Abstract
Alcoholic liver fatty disease (ALFD) is caused by excessive and chronic alcohol consumption. Alcohol consumption causes an imbalance in the intestinal microflora, leading to liver disease induced by the excessive release of endotoxins into the hepatic portal vein. Therefore, research on the intestinal microflora to identify treatments for ALFD is increasing. In this study, the protective effects of lactic acid bacteria (LAB) strains, including Levilactobacillus brevis, Limosilactobacillus reuteri, and Limosilactobacillus fermentum, were evaluated in ethanol-induced HepG2 cells. Among the evaluated LAB, nine strains increased aldehyde dehydrogenase (ALDH) levels and downregulated lipid peroxidation and liver transferase in the ethanol-induced HepG2 cells. Moreover, L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 protected against ethanol-induced HepG2 cell damage by regulating CYP2E1, antioxidant enzymes (SOD, CAT, and GPX), lipid synthesis factors (SREBP1C and FAS), and lipid oxidation factors (PPARα, ACO, and CPT-1). Moreover, five LAB were confirmed to be safe probiotics based on antibiotic susceptibility and hemolysis assays; their stability and adhesion ability in the gastrointestinal tract were also established. In conclusion, L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 may be useful as new probiotic candidates for ALFD prevention.
Keywords: probiotics, oxidative stress, alcohol, hepatoprotective, CYP2E1
1. Introduction
Alcoholic liver disease (ALD) is caused by chronic alcohol consumption and includes alcohol-induced liver cirrhosis, fibrosis, hepatitis, and liver cancer [1,2]. In particular, alcoholic fatty liver disease (AFLD), a common liver disease in many countries, is responsible for the death of at least three million people according to the World Health Organization (WHO) [3]. Three-month short-term mortality rate in patients with severe alcoholic steatohepatitis is very high, approaching 40–50% [4]. As only a few treatments are available for AFLD, the discovery of new useful treatments for AFLD is needed [2].
Alcohol is mainly metabolized via oxidation, catalyzed by alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) [5]. In this metabolism, cytochrome P450 2E1 (CYP2E1), which is activated in conjunction with ADH, induces oxidative stress, causing an imbalance between the production and elimination of reactive oxygen species (ROS) [6]. However, ROS levels are reduced by the expression of antioxidant enzymes, including superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPX) [7]. Alcohol intake also delays fatty acid oxidation by inhibiting peroxisome proliferator-activated receptor α (PPARα) and increases lipogenesis by activating sterol regulatory element-binding transcription factor 1C (SREBP1C), which may lead to fatty liver [1]. Therefore, a functional food that exhibits antioxidant activity and modulates lipid metabolism in hepatocytes could be a therapeutic agent for preventing AFLD.
Lactic acid bacteria (LAB), the most commonly used probiotics, are living microorganisms that provide health benefits by improving the balance in the host’s intestinal microbiota [8]. Recently, LAB, especially the Lactobacilaceae family, have proven to be therapeutic based on scientific research that revealed their range of health benefits, including diarrhea prevention, anti-allergy effects, and immune system modulation [9]. LAB can prevent AFLD by suppressing oxidative stress and improving the intestinal barrier function to reduce endotoxemia in the gut–liver axis [10]. For example, the amount of Bacteroides and Firmicutes is low in the gut microbiota of patients with ALD, leading to intestinal dysbiosis and pathogenic bacterial overgrowth. However, LAB can normalize the intestinal microflora [2]. In a previous study, Levilactobacillus brevis HY7410 and Limosilactobacillus fermentum MG590 lowered blood alcohol concentration by enhancing ADH and ALDH activity [11,12]. Moreover, L. brevis SBC8803, Limosilactobacillus reuteri DSM17938, and L. fermentum protected the liver of ethanol-fed mice [3,13,14]. Thus, LAB modulate the altered gut microbiota caused by alcohol and could thus be a promising treatment for the prevention of ALD. However, studies on the protective effects of LAB on AFLD are insufficient compared with those on non-AFLD.
Thus, we determined the ALDH activity and antioxidant and lipid metabolism of LAB, including L. brevis, L. reuteri, and L. fermentum isolated from humans and fermented food, in ethanol-induced HepG2 cells. Additionally, to confirm the properties of the probiotics, the safety and intestinal cell adhesion ability of those LAB having an ALFD inhibitory effect were determined.
2. Materials and Methods
2.1. Isolation of Bacterial Strains and Preparation of Cell-Free Extracts (CFEs)
All LAB strains used in this study were collected from MEDIOGEN (Jecheon, Korea). LAB used in this study were isolated from humans (MG4229, MG4296, MG4224, MG4231, MG4237, MG4244, MG4294, and MG4295) and fermented foods (MG5250, MG5280, MG5306, MG5311, MG5025, MG5149, and MG5458). In addition, L. fermentum MG590, a probiotic that alleviates AFLD, was used as a positive control [12]. All strains were cultured in MRS broth (de Man, Rogosa and Sharpe; Difco, Detroit, MI, USA) for 18 h at 37 °C under anaerobic chamber (Hanbaek Scientific Co., Gyeonggi-do, Korea).
CFEs were prepared according to the method of Park et al. [15]. For CFEs, each strain was collected by centrifugation (4000× g, 20 min at 4 °C). The collected pellet was lyophilized and resuspended in phosphate-buffered saline (PBS) at 10 mg/mL. The suspension was homogenized for 50 s using a sonicator (KFS-150N; Korea Process Technology Ltd., Seoul, Korea) and allowed to rest on ice for 1 min (repeated three times); the suspension was then centrifuged (4000× g) for 10 min at 4 °C. The supernatants were filter-sterilized using a 0.22 µm polytetrafluoroethylene membrane filter (ADVANTEC, Tokyo, Japan) and kept at −80 °C until use.
2.2. ALDH Activity
ALDH activity was determined as previously described [16]. In 10 µL of CFEs, 700 µL of distilled water, 375 µL of 1 M Tris–HCl buffer (pH 8.8, Sigma–Aldrich, St. Louis, MO, USA), and 150 µL of 25 mM NAD+ (Sigma–Aldrich) were reacted. After 10 min, ALDH (5 U/mL, Sigma–Aldrich) was added to the mixed samples. The optical density in kinetic mode (interval 10 min for 90 min) was determined at 340 nm using a microplate reader (EPOCH2, Biotek, Winooski, VT, USA). The protein content of the CFEs was determined using the Bradford assay (Bio-Rad, Hercules, CA, USA). The ALDH activity was calculated as the molar extinction coefficient of NADH (6.22 mM−1 cm−1) and expressed as units/mg protein/min.
2.3. Cell Culture
HepG2 cells (88065, KCLB, Seoul, Korea) were cultured in minimum essential media (MEM; Gibco, MT, USA) with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (PS; Gibco). HT-29 cells (30038, KCLB) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 10% FBS and 1% PS at 37 °C in a 5% CO2 incubator. The cells were subcultured at 70%–80% confluence.
2.4. Cell Viability
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [17]. HepG2 cells were seeded in 96-well plates at 4 × 105 cells/mL. After overnight growth, the cells were treated with CFEs for 1 h and then with or without ethanol for the next 24 h. The MTT solution (0.2 mg/mL) was added, and the cells were further cultured for 2–4 h. After incubation, the formazan crystals in each well were dissolved in DMSO. The absorbance at 550 nm was measured using a microplate reader.
2.5. Determination of Lipid Peroxidation and Glutathione (GSH) Content
Measurement of lipid peroxidation and GSH content was performed according to the manufacturer’s instructions (Cayman Chemical, Ann Arbor, MI, USA) and normalized to protein content using the Bradford assay.
For evaluation of lipid peroxidation, 5 × 106 cells in 100 mm plates were incubated in the presence or absence of CFEs for 1 h. Thereafter, the cells were stimulated with ethanol (3%) for 24 h. After incubation, lipid peroxidation was measured using a microplate reader at 540 nm and calculated using the malondialdehyde (MDA) calibration curve.
To measure GSH content, the cells (4 × 105 cells/mL) in a 6-well plate were incubated with or without CFEs for 1 h and then stimulated with ethanol (3%) for 24 h. After incubation, total glutathione was measured using a microplate reader at 405 nm.
2.6. Measurement of Liver Injury
Liver injury was measured by alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels using commercially available assay kits (Cayman Chemical) in accordance with the manufacturer’s instructions and normalized to protein content using the Bradford assay. Briefly, cells (5 × 105 cells/mL) in a 6-well plate were incubated in the presence or absence of CFEs for 1 h. Thereafter, the cells were stimulated with 3% ethanol for 24 h. After incubation, the ALT and AST levels of the cell lysates were measured using a microplate reader at 340 nm.
2.7. mRNA Extraction and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
The mRNA from HepG2 cells was isolated using 0.5 mL of NuceloZOL (MACHEREY–NAGEL GmbH & Co. KG, Dueren, Germany), according to the manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA using reverse transcriptase premix (Intron, Seongnam-si, Korea). qRT-PCR was performed using the CFX Connect Real-Time PCR Detection System (Bio-Rad), and target gene expression was assessed using the iQ™ SYBR® Green Supermix (Bio-Rad) by the standard metho (95 °C for 3 min, followed by 39 cycles at 95 °C for 10 s, 55–60 °C for 30 s, 72 °C for 30 s). The forward and reverse primers used are listed in Table S1. The relative expression of the target gene was normalized to that of GAPDH and analyzed by the 2−ΔΔCT method.
2.8. Probiotic Properties
2.8.1. Antibiotic Susceptibility and Hemolysis Assay
Antibiotic susceptibility was measured using antibiotic strips according to the manufacturer’s instructions (bioMérieux, Marcy-l’Étoile, France). Antibiotic resistance was confirmed according to the European Food Safety Authority (EFSA) guidelines [18].
The hemolysis assay was performed using tryptic soy agar (BD Bioscience, NJ, USA) plates containing 5% (w/v) sheep blood (MBCell, Seoul, Korea) [19]. The zone was observed as a green colony (α-hemolysis), a clean zone (β-hemolysis), and no color change (γ-hemolysis).
2.8.2. Gastrointestinal Tract (GIT) Stability and Adhesion
The survival rate in simulated GIT was evaluated according to the Maragkoudakis’s method with slight modifications [20]. Briefly, the LAB were cultured for 18 h and washed twice with PBS (pH 7.4) after centrifugation (4000× g for 5 min at 4 °C). The collected LAB were resuspended to 108 CFU/mL in simulated gastric fluid containing 3 g/L pepsin (adjusted to pH 3 and 4 with 1 N HCl) for 2 h and simulated intestinal fluid containing 1 g/L pancreatin adjusted to pH 7 and 8 with 1 N NaOH for 4 h, incubated at 37 °C. LAB were measured by counting viable cells using MRS agar plates.
The adhesion ability of LAB was evaluated using HT-29 colorectal cells, as described previously [21]. HT-29 colorectal cells (1 × 105 cells/mL) were incubated in 12-well plates in 5% CO2 at 37 °C for 24 h. The LAB were cultured in MRS broth at 37 °C for 24 h. The strains were resuspended at 1 × 108 CFU/mL in DMEM without FBS and PS and administered to cells. After 2 h, the cells were washed twice and then detached with PBS. The number of viable LAB was measured by plate counting on MRS agar and calculated by log CFU/mL.
2.9. Statistical Analysis
All experimental results are presented as the mean ± standard deviation (SD, n = 3). The statistical significance of differences was calculated using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test at p < 0.05 (SPSS, version 21; IBM Inc., Armonk, NY, USA).
3. Results
3.1. ALDH Activity of the LAB Strains
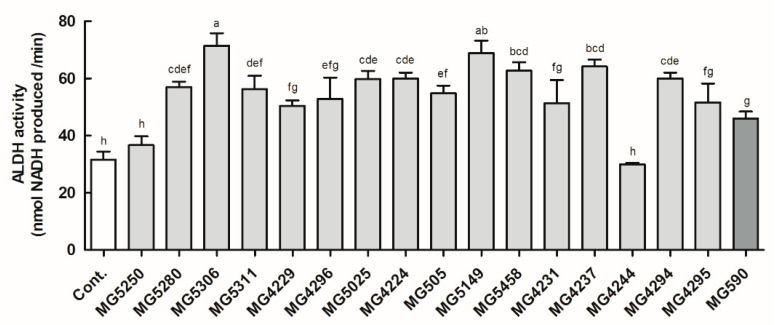
ALDH activity of all LAB strains, except MG5250 and MG4244, was increased compared with that of the control (Figure 1). In addition, nine LAB strains—MG5280 (1.83-fold of the control), MG5306 (2.29-fold of the control), MG5311 (1.80-fold of the control), MG4224 0 (1.60-fold of the control), MG505 (1.73-fold of the control), MG5149 (2.21-fold of the control), MG5458 (2.01-fold of the control), MG4237 (2.06-fold of the control), and MG4294 (1.92-fold of the control)—showed higher activity than MG590 (1.48-fold of the control), which was used as a positive control. Therefore, nine LAB strains with higher ALDH activity than the positive control were tested in HepG2 cells.
Figure 1.
Effect of CFEs (5 mg/mL) from LAB strains on ALDH activity. Data are expressed as the mean ± SD (n = 3). Different letters above the columns indicate significance at p < 0.05 based on Duncan’s test. Cont, control.
3.2. Protective Effect of LAB Strains on Ethanol-Induced HepG2 Cells
Prior to the assessment, the cytotoxic and protective effects of CFEs from LAB were confirmed in HepG2 cells with or without ethanol (Table 1). All LAB strains showed no cytotoxicity at 100 μg/mL (91.14 to 100.89%) in the HepG2 cells. After treatment with various concentrations of ethanol to induce cell injury, a significant cell death of less than 60% was confirmed when the HepG2 cells were stimulated with >3% ethanol (Figure S1). Thus, in subsequent experiments, HepG2 cell injury was induced by treatment with 3% ethanol. Viability of the ethanol-induced HepG2 cells was decreased by approximately 55% when compared with that of the non-ethanol-treated control. Nonetheless, all LAB strains were found to increase the viability of HepG2 cells treated with ethanol (73.27 to 91.47%).
Table 1.
Effect of CFEs from LAB strains on the viability of HepG2 cells exposed with or without ethanol.
| Lactic Acid Bacteria (μg/mL) | Cell Viability (%) | |||
|---|---|---|---|---|
| Control | + 3% Ethanol | |||
| Untreated | 100.00 ± 5.68 | 55.40 ± 2.07 | ||
| Levilactobacillus brevis | MG5280 | 50 | 99.13 ± 7.37 | 79.99 ± 5.68 *** |
| 100 | 97.26 ± 5.94 | 73.27 ± 4.13 *** | ||
| MG5306 | 50 | 96.91 ± 4.38 | 76.36 ± 3.47 *** | |
| 100 | 94.43 ± 2.04 | 75.04 ± 4.24 *** | ||
| MG5311 | 50 | 98.17 ± 5.43 | 83.47 ± 1.53 *** | |
| 100 | 99.63 ± 3.52 | 86.66 ± 2.17 *** | ||
| Limosilactobacillus reuteri | MG4224 | 50 | 96.26 ± 5.03 | 85.02 ± 1.84 *** |
| 100 | 99.49 ± 3.76 | 87.85 ± 0.89 *** | ||
| MG505 | 50 | 97.44 ± 5.55 | 85.02 ± 0.70 *** | |
| 100 | 96.58 ± 0.53 | 87.85 ± 1.91 *** | ||
| MG5149 | 50 | 92.51 ± 6.83 | 83.47 ± 8.61 *** | |
| 100 | 94.72 ± 11.25 | 86.66 ± 7.77 *** | ||
| MG5458 | 50 | 91.26 ± 1.18 | 82.60 ± 1.31 *** | |
| 100 | 91.14 ± 6.32 | 89.59 ± 0.96 *** | ||
| Limosilactobacillus fermentum | MG4237 | 50 | 95.37 ± 2.16 | 80.33 ± 2.04 *** |
| 100 | 91.71 ± 0.46 | 91.47 ± 1.23 *** | ||
| MG4294 | 50 | 93.89 ± 2.90 | 80.53 ± 1.92 *** | |
| 100 | 93.89 ± 3.59 | 80.53 ± 0.84 *** | ||
| MG590 | 50 | 100.89 ± 2.31 | 68.82 ± 1.46 ** | |
| 100 | 100.63 ± 2.29 | 75.00 ± 2.73 *** | ||
The results are expressed as mean ± SD (n = 3). Significance was based on Duncan’s test: ** p < 0.01, and *** p < 0.001 compared to the same column control.
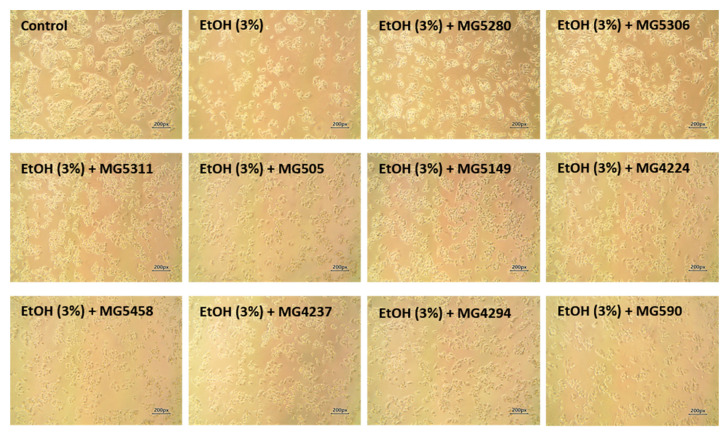
In terms of cell morphology, treatment of HepG2 cells with ethanol resulted in a change in epithelial cell shape and number (Figure 2). However, pretreatment with LAB strains averted cell damage caused by ethanol treatment by maintaining the original cell shape and number.
Figure 2.
Microscopic morphological images of ethanol-induced HepG2 cells treated with or without CFEs (100 μg/mL) from LAB strains. The images were captured at 10× magnification by phase-contrast light microscopy. EtOH, ethanol.
3.3. LAB Strains Regulate Oxidative Stress in Ethanol-Induced HepG2 Cells
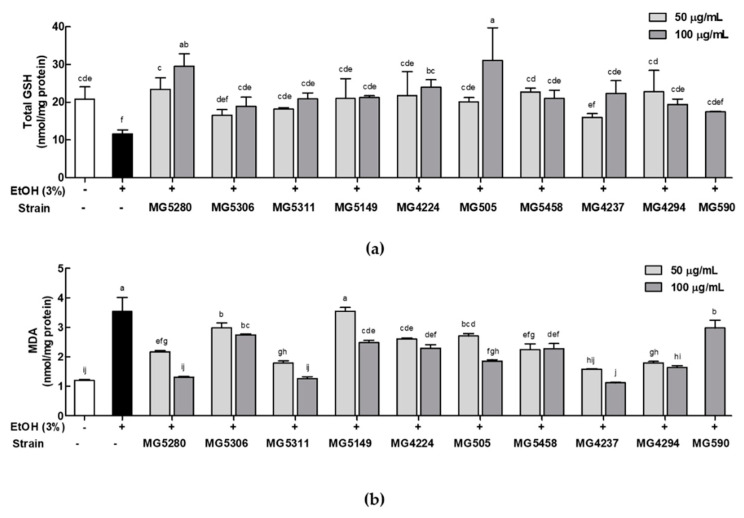
To confirm the protective effect of LAB against oxidative damage induced by ethanol treatment in HepG2 cells, total GSH and lipid peroxidation were measured. GSH content in cells induced by ethanol decreased by 0.56-fold when compared with that in cells induced by the control; however, when treated with LAB strains, GSH content was similar to or greater than that induced by the control (Figure 3a). In particular, MG5280 (29.50 ± 3.39 nmol/mg protein) and MG505 (31.04 ± 8.66 nmol/mg protein) showed significantly higher activity than the positive control MG590 (20.91 ± 0.45 nmol/mg protein).
Figure 3.
Effects of CFEs from LAB strains on (a) total GSH and (b) lipid peroxidation (determined by MDA levels) in ethanol-induced HepG2 cells. Data are expressed as mean ± SD (n = 3). Different letters at the column indicate significance at p < 0.05 based on Duncan’s test. EtOH, Ethanol.
Lipid peroxidation in HepG2 cells induced by ethanol was indicated by MDA levels (Figure 3b). The MDA levels of HepG2 cells treated only with ethanol was significantly increased by 2.94-fold compared with those in cells treated with the control; however, when pretreated with the nine LAB strains, the MDA levels of HepG2 cells were significantly decreased by approximately 0.38- to 0.99-fold compared with those of cells treated with the positive control L. fermentum MG590. Additionally, L. brevis MG5280 (1.31 ± 0.05 nmol/mg protein), L. brevis MG5311 (1.27 ± 0.12 nmol/mg protein), and L. fermentum MG4237 (1.12 ± 0.04 nmol/mg protein) markedly improved MDA levels as much as the control (1.20 ± 0.06 nmol/mg protein).
3.4. LAB Strains Protect against Liver Injury Induced by Ethanol in HepG2 Cells
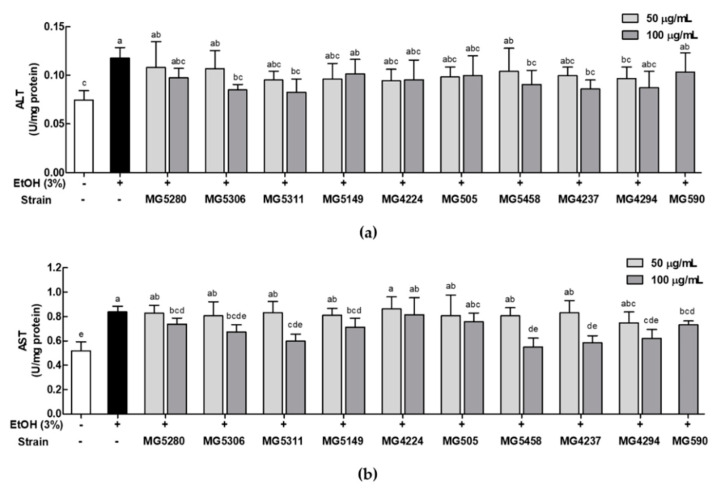
The protective effect of LAB strains against liver injury was assessed by the ALT and AST levels in ethanol-induced HepG2 cells. Ethanol significantly increased the levels of ALT and AST by 1.58- and 1.61-fold, respectively, compared with the control (Figure 4a,b). However, these increases were reduced upon treatment with the LAB strains by 0.66 to 0.99-fold in ALT and 0.70 to 0.92-fold in AST when compared with the ALT and AST levels in ethanol-induced HepG2 cells. In particular, L. brevis MG5311 (0.08 ± 0.01 and 0.60 ± 0.06 U/mg protein), L. reuteri MG5458 (0.09 ± 0.01 and 0.55 ± 0.07 U/mg protein), and L. fermentum MG4237 (0.09 ± 0.02 and 0.58 ± 0.06 U/mg protein) significantly suppressed the enzyme levels (ALT and AST, respectively) in HepG2 cells treated with ethanol relative to the control (0.07 ± 0.01 and 0.52 ± 0.07 U/mg protein).
Figure 4.
Inhibition of (a) ALT and (b) AST levels by CFEs from LAB strains in ethanol-induced HepG2 cells. Data are expressed as mean ± SD (n = 3). Different letters on the column indicate significance at p < 0.05 based on Duncan’s test. EtOH, ethanol.
Based on these results, L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 were selected as potential probiotics that can reduce damage to ethanol-induced HepG2 cells. Therefore, it was confirmed that these LAB had a protective effect against ALD. We then aimed to identify the underlying mRNA regulation related with ALFD.
3.5. LAB Strains Modulate Ethanol Metabolism by Enhancing mRNA Expression of Antioxidant Enzyme and Lipid Metabolism in Ethanol-Induced HepG2 Cells
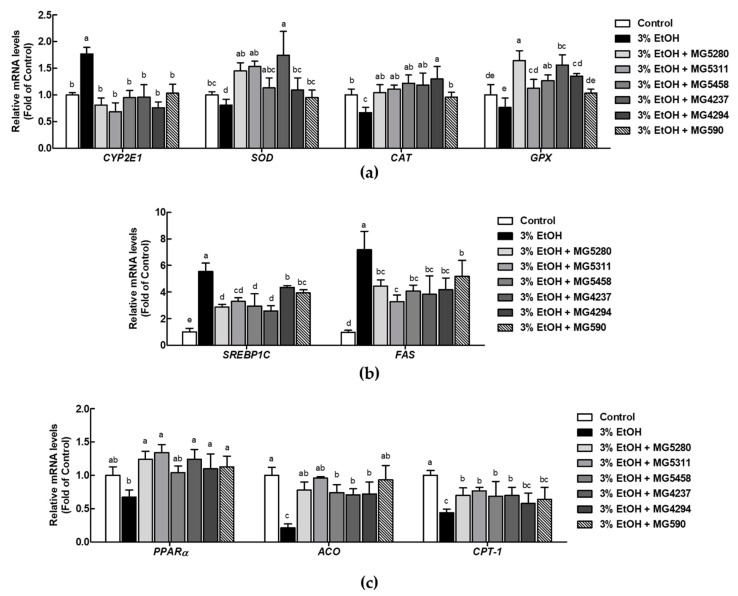
The effect of the LAB strains selected for the experiment on AFLD-related mRNA expression was investigated. Under oxidative stress in ethanol-induced HepG2 cells, expression level of CYP2E1 was significantly increased by 1.77-fold and that of SOD, CAT, and GPX was significantly reduced by 0.67-, 0.81-, and 0.76-fold, respectively, when compared with that in the control (Figure 5a). LAB treatment remarkably reversed the expression of these mRNA. In addition, to confirm the effect of LAB on ethanol-induced HepG2 cells, mRNA expression related to lipid metabolism was examined. Expression levels of SREBP1C and fatty acid synthase (FAS), which are lipogenesis-related factors, were increased by 5.54- and 7.18-fold after treatment of HepG2 cells with ethanol when compared with those after treatment with the control; however, LAB displayed a marked inhibition rate that ranged from 0.46 to 0.79-fold (Figure 5b). Expression levels of lipid oxidation factors, including PPAR, acyl–CoA oxidase (ACO), and carnitine palmitoyltransferase-1 (CPT-1), were significantly reduced in ethanol-induced HepG2 cells by 0.21-, 0.67-, and 0.44-fold compared with those in cells induced by the control. As expected, LAB exhibited significant upregulation of all tested lipid oxidation factors, with inhibition rates ranging from 1.34 to 4.50-fold (Figure 5c). In addition, LAB showed a better mRNA expression-modulating effect of all tested factors than L. fermentum MG590, the positive control.
Figure 5.
Effect of CFEs (100 μg/mL) from LAB strains on mRNA expression of (a) CYP2E1 and antioxidant enzymes (SOD, CAT and GPX), (b) lipid synthesis factors (SREBP1C and FAS), and (c) lipid oxidation factors (PPARα, ACO and CPT-1). Data are expressed as the mean ± SD (n = 3). Different letters on the column indicate significance at p < 0.05 based on Duncan’s test. EtOH, ethanol.
Taken together, all LAB have been shown to inactivate CYP2E1 to stimulate the antioxidant enzymes (SOD, CAT, and GPX) and modulate adipose metabolism-related factors (SREBP1C, FAS, PPAR, ACO, and CPT-1) in ethanol-induced HepG2 cells. L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 were also demonstrated to be effective at restoring ALFD through regulation of antioxidant enzyme expressions and lipid metabolism pathway.
3.6. Probiotic Properties of the LAB Strains
3.6.1. Safety of LAB as Probiotics
To confirm whether LAB can be used as a probiotic, safety tests using L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 were performed (Table 2). Antibiotic susceptibility was confirmed using the EFSA minimum inhibitory concentration (MIC) cutoff value [18]. All other antimicrobials, except tetracycline from L. brevis MG5311 and erythromycin from L. fermentum MG4237, were found to be below the standard indicated in Table S2. If the strain has hemolytic activity, the host’s red blood cells are destroyed [19]. By confirming the hemolytic activity, all strains were identified as γ-hemolytic—that is, no hemolytic activity was observed (Figure S2).
Table 2.
Results of antimicrobial test with the LAB strains.
| Antimicrobiotics 1 | L. brevis | L. reuteri | L. fermentum | ||
|---|---|---|---|---|---|
| MG5280 | MG5311 | MG5458 | MG4237 | MG4294 | |
| Ampicillin | S (0.75) |
S (0.5) |
S (0.75) |
S (0.094) |
S (0.094) |
| Gentamicin | S (0.094) |
S (0.047) |
S (2) |
S (0.19) |
S (0.19) |
| Kanamycin | S (3) |
S (3) |
S (6) |
S (4) |
S (4) |
| Streptomycin | S (4) |
S (6) |
S (24) |
S (3) |
S (6) |
| Tetracycline | S (6) |
R (>256) |
S (2) |
S (3) |
S (1.5) |
| Chloramphenicol | S (2) |
S (4) |
S (3) |
S (3) |
S (3) |
| Erythromycin | S (0.047) |
S (0.047) |
S (0.25) |
R (3) |
S (0.25) |
| Clindamycin | S (1.5) |
S (2) |
S (0.016) |
S (0.023) |
S (0.016) |
1 Susceptible (S) and resistant (R) strains according to the microbiology cutoff values from the EFSA guideline [18]. The minimum inhibitory concentrations are indicated in parentheses (µg/mL).
3.6.2. GIT Stability and Adhesion on HT-29 Colorectal Cells of L. brevis MG5311 and L. fermentum MG4237
The properties of L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 as probiotics were assessed in artificial GIT, including their adhesion ability to HT-29 colorectal cells (Table 3). In stimulated GIT, all strains had a survival rate of >98%. The cell number in the stimulated GIT ranged from 7.44 to 7.85 Log CFU/mL; the initial cell number ranged from 7.50 to 7.75 Log CFU/mL. Additionally, all strains could adhere to HT-29 colorectal cells, with an adhesion rate ranging from 55.36 to 84.77%. Among them, L. brevis MG5280 (84.21 ± 0.26%) and MG5311 (84.77 ± 0.45%) showed high adhesion rates.
Table 3.
Tolerance to artificial GI tract and adhesion of L. brevis MG5311 and L. fermentum MG4237 to HT-29 cells.
| Experiment | L. brevis | L. reuteri | L. fermentum | |||
|---|---|---|---|---|---|---|
| MG5280 | MG5311 | MG5458 | MG4237 | MG4294 | ||
| Stimulated gastrointestinal fluid (Log CFU/mL) |
Initial | 7.66 ± 0.02 | 7.50 ± 0.04 | 7.57 ± 0.04 | 7.75 ± 0.03 | 7.63 ± 0.01 |
| pH 3 | 7.65 ± 0.05 | 7.70 ± 0.02 | 7.58 ± 0.07 | 7.85 ± 0.03 | 7.71 ± 0.07 | |
| pH 4 | 7.64 ± 0.01 | 7.72 ± 0.06 | 7.58 ± 0.06 | 7.82 ± 0.08 | 7.72 ± 0.07 | |
| pH 7 | 7.65 ± 0.06 | 7.70 ± 0.04 | 7.51 ± 0.00 | 7.83 ± 0.01 | 7.56 ± 0.07 | |
| pH 8 | 7.63 ± 0.11 | 7.66 ± 0.10 | 7.44 ± 0.06 | 7.73 ± 0.01 | 7.55 ± 0.10 | |
| Adhesion ability | Initial | 8.52 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 | 7.23 ± 0.04 |
| (Log CFU/mL) | Adherent | 8.77 ± 0.02 | 6.96 ± 0.03 | 6.96 ± 0.03 | 6.96 ± 0.03 | 6.96 ± 0.03 |
| Cell adhesion (%) | 84.21 ± 0.26 | 84.77 ± 0.45 | 70.76 ± 0.865 | 79.29 ± 0.32 | 55.36 ± 1.00 | |
Data are expressed as the mean ± SD of duplicate experiments.
4. Discussion
Excessive alcohol consumption leads to liver damage [2]. ALD refers to a broad spectrum of alcohol-induced liver damage, including fatty liver, hepatitis, liver fibrosis, and cirrhosis [5]. Among these diseases, most people who frequently drink alcohol have fatty liver. Thus, the prevalence of ALFD is increasing [2]. The social costs, including the medical expenses of ALFD and its treatment, crimes, and accidents, are significant [22]. Abstinence from alcohol consumption may reverse mild ALD, but no effective drug has been found to treat ALFD [2]. Nonetheless, recent studies have reported that probiotics improve liver function [23]. In ethanol metabolism, ALDH plays an important role in converting highly toxic acetaldehyde decomposed by ADH into acetic acid, and oxidizing acetic acid to carbon dioxide and water, which are harmless to the human body [12]. Probiotics can contribute to ethanol metabolism by secreting their own ALDH to reduce aldehydes [12]. In this study, we demonstrated that LAB showed higher activity in ALDH, downregulated oxidative stress and lipogenesis genes, and upregulated lipid oxidation genes in ethanol-treated HepG2 cells. In addition, LAB, which have a protective function against ethanol-induced HepG2 cells, have been demonstrated to be valuable probiotics. Thus, this study was conducted to demonstrate the preventive efficacy of LAB, which can be used as a health functional food and a therapeutic alternative, against ALFD.
Oxidative stress is one of the factors that significantly contribute to the pathogenesis of ALD [24]. Oxidative stress, particularly ROS, is known to cause the oxidation of unsaturated fatty acids to produce lipid peroxides that induce fatty acid side chain reactions and MDA to damage cell structure, function, and DNA [2]. CYP2E1, which is expressed due to ROS generation, induces ALD and progresses to an advanced disease stage [25]. The increase in these factors is known to be reduced by GSH and antioxidant enzymes, such as SOD, CAT, and GPX, which convert O2− to H2O [26,27]. Therefore, the inhibition of CYP2E1 expression and an increase in antioxidant enzymes can effectively block the progression of ALD. By investigating LAB, L. brevis MG5280 and MG5311, L. reuteri MG5458, L. fermentum MG4237, and MG4294 were found to show excellent efficacy at relieving oxidative stress by elevating GSH and antioxidant enzymes. Our results were similar to those obtained with Probiotic V, a product that includes various LAB, which was found to reduce oxidative stress by inhibiting lipid peroxidation and the expression of CYP2E1 in HepG2 cells exposed to ethanol [28]. Moreover, the activity of enzymes, such as AST and ALT, has been reported as one of the most sensitive markers of hepatotoxicity [29]. In this study, the five LAB mentioned above were confirmed to alleviate ethanol-induced hepatotoxicity.
Alcohol consumption can affect lipid metabolism in the liver and cause hepatic steatosis [4]. The early growth response-1 (Erg-1) transcription factor involved in cellular stress is expressed by the aldehyde produced by CYP2E1, which stimulates SREPB-1C, a transcription factor that regulates hepatic cholesterol metabolism [30]. SREBP-1C induces lipid and cholesterol synthesis by promoting FAS [31]. Treatment with five LAB led to the inhibition of SREBP1C and FAS mRNA expression. Our results are consistent with a report by Farhin et al. who revealed that probiotics downregulated srebp1c and FAS in HepG2 cells exposed to ethanol [28]. Chronic alcohol consumption impedes lipid oxidation due to erroneous lysosomal biosynthesis, thereby delaying lipid degradation [30]. The transcription factor, PPARα, is increased by alcohol and affects the expression of subfactors ACO and CPT-1, which contribute to lipid oxidation in the mitochondria [32]. In our study, five LAB upregulated the mRNA expression of PPARα, ACO, and CPT-1 in ethanol-induced HepG2 cells. Hong et al. and Chu et al. reported that LAB treatment enhanced lipid oxidation by increasing the expression of PPARα, ACO, and CPT-1 in HepG2 cells [33,34]. Therefore, L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 could be a strategy to protect against ALFD by regulating lipid synthesis and oxidation in alcohol-induced HepG2 cells.
The term probiotics was derived as a comparative concept for the risk of antibiotics against LAB, which are microorganisms that play a beneficial role in the human body [35]. For LAB to be used as a probiotic, they must undergo safety and stability verification [36]. Thus, to use the five selected LAB as probiotics, safety and stability must be demonstrated. By conducting hemolytic and antibiotic resistance tests to confirm the safety of probiotics, we found that there were no hemolytic properties in any strain. However, L. brevis MG5311 was confirmed to be resistant to tetracycline; thus, further studies such as plasmid association are required. As probiotics play a role in maintaining health and regulating human GIT, their ability to grow at low and high pH by pepsin and bile should be confirmed [37]. In this study, all five LABs survived more than 98% in stimulated GIT. HT-29 colorectal cells are used extensively in adhesion studies because they best represent the morphological and physiological properties of human enterocytes [37]. Compared to other LAB, such as Lacticaseibacillus rhamnosus GG (10.33%), Lactiplantibacillus plantarum AdF10 (12.88%), and L. reuteri E (23.83%), the adhesion ability of all strains was significantly higher than 55.36% [38,39]. L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294 had better survival and adhesion than the above-mentioned probiotics, suggesting that they can be used as probiotics with improved efficacy. To more conclusively demonstrate the efficacy of reducing ALFD using a probiotic, further studies are needed to determine whether the same efficacy appears in vivo.
5. Conclusions
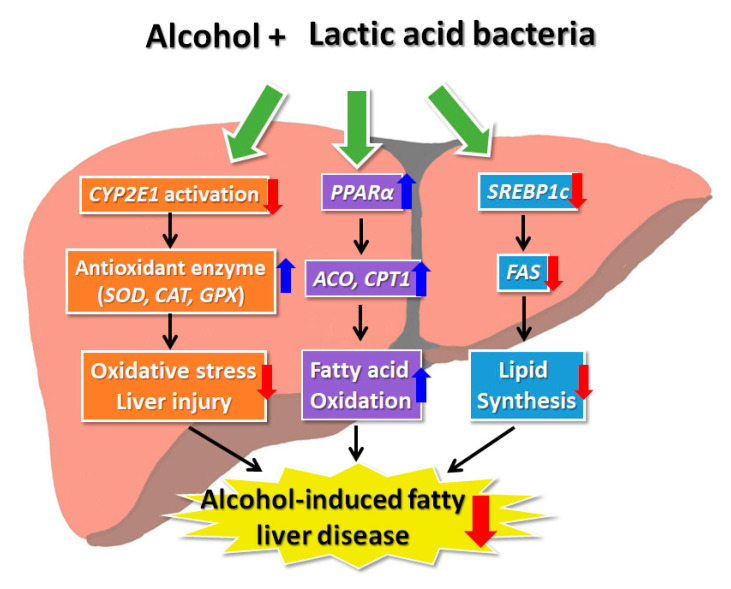
In conclusion, our findings suggest that LAB are effective at ameliorating damage in ethanol-induced HepG2 cells. In the current study, treatment with LAB reduced the expression level of CYP2E1 and increased the levels of antioxidant enzymes (SOD, CAT, and GPX) in ethanol-induced HepG2 cells to prevent cell injury. Further, LAB were found to possess a mechanism that contributes to AFLD prevention by relieving steatohepatitis through the regulation of abnormal lipid metabolism by ethanol (Figure 6). Therefore, LAB, including L. brevis MG5280 and MG5311, L. reuteri MG5458, and L. fermentum MG4237 and MG4294, as probiotics could serve as a functional food and a therapeutic agent for preventing ALFD.
Figure 6.
Lactic acid bacteria as probiotics exert a hepatoprotective effect by modulating antioxidant and lipid metabolism in ethanol-induced HepG2 cells.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9091844/s1, Table S1: Primers sequence used for qRT-PCR.; Table S2: The minimum inhibitory concentrations (MIC) cut-off value from EFSA. Figure S1: The viability of ethanol (EtOH) on HepG2 cells; Figure S2: The hemolytic activity of LAB strains.
Author Contributions
Conceptualization, C.-H.K.; methodology, J.Y.L., H.K. and Y.J.; investigation, J.Y.L. and Y.J.; resources, C.-H.K.; data curation, J.Y.L.; writing—original draft preparation, J.Y.L.; writing—review and editing, J.Y.L., H.K. and C.-H.K.; project administration, C.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available in a publicly accessible repository/data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang L., Yang C., Thomes P.G., Kharbanda K.K., Casey C.A., McNiven M.A., Donohue T.M., Jr. Lipophagy and alcohol-induced fatty liver. Front. Pharmacol. 2019;10:495. doi: 10.3389/fphar.2019.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu Z., Liu Y., Hu S., You Y., Wen J., Li W., Wang Y. Probiotics for alleviating alcoholic liver injury. Gastroenterol. Res. Pract. 2019;2019:9097276. doi: 10.1155/2019/9097276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng T.-X., Pu S.-L., Tan P., Du Y.-C., Qian B.-L., Chen H., Fu W.-G., Huang M.-Z. Liver metabolomics reveals the effect of Lactobacillus reuteri on alcoholic liver disease. Front. Physiol. 2020;11:1494. doi: 10.3389/fphys.2020.595382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim W. Diagnostic and therapeutic strategies for severe alcoholic hepatitis. Korean J. Gastroenterol. 2015;65:4–11. doi: 10.4166/kjg.2015.65.1.4. [DOI] [PubMed] [Google Scholar]
- 5.Orywal K., Szmitkowski M. Alcohol dehydrogenase and aldehyde dehydrogenase in malignant neoplasms. Clin. Exp. Pediatr. 2017;17:131–139. doi: 10.1007/s10238-016-0408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S.E., Koh H., Joo D.J., Nedumaran B., Jeon H.J., Park C.S., Harris R.A., Kim Y.D. Induction of SIRT1 by melatonin improves alcohol-mediated oxidative liver injury by disrupting the CRBN-YY1-CYP2E1 signaling pathway. J. Pineal Res. 2020;68:e12638. doi: 10.1111/jpi.12638. [DOI] [PubMed] [Google Scholar]
- 7.Halliwell B., Gutteridge J.C., Cross C.E. Free radicals, antioxidants, and human disease: Where are we now? J. Lab. Clin. Med. 1992;119:598–620. [PubMed] [Google Scholar]
- 8.Ducatelle R., Eeckhaut V., Haesebrouck F., Van Immerseel F. A review on prebiotics and probiotics for the control of dysbiosis: Present status and future perspectives. Animal. 2015;9:43–48. doi: 10.1017/S1751731114002584. [DOI] [PubMed] [Google Scholar]
- 9.Soccol C.R., Vandenberghe L.P.d.S., Spier M.R., Medeiros A.B.P., Yamaguishi C.T., Lindner J.D.D., Pandey A., Thomaz-Soccol V. The potential of probiotics: A review. Food Sci. Biotechnol. 2010;48:413–434. [Google Scholar]
- 10.Li F., Duan K., Wang C., McClain C., Feng W. Probiotics and alcoholic liver disease: Treatment and potential mechanisms. Gastroenterol. Res. Pract. 2016;2016:1–11. doi: 10.1155/2016/5491465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn Y.-T., Kim Y.-H., Bae J.-S., Lim K.-S., Huh C.-S., Yang W.-Y., Kim H.-S., Baek Y.-J. Effect of Lactobacillus brevis HY7401 intake on the serum ethanol concentration in rats. Korean J. Food Sci. Technol. 2004;36:604–608. [Google Scholar]
- 12.Kim J.-H., Kim H.-J., Son J.-H., Chun H.-N., Yang J.-O., Choi S.-J., Paek N.-S., Choi G.-H., Kim S.-K. Effect of Lactobacillus fermentum MG590 on alcohol metabolism and liver function in rats. J. Microbiol. Biotechnol. 2003;13:919–925. [Google Scholar]
- 13.Segawa S., Wakita Y., Hirata H., Watari J. Oral administration of heat-killed Lactobacillus brevis SBC8803 ameliorates alcoholic liver disease in ethanol-containing diet-fed C57BL/6N mice. Int. J. Food Microbiol. 2008;128:371–377. doi: 10.1016/j.ijfoodmicro.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Barone R., Rappa F., Macaluso F., Bavisotto C.C., Sangiorgi C., Di Paola G., Tomasello G., Di Felice V., Marcianò V., Farina F. Alcoholic liver disease: A mouse model reveals protection by Lactobacillus Fermentum. Clin. Transl. Gastroenterol. 2016;7:e138. doi: 10.1038/ctg.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J.-E., Oh S.-H., Cha Y.-S. Lactobacillus plantarum LG42 isolated from gajami sik-hae inhibits adipogenesis in 3T3-L1 adipocyte. Biomed Res. Int. 2013;2013:460927. doi: 10.1155/2013/460927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blandino A., Caro I., Cantero D. Comparative study of alcohol dehydrogenase activity in flor yeast extracts. Biotechnol. Lett. 1997;19:651–654. doi: 10.1023/A:1018386731116. [DOI] [Google Scholar]
- 17.Kumar P., Nagarajan A., Uchil P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018;2018:95505. doi: 10.1101/pdb.prot095505. [DOI] [PubMed] [Google Scholar]
- 18.EFSA Panel on Additives and Products. Substances Used in Animal Feed (FEEDAP) Rychen G., Aquilina G., Azimonti G., Bampidis V., Bastos M.d.L., Bories G., Chesson A., Cocconcelli P.S., et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16:e05206. doi: 10.2903/j.efsa.2018.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buxton R. Blood agar plates and hemolysis protocols. Am. Soc. Microbiol. 2005;30:1–9. [Google Scholar]
- 20.Jeong Y., Kim H., Lee J.Y., Won G., Choi S.-I., Kim G.-H., Kang C.-H. The antioxidant, anti-diabetic, and anti-adipogenesis potential and probiotic properties of lactic acid bacteria isolated from human and fermented foods. Fermentation. 2021;7:123. doi: 10.3390/fermentation7030123. [DOI] [Google Scholar]
- 21.Kim S., Choi S.-I., Jang M., Jeong Y., Kang C.-H., Kim G.-H. Anti-adipogenic effect of Lactobacillus fermentum MG4231 and MG4244 through AMPK pathway in 3T3-L1 preadipocytes. Food Sci. Biotechnol. 2020;29:1541–1551. doi: 10.1007/s10068-020-00819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang J.Y., Kim D.J. Epidemiology of alcoholic liver disease in Korea. Clin. Mol. Hepatol. 2018;24:93–99. doi: 10.3350/cmh.2017.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chávez-Tapia N.C., González-Rodríguez L., Jeong M., López-Ramírez Y., Barbero-Becerra V., Juárez-Hernández E., Romero-Flores J.L., Arrese M., Méndez-Sánchez N., Uribe M. Current evidence on the use of probiotics in liver diseases. J. Funct. Foods. 2015;17:137–151. doi: 10.1016/j.jff.2015.05.009. [DOI] [Google Scholar]
- 24.Cederbaum A.I., Lu Y., Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch. Toxicol. 2009;83:519–548. doi: 10.1007/s00204-009-0432-0. [DOI] [PubMed] [Google Scholar]
- 25.Nagappan A., Jung D.Y., Kim J.-H., Lee H., Jung M.H. Gomisin N alleviates ethanol-induced liver injury through ameliorating lipid metabolism and oxidative stress. Int. J. Mol. Sci. 2018;19:2601. doi: 10.3390/ijms19092601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apel K., Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y., Cederbaum A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel F., Parwani K., Patel D., Mandal P. Metformin and probiotics interplay in amelioration of ethanol-induced oxidative stress and inflammatory response in an in vitro and in vivo model of hepatic injury. Mediat. Inflamm. 2021;2021:6636152. doi: 10.1155/2021/6636152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutlu S., Colakoglu N., Halifeoglu I., Sandal S., Seyran A.D., Aydin M., Yılmaz B. Comparative evaluation of hepatotoxic and nephrotoxic effects of aroclors 1221 and 1254 in female rats. Cell Biochem. Funct. 2007;25:167–172. doi: 10.1002/cbf.1289. [DOI] [PubMed] [Google Scholar]
- 30.Osna N.A., Donohue T.M., Jr., Kharbanda K.K. Alcoholic liver disease: Pathogenesis and current management. Alcohol Res. Curr. Rev. 2017;38:147–161. doi: 10.35946/arcr.v38.2.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali A.T., Hochfeld W.E., Myburgh R., Pepper M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013;92:229–236. doi: 10.1016/j.ejcb.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Ghazali R. Ph.D. Thesis. University of Westminster; London, UK: 2017. Mechanisms into the development of fatty liver disease: Role of free fatty acids and alcohol. [Google Scholar]
- 33.Chu J., Joung H., Kim B.-K., Choi I.-S., Park T.-S. Inhibitory effects of Lactobacillus plantarum Q180 on lipid accumulation in HepG2 cells. Korean J. Food Nutr. 2019;32:738–744. [Google Scholar]
- 34.Hong S.-M., Chung E.-C., Kim C.-H. Anti-obesity effect of fermented whey beverage using lactic acid bacteria in diet-induced obese rats. Korean J. Food Sci. Anim. 2015;35:653–659. doi: 10.5851/kosfa.2015.35.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon H., Lee Y., Kang H.J., Ju J., Ji Y., Park H., Park H., Lee H., Holzapfel W.H. Two putative probiotic strains improve diet-induced hypercholesterolemia through modulating intestinal cholesterol uptake and hepatic cholesterol efflux. J. Appl. Microbiol. 2021;16:1–9. doi: 10.1111/jam.15181. [DOI] [PubMed] [Google Scholar]
- 36.Bang J.-H., Shin H.-J., Choi H.-J., Kim D.-W., Ahn C.-S., Jeong Y.-K., Joo W.-H. Probiotic potential of Lactobacillus isolates. J. Life Sci. 2012;22:251–258. doi: 10.5352/JLS.2012.22.2.251. [DOI] [Google Scholar]
- 37.Delgado S., O’sullivan E., Fitzgerald G., Mayo B. Subtractive screening for probiotic properties of Lactobacillus species from the human gastrointestinal tract in the search for new probiotics. J. Food Sci. Technol. 2007;72:M310–M315. doi: 10.1111/j.1750-3841.2007.00479.x. [DOI] [PubMed] [Google Scholar]
- 38.Sharma S., Kanwar S.S. Adherence potential of indigenous lactic acid bacterial isolates obtained from fermented foods of Western Himalayas to intestinal epithelial Caco-2 and HT-29 cell lines. J. Food Sci. Technol. 2017;54:3504–3511. doi: 10.1007/s13197-017-2807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudík B., Kiňová Sepová H., Bilka F., Pašková Ľ., Bilková A. Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie Leeuwenhoek. 2020;113:1191–1200. doi: 10.1007/s10482-020-01426-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a publicly accessible repository/data are contained within the article or Supplementary Materials.