Abstract
Background
The current coronavirus disease-19 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a global outbreak of a disease from a new coronavirus. Several databases have been published on this pandemic, but the research community still needs an easy way to get comprehensive information on COVID-19.
Objectives
COVID-19 pandemic database (CO-19 PDB) aims to provide wonderful insights for COVID-19 researchers with the well-gathered of all the COVID-19 data to one platform, which is a global challenge for the research community these days.
Methods
We gathered 59 updated databases since December-2019 until May 2021 and divided them into six categories: digital image database, genomic database, literature database, visualization tools database, chemical structure database, and social science database. These categories focus on taking number of functions from the images, information from gene sequences, updates from relevant papers, essays, reports, articles, and books, the data or information in the form of maps, graphs, and charts, information of bonds between atoms, and updates about events of the physical and social environment, respectively.
Results
Users can search the information of interest in two ways including typing the name of the database in the search bar or by clicking the right category directly. Computer languages such as CSS, PHP, HTML, Java, etc. are utilized to construct CO-19 PDB.
Conclusion
This article attempts to compile up-to-date appropriate COVID-19 datasets and resources that have not been compiled and given in such an accessible and user-friendly manner. As a result, the CO-19 PDB offers extensive open data sharing for both worldwide research communities and local people. Further, we have planned future development of new features, that will be awesome for future study.
Keywords: CO-19 PDB, COVID-19, Digital image, Genomic, Chemical structure
Introduction
Background
COVID-19 pandemic broke out in the end of the year 2019 [1], and spread to 211 countries quickly [2,3]. It is caused by a novel virus known as severe acute respiratory syndrome coronavirus 2. (SARS-CoV-2) [4], which is responsible for the violent (COVID-19) pandemic [5]. Researchers recently clearly stated that COVID-19 patients getting angiotensin-converting-enzyme inhibitors (ACEI) could experience better health [5,6]. The COVID-19 vaccine, considered to be an important prophylactic safety and prevention technique, is being manufactured in about 90 institutions around the world [2]. Previously we have published several databases on different research areas in well-cited international journals, such as, databases on phosphorylation animal and fungi (DBPAF), in Scientific Report journal [7], a database of circadian genes in eukaryotes (CGDB), in Nucleic Acids Research journal [8], database for protein phosphorylation sites in prokaryotes (dbPSP), in Oxford Database journal [9], Emergency Data Base of COVID-19 (EDBCO-19) [10], Database of plant Research (DBPR) [11] and so on. Taken together, we have provided a huge platform named HABDSK to scientific community in the form of eleven databases of a different research field that can be freely accessed [12] with timely updates.
Aim of the study
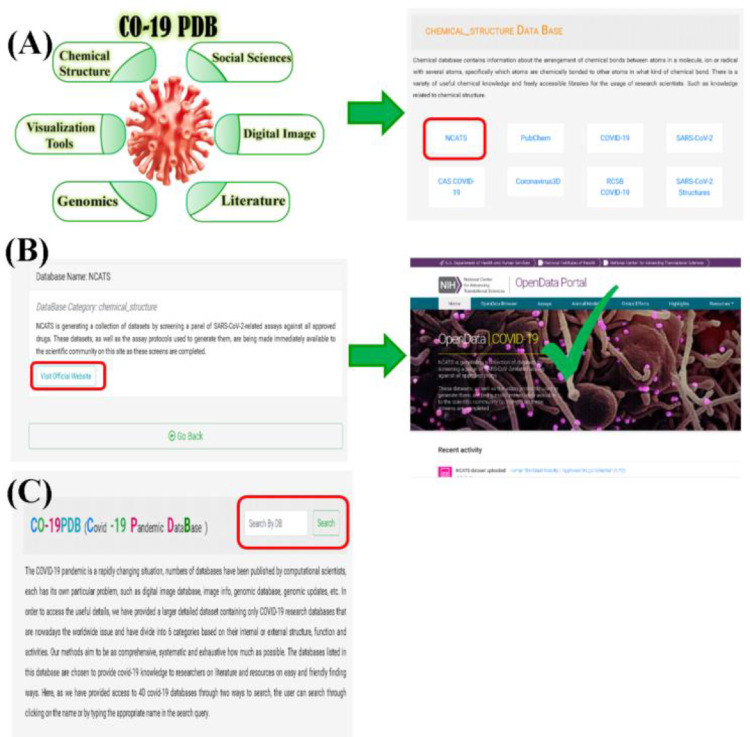
This article attempts to gather all the knowledge needed for the COVID-19 pandemic worldwide by analyzing papers and databases that have been published by a recent global scientists, Like the regular publishing of articles reported in the Centers for Disease Control and Prevention (CDC) database [13], statistics are reported by World Health Organization (WHO) [14], and several other databases have been online on the COVID-19 pandemic in various study areas and facets of COVID-19, each and every one has its own information and importance, however, in this crucial time, we have gathered all this data on a friendly and faster-finding platform for the global scientific community, and have grouped them into 6 categories so researchers can query for the knowledge they need in a short time. In short, we have provided a new way of searching in the form of a new feature (Fig. 2A), and will be useful for future research.
Fig. 2.
The usage and statistic of CO-19 PDB, (A) Searching by clicking the name of the category. (B) Can search the name of needed database. (C) The percentage of the collected data.
Materials and methods
Construction of database
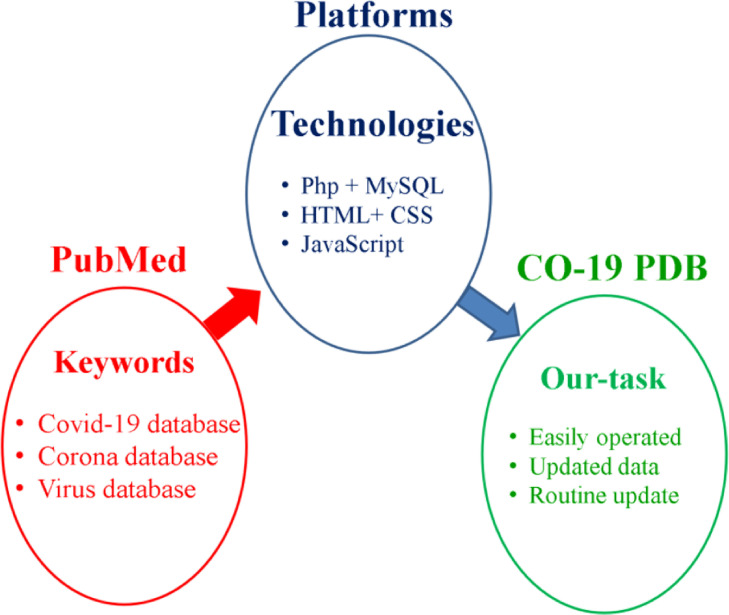
CO-19 PDB, data is collected and constructed on the friendly finding way (Fig. 1 ), many keywords have been used for searching the data such as; COVID-19 database, Corona database, Virus database, etc. in several searching engines like Google, Google Scholar and especially PubMed. Computer programming platforms have been used in the construction of the database. Finally, CO-19 PDB is available for researchers to be operated easily.
Fig. 1.
The data collection and construction procedure of the CO-19 PDB.
Use of the CO-19 PDB
Our database offers two ways to search. Users can search by clicking on the categories that lead to the proper table of the categories with small description which is shown in the (Fig. 2 A). Further clicking will lead to the official link with a small description of the needed query, shown in (Fig. 2B), furthermore clicking will give the new window of needed database which we have placed the tickmark sign as an example. In addition, insert the required database name in the search bar that is mentioned at the top of the main page and has been highlighted in the (Fig. 2C).
Results
Statistic of the database
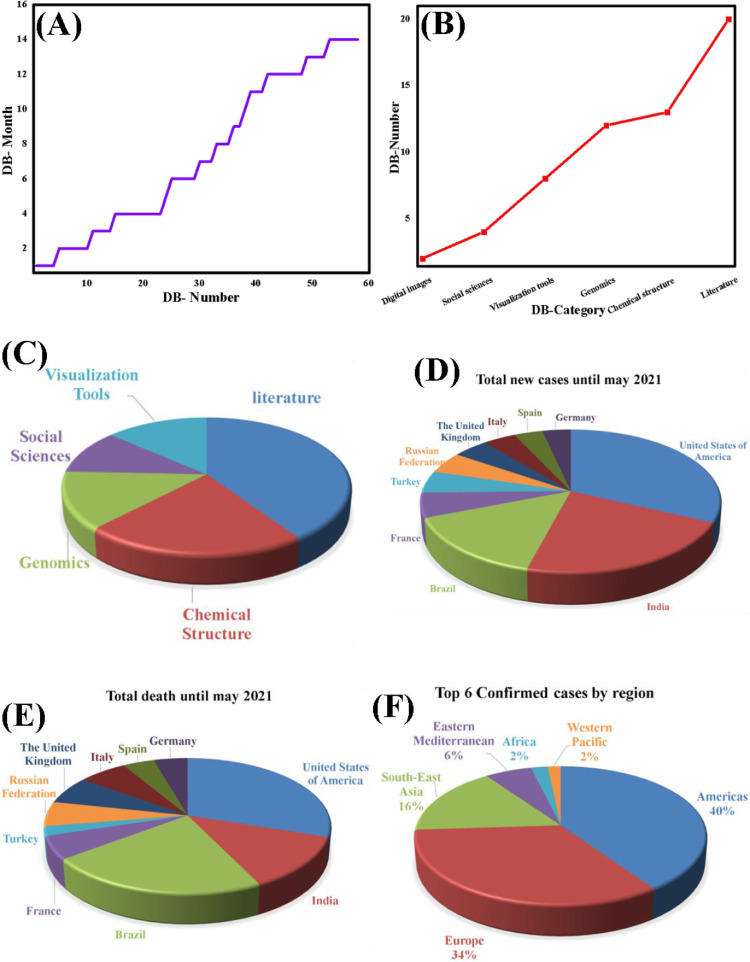
In this work, we have focused on COVID-19 databases, and have collected all the database from December 2019 till May 2021, (Fig. 3 A) is the growth of monthly wise databases, which show the tremendous growth with the passage of time, while (Fig. 3B) is the category wise growth of databases, in which, the literature category shown the highest value, due to vast amount of globally published literature in COVID-19 pandemic, in (Fig. 3C) the clear percentile of the data is shown, which can be of great help to further researchers, (Fig. 3D) depicts the overall number of new cases, while (Fig. 3E) depicts the overall number of deaths from December 2019 till 5 May 2021, in which the United States and India are at the top, and (Fig. 3F) depicts the top 6 confirmed cases by area, with America and Europe at the top. For more updated information and research we have provided this separate platform. It is to be noted, that all the redundant, disabled, and non-assessable database links have been updated or removed and provided new and updated COVID-19 databases which are in the form of database and table, (Table S1).
Fig. 3.
The statistic of CO-19 PDB, (A) Monthly wise growth, (B) Category-wise growth, (C) The percentage of the collected data, (D) Total new cases, (E) Total death, (F) Top 6 confirmed cases by region.
Previous published work and development of new features
Previously many articles have been published in well-known journals [15], [16], [17], [18] (Tab: 1), which have collected the databases of different organisms and different research area, e.g. Biological databases for human research [19] which have collected 74 human databases, “Online Databases for Taxonomy and Identification of Pathogenic Fungi and Proposal for a Cloud-Based Dynamic Data Network Platform [20]” have collected 24 fungi databases, so that a well comprehensive COVID-19 database is also needed for the research community to sort and save all the COVID-19 data for future researchers [21], because nowadays database of COVID-19 has been an integral part of modern biology. Further, published work has collected the databases and has presented them in the form of a Table, while in our work we have provided the table as well as the database of the databases, named COVID-19 PD, the Comparison is given in (Table 1 ). Furthermore, to make easier and clearer, we have categorized the databases on the basis of their properties such as, digital image database, genomic database, literature database, visualization tools database, chemical structure database, and social science database, and have planned future development of new features, which are shown in (Fig. 2A). that have not been provided before.
Table 1.
Comparison of CO-19 PDB with other published work.
| Authors | Year | Category | Form of | DB. No | Journal name | Ref. |
|---|---|---|---|---|---|---|
| Our work | 2021 | COVID-19 | DB+Table | 59 | ……. | …. |
| Rigden and Fernández 2021 | 2021 | Covid+other | Table | 89 | Nucleic Acids Res. | [22] |
| Rigden and Fernández 2020 | 2020 | Nucleic acid | Table | 70 | Nucleic Acids Res. | [18] |
| Xu 2012 | 2012 | Protein | Table | 121 | Curr. Protoc. Mol. Biol. | [15] |
| Zou, Ma et al. 2015 | 2015 | Human | Table | 74 | Genomics, Proteomics Bioinf. | [19] |
| Harper 1994 | 1994 | DNA+Protein | Table | 50 | Curr. Opin. Biotechnol. | [17] |
Discussion
CO-19 PDB classification
The classification of various viruses is based on the selection and contrast displayed by the viruses, them to distinguish [23] based on sequence similarity [24], the molecular structure of the genome [25], pathogenicity [26], structural similarities [27,28] and host range [29]. Huge research has been done on different viruses and has been saved in the form of published literature or databases, for an easy access and using this huge data, several databases have been released previously such as NIH, COVID-19 Data Portal and EDBCO-19. Currently on COVID-19 Pandemic, number of databases have been published, but a simplified and easier-to-use research is still needed for the conformant of the scientific community, so we have gathered revised datasets and grouped them into several categories based on their external and internal structure and function as described below, that can be access through this link https://www.habdsk.org/co-19pdb.php
Chemical structure database
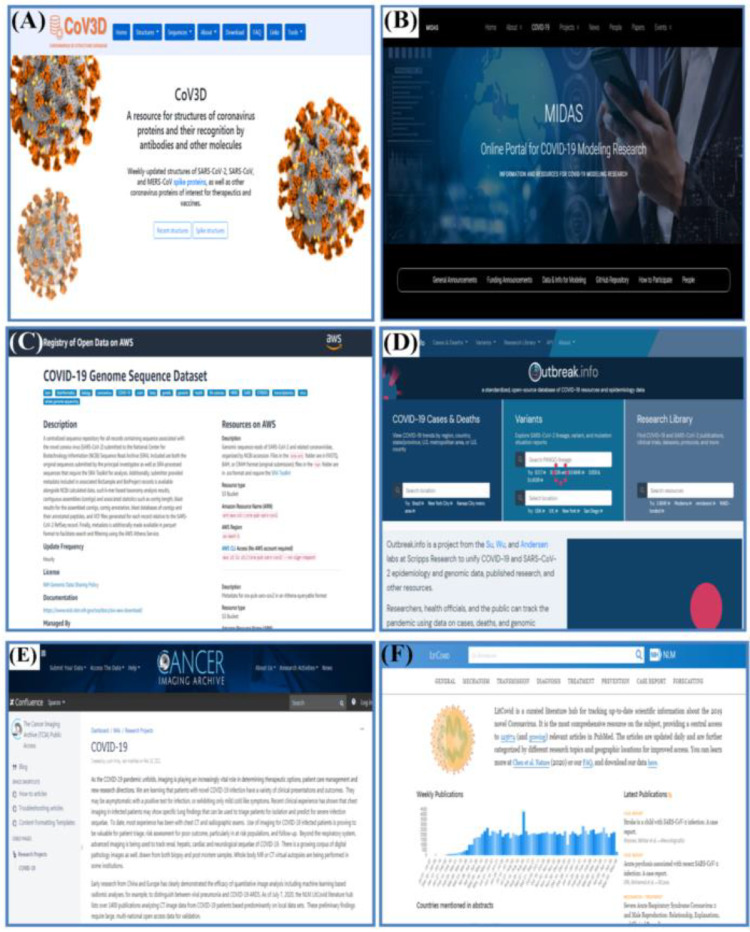
A chemical database contains information about the arrangement of chemical bonds between atoms in a molecule, ion, or radical with several atoms, specifically which atoms are chemically bonded to other atoms in what kind of chemical bond [30].There is a variety of useful chemical knowledge and freely accessible libraries for the usage of research scientists. Such as knowledge related to the chemical structure [31] (Fig. 4 A) shown COV3D is a weekly modified database that provides a detailed annotated collection of coronavirus protein structures and their identification by antibodies and other molecules.
Fig. 4.
The main pages of some common using databases in COVID-19 pandemic.
Visualization tools database
Visualization tool database contains a graphical representation of any data or information. Visual elements such as maps, graphs, and charts are some of the few data visualization tools that provide the audience with an easy and accessible way of understanding the represented information. Using Visualization tools, the COVID-19 data and information can be read and generated easily and quickly [32].(Fig. 4B) shown MIDAS is a global network of scientists who developed and applied theoretical, methodological, and mathematical models to help explain the complexities of infectious diseases in terms of pathogenesis, dissemination, efficient management methods, and forecasting.
Genomic database
Genomics is an interdisciplinary biological field that focuses on the structure, work, development, and edition of the genome. It is a complete set of DNA of an organism including all its genes [33]. Mostly it includes gene sequencing and analysis by using high-volume DNA sequencing and bioinformatics to evaluate the role and composition of whole genomes [34]. (Fig. 4C) shown COVID-19 genomic sequence database is a consolidated sequence database for all records containing sequences associated with the novel corona virus (SARS-CoV-2) that have been sent to the Sequence Read Archive (SRA) at the National Center for Biotechnology Information.
Social science database
Social science is the study of the physical and modern sphere. It is basically an analysis of international relations or the scientific study of human culture. Simply, it deals with the humans-their development, behavior, relationship, and resources they use and many kinds of organizations such as family, school, workplace, etc. it has brought awareness among the people about our environments and the incidents that happened in the past [35]. (Fig. 4D) shown main page named “Outbreak.info” is a web initiative that seeks to collect COVID-19 and SARS-CoV-2 epidemiology and genomic evidence, as well as published research and other materials.
Digital image database
A digital picture is consisting of graphic components, also known as pixels, each with a limited, distinct quantity of numerical representation due to its strength or gray color from its two-dimensional functions [36]. A digital image has the potential to improve a number of functions, such as the understanding of information for low-contrast films, the electronic transfer of images to remote installations, and the storage space requirements for archiving once treatment has been completed [37]. (Fig. 4E) shown, the cancer image archive (TCIA) is a service that de-identifies and hosts a large archive of medical image of cancer, that plays an important role in Covid-19 pandemic.
Literature database
The scientific literature contains publications that report a novel, experimental and theoretical work in the natural and social sciences, and is often abbreviated as literature within a scientific field [38]. Literature Database is an on-line, searchable bibliographic database of selected papers, essays, report, articles, and books relevant to the study of arts and cultural policy [39]. (Fig. 4F) shown “LitCovid” is a curated literature hub for monitoring up-to-date research knowledge about the newly discovered Corona virus in 2019. It is the most extensive resource on the topic, providing unified access to 125674 related PubMed articles.
Conclusion
People around the world are widely infected with COVID-19. Global scientists are working on COVID-19 and nearly 90 well-known research institutions are on record. Computational work has been done, and many datasets have been released and updated on a regular basis. Therefore, we have created a database of databases that contains all updated COVID-19 data on an easy platform and is accessible to all global researchers to aid in their research and study. To prevent wastage of time and to make it easier to find, we have classified the updated data into six categories according to their physical and chemical properties and have two ways of accessing to it, users can search by clicking on the category or enter the name of the database they need in the search bar. In short, the goal of this article is to put together up-to-date applicable COVID-19 datasets and resources that have not been gathered and provided before in such an easy and friendly finding ways. As a result, CO-19 PDB provides wide-ranging open data sharing for both global research communities and local people.
Author's contribution
Dr. Shahid Ullah and Prof. Tianshun Gao supervised the project. Dr. Anees Ullah, Ms. Wajeeha Rahman, Mr. Farhan Ullah, Dr. Sher Bahadar Khan, Mr. Gulzar Ahmad, and Mr. Muhammad Ijaz collected and verified the data carefully, all authors reviewed the manuscript and agreed to submit.
Declaration of Competing Interest
To avoid future conflict, Co-19PDB database is uploaded on (http://www.habdsk.org/co-19pdb.php) so that we have provided some content in this article.
Acknowledgment
To avoid future conflict, CO-19 PDB database is uploaded on (http://www.habdsk.org/co-19pdb.php) so that we have provided some content in this article. This project is supported by National Natural Science Foundation of China [32100434] and Shenzhen's introduction of talents and research start-up [392020].
Biographies
Anees Ullah: Bachelor in medicine: Kyrgyz state medical University, Email: anesullah923@gmail.com
Wajeeha Rahman: done Master: now working as a researcher in S-Khan Lab Mardan KPK, Pakistan http://www.habdsk.org/students.php, Email: wajeeharehman7464@gmail.com
Farhan Ullah: done Master: now working as a researcher in S-Khan Lab Mardan KPK, Pakistan http://www.habdsk.org/students.php, Email: Farhanu686@gmail.com
Sher Bahadr khan: Assistant professor: Department of Animal Health, The University of Agriculture, Peshawar, Pakistan, Email: drsher@aup.edu.pk
Gulzar Ahmad: done Master: now working as a researcher in S-Khan Lab Mardan KPK, Pakistan http://www.habdsk.org/students.php, Email: gulzarbiotech@gmail.com
Muhammad Ijaz: done Master: now working as a researcher in S-Khan Lab Mardan KPK, Pakistan http://www.habdsk.org/students.php, Email: m.ijaz.biotech@gmail.com
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpbup.2021.100031.
Appendix. Supplementary materials
References
- 1.Li H., Liu S.M., Yu X.H., Tang S.L., Tang C.K. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105951. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L., Tian D., Liu W. Strategies for vaccine development of COVID-19. Sheng Wu Gong Cheng Xue Bao. 2020:593–604. doi: 10.13345/j.cjb.200094. [DOI] [PubMed] [Google Scholar]
- 3.M. Gbetuwa, U. Iqbal, RE: COVID19 impact on human behaviour in Sierra Leone, (2020).
- 4.Contini C., di Nuzzo M., Barp N., Bonazza A., de Giorgio R., Tognon M., Rubino S. The novel zoonotic COVID-19 pandemic: an expected global health concern. J. Infect. Dev. Countries. 2020;14:254–264. doi: 10.3855/jidc.12671. [DOI] [PubMed] [Google Scholar]
- 5.Rico-Mesa J.S., White A., Anderson A.S. Outcomes in patients with COVID-19 infection taking ACEI/ARB. Curr. Cardiol. Rep. 2020;22:1–4. doi: 10.1007/s11886-020-01291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humayun A., Anwar M.I., Alam S.S., Iqbal U. Proceedings. 2020. Epidemic to Pandemic: Just a Matter of Time; pp. 1–3. [Google Scholar]
- 7.Ullah S., Lin S., Xu Y., Deng W., Ma L., Zhang Y., Liu Z., Xue Y. dbPAF: an integrative database of protein phosphorylation in animals and fungi. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Shui K., Zhang Y., Lv Y., Deng W., Ullah S., Zhang L., Xue Y. CGDB: a database of circadian genes in eukaryotes. Nucleic Acids Res. 2016:gkw1028. doi: 10.1093/nar/gkw1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Z., Wang B., Zhang Y., Wang Y., Ullah S., Jian R., Liu Z., Xue Y. dbPSP: a curated database for protein phosphorylation sites in prokaryotes. Database. 2015:2015. doi: 10.1093/database/bav031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullah S., Ullah F., RahmanW I.M., Ahmad G., Ullah W. EDBCO-19: Emergency Data Base of COVID-19. J. Clin. Med. Res. 2020;2:1–4. [Google Scholar]
- 11.S. Ullah, W. Rahman, F. Ullah, G. Ahmad, M. Ijaz, T. Gao, DBPR: DataBase of Plant Research, (2021).
- 12.KhanSA J.M., Anand V., Soomro K.A., Junaid M. Tremendous Contribution of Dr. Shahid Ullah to Scientific Community during COVID-19 Pandemic in the Form of Scientific Research. J. Clin. Med. Res. 2020;2:1–7. [Google Scholar]
- 13.S.B. Thacker, C.D.C. Library. COVID-19 Research Articles Downloadable Database., (2020). https://www.cdc.gov/library/researchguides/2019novelcoronavirus/researcharticles.html. (Accessed September 14, 2021).
- 14.World Health Organization, Global research on coronavirus disease (COVID-19)., (2021). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov. (Accessed September 14, 2021).
- 15.Xu D., Xu Y. Protein databases on the internet. Curr. Protocols Mol. Biol. 2004;68:19.4.1–19.4.15. doi: 10.1002/0471142727.mb1904s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bader G.D., Cary M.P., Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper R. Access to DNA and protein databases on the Internet. Curr. Opin. Biotechnol. 1994;5:4–18. doi: 10.1016/s0958-1669(05)80063-1. [DOI] [PubMed] [Google Scholar]
- 18.Rigden D.J., Fernández X.M. The 27th annual Nucleic Acids Research database issue and molecular biology database collection. Nucleic Acids Res. 2020;48:D1–D8. doi: 10.1093/nar/gkz1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou D., Ma L., Yu J., Zhang Z. Biological databases for human research. Genomics Proteomics Bioinformatics. 2015;13:55–63. doi: 10.1016/j.gpb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prakash P.Y., Irinyi L., Halliday C., Chen S., Robert V., Meyer W. Online databases for taxonomy and identification of pathogenic fungi and proposal for a cloud-based dynamic data network platform. J. Clin. Microbiol. 2017;55:1011–1024. doi: 10.1128/JCM.02084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M.-C. Lin, U. Iqbal, Y.-C. Li, AI in medicine: big data remains a challenge, (2018). [DOI] [PubMed]
- 22.Rigden D.J., Fernández X.M. The 2021 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic Acids Res. 2021;49:D1–D9. doi: 10.1093/nar/gkaa1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A.M.Q. King, E. Lefkowitz, M.J. Adams, E.B. Carstens, Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses, Elsevier, 2011.
- 24.Simmonds P. Methods for virus classification and the challenge of incorporating metagenomic sequence data. J. Gen. Virol. 2015;96:1193–1206. doi: 10.1099/jgv.0.000016. [DOI] [PubMed] [Google Scholar]
- 25.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.W.S.M. Wold, M.G. Ison, Adenovirus, in: Fields Virology 6th Edition, Lippincott, Williams & Wilkins, 2013: pp. 1732–1767.
- 27.Büchen-Osmond C. Further progress in ICTVdB, a universal virus database. Arch. Virol. 1997;142:1734–1739. [PubMed] [Google Scholar]
- 28.Buechen-Osmond C., Dallwitz M. Towards a universal virus database—progress in the ICTVdB. Arch. Virol. 1996;141:392–399. doi: 10.1007/BF01718409. [DOI] [PubMed] [Google Scholar]
- 29.Hare J.D., Short H.B. Manual of clinical microbiology. Am. J. Dis. Child. 1975;129:754. [Google Scholar]
- 30.Gross L., Mohn F., Moll N., Liljeroth P., Meyer G. The chemical structure of a molecule resolved by atomic force microscopy. Science. 2009;325:1110–1114. doi: 10.1126/science.1176210. [DOI] [PubMed] [Google Scholar]
- 31.Jick S.S., Kaye J.A., Vasilakis-Scaramozza C., Rodríguez L.A.G., Ruigómez A., Meier C.R., Schlienger R.G., Black C., Jick H. Validity of the general practice research database. Pharmacotherapy. 2003;23:686–689. doi: 10.1592/phco.23.5.686.32205. [DOI] [PubMed] [Google Scholar]
- 32.E. Graham, Introduction: Data visualisation and the Humanities, (2017).
- 33.O.M. de la S. Advisory, W.H. Organization, Genomics and world health: report of the Advisory Committee on Health Research, World Health Organization, 2002.
- 34.Ogbe R.J., Ochalefu D.O., Olaniru O.B. Bioinformatics advances in genomics-a review. Int. J. Curr. Res. Rev. 2016;8:5–11. [Google Scholar]
- 35.Giorgi A. The relationships among level, type, and structure and their importance for social science theorizing: a dialogue with Schütz. Duquesne Stud. Phenomenol. Psychol. 1979;3:81–92. [Google Scholar]
- 36.F. Patin, An introduction to digital image processing. online]: http://www.programmersheaven.com/articles/patin,ImageProc.Pdf. (2003).
- 37.Cumberlin R.L., Rodgers J.E., Fahey F.H. Digital image processing of radiation therapy portal films. Comput. Med. Imaging Graph. 1989;13:227–233. doi: 10.1016/0895-6111(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 38.Renear A.H., Sacchi S., Wickett K.M. Definitions of dataset in the scientific and technical literature. Proc. Am. Soc. Inf. Sci. Technol. 2010;47:1–4. [Google Scholar]
- 39.Lin B.K., Clyne M., Walsh M., Gomez O., Yu W., Gwinn M., Khoury M.J. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am. J. Epidemiol. 2006;164:1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.