Abstract
Lactobacillus acidophilus is a common kind of lactic acid bacteria usually found in the human gastrointestinal tract, oral cavity, vagina, and various fermented foods. At present, many studies have focused on the probiotic function and industrial application of L. acidophilus. Additionally, dozens of L. acidophilus strains have been genome sequenced, but there has been no research to compare them at the genomic level. In this study, 46 strains of L. acidophilus were performed comparative analyses to explore their genetic diversity. The results showed that all the L. acidophilus strains were divided into two clusters based on ANI values, phylogenetic analysis and whole genome comparison, due to the difference of their predicted gene composition of bacteriocin operon, CRISPR-Cas systems and prophages mainly. Additionally, L. acidophilus was a pan-genome open species with a difference in carbohydrates utilization, antibiotic resistance, EPS operon, surface layer protein operon and other functional gene composition. This work provides a better understanding of L. acidophilus from a genetic perspective, and offers a frame for the biotechnological potentiality of this species.
Keywords: Lactobacillus acidophilus, comparative genomics, CRISPR-Cas, bacteriocin, antibiotic resistance
1. Introduction
Lactobacillus acidophilus, a Gram-positive bacterium with low GC content (34–37%), belongs to phylum Firmicutes, class Bacilli, order Lactobacillales, Family Lactobacillaceae, and Genus Lactobacillus. L. acidophilus, originally isolated from the infant feces in the 1900s and mostly found in the human gastrointestinal tract, oral cavity, vagina, and various fermented foods. Additionally, NCFM (a typical L. acidophilus strain) is one of the most well commercially and clinically well-researched probiotics. Since it was isolated, it has been studied and found to have a variety of beneficial properties for gastrointestinal and general health. Alleviating inflammatory bowel disease through reducing cytokines is the most well-known benefit of L. acidophilus [1,2,3]. Additionally, other health-associated functions of L. acidophilus have also attracted much attention, such as in alleviating cancer [4,5], regulating immunity [6,7], reducing cholesterol [8], and relieving diarrhea [9]. According to recent reports, it had been found that L. acidophilus could directly or indirectly interfere with the intestinal microbiota or host metabolism by some active substances, such as extracellular polysaccharides (EPS), surface layer protein (SLP) and bacteriocin.
With the development of high-throughput sequencing technology and comparative genomics approaches, numerous reports focused on genomes of lactobacilli, such as Lacticaseibacillus rhamnosus [10], Lacticaseibacillus paracasei [11], Limosilactobacillus reuteri [12,13], Ligilactobacillus salivarius [14], Limosilactobacillus fermentum [15], Limosilactobacillus mucosae [16], and Ligilactobacillus ruminis [17]. These studies used comparative genomics methods to reveal general genome characteristics, phylogenetic relationships and functional genes related to niche adaptation and probiotic activity of different strains, which let researchers have a new understanding of the metabolic capabilities and function roles of lactobacilli at the genetic level.
Although L. acidophilus possesses many probiotic characteristics, for instance, L. acidophilus NCFM, ATCC 4356 and ATCC 53544 were well-known probiotics and had been genomic sequenced; rare research on comparative genomics of this species has been reported [18,19].
In current researches, the probiotic functions of different L. acidophilus strains showed both similarities and differences which might be resulted from the diversification of L. acidophilus. Analysis of comparative genomes is possible to excavate the diversification of this species to expose the correlation between genotypes and phenotypes, and to predict the potential probiotic functions for new L. acidophilus isolates strains based on past researches. Hence, the purpose of this study was to sequence more genomes of L. acidophilus and to carry out comparative genomics analyses. These works will consolidate the foundation for future exploration of L. acidophilus, especially some typical physiological characteristics and synthetic beneficial metabolites.
2. Materials and Methods
2.1. Bacterial Strains Culturing and Genome Sequencing
Eleven strains of L. acidophilus were isolated from healthy human feces from different regions of China (Table 1), then 16S rRNA genes were sequenced for species identification. Additionally, L. acidophilus CCFM137 is another strain deposited at the Culture Collection of Food Microorganisms in Jiangnan University, Wuxi, China (CCFM). These 12 determined L. acidophilus strains were cultured with de Man, Rogosa and Sharpe (MRS) medium in an anaerobic workstation for 24 h [20] The draft genomes of these 12 strains were sequenced by Majorbio BioTech Co. (Shanghai, China), and SOAPde novo and GapCloser were used to assemble and fill the reads of draft genomes [21]. Additionally, another thirty four publicly available genomes of L. acidophilus from the National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) (accessed on 15 August 2021) were used for comparison (Table 1).
Table 1.
Source information, BioSample and general genome features of 46 L. acidophilus strains.
| Strain | BioSample | Size(Mb) | GC(%) | CDS no. | Source |
|---|---|---|---|---|---|
| ATCC 4356 | SAMN03105773 | 1.9567 | 34.6 | 1730 | Human |
| ATCC 4796 | SAMN00001471 | 2.0205 | 34.7 | 1747 | Human |
| ATCC 53544 | SAMN07357495 | 1.99191 | 34.7 | 1773 | Human |
| BIO6307 | SAMN12856535 | 1.96977 | 34.6 | 1790 | Unknown |
| CCFM137 | SAMN19655193 | 1.951495 | 34.58 | 1904 | Human |
| CIP 76.13 | SAMEA2272342 | 1.95182 | 34.6 | 1741 | Human |
| CIRM-BIA 442 | SAMEA2272381 | 1.98699 | 34.7 | 1787 | Dairy product |
| CIRM-BIA 445 | SAMEA2272655 | 2.00201 | 34.6 | 1782 | Dairy product |
| DS10_1A | SAMN05583778 | 1.97072 | 34.6 | 1787 | Commercial Probiotic Products |
| DS13_1A | SAMN05583782 | 1.96427 | 34.6 | 1780 | Commercial Probiotic Products |
| DS13_1B | SAMN05583783 | 1.96393 | 34.6 | 1785 | Commercial Probiotic Products |
| DS2_1A | SAMN05583785 | 1.98202 | 34.6 | 1812 | Commercial Probiotic Products |
| DS20_1 | SAMN06464087 | 1.96947 | 34.6 | 1795 | Commercial Probiotic Products |
| DS24_1 | SAMN06464090 | 1.96837 | 34.6 | 1790 | Commercial Probiotic Products |
| DS5_1A | SAMN05583788 | 1.96883 | 34.6 | 1784 | Commercial Probiotic Products |
| DS8_1A | SAMN05583791 | 1.96757 | 34.6 | 1787 | Commercial Probiotic Products |
| DS9_1A | SAMN05583792 | 1.96901 | 34.6 | 1792 | Commercial Probiotic Products |
| DSM 20079 | SAMN06606133 | 2.00997 | 34.7 | 1760 | Human |
| DSM 20242 | SAMEA2272474 | 2.04786 | 34.7 | 1814 | Unknown |
| DSM 9126 | SAMEA2272239 | 1.99176 | 34.6 | 1790 | Unknown |
| FAHWH11L56 | SAMN19655182 | 1.975822 | 34.591 | 1881 | Human |
| FCQHC4LH1 | SAMN19655183 | 1.962741 | 34.569 | 1877 | Human |
| FFJND6L5 | SAMN19655184 | 1.960445 | 34.562 | 1878 | Human |
| FFJND7L5 | SAMN19655185 | 1.989652 | 34.581 | 1905 | Human |
| FGSYC48L79 | SAMN19655186 | 1.990393 | 34.662 | 1984 | Human |
| FHNXY41L162 | SAMN19655187 | 2.051142 | 34.733 | 2112 | Human |
| FNMGHHHT12L40 | SAMN19655188 | 1.992905 | 34.658 | 1894 | Human |
| FSHXBX32L130 | SAMN19655189 | 2.140788 | 34.893 | 2218 | Human |
| FSI4 | SAMN03274004 | 1.99197 | 34.7 | 1790 | Fermented dairy product |
| FXJSW24L139 | SAMN19655190 | 2.115801 | 34.647 | 2059 | Human |
| FXJSW48L59 | SAMN19655191 | 2.048808 | 34.753 | 2100 | Human |
| FZJTZ18L25 | SAMN19655192 | 2.115442 | 34.846 | 2170 | Human |
| LA_AVK1 | SAMN13198235 | 1.96274 | 34.6 | 1690 | Unknown |
| LA_AVK2 | SAMN13198280 | 1.96264 | 34.6 | 1690 | Unknown |
| LA1 | SAMN05631052 | 1.9912 | 34.7 | 1787 | Fermented dairy product |
| La-14 | SAMN02603216 | 1.99158 | 34.7 | 1781 | Human |
| LA-G80-111 | SAMN15165794 | 1.99198 | 34.7 | 1788 | Unknown |
| LMG P-21904 | SAMN07187785 | 1.96566 | 34.6 | 1780 | Commercial Probiotic Products |
| NCFM | SAMN02603047 | 1.99356 | 34.7 | 1760 | Human |
| P2 | SAMN07665576 | 2.04684 | 35.7 | 1904 | Commercial Probiotic Products |
| s-13 | SAMN15579847 | 1.96575 | 34.6 | 1778 | Unknown |
| s-4 | SAMN15579838 | 1.95327 | 34.6 | 1724 | Unknown |
| UBLA-34 | SAMN10136005 | 1.95104 | 34.6 | 1762 | Fermented foods |
| WG-LB-IV | SAMN04628015 | 1.95169 | 34.6 | 1780 | Fermented dairy product |
| YT1 | SAMN08142761 | 2.09254 | 34.7 | 1878 | Unknown |
The underline represented that draft genomes of these strains were sequenced in this study.
2.2. The Average Nucleotide Identity (ANI) Values and Phylogenetic Analyses
The ANI values of any two genomes were calculated through a python script (https://github.com/widdowquinn/pyani) (accessed on 19 January 2021)and the resulting matrix was clustered and visualized using Seaborn, a Python data visualization library [22].
2.3. Pan-Genome and Core-Genome Analysis
Pan-genome and core-genome calculation of the L. acidophilus was performed using PGAP-1.2.1 [23]. Curve fitting of the pan-genome and core-genome were performed using a power-law regression based on Heaps’ law and an exponential regression model, respectively [24,25]. PanGP software was used to conduct fitting and visualize the core- and pan-genomes [26]. Venn diagram of core-genome and specific genome of those strains was made by using Orthomcl software [27].
2.4. Phylogenetic Comparison
Orthologous sequences of those forty-six strains were extracted by Orthomcl-v2.0.9 software for clustering orthologous genes [28]. A phylogenetic tree was constructed using the neighbor-joining (NJ) algorithm with default parameters in MEGA 7.0 software [29], and the phylogenetic tree was decorated with Evolgenius (http://www.evolgenius.info/evolview/) (accessed on 10 February 2021) [30].
2.5. Whole Genome Comparison
Genome wide visualization of coding sequences identity among all strains was used BLAST ring image to perform generator (BRIG). Additionally, L. acidophilus ATCC33323 was taken as a reference genome [31].
2.6. Genotype Analysis of Carbohydrate Metabolism
Those L. acidophilus genomes were annotated by HMMER-3.1, and the carbohydrate active enzymes were analyzed by Carbohydrate Active Enzymes Database7 (CAZy) (http://www.cazy.org/) (accessed on 10 February 2021) [32,33]. Additionally, L. acidophilus strains were clustered by software HEMI version 1.0.
2.7. Genotype Analysis of Antibiotic Resistance
The resistance gene identifier (RGI) software based on a comprehensive antibiotic resistance database (CARD) was used to analyze the antibiotic resistance of all L. acidophilus strains [34].
2.8. Prediction of the EPS and Surface Layer Protein Gene Operon
The sequences of those L. acidophilus strains were aligned with the EPS-encoding operon by the basic local alignment search tool (BLAST) program [35]. The presence of genes was determined based on the alignment fragment size and identity [36]. Easy sequencing in PostScript (ESPript) online program was used to display the aligned sequences [37].
2.9. Prediction of Bacteriocin Operon
The genes for potential bacteriocin operons were mined by using BAGEL4 webserver [38]. Additionally, BLASTP was used to analyze the domains of bacteriocin against the non-redundant protein databases created by BLASTP based on the National Center for Biotechnology Information (NCBI).
2.10. Identification of CRISPR-Cas Systems and Prophage
The CRISPRCasFinder with default parameters was used to identify the clustered regularly interspaced short palindromic repeats (CRISPR) regions and CRISPR-associated (Cas) proteins [39]. The PHASTER (PHAge Search Tool Enhanced Release) webserver was used to identify and annotate the prophages within all the strains [40].
2.11. Ethics Statement
This study was approved by the Ethics Committee in Jiangnan University, China (SYXK 2012-0002). All the fecal samples from healthy persons were for public health purposes and these were the only human materials used in the present study. Written informed consent for the use of their fecal samples was obtained from the participants or their legal guardians. All of them conducted health questionnaires before sampling and no human experiments were involved. The collection of the fecal sample had no risk of predictable harm or discomfort to the participants.
3. Results
3.1. Genome Characteristics of L. acidophilus
In this study, 11 strains of L. acidophilus were isolated from human feces in different areas in China (Table 2), and in total 46 genomes were compared in this study (Table 1). The genome size of all the strains ranged from 1.95 Mb to 2.09 Mb, and the average size was 1.98 Mb. The average G + C content and number of coding sequences was 34.66% and 1780, respectively. Compared with the strains isolated from humans, those from fermented foods and commercial probiotic products possessed more coding sequences.
Table 2.
Host Information of 11 New Isolated L. acidophilus.
| Strain | Age | Gender | Modernization |
|---|---|---|---|
| FAHWH11L56 | 3 | Female | City |
| FCQHC4LH1 | 20 | Male | Rural |
| FFJND6L5 | 24 | Male | City |
| FFJND7L5 | 24 | Female | Rural |
| FGSYC48L79 | NA | NA | Rural |
| FHNXY41L162 | 80 | Female | Rural |
| FNMGHHHT12L40 | 23 | Female | City |
| FSHXBX32L130 | 59 | Female | Rural |
| FXJSW24L139 | 2.3 | Female | Rural |
| FXJSW48L59 | 12 | Female | Rural |
| FZJTZ18L25 | 79 | Male | Rural |
3.2. Pan-Genome and Core-Genome of L. acidophilus
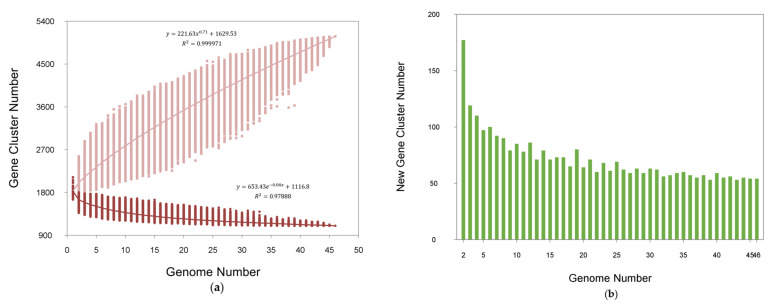
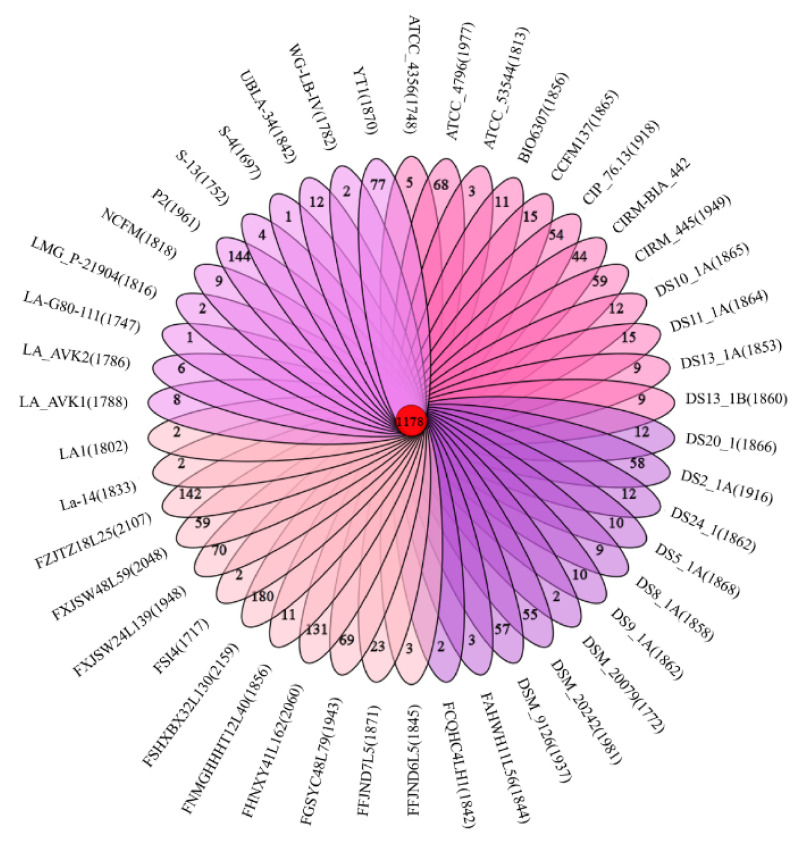
Pan-genome is the general term for all the genes of a species. In this study, the genetic diversity of L. acidophilus was shown by the core- and pan-genome curves. Additionally, the curve presented an asymptotic trend (Figure 1a). The number of new gene cluster increments gradually decreased from 141 to 13 (Figure 1b). According to the regression equation of the pan-genome, its size increased infinitely as new genomes were added sequentially, which indicated the pan-genome of L. acidophilus could be considered as an open state, which meant that the pan-genome size of species increased with the number of sequenced genomes. With the equation of core-genome, there were approximately 1117 genes harbored in the core-genome of L. acidophilus (Figure 1a). The specific and homologous core genes were represented by the Venn diagram among all the 46 L. acidophilus strains, which showed that there were 1178 shared genes among all the strains. Additionally, the number of distinctive genes for each L. acidophilus strain ranged from 1 to 180 (Figure 2).
Figure 1.
Pan-genome and core-genome of L. acidophilus. (a) Pan-genome plot is represented by the accumulated number of new genes against the number of genomes added. Core-genome plot is represented by the accumulated number of genes attributed to the core-genomes against the number of added genomes. (b) The new gene cluster number plot shows the changes in new genes of all strains genomes as the number of L. acidophilus strains increases.
Figure 2.
Venn diagram of L. acidophilus. The middle circle of the Venn diagram shows the number of homologous genes of these 46 L. acidophilus. Additionally, every branch shows the number of their unique genes.
3.3. ANI, Phylogenetic Analyses and Whole Genome Comparison of L. acidophilus
ANI analysis is a common standard for species classification and clustering. A previous study found that in most lineages, there was a clear dividing line of ANI within and between species, in which the ANI value of the same species was higher than 95%, and the ANI value of different species was less than 95% [41]. The results showed that all the 46 strains with ANI values above 97% belonged to L. acidophilus. However, all the strains were significantly divided into two regions, in which a small region consisted of FZJTZ18L25, FSHXBX32L130, FGSYC48L79, FHNXY41L162 and FXJSW28L59 and a large region consisted of all the other 41 strains (Figure 3).
Figure 3.
ANI Heatmap of L. acidophilus. Proposed species cut-off boundary is above 97%, showing identity within these strains.
A phylogenetic tree was performed based on the homologous genes of 46 L. acidophilus genomes in order to analyze the phylogenetic relationship of L. acidophilus strains (Figure 4). The phylogenetic tree revealed that all the 46 L. acidophilus strains formed three branches, which were rooted by L. gasseri ATCC33323 as an outgroup, and the L. acidophilus strains were divided into three clades (clade A, B and C). Clade A and clade B included six strains which could synthesize Helveticin-J, while all the strains in clade C predictably had the Operon Acidocin_J1132_beta_peptide_N-terminal. Interestingly, the composition of clade a on phylogenetic tree was the same as the small region in the ANI heatmap.
Figure 4.
Phylogenetic analysis of L. acidophilus. Phylogenetic tree showing the relationship among 46 L. acidophilus strains with L. gasseri ATCC33323 as outgroup. The tree was grouped into three clades signed by different colors of lines (red represented clade A, blue represented clade B and black represented clade C). The dots represented strains that contained the operon of bacteriocins. In specific, dark purple dots represented Acidocin_J1132_beta_peptide_N-terminal, claret dots represented Bacteriocin_helveticin_J, orange dots Enterolysin_A, mazarine dots Helveticin-J, green dots Lanthipeptide. Additionally, circular dots represented one bacteriocin gene operon while square dots represented two. Orange font represented strains isolated in this study, black represented obtained from NCBI. Pink frames represented strains that contained a complete CRISPR/Cas system. Wathet highlight represented strains contained intact prophage.
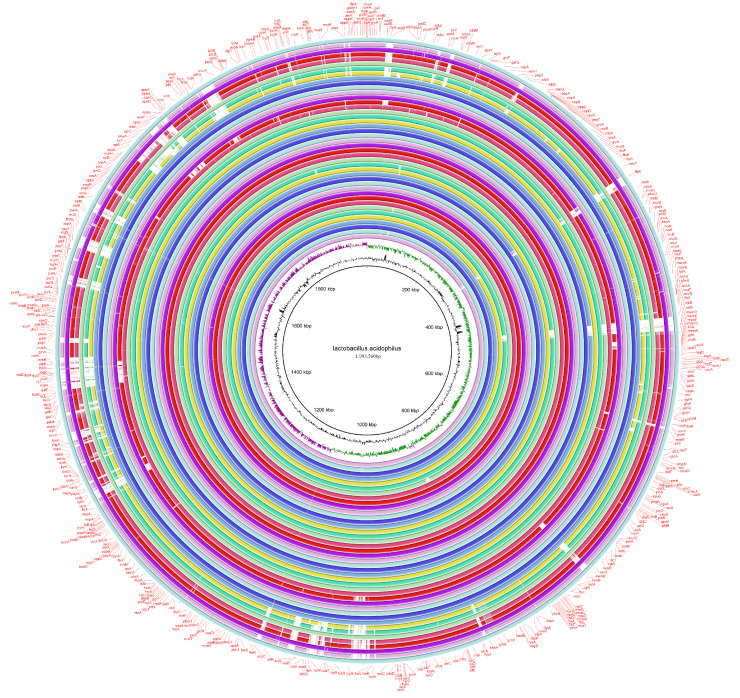
With L. acidophilus NCFM as a referenced genome, the whole genomes of all the 46 strains were analyzed through BRIG software. Most strains had no significant difference in their genome composition, apart from the clade A and small region clustered strains FZJTZ18L25, FXJSW28L59, FSHXBX32L130, FHNXY41L162, FGSYC48L79 and YT1, whose genome sequences located at rings 2, 4, 5, 7, 8 and 14, counting from the outside in the ring image (Figure 5). Those six genetically similar strains had similar gene deletion, such as ftsK, celB, repB and marR, which encoded putative cell division proteins, cellobiose-specific PTS IIC, putative replication initiator proteins and transcriptional regulators, respectively. Additionally, some genes related to ribose encoding including rbsB, rbsK and rbsR were absent.
Figure 5.
The whole genome comparison of L. acidophilus.
3.4. Genotype Analysis for Carbohydrates Utilization in L. acidophilus
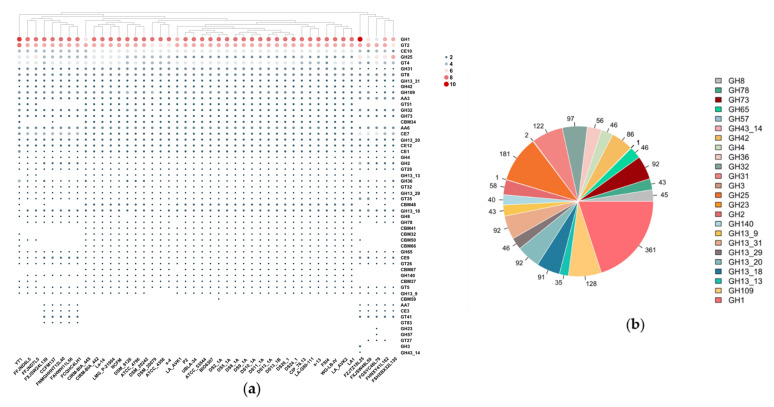
In this study, the CAZy database was utilized to analyze the 46 L. acidophilus genomes in order to explore how those strains using different carbohydrates. The results revealed that there were 54 genes encoding predicted enzymes which could activate carbohydrates, including 24 glycoside hydrolases (GH) families, 12 glycosyl transferases (GT) families, three auxiliary activities (AA) families, 9 carbohydrate-binding modules (CBM) families and six carbohydrate esterases (CE). GH1 (β-glucosidase, EC 3.2.1.21), GH3 (β-glucosidase, EC 3.2.1.21), GH13_20 (α-amylase, EC 3.2.1.1), GH13_31 (α-amylase, EC 3.2.1.1), GH25 (lysozyme, EC 3.2.1.17), GH73 (lysozyme, EC 3.2.1.17), GT2 (cellulose synthase, EC 2.4.1.12), GT4 (sucrose synthase, EC 2.4.1.13), GT8 (lipopolysaccharide α-1,3-galactosyltransferase, EC 2.4.1.44), GT41 (lipopolysaccharide α-1,3-galactosyltransferase, EC 2.4.1.44), GT51 (murein polymerase, EC 2.4.1.129), AA3 (cellobiose dehydrogenase, EC 1.1.99.18), AA6 (1,4-benzoquinone reductase, EC. 1.6.5.6), CE7 (acetyl xylan esterase, EC 3.1.1.72), CE10 (arylesterase, EC 3.1.1.-) and CE12 (pectin acetylesterase, EC 3.1.1.-) were distributed among all the L. acidophilus strains, while the remaining families had different distribution in those strains (Figure 6a).
Figure 6.
Carbohydrate utilization families identified in L. acidophilus. (a) Heatmap showing the distribution of carbohydrates utilization families. (b) Pie chart indicating the number of each GH family identified.
In carbohydrate metabolism, glycosyl hydrolases are the key enzymes. Among all the 24 predicted GH families, 15 of them were present in each strain. Additionally, those 15 families were involved in the metabolism of common carbohydrates, such as glucose, galactose, fructose, sucrose, starch and maltose in the human diet. In all GH families, GH3 (β-glucosidase, EC 3.2.1.21), GH23 (lysozyme type G, EC 3.2.1.17), GH43_14 (β-xylosidase, EC 3.2.1.37) and GH57 (α-amylase, EC 3.2.1.1) were the least four GH families (Figure 6b).
3.5. Genotype Analysis for Antibiotic Resistance of L. acidophilus
The number of different genes in all L. acidophilus was analyzed and annotated by RGI software. Genetically, macB and lmrB were the top two genes of antibiotic resistance in those strains whose numbers were nine to ten and four to five, respectively, while the number of other genes was less than four. In contrast, vanRM, ugd, poxtA, telT, ErmB, mel and mef(B) genes were the least seven genes related to antibiotic resistance in L. acidophilus (Figure 7a).
Figure 7.
Antibiotic resistance gene of L. acidophilus. (a) Heatmap showing number of antibiotic resistance gene. (b) Pie chart indicating the classes of antibiotics. (c) Bar plot showing gene number of different kinds of resistance mechanism.
With all the genes of antibiotic resistance, there were 18 different classes of antibiotic (acridine dye, aminocoumarin antibiotic, cephamycin, elfamycin antibiotic, fluoroquinolone antibiotic, fosfomycin, fusidic acid, glycopeptide antibiotic, lincosamide antibiotic, macrolide antibiotic, mupirocin, nitroimidazole antibiotic, oxazolidinone antibiotic, peptide antibiotic, pleuromutilin antibiotic, rifamycin antibiotic, streptogramin antibiotic and tetracycline antibiotic) resistant genes in L. acidophilus. Fluoroquinolone, glycopeptide, lincosamide, macrolide and tetracycline antibiotics were the most five classes of antibiotics that L. acidophilus could tolerate. There were more than 300 relevant genes of these five antibiotics distributed in L. acidophilus. (Figure 7b). For the resistance pattern, L. acidophilus could resist antibiotics through antibiotic efflux, antibiotic target alteration and antibiotic target protection (Figure 7c).
3.6. Comparative Analysis of Functional Gene Composition of L. acidophilus
The Clusters of Orthologous Groups (COG) database is a database for identifying orthologous genes. After COG comparison and analysis of the difference in the number of functional genes, the results showed that there was a significant difference in some functional genes, such as amino acid transport genes, carbohydrate transport and metabolism genes, cell wall/membrane/envelope biogenesis genes, coenzyme transport genes, defense mechanisms genes, energy production genes, mobilome (prophage and transposons) genes and transcription genes in L. acidophilus (Figure 8a–i). Furthermore, from PCA analysis, most L. acidophilus strains were clustered together except a small clustered group consisted of five strains including YT1, FZJTZ18L25, FSHXBX32L130, FGSYC48L79, FHNXY41L162 and FXJSW28L59, which was consistent with the phylogenetic tree clustering result (Figure 8j).
Figure 8.
COG analysis of L. acidophilus. (a) Composition of COG function categories. (b–i) Number of amino acid transport genes; carbohydrate transport and metabolism genes; cell wall/membrane/envelope biogenesis genes; coenzyme transport genes; defense mechanisms genes; energy production genes; energy production genes; mobilome (prophage and transposons) genes; transcription genes in clade A and C L. acidophilus strains. ***: p < 0.001, ****: p < 0.0001. All data are presented as mean ± SEM. (j) PCA analysis of composition of COG function categories in different L. acidophilus strains.
3.7. Prediction of the EPS Operon in L. acidophilus
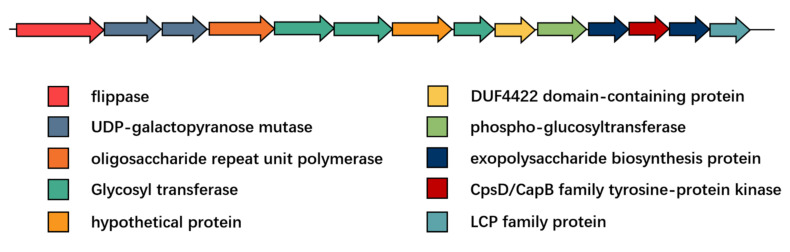
EPS is a key structural and functional composition in L. acidophilus. To figure out whether the newly genome-sequenced L. acidophilus could produce EPS, Orthomcl software and BlastN were used to predict the gene operon of EPS. All the protein sequences of EPS gene clusters were integrated together according to NCFM after Orthomcl analysis. For NCFM, the EPS gene cluster mainly composed of 14 genes including the highly conserved proteins LCP family protein (EpsA), exopolysaccharide biosynthesis protein (EpsB), CpsD/CapB family tyrosine-protein kinase (EpsC), exopolysaccharide biosynthesis protein (EpsD), phospho-glucosyltransferase (EpsE), DUF4422 domain-containing protein (EpsF), flippase (EpsI), UDP-galactopyranose mutase (EpsJ) and six variable proteins representing glycosyltransferases and polysaccharide polymerases (Figure 9). Among all the 46 L. acidophilus strains, there were only nine strains (CIRM_BIA_442, FAHWH11L56, FCQHC4LH1, FFJND6L5, FNMGHHHT12L40, FXJSW24L139, La-14 and NCFM) consisting of whole EPS-producing operons (Figure 10).
Figure 9.
EPS gene cluster in L. acidophilus NCFM.
Figure 10.
Heatmap of EPS gene cluster in L. acidophilus. Black green regions represented the specific genes exist in strains. The orange regions represented the specific genes do not exist in strains.
3.8. Prediction of the Surface Layer Protein Operon in L. acidophilus
Surface layer protein was one of the contents that could possess the biological properties of L. acidophilus. To explore whether the L. acidophilus newly isolated in this study could produce surface layer protein, the homologous gene operon that could encode this protein was predicted by Orthomcl software and BlastN. In most L. acidophilus strains, there were three independent genes relevant to surface layer protein, slpA could encode for the pore-forming S-layer protein SlpA (44,884 Da), and other two genes were identified as absent genes called slpB (43,636 Da), which could encode hypothetical SLAP domain-containing protein SlpB with 53% similarity to SlpA in the N-terminal and middle parts and only one amino acid residue difference in the C-terminal if this silent gene could express. With homologous gene analysis and multiple alignments of protein sequences, eight L. acidophilus strains including CCFM137, CIP_76.13, CIRM_BIA_445, DSM_9126, FCQHC4LH1, FGSYC48L79, FSHXBX32L130 and FZJTZ18L25 could not encode SlpA protein while other thirty-eight strains could encode it predictively. For putative protein SlpB, each strain had the corresponding genes (Figure 11).
Figure 11.
Heatmap of surface layer protein gene in L. acidophilus. Black green regions represented the specific genes exist in strains. The orange regions represented the specific genes do not exist in strains.
3.9. Prediction of the Bacteriocin Operon in L. acidophilus
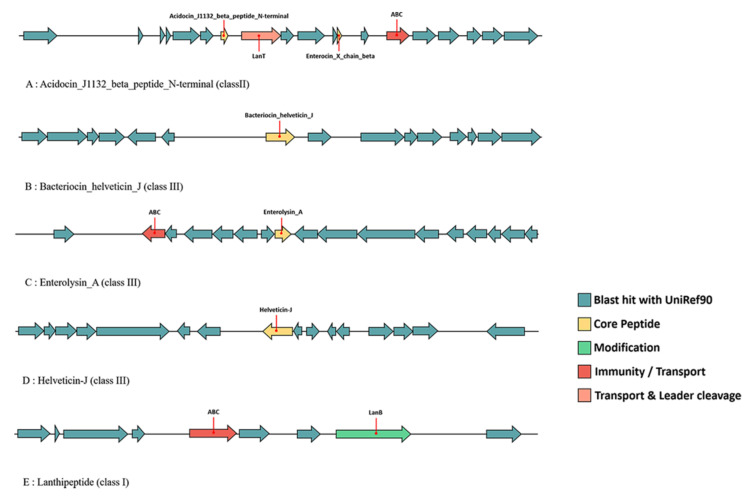
The potential operon of bacteriocin in the 46 L. acidophilus strains was mined using the BAGEL4 webserver in this study. The results showed that in total five different bacteriocins including acidocin_J1132_beta_peptide_N-terminal (6.2), bacteriocin_helveticin_J (6.3), enterolysin_A (64.3), helveticin-J (70.3) and lanthipeptide were predicted (Figure 4). In general, the bacteriocins synthesized by L. acidophilus covered all three classes, of which class II was the main one. Each strain was predicted that they could synthesize bacteriocin_helveticin_J and enterolysin_A, while only six and three strains could synthesize helveticin-J and lanthipeptide, respectively. Acidocin_J1132_beta_peptide_N-terminal was another common bacteriocin, which was identified in forty strains. Among all the 46 L. acidophilus, La-14 and WG-LB-IV were two special strains which could synthesize acidocin_J1132_beta_peptide_N-terminal, bacteriocin_helveticin_J and enterolysin_A and with two operons, respectively, while the other 44 strains only possessed one operon. Each bacteriocin gene operon in L. acidophilus contained its core peptides (Figure 12).
Figure 12.
Predicted bacteriocin gene operon in L. acidophilus.
3.10. Prediction of Prophages and CRISPR-Cas Systems in L. acidophilus
The prophages of L. acidophilus were predicted by PHASTER, and only six strains (FGSYC48L79, FHNXY41L162, FSHXBX32L130, FXJSW48L59, FZJTZ18L25 and YT1) contained intact prophages sequences (Table 3). Among them, FGSYC48L79 and FHNXY41L162 carried out two prophages, FSHXBX32L130 carried out three prophages, while FXJSW48L59, FZJTZ18L25 and YT1 only possessed one prophage, respectively. Interestingly, FGSYC48L79, FHNXY41L162, FSHXBX32L130, FXJSW48L59 and FZJTZ18L25 were clustered into same clade (clade A), while YT1 was clustered into their neighboring clade (clade B) on the phylogenetic tree (Figure 4).
Table 3.
CRISPR-Cas systems and prophages in L. acidophilus.
| Strain | CRISPR-Cas | Prophage |
|---|---|---|
| ATCC 4356 | incomplete | incomplete |
| ATCC 4796 | incomplete | incomplete |
| ATCC 53544 | incomplete | incomplete |
| BIO6307 | incomplete | incomplete |
| CCFM137 | incomplete | incomplete |
| CIP 76.13 | incomplete | incomplete |
| CIRM-BIA 442 | incomplete | incomplete |
| CIRM-BIA 445 | incomplete | incomplete |
| DS10_1A | incomplete | incomplete |
| DS11_1A | incomplete | incomplete |
| DS13_1A | incomplete | incomplete |
| DS13_1B | incomplete | incomplete |
| DS2_1A | incomplete | incomplete |
| DS20_1 | incomplete | incomplete |
| DS24_1 | incomplete | incomplete |
| DS5_1A | incomplete | incomplete |
| DS8_1A | incomplete | incomplete |
| DS9_1A | incomplete | incomplete |
| DSM 20079 | incomplete | incomplete |
| DSM 20242 | incomplete | incomplete |
| DSM 9126 | incomplete | incomplete |
| FAHWH11L56 | incomplete | incomplete |
| FCQHC4LH1 | incomplete | incomplete |
| FFJND6L5 | incomplete | incomplete |
| FFJND7L5 | incomplete | incomplete |
| FGSYC48L79 | incomplete | intact |
| FHNXY41L162 | complete | intact |
| FNMGHHHT12L40 | incomplete | incomplete |
| FSHXBX42L130 | incomplete | intact |
| FSI4 | incomplete | incomplete |
| FXJSW24L139 | incomplete | incomplete |
| FXJSW48L59 | complete | intact |
| FZJTZ18L25 | incomplete | intact |
| LA_AVK1 | incomplete | incomplete |
| LA_AVK2 | incomplete | incomplete |
| LA1 | incomplete | incomplete |
| La-14 | incomplete | incomplete |
| LA-G80-111 | incomplete | incomplete |
| LMG P-21904 | incomplete | incomplete |
| NCFM | incomplete | incomplete |
| P2 | incomplete | incomplete |
| s-13 | incomplete | incomplete |
| s-4 | incomplete | incomplete |
| UBLA-34 | incomplete | incomplete |
| WG-LB-IV | incomplete | incomplete |
| YT1 | complete | intact |
All the 46 genome sequences were uploaded to CRISPRCasFinder and predicted by orphan CRISPRs, which had no Cas proteins. Only three strains (FHNXY41L162, FXJSW48L59 and YT1) had complete CRISPR-Cas systems and all of them contained intact prophages (Figure 4). However, none of their spacers in CRISPR could correspond to the prophages’ sequences within them.
4. Discussion
L. acidophilus is a kind of lactic acid bacteria that has been widely used in industry for a long time and has excellent health-associated benefits. In previous studies, L. acidophilus was reported as a member of L. acidophilus group for phylogeny and comparative genome analysis [19]. However, due to the limitation of sequencing technology development and genome predictive analysis tools at that time, the results of gene prediction and annotation have an era limitation. With the development of sequencing technology and bioinformatics tools, researchers have the possibility of deeper analyzing the genomes. In this study, for the first time, L. acidophilus was analyzed to figure out the differences among strains through the comparative genomics approaches, including generally genomic characteristics, phylogenetic analysis, and prediction of some functional genes.
The average G + C content of L. acidophilus, one of the standard features in bacterial taxonomy, was 34.66%, and this value could reflect the genetic relationship in evolution to some extent. This value was consistent with that in Bergey’s Manual of Systematics of Archaea and Bacteria [42], but it was lower compared to other lactobacilli (with more than 40% G + C content). Additionally, its average size of genome was 1.98 Mb with ~1800 CDSs. Combined with COG analysis, the core genes of those strains were mainly carbohydrate transport and metabolism, defense mechanisms, translation, ribosomal structure and biogenesis and other basic functions.
The pan-genome consists of the core genome and the dispensable genome. The genetic plasticity and environmental adaptation potential of a species could be indicated from the relative size and content of the pan-genome. In the context of rapid development of high-throughput sequencing technology, it is convenient and quick to generate the whole genome for a strain of bacteria. Additionally, it is valuable to sequence its pan-genome to estimate the size of the entire gene repertoire and the diversity of this species [25]. Through the prediction of mathematical models, even if hundreds of genomes of each species were sequenced, there would be newly discovered genes in subsequent studies [43]. However, compared with the pan-genome of other lactobacilli, such as Limosilactobacillus mucosae (8100 genes) [16], Ligilactobacillus ruminis (10,000 genes) [17], Lacticaseibacillus rhamnosus (8200 genes) [44] and L. gasseri (6500 genes) [45], the pan-genome of L. acidophilus was smaller (5200 genes) and open. From the perspective of habitat, with Limosilactobacillus mucosae as an example, in addition to human feces and fermented dairy products, they had different habitat sources such as piglets, dogs and cattles. Their wide habitat range made them have larger gene pools for lateral gene transfer. The source of L. acidophilus used in this study was not sundry, and those strains were isolated from human feces and fermented products mainly. Hence, the open and narrow pan-genome status of L. acidophilus indicated that its genetic diversity could be further enriched, and it also had the ability to continue to adapt to various ecological niches. The similar inference had also been mentioned in previous study of Streptococcus [46]. From another point of view, the pan-genome of L. acidophilus may be due to its relatively stable intestinal niche, the small intestine environment, which was different from other lactobacilli colinized in the large intestine, and the living conditions abundance of the microflora of small intestine were lower than that in the large intestine, therefore, the living environment and niche for L. acidophilus was more stable, and there were fewer external changes and interferences, which contributed to that L. acidophilus did not need a wider pan-genome range to adapt to its niche.
ANI has been used to substitute DNA-DNA hybridization as the gold standard for prokaryotic species genetical circumscriptions [22]. Based on ANI, some phenotypically and genotypically closely related species, such as L. casei, Lacticaseibacillus paracasei and Lacticaseibacillus rhamnosus, that were difficult to distinguish on taxonomic level, had a new method to be identified [47,48,49]. Therefore, this study followed previous methods and carried out ANI analysis on L. acidophilus with the aim to explore the diversity of the species and the existence possibility of subspecies. All the 46 strains with ANI value above 97% were classified into two clusters, with five strains showing an ANI value over 99%, and the other group consisted of 41 strains including L. acidophilus NCFM with an ANI range 97–98% compared with another group. The ANI value of L. acidophilus was relatively narrow, compared with Limosilactobacillus mucosae (95.5%) [16] and Ligilactobacillus ruminis (96%) [17], representing that the proportion of variable genes was less, and the diversity was not rich. Combining the results of pan-genome analysis, it was speculated that L. acidophilus may not need many genes to adapt to different niches. Phylogenetic analyses and whole genome comparison of L. acidophilus were performed to further mine the relationship of the two classified groups. Interestingly, the five strains gathered in ANI analysis were also clustered in the branch on the phylogenetic tree. Meanwhile, they had similar levels of gene deletion. It was speculated that those five strains may be potential subspecies in L. acidophilus. However, due to the small sample size, this hypothesis still needs further verification.
For bacteria living in the intestine, their ability to use nutrients partly determined their ability to reside and survive in the intestine. In different nutrient environments, the carbohydrate utilization genes in L. acidophilus will be differentially expressed [50]. In silico, there were 24 GH series involving carbohydrate metabolism showed that GH1 accounted for a relatively high proportion of GH, showing that the main carbon source of L. acidophilus was glucose. Additionally, the number of genes in carbohydrate transport and metabolism in COG analysis showed that there was a significant difference between clade A and C, which meant that in L. acidophilus there might be two branches with different carbohydrate utilization.
Similar to other probiotics, L. acidophilus can easily obtain different antibiotic resistance genes in the intestine through mobile genetic elements. If it is to be added to food as a food additive, it needs to undergo safety verification [51]. In this study, predictions of antibiotic resistance genes have been made for each strain. Additionally, L. acidophilus has the most resistance genes of fluoroquinolone, glycopeptide, lincosamide, macrolide and tetracycline, which was consistent with previous reports [52,53,54,55].
The lactobacilli EPS had the effect of regulating intestinal immunity. Additionally, L. acidophilus NCFM had been reported that its EPS induced genes expression related to immunity both in vitro and in vivo [56]. Additionally, the EPS (LA-EPS-20079) of L. acidophilus DSM20079 had been proved that exerted a direct cytotoxic action on the tumors cells in addition to stimulating the immune response and inflammatory pathway [4]. Referring to NCFM as the standard [2005NCFM], the EPS cluster showed high synteny to EPS cluster of L. gasseri [45], L. johnsonii [57], Lacticaseibacillus casei [58] and streptococci [59]. The relationship between EPS gene clusters and EPS synthesis in Lacticaseibacillus casei was investigated, and that glucose-1-phosphate thymidyltranseferase gene (LC2W_2179), uncharacterized EPS biosynthesis protein (LC2W_2188), and EPS biosynthesis protein (LC2W_2189) were related to EPS biosynthesis. According to these results, epsB (exopolysaccharide biosynthesis protein gene) and epsD (exopolysaccharide biosynthesis protein gene) may exert the same effect in L. acidophilus NCFM. Although not all the L. acidophilus strains had a complete EPS gene cluster composing of 14 genes, similar to NCFM, each strain had epsB and epsD, indicating that all the 46 strains may have the potential to synthesize EPS.
Surface layer protein A (SlpA) of L. acidophilus is a key factor in probiotic–host crosstalk and could trigger immunomodulation in the host [2,60,61]. Different from EPS, SlpA is a protein regulated by a single gene. There were eight out of all the L. acidophilus strains that possessed no SlpA encoding gene. However, the expression of surface layer protein is related to some environmental factors, such as anaerobic conditions [62], bile salt concentrations [63], mucin, pancreatin and pH [64]. Therefore, whether L. acidophilus could produce SlpA or not need more experiments to verify.
Since the discovery of antimicrobials, compounds that could kill or inhibit the growth of bacteria, human life expectancy was improved. Nevertheless, antibiotic resistance has become a major threat. The bacteriocin derived from the intestinal microflora has shown great potential in maintaining intestinal homeostasis and biological control of pathogenic bacteria [65]. Bacteriocin production is one of the characteristics of L. acidophilus. For example, L. acidophilus JCM 1132 produced acidocin J1132 that had a narrow inhibitory spectrum [66], in addition, TK9201 produced acidocin A [67], DSM20079 generated acidocin D20079, and PNW3 produced a bacteriocin predicted to be helveticin J [68]. Additionally, a novel class III bacteriocin gene (NX371) was mined by bioinformatic analysis in L. acidophilus NX2-6, which had 98.15% homology of helveticin J [69]. Combining the results of previous researches and bioinformatics predictions in this research, it can be inferred that, L. acidophilus mainly produce a variety of class II (acidocin) and class III (helveticin J) bacteriocins. Acidocin was the most common bacteriocin in L. acidophilus [66,69,70,71,72,73,74,75]. However, in the prediction of bacteriocin operon, six strains did not have the gene cluster of acidocin-like bacteriocin. Coincidentally, all of them had hypothetical operon of helveticin-J. It was inferred that they may inhibit other bacteria by encoding helveticin-J instead of acidocin-like bacteriocin. Gene cluster of enterolysin A was predicted in all strains, and there were no relevant reports that it can be purified from L. acidophilus. Enterolysin A, an antimicrobial protein that could inhibit the growth of specific bacteria, was purified from an Enterococcus faecalis LMG 2333 [76]. Enterococcus has a high tendency to acquire and express new determinants of resistance, and the acquired resistance can then be transferred to other bacteria through mobile genetic elements [77]. That may be the reason why some L. acidophilus had the operon of enterolysin A. Lanthipeptide is not a typical bacteriocin in lactobacilli, but three L. acidophilus strains possessed the potentially synthetic genes.
In this study, only six strains carried out intact prophages sequences, and the number was very few compared to other lactobacilli [16,17,44,46]. Moreover, as the result of COG analysis, the number of prophages and transposons genes of those six strains clustered in clade A were more than twice as much as clade C. The reason could be due to the niche of L. acidophilus, which is the small intestine, a place where the living environment is much worse than the colon and cecum. This will result in not many prophages being able to exist in such a niche and L. acidophilus may not integrate the sequences of the prophages into their own genomes. Similarly, only three strains had complete CRISPR-Cas systems, which meant that they did not need the existence of CRISPR-Cas system, whose possession could be non-adaptive for strains [78] to resist foreign DNA invasion.
5. Conclusions
Based on ANI values, phylogenetic analysis and whole genome comparison, 46 L. acidophilus strains could be divided into two parts, which could be caused by the difference of predicted gene composition of bacteriocin operon, CRISPR-Cas systems and prophages mainly. In addition, predictably, genes related to carbohydrate utilization, EPS production, surface layer protein production, and antibiotic resistance of different L. acidophilus strains were all different. Hence, genome sequencing and genetic analysis enabled this research to deeply understand and exploit the biotechnology potential of L. acidophilus.
Author Contributions
Conceptualization, B.Y., H.Z. and W.C.; methodology, Z.H. and X.Z.; soft-ware, Z.H. and X.Z.; validation, Z.H., J.Z. and B.Y.; formal analysis, Z.H. and X.Z.; investigation, Z.H. and X.Z.; resources, J.Z.; data curation, Z.H. and B.Y.; writing—original draft preparation, Z.H. and B.Y.; writing—review and editing, R.P.R., C.S. and B.Y.; visualization, Z.H.; supervision, J.Z., H.Z. and B.Y.; project administration, J.Z.; funding acquisition, W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Nos. 32021005, 31820103010), National First-Class Discipline Program of Food Science and Technology (JUFSTR20180102), the Fundamental Research Funds for the Central Universities (JUSRP52003B), and Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
Institutional Review Board Statement
This study was approved by the Ethics Committee in Jiangnan University, China (SYXK 2012-0002) (15 February 2012).
Informed Consent Statement
Written informed consent for the use of their fecal samples was obtained from the participants or their legal guardians. All of them conducted health questionnaires before sampling and no human experiments were involved. The collection of the fecal sample had no risk of predictable harm or discomfort to the participants.
Data Availability Statement
All raw sequencing data analysed in this study have been submitted to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) (accessed on 28 June 2021) under the BioProject PRJNA736624.
Conflicts of Interest
All authors declared no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vemuri R., Gundamaraju R., Shinde T., Perera A.P., Basheer W., Southam B., Gondalia S.V., Karpe A.V., Beale D.J., Tristram S., et al. Lactobacillus acidophilus DDS-1 Modulates Intestinal-Specific Microbiota, Short-Chain Fatty Acid and Immunological Profiles in Aging Mice. Nutrients. 2019;11:1297. doi: 10.3390/nu11061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lightfoot Y.L., Selle K., Yang T., Goh Y.J., Sahay B., Zadeh M., Owen J.L., Colliou N., Li E., Johannssen T., et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 2015;34:881–895. doi: 10.15252/embj.201490296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Gu Y., Fang K., Mao K., Dou J., Fan H., Zhou C., Wang H. Lactobacillus acidophilus and Clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef. Microbes. 2018;9:775–787. doi: 10.3920/BM2017.0035. [DOI] [PubMed] [Google Scholar]
- 4.El-Deeb N.M., Yassin A.M., Al-Madboly L.A., El-Hawiet A. A novel purified Lactobacillus acidophilus 20079 exopolysaccharide, LA-EPS-20079, molecularly regulates both apoptotic and NF-kappa B inflammatory pathways in human colon cancer. Microb. Cell Factories. 2018;17:29. doi: 10.1186/s12934-018-0877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuo Q., Yu B.H., Zhou J., Zhang J.Y., Zhang R.L., Xie J.Y., Wang Q.L., Zhao S.L. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci. Rep. 2019;9:20128. doi: 10.1038/s41598-019-56661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paineau D., Carcano D., Leyer G., Darquy S., Alyanakian M.-A., Simoneau G., Bergmann J.-F., Brassart D., Bornet F., Ouwehand A.C. Effects of seven potential probiotic strains on specific immune responses in healthy adults: A double-blind, randomized, controlled trial. FEMS Immunol. Med. Microbiol. 2008;53:107–113. doi: 10.1111/j.1574-695X.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 7.Ouwehand A.C., Tiihonen K., Saarinen M., Putaala H., Rautonen N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: Intestinal and immune parameters. Br. J. Nutr. 2009;101:367–375. doi: 10.1017/S0007114508003097. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y., Wang J.F., Quan G.H., Wang X.J., Yang L.F., Zhong L.L. Lactobacillus acidophilus ATCC 4356 Prevents Atherosclerosis via Inhibition of Intestinal Cholesterol Absorption in Apolipoprotein E-Knockout Mice. Appl. Environ. Microbiol. 2014;80:7496–7504. doi: 10.1128/AEM.02926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitapanarux I., Chitapanarux T., Traisathit P., Kudumpee S., Tharavichitkul E., Lorvidhaya V. Randomized controlled trial of live Lactobacillus acidophilus plus bifidobacterium bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients. Radiat. Oncol. 2010;5:31. doi: 10.1186/1748-717X-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douillard F.P., Ribbera A., Kant R., Pietila T.E., Jarvinen H.M., Messing M., Randazzo C.L., Paulin L., Laine P., Ritari J., et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013;9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smokvina T., Wels M., Polka J., Chervaux C., Brisse S., Boekhorst J., van Hylckama Vlieg J.E., Siezen R.J. Lactobacillus paracasei comparative genomics: Towards species pan-genome definition and exploitation of diversity. PLoS ONE. 2013;8:e68731. doi: 10.1371/journal.pone.0068731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J., Zhao X., Lin X.B., Gänzle M. Comparative genomics Lactobacillus reuteri from sourdough reveals adaptation of an intestinal symbiont to food fermentations. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Jiang X., Yang Y., Zhang J., Zeng X., Wu Z., Sun Y., Pan D. Prevention of necrotizing enterocolitis through surface layer protein of Lactobacillus acidophilus CICC6074 reducing intestinal epithelial apoptosis. J. Funct. Foods. 2018;47:91–99. doi: 10.1016/j.jff.2018.05.045. [DOI] [Google Scholar]
- 14.Lee J.-Y., Han G.G., Kim E.B., Choi Y.-J. Comparative genomics of Lactobacillus salivarius strains focusing on their host adaptation. Microbiol. Res. 2017;205:48–58. doi: 10.1016/j.micres.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Verce M., De Vuyst L., Weckx S. Comparative genomics of Lactobacillus fermentum suggests a free-living lifestyle of this lactic acid bacterial species. Food Microbiol. 2020;89:10. doi: 10.1016/j.fm.2020.103448. [DOI] [PubMed] [Google Scholar]
- 16.Jia Y., Yang B., Ross P., Stanton C., Zhang H., Zhao J., Chen W. Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches. Genes. 2020;11:95. doi: 10.3390/genes11010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., Yang B., Ross R.P., Stanton C., Zhao J., Zhang H., Chen W. Comparative Genomics Analysis of Lactobacillus ruminis from Different Niches. Genes. 2020;11:70. doi: 10.3390/genes11010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrangou R., Altermann E., Hutkins R., Cano R., Klaenhammer T.R. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA. 2003;100:8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger B., Pridmore R.D., Barretto C., Delmas-Julien F., Schreiber K., Arigoni F., Bruessow H. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 2007;189:1311–1321. doi: 10.1128/JB.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donnell M.M., Harris H.M.B., Jeffery I.B., Claesson M.J., Younge B., O’Toole P.W., Ross R.P. The core faecal bacterial microbiome of Irish Thoroughbred racehorses. Lett. Appl. Microbiol. 2013;57:492–501. doi: 10.1111/lam.12137. [DOI] [PubMed] [Google Scholar]
- 21.Luo R.B., Liu B.H., Xie Y.L., Li Z.Y., Huang W.H., Yuan J.Y., He G.Z., Chen Y.X., Pan Q., Liu Y.J., et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:6. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter M., Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y.B., Wu J.Y., Yang J.H., Sun S.X., Xiao J.F., Yu J. PGAP: Pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasko D.A., Rosovitz M., Myers G.S., Mongodin E.F., Fricke W.F., Gajer P., Crabtree J., Sebaihia M., Thomson N.R., Chaudhuri R. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin H., Riley D., Cattuto C., Medini D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y., Jia X., Yang J., Ling Y., Zhang Z., Yu J., Wu J., Xiao J. PanGP: A tool for quickly analyzing bacterial pan-genome profile. Bioinformatics. 2014;30:1297–1299. doi: 10.1093/bioinformatics/btu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Stoeckert C.J., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelleher P., Bottacini F., Mahony J., Kilcawley K.N., van Sinderen D. Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genom. 2017;18:20. doi: 10.1186/s12864-017-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailund T., Brodal G.S., Fagerberg R., Pedersen C.N.S., Phillips D. Recrafting the neighbor-joining method. BMC Bioinform. 2006;7:8. doi: 10.1186/1471-2105-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian B., Gao S.H., Lercher M.J., Hu S.N., Chen W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombard V., Ramulu H.G., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.L.V., Cheng A.A., Liu S.H., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J.R., Tang W., Zheng Y.N., Xing Z.Q., Wang Y.P. Functional and bioinformatics analysis of an exopolysaccharide-related gene (epsN) from Lactobacillus kefiranofaciens ZW3. Arch. Microbiol. 2016;198:611–618. doi: 10.1007/s00203-016-1217-5. [DOI] [PubMed] [Google Scholar]
- 36.Petrova M.I., Macklaim J.M., Wuyts S., Verhoeven T., Vanderleyden J., Gloor G.B., Lebeer S., Reid G. Comparative Genomic and Phenotypic Analysis of the Vaginal Probiotic Lactobacillus rhamnosus GR-1. Front. Microbiol. 2018;9:1278. doi: 10.3389/fmicb.2018.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Heel A.J., de Jong A., Song C.X., Viel J.H., Kok J., Kuipers O.P. BAGEL4: A user-friendly webserver to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Neron B., Rocha E.P.C., Vergnaud G., Gautheret D., Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y.J., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:8. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitman W.B., Rainey F., Kämpfer P., Trujillo M., Chun J., DeVos P., Hedlund B., Dedysh S., Nedashkovskaya O. Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley; Hoboken, NJ, USA: 2016. [Google Scholar]
- 43.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Huang D., Yang B., Chen Y., Stanton C., Ross R.P., Zhao J., Zhang H., Chen W. Comparative genomic analyses of Lactobacillus rhamnosus isolated from Chinese subjects. Food Biosci. 2020;36:100659. doi: 10.1016/j.fbio.2020.100659. [DOI] [Google Scholar]
- 45.Zhou X., Yang B., Stanton C., Ross R.P., Zhao J., Zhang H., Chen W. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genom. 2020;21:1–16. doi: 10.1186/s12864-020-6527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefebure T., Stanhope M.J. Evolution of the core and pan-genome of Strepococcus: Positive selection, recombination, and genome composition. Genome Biol. 2007;8:30. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang C.H., Li S.W., Huang L., Watanabe K. Identification and Classification for the Lactobacillus casei Group. Front. Microbiol. 2018;9:1974. doi: 10.3389/fmicb.2018.01974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D.D., Gu C.T. Lactobacillus pingfangensis sp. nov., Lactobacillus daoliensis sp. nov., Lactobacillus nangangensis sp. nov., Lactobacillus daowaiensis sp. nov., Lactobacillus dongliensis sp. nov., Lactobacillus songbeiensis sp. nov. and Lactobacillus kaifaensis sp. nov., isolated from traditional Chinese pickle. Int. J. Syst. Evol. Microbiol. 2019;69:3251–3261. doi: 10.1099/ijsem.0.003619. [DOI] [PubMed] [Google Scholar]
- 49.Rocha J., Botelho J., Ksiezarek M., Perovic S.U., Machado M., Carrico J.A., Pimentel L.L., Salsinha S., Rodriguez-Alcala L.M., Pintado M., et al. Lactobacillus mulieris sp. nov., a new species of Lactobacillus delbrueckii group. Int. J. Syst. Evol. Microbiol. 2020;70:1522–1527. doi: 10.1099/ijsem.0.003901. [DOI] [PubMed] [Google Scholar]
- 50.Barrangou R., Azcarate-Peril M.A., Duong T., Conners S.B., Kelly R.M., Klaenhammer T.R. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA. 2006;103:3816–3821. doi: 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Granato D., Barba F.J., Kovacevic D.B., Lorenzo J.M., Cruz A.G., Putnik P. Functional Foods: Product Development, Technological Trends, Efficacy Testing, and Safety. Annu. Rev. Food Sci. Technol. 2020;11:93–118. doi: 10.1146/annurev-food-032519-051708. [DOI] [PubMed] [Google Scholar]
- 52.Gupta P.K., Mital B.K., Gupta R.S. Antibiotic sensitivity pattern of various Lactobacillus acidophilus strains. Indian J. Exp. Biol. 1995;33:620–621. [PubMed] [Google Scholar]
- 53.Cui Y., Qu X. Comparative Analysis of Two Component Signal Transduction Systems of The Lactobacillus Acidophilus Group. Braz. J. Microbiol. 2011;42:147–157. doi: 10.1590/S1517-83822011000100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Georgieva R., Yocheva L., Tserovska L., Zhelezova G., Stefanova N., Atanasova A., Danguleva A., Ivanova G., Karapetkov N., Rumyan N., et al. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 2015;29:84–91. doi: 10.1080/13102818.2014.987450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayrhofer S., van Hoek A., Mair C., Huys G., Aarts H.J.M., Kneifel W., Domig K.J. Antibiotic susceptibility of members of the Lactobacillus acidophilus group using broth microdilution and molecular identification of their resistance determinants. Int. J. Food Microbiol. 2010;144:81–87. doi: 10.1016/j.ijfoodmicro.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 56.Li L., Jiang Y.-J., Yang X.-Y., Liu Y., Wang J.-Y., Man C.-X. Immunoregulatory effects on Caco-2 cells and mice of exopolysaccharides isolated from Lactobacillus acidophilus NCFM. Food Funct. 2014;5:3261–3268. doi: 10.1039/C4FO00565A. [DOI] [PubMed] [Google Scholar]
- 57.Pridmore R.D., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song X., Xiong Z.Q., Kong L.H., Wang G.Q., Ai L.Z. Relationship Between Putative eps Genes and Production of Exopolysaccharide in Lactobacillus casei LC2W. Front. Microbiol. 2018;9:1882. doi: 10.3389/fmicb.2018.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stingele F., Neeser J.R., Mollet B. Identification and characterization of the eps (Exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Z., Xu P., Wu Z., Pan D. Anti-inflammatory activity of surface layer protein SlpA of Lactobacillus acidophilus CICC 6074 in LPS-induced RAW 264.7 cells and DSS-induced mice colitis. J. Funct. Foods. 2018;51:16–27. doi: 10.1016/j.jff.2018.10.008. [DOI] [Google Scholar]
- 61.Wang H., Zhang Q., Niu Y., Zhang X., Lu R. Surface-layer protein from Lactobacillus acidophilus NCFM attenuates tumor necrosis factor-alpha-induced intestinal barrier dysfunction and inflammation. Int. J. Biol. Macromol. 2019;136:27–34. doi: 10.1016/j.ijbiomac.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 62.Jakava-Viljanen M., Avall-Jaaskelainen S., Messner P., Sleytr U.B., Palva A. Isolation of three new surface layer protein genes (slp) from Lactobacillus brevis ATCC 14869 and characterization of the change in their expression under aerated and anaerobic conditions. J. Bacteriol. 2002;184:6786–6795. doi: 10.1128/JB.184.24.6786-6795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khaleghi M., Kermanshahi R.K., Yaghoobi M.M., Zarkesh-Esfahani S.H., Baghizadeh A. Assessment of Bile Salt Effects on S-Layer Production, slp Gene Expression and, Some Physicochemical Properties of Lactobacillus acidophilus ATCC 4356. J. Microbiol. Biotechnol. 2010;20:749–756. doi: 10.4014/jmb.0906.06050. [DOI] [PubMed] [Google Scholar]
- 64.Ramiah K., van Reenen C.A., Dicks L.M.T. Expression of the Mucus Adhesion Gene Mub, Surface Layer Protein Slp and Adhesion-Like Factor EF-TU of Lactobacillus acidophilus ATCC 4356 Under Digestive Stress Conditions, as Monitored with Real-Time PCR. Probiotics Antimicrob. Proteins. 2009;1:91–95. doi: 10.1007/s12602-009-9009-8. [DOI] [PubMed] [Google Scholar]
- 65.Garcia-Gutierrez E., Mayer M.J., Cotter P.D., Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10:1–21. doi: 10.1080/19490976.2018.1455790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tahara T., Oshimura M., Umezawa C., Kanatani K. Isolation, partial characterization, and mode of action of acidocin J1132, a two-component bacteriocin produced by Lactobacillus acidophilus JCM 1132. Appl. Environ. Microbiol. 1996;62:892–897. doi: 10.1128/aem.62.3.892-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanatani K., Oshimura M., Sano K. Isolation and characterization of acidocin a and cloning of the bacteriocin gene from lactobacillus-acidophilus. Appl. Environ. Microbiol. 1995;61:1061–1067. doi: 10.1128/aem.61.3.1061-1067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alayande K.A., Aiyegoro O.A., Ateba C.N. Distribution of Important Probiotic Genes and Identification of the Biogenic Amines Produced by Lactobacillus acidophilus PNW3. Foods. 2020;9:1840. doi: 10.3390/foods9121840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng F.Q., Zhu X.Y., Zhao H.Z., Nie T., Lu F.X., Lu Z.X., Lu Y.J. A class III bacteriocin with broad-spectrum antibacterial activity from Lactobacillus acidophilus NX2-6 and its preservation in milk and cheese. Food Control. 2021;121:107597. doi: 10.1016/j.foodcont.2020.107597. [DOI] [Google Scholar]
- 70.Tenbrink B., Minekus M., Vandervossen J., Leer R.J., Huisintveld J.H.J. Antimicrobial Activity of Lactobacilli-Preliminary Characterization and Optimization of Production of Acidocin-B, A Novel Bacteriocin Produced by Lactobacillus-Acidophilus-M46. J. Appl. Bacteriol. 1994;77:140–148. doi: 10.1111/j.1365-2672.1994.tb03057.x. [DOI] [PubMed] [Google Scholar]
- 71.Leer R.J., Vandervossen J., Vangiezen M., Vannoort J.M., Pouwels P.H. Genetic-Analysis of Acidocin-B, A Novel Bacteriocin Produced by Lactobacillus Acidophilus. Microbiology. 1995;141:1629–1635. doi: 10.1099/13500872-141-7-1629. [DOI] [PubMed] [Google Scholar]
- 72.Chumchalova J., Stiles J., Josephsen J., Plockova M. Characterization and purification of acidocin CH5, a bacteriocin produced by Lactobacillus acidophilus CH5. J. Appl. Microbiol. 2004;96:1082–1089. doi: 10.1111/j.1365-2672.2004.02237.x. [DOI] [PubMed] [Google Scholar]
- 73.Majhenic A.C., Venema K., Allison G.E., Matijasic B.B., Rogelj I., Klaenhammer T.R. DNA analysis of the genes encoding acidocin LF221 A and acidocin LF221 B, two bacteriocins produced by Lactobacillus gasseri LF221. Appl. Microbiol. Biotechnol. 2004;63:705–714. doi: 10.1007/s00253-003-1424-2. [DOI] [PubMed] [Google Scholar]
- 74.Deraz S.F., Karlsson E.N., Hedstrom M., Andersson M.M., Mattiasson B. Purification and characterisation of acidocin D20079, a bacteriocin produced by Lactobacillus acidophilus DSM 20079. J. Biotechnol. 2005;117:343–354. doi: 10.1016/j.jbiotec.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Deraz S.F., Karlsson E.N., Khalil A.A., Mattiasson B. Mode of action of acidocin D20079, a bacteriocin produced by the potential probiotic strain, Lactobacillus acidophilus DSM 20079. J. Ind. Microbiol. Biotechnol. 2007;34:373–379. doi: 10.1007/s10295-007-0206-8. [DOI] [PubMed] [Google Scholar]
- 76.Nilsen T., Nes I.F., Holo H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003;69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hegstad K., Mikalsen T., Coque T.M., Werner G., Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 78.Bondy-Denomy J., Davidson A.R. To acquire or resist: The complex biological effects of CRISPR-Cas systems. Trends Microbiol. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw sequencing data analysed in this study have been submitted to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) (accessed on 28 June 2021) under the BioProject PRJNA736624.