to define how B cells produce DSA in response to organ transplantation.
Germinal centers (GC) are formed when activated B cells enter lymphoid follicles. Activated B cells in GC (termed GC B cells) undergo a dynamic process and differentiate into either plasma cells that secret high-affinity antibodies or memory B cells.11 Several key transcription factors regulate the generation of GC B cells and the differentiation of plasma cells. B cell lymphoma 6 (Bcl6) is highly expressed in GC B cells and controls their generation. In the absence of Bcl6, GC formation and Ig affinity maturation are significantly impaired.12 B lymphocyte-induced maturation protein 1(Blimp-1) is not required for the generation of GC B cells but is essential for the differentiation of plasma cells.13 Interferon regulatory factor 4 (IRF4) belongs to the IRF family of transcription factors. IRF4 is preferentially expressed in immune cells and controls their differentiation and function. In B cells, IRF4 is essential for both the generation of GC B cells and the differentiation into plasma cells.14, 15 In the setting of transplantation, it is unknown how IRF4 regulates GC B and plasma cells, and the subsequent development of DSA and chronic rejection.
Here we generated and characterized the Irf4gfp reporter mice and found that IRF4 was dynamically expressed at different stages of B cell development. We then investigated the role of IRF4 in regulating B cell response during chronic rejection. We found that in the presence of transient CTLA4-Ig treatment, IRF4 deletion in B cells inhibited chronic rejection and induced long-term heart allograft survival. Mechanistically, IRF4 deletion in B cells abolished Bcl6 expression in GC B cells, eliminated GC B and plasma cells, abrogated the production of de novo IgG1 DSA, and impaired Th1 and follicular helper T (Tfh) cell response. Hence, IRF4 is required for B cells to mediate chronic allograft rejection.
Material and Methods
Mice
Balb/c, Irf4fl/fl, and Cd19-Cre mice were purchased from Jackson Laboratory (Bar Harbor, ME). Irf4fl/fl mice were crossed to Cd19-Cre mice to generate Irf4fl/flCd19-Cre+/– mice (indicated as Irf4fl/flCd19-Cre mice throughout the text). We designed the Irf4gfp reporter mice and requested Jackson Laboratory Model Generation Services to generate this mouse strain. All animal experiments were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute.
Flow cytometric analysis
Flow cytometric analysis of lymphocytes has been previously described.16 Detailed information is provided in Supplementary Material.
NP-KLH immunization and ELISA
Irf4fl/fl control and Irf4fl/flCd19-Cre mice were immunized with NP-KLH. Host immune response to NP-KLH was determined by ELISA17, 18 and flow cytometry analysis. Detailed information is provided in Supplementary Material.
Heart and skin transplantation
Irf4fl/fl and Irf4fl/flCd19-Cre mice were transplanted with Balb/c skins and hearts as previously described.16, 19 Detailed information is provided in Supplementary Material.
Tissue histology and morphometric analysis of graft arteries
Hematoxylin & eosin (H&E) staining and Verhoeff-Van Gieson (VVG) staining were performed on paraffin sections of heart allografts. For morphometric analysis of coronary arteries from the allograft sections, images of arteries were captured digitally with a light microscope (Nikon Eclipse 80i, Nikon, Japan). ImageJ software (NIH, Bethesda, MD) was used to calculate areas of lumen and intima of each artery in VVG-stained images. Neointimal index was used to indicate the degree of lumen occlusion of each artery, which was calculated by neointimal volume (intimal area value – luminal area value)/stent volume (intimal area value) × 100.20
Detection of DSA in serum
Serum samples from transplant recipients were used for detecting DSA. Detailed information is provided in Supplementary Material.
Statistics
Data were represented as mean ± SD and analyzed with Prism version 8 (GraphPad Software). The P values of heart graft survival were determined by the log-rank test. Other measurements were performed using unpaired Student’s t-test. Differences were considered significant when P < 0.05.
Results
IRF4 is dynamically expressed at different stages of B cell development
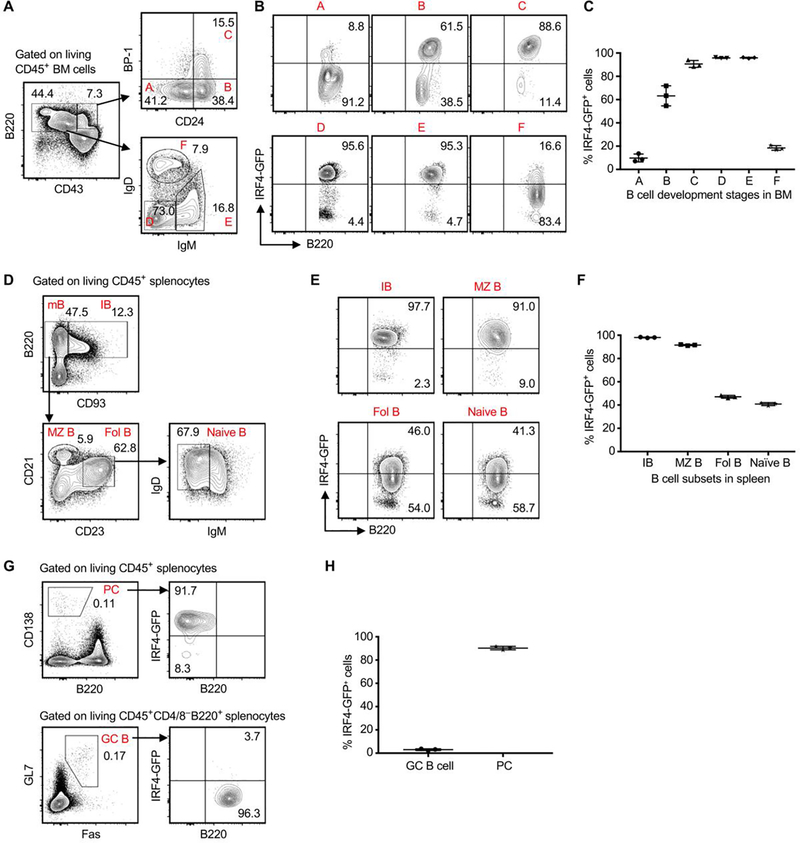
To determine the role of IRF4 in allogeneic B cell responses, we first assessed the expression levels of IRF4 in the B cell lineage. To this end, we generated the Irf4gfp reporter mice, in which a P2A.GFP_P2A.DTR_stop cassette was inserted immediately after the last exon of WT B6 mouse Irf4 by using the CRISPR/Cas9 technique (Supplemental Figure 1).
Irf4gfp reporter mice at 8 weeks of age were used to determine IRF4 expression at different stages of B cell development. Living CD45+ cells from bone marrows were analyzed by flow cytometry according to the Hardy Fraction gating strategy.21, 22 As shown in Figure 1A, CD43highB220+ cells were segregated into Hardy fractions A (Pre-Pro-B), B (pro-B), and C (early Pre-B) based on different expressions of CD24 and BP-1. CD43mid/lowB220+ cells were segregated into Hardy fractions D (late Pre-B), E (immature B), and F (mature B) based on different expressions of IgD and IgM (Figure 1A). By gating on each Hardy fraction, we found that the expression level of IRF4-GFP was low in Pre-Pro-B cells, gradually increased in pro-B and early Pre-B cells, peaked in late Pre-B and immature B cells, and then declined in mature B cells (Figure 1B,C).
Figure 1. IRF4 is dynamically expressed during B cell development.
Splenocytes and bone marrow (BM) cells were obtained from Irf4gfp reporter mice at 8 weeks of age, followed by flow cytometry analysis. (A-C) Analysis of IRF4 expression during B cell development in BM. (A) The gating strategy for identifying Hardy fractions A through F, highlighted in red font. (B and C) Representative contour plots (gated on each indicated Hardy fraction) and the graph display % IRF4-GFP+ cells in each Hardy fraction. (D-H) Analysis of IRF4 expression in B cell subsets in spleen. (D) The gating strategy for detecting immature B (IB), mature B (mB), marginal zone B (MZ B), follicular B (Fol B), and naïve B cells. (E and F) Representative plots (gated on each indicated B cell subset) and the graph show % IRF4-GFP+ cells in each B cell subset. (G) Representative plots display the gating of plasma cells (PC) and GC B cells (left panels), and % IRF4-GFP+ cells in PC and GC B cells (right panels). (H) The graph shows % IRF4-GFP+ cells in PC and GC B cells. Data in C, F and H are shown as mean ± SD (n = 3) and are from one experiment that is representative of two independent experiments.
Living CD45+ splenocytes were analyzed by using the gating strategy depicted in Figure 1D. Immature B (IB) cells in spleens were CD93+B220+, whereas mature B (mB) cells were CD93–B220+. Mature B cells were segregated into follicular B (Fol B) and marginal zone B (MZ B) cells based on different expressions of CD21 and CD23. Fol B cells contained IgD+IgM– naïve B cells (Figure 1D). By gating on each B cell subset, we found that most immature B cells highly expressed IRF4-GFP. The expression level of IRF4-GFP was then gradually decreased in MZ B, Fol B, and naïve B cells (Figure 1E,F). Moreover, CD45+ splenocytes of naïve mice contained low frequencies of B220–CD138+ plasma cells (PCs) and B220+GL7+Fas+ germinal center (GC) B cells. We found that GC B cells almost completely lost IRF4-GFP expression, whereas PCs highly expressed IRF4-GFP (Figure 1G,H).
We briefly analyzed B cell subsets in peritoneal cavity. Supplemental Figure 2A shows the gating strategy for detecting IRF4-GFP expression in B1a, B1b and B2 cell subsets. The expression level of IRF4-GFP in B2 cells was significantly lower than those of B1a and B1b cells (Supplemental Figure 2). Taken together, IRF4 is dynamically expressed at different stages of B cell development.
Ablation of IRF4 eliminates GC B cells
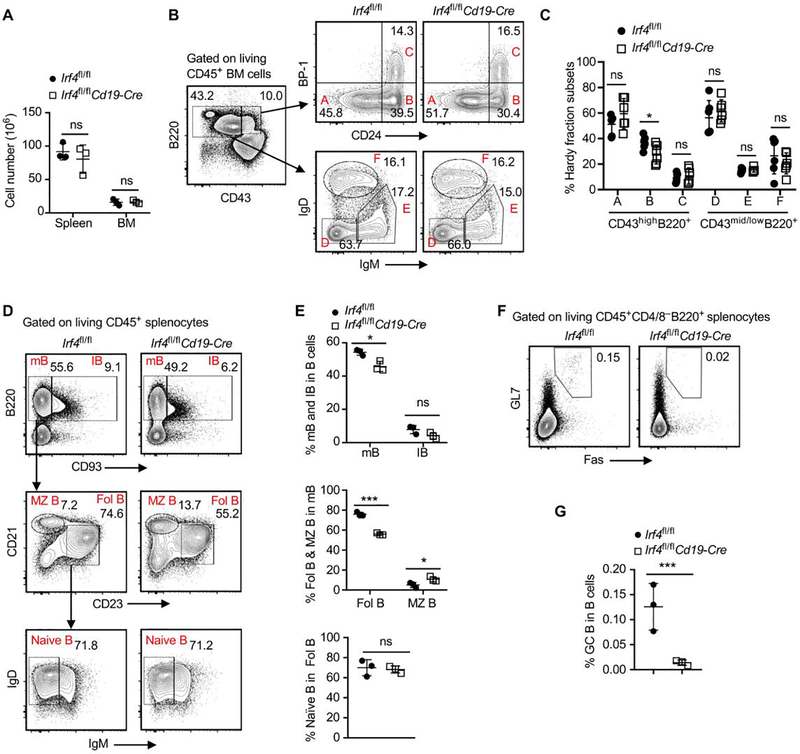
To determine whether IRF4 controls B cell development, we generated the Irf4fl/flCd19-Cre mice, in which IRF4 was conditionally deleted only in B cells. Irf4fl/fl control and Irf4fl/flCd19-Cre mice at 8 weeks of age were used for flow cytometry analysis. Compared with Irf4fl/fl control mice, IRF4 deletion in B cells (Irf4fl/flCd19-Cre mice) did not affect the absolute numbers of CD45+ immune cells in spleens and bone marrows (Figure 2A). By comparing Hardy fractions A-F in bone marrows between Irf4fl/fl control and Irf4fl/flCd19-Cre mice, we found that IRF4 deletion in B cells significantly reduced the frequency of the pro-B cell subset (Hardy fraction B), but did not affect the frequencies of other developmental B cell subsets (Figure 2B,C).
Figure 2. Ablation of IRF4 in B cells eliminates GC B cells.
Splenocytes and bone marrow (BM) cells were obtained from Irf4fl/flCd19-Cre or Irf4fl/fl control mice at 8 weeks of age, followed by flow cytometry analysis. (A) Total numbers of CD45+ splenocytes and femur BM cells in indicated mouse groups. Data are shown as mean ± SD and are from one experiment that is representative of two independent experiments. (B and C) Representative contour plots and the bar graph display % Hardy fraction subsets A to C in CD43highB220+ BM cells and % Hardy fraction subsets D to F in CD43mid/lowB220+ BM cells. Data in C are shown as mean ± SD and are from one experiment that is representative of two independent experiments. (D and E) Representative plots and bar graphs show % mB and IB cells in splenic B cells, % Fol B and MZ B cells in mB cells, and % naïve B cells in Fol B cells. (F and G) Representative plots and the bar graph show % GC B cells in CD45+CD4–CD8–B220+ splenic B cells. Data in E and G are shown as mean ± SD and are from one experiment that is representative of two independent experiments. ns, P > 0.05; *P < 0.05; ***P < 0.001 by unpaired Student’s t test.
By comparing B cell subsets in spleens between Irf4fl/fl control and Irf4fl/flCd19-Cre mice, we found that IRF4 deletion in B cells significantly reduced the frequencies of mature B and Fol B cells, and increased the frequency of MZ B cells (Figure 2D,E). Figure 2F shows the representative plots for detecting GC B cells in spleens. Importantly, deletion of IRF4 in B cells almost completely eliminated the GC B cell population (Figure 2F,G).
Moreover, IRF4 deletion in B cells significantly reduced B1a cell frequency and increased B2 cell frequency in peritoneal cavity (Supplemental Figure 3). Taken together, ablation of IRF4 in B cells affects the frequencies of several B cell subsets, and particularly eliminates GC B cells.
Ablation of IRF4 in B cells abrogates the production of antigen-specific antibodies in response to NP-KLH immunization
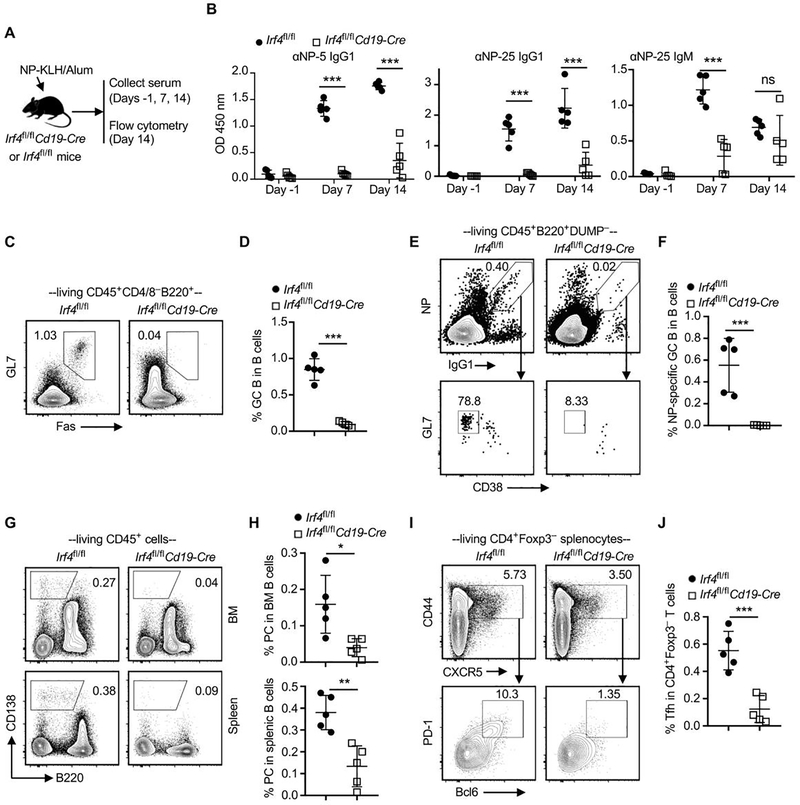
To investigate whether deletion of IRF4 impairs B cell function, Irf4fl/fl control and Irf4fl/flCd19-Cre mice were immunized with NP-KLH in alum on day 0. Serum samples were collected on days −1, 7, and 14 for detecting NP-KLH–specific antibodies using ELISA (Figure 3A). Prior to immunization (day −1), NP-specific antibodies were undetectable in serum samples of both mouse groups. Post immunization (days 7 and 14), serum samples from Irf4fl/flCd19-Cre mice contained significantly lower amounts of anti-NP-25 IgG1 (total anti-NP-KLH IgG1 antibodies) and anti-NP-5 IgG1 (high-affinity anti-NP-KLH IgG1 antibodies) than those of Irf4fl/fl control mice. Indeed, NP-KLH/Alum immunization largely failed to induce the production of anti-NP-25 IgG1 and anti-NP-5 IgG1 in Irf4fl/flCd19-Cre mice. Serum samples from Irf4fl/flCd19-Cre mice also contained significantly lower amounts of anti-NP-25 IgM than those of Irf4fl/fl control mice at day 7 post immunization (Figure 3B).
Figure 3. Ablation of IRF4 in B cells abrogates the production of antigen-specific antibody in response to NP-KLH immunization.
Irf4fl/fl control and Irf4fl/flCd19-Cre mice were immunized with NP-KLH in alum on day 0. Serum samples were collected on days −1, 7, and 14 for ELISA. Splenocytes and BM cells were obtained on day 14 for flow cytometry analysis. (A) Schematic of the experimental design. (B) Optical density (OD) values indicate the presence of anti-NP-5 (αNP-5) IgG1, anti-NP-25 (αNP-25) IgG1, and anti-NP-25 (αNP-25) IgM in sera of indicated recipient groups. Test sera were diluted 1:8100. (C and D) Representative contour plots and the bar graph show % Fas+GL7+ GC B cells in splenic B cells. (E and F) % NP-specific GC B cells (NP+IgG1+GL7+CD38–) in B220+DUMP– splenic B cells. DUMP includes antibodies against IgM, IgD, CD3, CD11b, CD11c, CD138, and Gr-1. (G and H) % CD138+B220– plasma cells (PC) in splenocytes and BM cells. (I and J) % CD44+CXCR5+PD-1+Bcl6+ Tfh cells in CD4+Foxp3– splenocytes. Data in B, D, F, H, and J are shown as mean ± SD (n = 5) and are from one experiment that is representative of two independent experiments. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired Student’s t test.
Splenocytes and bone marrow cells were obtained on day 14 for flow cytometry analysis. The gating strategy for detecting NP-specific GC B cells (NP+IgG1+GL7+CD38– cells among B220+DUMP– cells) was shown in Figure 3E. We found that the frequencies of total GC B cells (Figure 3C,D), NP-specific GC B cells (Figure 3E,F), and plasma cells (Figure 3G,H) in Irf4fl/flCd19-Cre mice were significantly lower than those of Irf4fl/fl control mice. In addition, Irf4fl/flCd19-Cre mice also contained significantly less CD44+CXCR5+Bcl6+PD-1+ follicular helper T (Tfh) cells in spleens than that of Irf4fl/fl control mice (Figure 3I,J). Therefore, ablation of IRF4 abrogates B cell response to NP-KLH.
Ablation of IRF4 in B cells alone fails to prevent acute allograft rejection
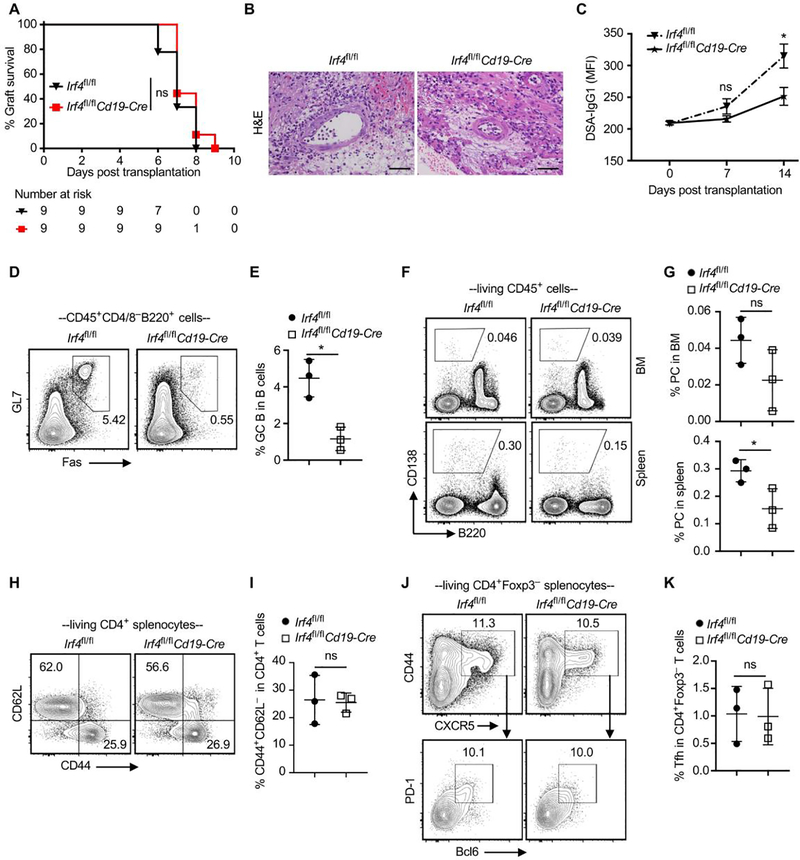
To determine whether IRF4 expression in B cells regulates DSA production and affects transplant outcomes, fully MHC-mismatched Balb/c heart allografts were transplanted into either Irf4fl/fl control or Irf4fl/flCd19-Cre mice on day 0. We found that IRF4 deletion in B cells (Irf4fl/flCd19-Cre mice) did not prolong cardiac allograft survival (Figure 4A). At day 7 post-transplant, heart allografts from both groups exhibited massive cellular infiltration and severe tissue damage (Figure 4B). Serum samples were collected on days 0 (prior to grafting), 7, and 14 for assessing DSA levels. The basal IgG1 DSA serum level prior to grafting was comparable in the two groups. However, IRF4 deletion in B cells significantly reduced the production of de novo IgG1 DSA at day 14 post-transplant (Figure 4C).
Figure 4. Ablation of IRF4 in B cells alone impairs IgG1 DSA production but fails to prevent acute allograft rejection.
Balb/c hearts were transplanted into Irf4fl/fl control or Irf4fl/flCd19-Cre mice. (A) % heart allograft survival (n = 9). ns, P > 0.05 by log-rank test. (B) H&E-stained sections (x200 magnification) of heart allografts harvested at day 7 post-transplant. Scale bars indicate 50 μm. (C) Serum samples were collected from transplant recipients at indicated days, and then incubated with Balb/c donor cells. The graph shows IgG1 MFI of CD45+ Balb/c donor cells. Data are shown as mean ± SD (n = 5) and are from one experiment that is representative of two independent experiments. (D and E) Representative contour plots and bar graphs show % GC B cells in splenic B cells. (F and G) % PC in splenocytes and BM cells. (H and I) % CD62L–CD44+ cells in CD4+ splenocytes. (J and K) % Tfh cells in Foxp3–CD4+ splenocytes. Data in E, G, I and K are shown as mean ± SD (n = 3) and are from one experiment that is representative of two independent experiments. ns, P > 0.05; *P < 0.05 by unpaired Student’s t test.
Splenocytes and bone marrow cells were analyzed on day 14. IRF4 deletion in B cells did not alter the number of splenocytes in heart allograft recipients (Supplemental Figure 4). In line with the impaired production of de novo IgG1 DSA, Irf4fl/flCd19-Cre recipients had significantly lower frequencies of GL7+Fas+ GC B cells (Figure 4D,E) and CD138+B220– plasma cells (Figure 4F,G) in spleens than did Irf4fl/fl recipients. IRF4 deletion in B cells did not affect the frequencies of CD4+CD62L–CD44+ T cells (Figure 4H,I) and CD44+CXCR5+Bcl6+PD1+ Tfh cells (Figure 4J,K) in transplant recipients. Therefore, IRF4 deletion in B cells alone impairs IgG1 DSA production but fails to prevent acute allograft rejection.
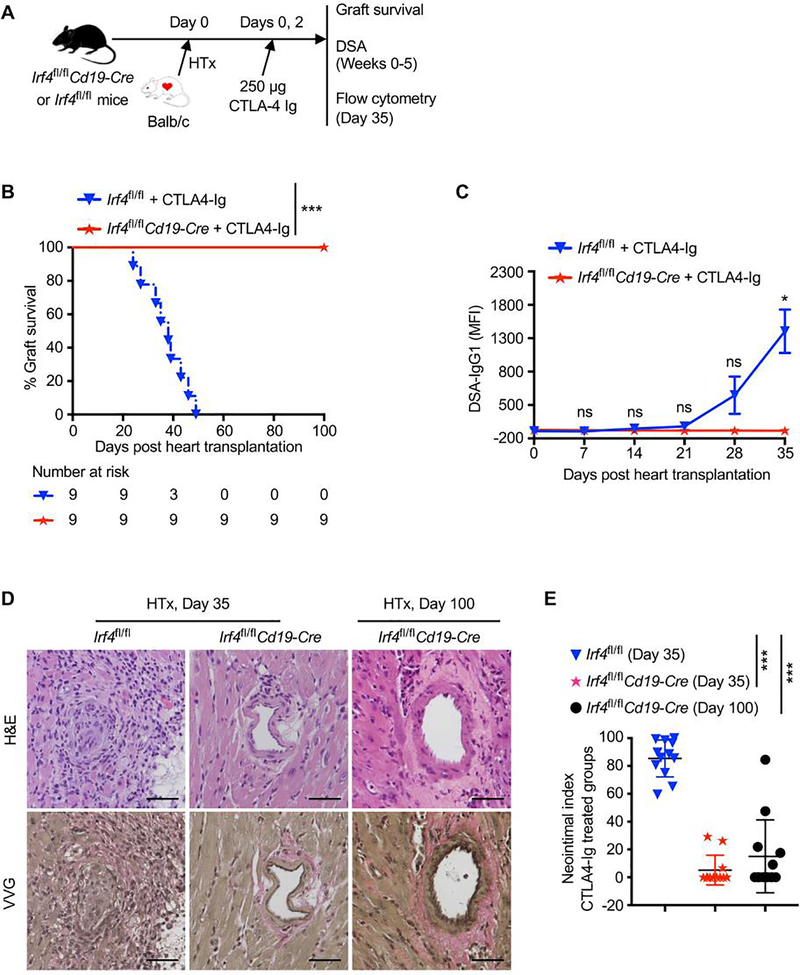
Deletion of IRF4 in B cells abrogates DSA production and inhibits chronic allograft rejection in CTLA4-Ig treated mice
We investigated whether deletion of IRF4 in B cells would affect transplant outcomes when acute cellular rejection is inhibited. Irf4fl/fl control or Irf4fl/flCd19-Cre mice were transplanted with Balb/c hearts on day 0, and were treated with 250 μg CTLA4-Ig on days 0 and 2 (Figure 5A). All heart allografts were chronically rejected within 50 days in CTLA4-Ig–treated Irf4fl/fl recipients (MST = 37.1 ± 8.32 days). By contrast, none of the CTLA4-Ig–treated Irf4fl/flCd19-Cre recipients rejected the heart allografts (MST >100 days) (Figure 5B). Hence, ablation of IRF4 in B cells induces transplant acceptance in CTLA4-Ig treated mice.
Figure 5. Deletion of IRF4 in B cells abrogates de novo DSA production and prevents chronic allograft rejection in CTLA4-Ig treated mice.
Irf4fl/fl control or Irf4fl/flCd19-Cre mice were transplanted with Balb/c hearts on day 0, and treated with 250 μg CTLA4-Ig on days 0 and 2. (A) Schematic of the experimental design. (B) % allograft survival after heart transplantation (HTx) (n = 9). ***P < 0.001 by log-rank test. (C) Serum samples were collected from transplant recipients at indicated days, and then incubated with Balb/c donor cells. The graph shows IgG1 MFI of CD45+ Balb/c donor cells. Data are shown as mean ± SD (n = 3) and are from one experiment that is representative of two independent experiments. ns, P > 0.05; *P < 0.05 by unpaired Student’s t test. (D and E) Heart allografts were harvested from CTLA4-Ig–treated groups at indicated days post-transplant. (D) Representative images show the H&E- and VVG-stained sections (x200 magnification) of heart allografts. Scale bars indicate 50 μm. (E) Morphometric quantification (neointimal index) of cardiac allograft vasculopathy. Arteries in VVG-stained sections (four grafts per group) were analyzed. ***P < 0.001 by unpaired Student’s t test.
Serum samples were collected from recipients on day 0 (prior to transplant) and then weekly for 5 weeks post-transplant. The serum IgG1 level remained low in both groups for 3 weeks, and was increased at weeks 4 and 5 in CTLA4-Ig–treated Irf4fl/fl control but not Irf4fl/flCd19-Cre recipients (Figure 5C). Thus, ablation of IRF4 in B cells abrogates the production of de novo DSA in CTLA4-Ig treated recipients.
Histologically, heart allografts from CTLA4-Ig–treated Irf4fl/fl recipients at day 35 post-transplant developed severe chronic rejection, characterized by perivascular cellular infiltration and severe neointima formation. By contrast, heart allografts from CTLA4-Ig–treated Irf4fl/flCd19-Cre recipients displayed minimal vascular injury and near absence of neointima formation at day 35 post-transplant, and also displayed significantly less signs of chronic rejection even at day 100 post-transplant (Figure 5D,E). Therefore, ablation of IRF4 in B cells inhibits chronic rejection in CTLA4-Ig treated recipients.
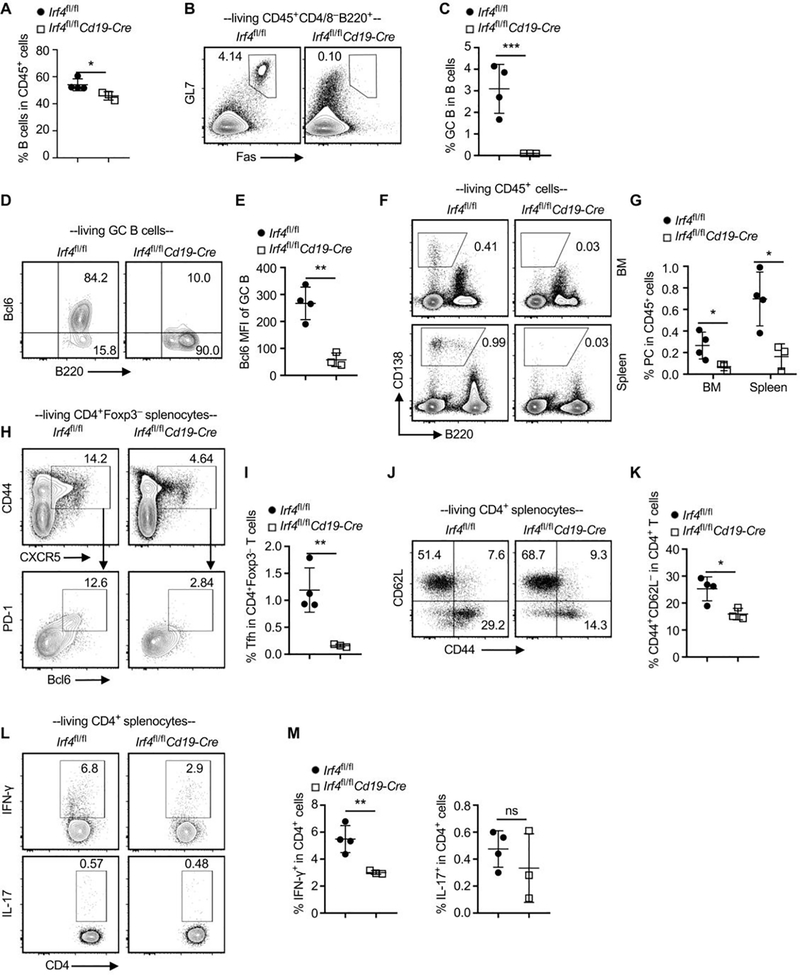
Ablation of IRF4 in B cells inhibits allogeneic B cell and T cell responses in CTLA4-Ig treated recipients
To investigate the cellular mechanisms of allograft acceptance in our model, splenocytes and bone marrow cells were obtained from the CTLA4-Ig–treated Irf4fl/fl control or Irf4fl/flCd19-Cre recipients at day 35 post heart grafting, followed by flow cytometry analysis. Irf4fl/flCd19-Cre recipients displayed significant decreases in splenocyte number and B cell frequency than did Irf4fl/fl recipients (Figure 6A and Supplemental Figure 5). Importantly, compared with Irf4fl/fl recipients, Irf4fl/flCd19-Cre recipients contained significantly lower frequency of GC B cells (Figure 6B,C) and their GC B cells barely expressed Bcl6 (Figure 6D,E). Irf4fl/flCd19-Cre recipients also contained significantly lower frequency of plasma cells than did Irf4fl/fl recipients (Figure 6F,G). Expression levels of Bcl6 in plasma cells, as well as Blimp-1 in GC B and plasma cells, were not significantly different between the two recipient groups (Supplemental Figure 6).
Figure 6. Ablation of IRF4 in B cells abrogates allogeneic B and T cell response in CTLA4-Ig treated recipients.
Splenocytes and BM cells were obtained from CTLA4-Ig–treated Irf4fl/fl control (n = 4) or Irf4fl/flCd19-Cre recipients (n = 3) at day 35 after Balb/c heart transplantation, followed by flow cytometry analysis. (A) The bar graph shows % B cells in CD45+ splenocytes. (B and C) Representative contour plots and the bar graph show % GC B cells in splenic B cells. (D and E) % Bcl6+ cells (D) and Bcl6 MFI (E) in splenic GC B cells. (F and G) % PC in BM and splenic CD45+ cells. (H and I) % Tfh cells in Foxp3–CD4+ splenocytes. (J and K) % CD62L–CD44+ T cells in CD4+ splenocytes. (L and M) % IFN-γ+ and IL-17+ T cells in CD4+ T cells. Data in A, C, E, G, I, K, and M are shown as mean ± SD and are from one experiment that is representative of two independent experiments. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired Student’s t test.
Compared with Irf4fl/fl control recipients, Irf4fl/flCd19-Cre recipients displayed significantly lower frequencies of CD44+CXCR5+Bcl6+PD-1+ Tfh cells (Figure 6H,I), CD4+CD62L–CD44+ T cells (Figure 6J,K), and IFN-γ producing Th1 cells (Figure 6L,M). IL-17A producing Th17 cells were barely detectable in both groups (Figure 6L,M). Taken together, ablation of IRF4 in B cells inhibits allogeneic B cell and T cell responses in CTLA4-Ig treated recipients.
Ablation of IRF4 in B cells fails to prevent heart allograft rejection in skin-sensitized mice
To determine whether IRF4 deletion in B cells affects heart allograft survival in recipients that are pre-sensitized to donor antigens, Irf4fl/flCd19-Cre and Irf4fl/fl control recipients were transplanted with Balb/c skin allografts. Thirty days later, recipient mice were transplanted again with Balb/c heart allografts and treated with four doses of 250 μg CTLA4-Ig on days 0, 2, 4, and 6 post-heart transplantation (Supplemental Figure 7A). All those Balb/c heart allografts were acutely rejected in both Irf4fl/flCd19-Cre and control recipients (Supplemental Figure 7B). Compared with Irf4fl/fl recipients, Irf4fl/flCd19-Cre recipients contained significantly lower frequencies of GC B cells (Supplemental Figure 7C,D) and plasma cells (Supplemental Figure 7E,F). Nevertheless, both recipient groups have more than 30% CD62L–CD44+ effector memory-like cells within splenic CD4+ T cells (Supplemental Figure 7G,H). These findings suggest that pre-sensitization with Balb/c skin allografts generates potent memory T cell response to acutely reject the later transplanted Balb/c heart allografts, which could not be effectively inhibited by IRF4 deletion in B cells plus CTLA4-Ig.
Discussion
We found that IRF4 was most highly expressed in immature B cells and was barely expressed in GC B cells. However, deletion of IRF4 largely spared immature B cells but most significantly eliminated GC B cells. We further found that IRF4 deletion abrogated the expression of GC regulator Bcl6.12 This possibly explains why IRF4 is barely expressed in GC B cells but controls their generation. In consistent with our findings, Willis S et al. have indicated that although IRF4 is absent in GC B cells, it is required for the development of early GC B cells.23 Moreover, Ochiai K et al. have reported that transient IRF4 expression is sufficient to activate Bcl6 gene and support the later generation of GC B cells.24
We found that IRF4 deletion in B cells largely eliminated plasma cells in transplant recipients. This elimination of plasma cells may be due to the lack of GC B cells upon IRF4 deletion. Of note, IRF4 itself is required for the differentiation of plasma cells, as deletion of IRF4 until after the generation of GC B cells still eliminates post-GC plasma cells.25 Overall, the deletion of IRF4 in B cells eliminates GC B/plasma cells and subsequently the production of IgG1 DSA, which correlates well with the inhibition of chronic rejection in our model.
Compared with CTLA4-Ig–treated Irf4fl/fl control recipients, CTLA4-Ig–treated Irf4fl/flCd19-Cre recipients contained significantly lower frequencies of Tfh cells, IFN-γ producing Th1 cells, and CD4+CD62L–CD44+ T cells at day 35 post-transplant. Thus, deletion of IRF4 in B cells inhibits the recovery of T cell response after the transient CTLA4-Ig treatment. Indeed, B cells can function as antigen-presenting cells and are involved in T-cell mediated transplant rejection.26–29 Ng Y et al. showed that B cell deficiency does not affect acute skin allograft rejection but reduces the frequency of IFN-γ producing T cells during the memory phase.26 Tfh cell differentiation is a multistage and multifactorial process. The late stage of Tfh cell differentiation involves GC.30 Lack of GC B cells may be one of the reasons why graft-accepted Irf4fl/flCd19-Cre recipients contained fewer Tfh cells.
B cell response in transplantation is unique, as allogeneic B cells remain chronically alloantigen-exposed, still responsive to alloantigens under T cell-targeted immunosuppression.8 Detailed investigation into the molecular mechanisms of this distinctive B cell response may provide insight for better B cell targeting in transplant therapy. This study probed the transcriptional regulation in B cells, focusing on a heart transplantation model in which CTLA4-Ig inhibited T cell-mediated acute rejection. We demonstrated the importance of B cells in promoting chronic rejection by deleting a single transcription factor IRF4 in B cells. Mechanistically, IRF4 is intrinsically required for allogeneic B cell response at multiple levels, including GC B cell generation, plasma cell differentiation, DSA production, and support of T cell response. Further analysis of the IRF4-controlled gene networks in B cells will open a new avenue to unveil the anti-graft B cell response at the molecular level.
In summary, B cells require IRF4 to mediate chronic transplant rejection. We have previously reported that IRF4 in T cells dictates T cell responses against allografts.16, 19 Hence, targeting IRF4 has the potential to eliminate both B and T cell responses to transplants, and represents an attractive therapeutic strategy for promoting transplant acceptance.
Supplementary Material
Acknowledgements
This study was supported by the US National Institutes of Health grant (#NIH R01AI132492 to WC) and the J.C. Walter Jr. Transplant Center Foundation (to AOG). The authors would like to thank the Jackson Laboratory Model Generation Services and the Houston Methodist Flow Cytometry Core Facility for excellent services. The authors appreciate Preston Arnold for helpful scientific discussion.
Footnotes
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lechler RI, Sykes M, Thomson AW, Turka LA. Organ transplantation--how much of the promise has been realized? Nat Med. 2005;11:605–13. [DOI] [PubMed] [Google Scholar]
- 2.Le Moine A, Goldman M, Abramowicz D. Multiple pathways to allograft rejection. Transplantation. 2002;73:1373–81. [DOI] [PubMed] [Google Scholar]
- 3.Kwun J, Bulut P, Kim E, et al. The role of B cells in solid organ transplantation. Semin Immunol. 2012;24:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libby P, Pober JS. Chronic rejection. Immunity. 2001;14:387–97. [DOI] [PubMed] [Google Scholar]
- 5.Kim KS, Denton MD, Chandraker A, et al. CD28-B7-mediated T cell costimulation in chronic cardiac allograft rejection: differential role of B7–1 in initiation versus progression of graft arteriosclerosis. Am J Pathol. 2001;158:977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Q, Yang Y. Late and chronic antibody-mediated rejection: main barrier to long term graft survival. Clin Dev Immunol. 2013;2013:859761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platt JL. Antibodies in transplantation. Discov Med. 2010;10:125–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Karahan GE, Claas FH, Heidt S. B Cell Immunity in Solid Organ Transplantation. Front Immunol. 2016;7:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loupy A, Hill GS, Jordan SC. The impact of donor-specific anti-HLA antibodies on late kidney allograft failure. Nat Rev Nephrol. 2012;8:348–57. [DOI] [PubMed] [Google Scholar]
- 10.Mao Q, Terasaki PI, Cai J, et al. Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five-year longitudinal study. Am J Transplant. 2007;7:864–71. [DOI] [PubMed] [Google Scholar]
- 11.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–70. [DOI] [PubMed] [Google Scholar]
- 13.Minnich M, Tagoh H, Bonelt P, et al. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17:331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook SL, Franke MC, Sievert EP, Sciammas R. A Synchronous IRF4-Dependent Gene Regulatory Network in B and Helper T Cells Orchestrating the Antibody Response. Trends Immunol. 2020;41:614–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Zhang HD, Shi XM, et al. Ablation of Transcription Factor IRF4 Promotes Transplant Acceptance by Driving Allogenic CD4(+) T Cell Dysfunction. Immunity. 2017;47:1114–28.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrio L, Roman-Garcia S, Diaz-Mora E, et al. B Cell Development and T-Dependent Antibody Response Are Regulated by p38gamma and p38delta. Front Cell Dev Biol. 2020;8:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rydyznski CE, Cranert SA, Zhou JQ, et al. Affinity Maturation Is Impaired by Natural Killer Cell Suppression of Germinal Centers. Cell Rep. 2018;24:3367–73 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang HD, Wu J, Zou DW, et al. Ablation of interferon regulatory factor 4 in T cells induces “memory” of transplant tolerance that is irreversible by immune checkpoint blockade. Am J Transplant. 2019;19:884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Chen S, Lan P, et al. Macrophage subpopulations and their impact on chronic allograft rejection versus graft acceptance in a mouse heart transplant model. Am J Transplant. 2018;18:604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy RR, Li YS, Allman D, Asano M, Gui M, Hayakawa K. B-cell commitment, development and selection. Immunol Rev. 2000;175:23–32. [PubMed] [Google Scholar]
- 23.Willis SN, Good-Jacobson KL, Curtis J, et al. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol. 2014;192:3200–6. [DOI] [PubMed] [Google Scholar]
- 24.Ochiai K, Maienschein-Cline M, Simonetti G, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein U, Casola S, Cattoretti G, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–82. [DOI] [PubMed] [Google Scholar]
- 26.Ng YH, Oberbarnscheidt MH, Chandramoorthy HC, Hoffman R, Chalasani G. B cells help alloreactive T cells differentiate into memory T cells. Am J Transplant. 2010;10:1970–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noorchashm H, Reed AJ, Rostami SY, et al. B cell-mediated antigen presentation is required for the pathogenesis of acute cardiac allograft rejection. J Immunol. 2006;177:7715–22. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Q, Ng YH, Singh T, et al. B cells mediate chronic allograft rejection independently of antibody production. J Clin Invest. 2014;124:1052–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong AS. B cells as antigen-presenting cells in transplantation rejection and tolerance. Cell Immunol. 2020;349:104061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crotty S T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.