Abstract
The fungal genus Myrothecium was once polyphyletic but a recent reconsideration of the family Stachybotryaceae spilt it into several genera. The ex-neotype specimen of the species Myrothecium verrucaria is now recognized as Albifimbria verrucaria. The well-studied plant pathogen and candidate bioherbicide CABI-IMI 368023, previously identified as M. verrucaria, was analyzed morphologically and genetically and found to be most consistently aligned with the other representatives of A. verrucaria.
Keywords: Myrothecium verrucaria, Albifimbria verrucaria, taxonomic classification, bioherbicide
1. Introduction
The genus Myrothecium was named by Tode in 1790 and has been revised and reconsidered numerous times since then. Tulloch [1] provided a thorough examination of the genus drawing from direct examination of materials from several collections and developed a taxonomic key. However, substantial difficulty was noted in differentiating among species within the genus and even between closely related genera, often relying on admittedly superficial characteristics. Over the years, the members of this genus have attracted attention due to their abundant production of lytic enzymes and the presence of highly unique mycotoxins [2,3,4,5]. While relatively few members of the genus are reported as pathogenic on plants and generally as ‘minor’ or ‘weak’ pathogens, a few isolates are known to be highly virulent on several weedy plants [6,7,8]. One isolate, collected from a diseased sicklepod (Senna obtusfolia) in DeSoto County (Mississippi, USA) morphologically identified as M. verrucaria and deposited as CABI-IMI 368,023 [9], has been thoroughly investigated as a bioherbicide for some difficult-to-control weeds in aquatic, agronomic and non-agronomic settings [10,11,12,13,14].
DNA sequencing offers a powerful tool in taxonomy and can confirm or dispel long-standing taxonomic understanding. Chen et al. [15] reviewed the existing morphologic descriptions of Myrothecium coupled with ITS and EF1-α sequence data, as well as several related genera for which sequence data were publicly available. They concluded that Myrothecium was polyphyletic and that the species concept was not well resolved. Working at the same time, Lombard et al. [16] sequenced six genes from nearly one hundred type specimens (ex-type or ex-epitype) and several hundred other isolates from public and private collections. These sequence data were used to revise the family Stachybotriaceae (Hypocreomycetidae, Sordariomycetes, Pezizomycotina and Ascomycota). In that examination of Stachybotriaceae, the genus name Albifimbria (a reference to the white fringe around the sporodochia) was introduced and representatives previously described as M. verrucaria received the epithet A. verrucaria as the typified representative. In addition, several other former members of Myrothecium were placed in Albifimbira, including A. lateralis, A. terrestris and A. viridis.
The recognition of CABI-IMI 368,023 as M. verrucaria pre-dated DNA-based taxonomic consideration. Therefore, in the context of the substantial revisions within Stachybotriaceae and the importance of this particular isolate, we compared six genes and photomicrographs of CABI-IMI 368,023 with published sequences and images of Albifimbria spp. and closely related taxa.
2. Materials and Methods
2.1. Phenotypic Characterization
CABI-IMI 368,023 was originally isolated from diseased sicklepod (Senna obtusfolia) in DeSoto County (Mississippi, USA) and has been maintained at the USDA-ARS in Stoneville, MS, USA. After seven days of growth on potato dextrose agar (PDA) at 28 degrees, photomicrographs were taken of CABI-IMI 368,023 with a Keyence VHX5000 light microscope from 20× to 2500× magnification with integrated digital measurements. Digital images were recorded, and morphology was compared to representative images from Lombard et al. [16].
2.2. Genetic Characterization
Genomic DNA was extracted from the fungal isolate CABI-IMI 368,023 with the Zymo Fungal Miniprep Kit (#D6005) from approximately 108 conidia. The genome was sequenced using one Spot-ON Flow Cell (R9) on the Oxford Nanopore Technologies (ONT) MinION platform following ligations using standard nanopore protocols (Ligation Kit SQK-KSK109). Sequencing was conducted over 72 h, and base calling was conducted with the Fast Basecalling implementation by using the innate MinKNOW GUI with a minimum Q-score = 7 for sequence inclusion. This resulted in 8.5 × 109 called bases, 1.3 × 106 sequences, average sequence length = 6154 bp and longest read = 75,816 bp with raw data of N50 = 8438. Since the true identity of this isolate was unknown, we extracted the Internal Transcribed Spacer (ITS) regions within silico PCR within the program Geneious (v.11.1.5) by using the primers detailed in Lombard et al. [16]. A BLASTn query of the ITS sequence against GenBank revealed >95% identity with the type specimen of Albifimbria verrucaria as well as with A. lateralis and A. virdis. Since the ITS sequence identity alone has limited power to differentiate Albifimbria, additional queries were made against 4 Albifimbira species and 20 other Stachybotriaceae using the cmdA, ITS, LSU, rpb2, tef1 and tub2 genes, as in Lombard et al. [16]. These gene sequences, from all 19 Albifimbria isolates (12 A. verrucaria, 3 A. terrestris, 3 A. viridis and 1 A. lateralis strain) used by Lombard et al. [16] as well the sister genera Dimorphiseta, Smaragdiniseta, Parvothecium, Inaequalispora, Virgatospora, Peethambara, Septomyrothecium and Paramyrothecium roridum NRRL 2183 (the only closely related species that is entirely sequenced), were concatenated and aligned using MUSCLE (v.3.8.425) as implemented in Geneious (v.11.1.5), and a Neighbor-Joining Tree was generated (Tamura-Nei distance model) by using Paramyrothecium roridum as an outgroup and resamples using bootstrapping with 1000 replicates.
3. Results and Discussion
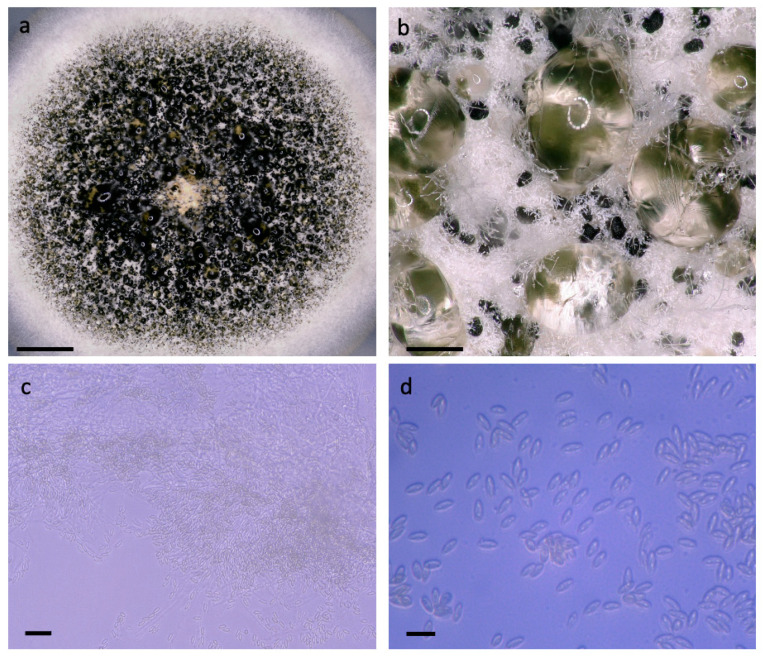
Figure 1 presents microscopic observations that support the placement of this isolate within A. verrucaria. Measurements of ten conidia yielded an average length of 7.0 µm ± 0.9 and width of 2.7 µm ± 0.5 µm. When grown on PDA, the mycelium is white with a buff reverse. Masses of spores range from very dark olive to nearly black and clustered in moist masses. These features are as described in figures and text by Tulloch [1], with the exception of the lack of fantailed appendages on the conidia.
Figure 1.
Photomicrographs of CABI-IMI 368023. Images (a–d) are captured on Keyence VHX5000 light microscope at 20, 200, 1000 and 2500× magnification, respectively. (a) Culture morphology with dark region of sporulation surrounded by white mycelial margin, bar = 5 mm. (b) Dark spore masses embedded in white arial mycelia and droplets of condensation, bar = 400 µm. (c) Closer examination of free conidia and dense hyphal mass, bar = 25 µm. (d) Elongated conidia, lacking fantail appendage, bar = 10 µm.
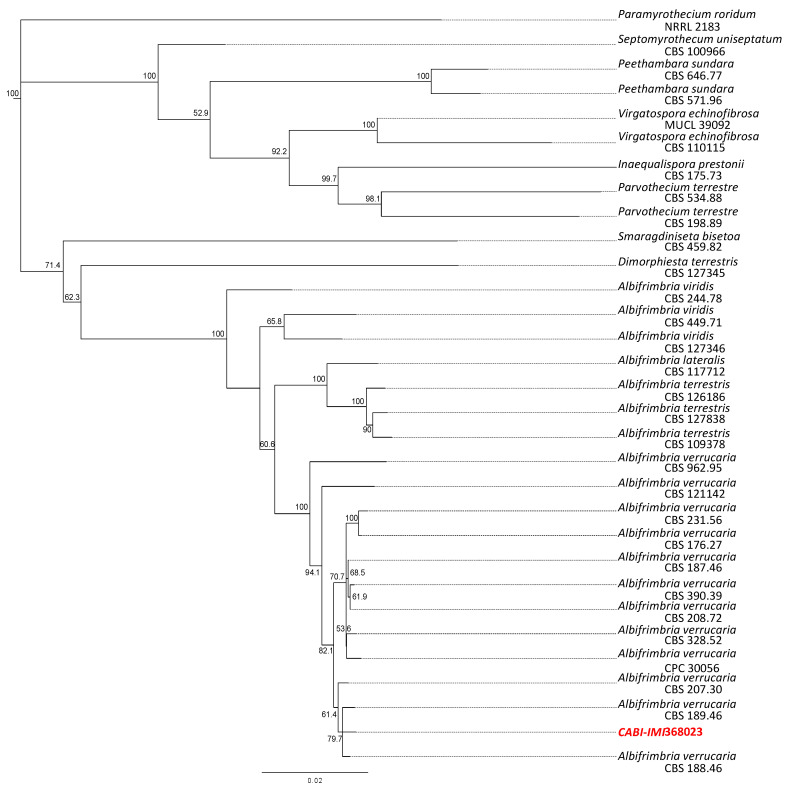
Generic sequences of the cmdA, ITS, LSU, rpb2, tef1 and tub2 genes from CABI-IMI 368,023 are deposited in GenBank (MZ673262–MZ673264). The type specimen of Albifimbria verrucaria was the closest match to CABI-IMI 368,023 for four of the six genes examined (cmdA, LSU, rbpb2 and tub2), with identity of 99.2% to 99.9% between the two isolates (Table 1). The other two genes (ITS and tef1) showed 99.8% and 96.2% identity, respectively, to A. verrucaria. The ITS and LSU genes had high homology relative to many isolates of other genera, indicating that they had poor taxonomic resolution. The cmdA and rbp2 genes appeared, in the context of these isolates, to best differentiate A. verrucaria. In order to further examine the placement of CABI-IMI 368,023 within this clade of Stachybotriaceae, we constructed a phylogenetic tree of likely close relatives, including the Paramyrothecium roridum as the outgroup (Figure 2). With the recent reconsiderations of the family, due to emerging genetic resources [15,16], this clade appears to be well-resolved with the monophyletic branches. CABI-IMI 368,023 is present in a division with only other A. verrucaria with very low genetic differences separating represented members of species.
Table 1.
Genetic similarity of Albifimbria verrucaria strain CABI-IMI 368,023 to other Albifimbria taxa and select Stachybotriaceae. Presented are percent identical sequences with the number of exact matches/total length presented parenthetically following BLASTn comparisons for the genes calmodulin (cmdA), internal transcribed spacer 5.8S (ITS), 28S RNA large subunit (LSU), RNA polymerase II second subunit (rpb2), translation elongation factor 1-alpha (tef1) and beta-tubulin (tub2). Accession numbers for the gene sequences used are presented in Lombard et al. (2016). Instances of >98% identity are highlighted in grey.
| Species and Isolate Number | cmdA | ITS | LSU | rpb2 | tef1 | tub2 |
|---|---|---|---|---|---|---|
| Achroiostachys humicola CBS 868.73 *,† | 73.4% (300/409) | 73.6% (521/708) | 97.0% (801/826) | 76.2% (550/722) | 90.1% (127/141) | 84.3% (305/362) |
| Albifimbria lateralisCBS 117,712 *,† | 91.3% (608/666) | 99.4% (666/670) | 99.3% (820/836) | 94.6% (682/721) | 89.4% (405/453) | 96.7% (352/364) |
| A. terrestris CBS 126,186 *,† | 91.3% (605/663) | 99.4% (666/650) | 99.2% (819/826) | 93.9% (675/719) | 89.2% (405/454) | 96.7% (551/363) |
| A. verrucaria NRRL 2003 *,†,‡ | 98.5% (649/659) | 99.8% (633/643) | 99.9% (825/826) | 99.3% (716/721) | 96.2% (430/447) | 99.2% (356/359) |
| A. virdis CBS 449.71 *,† | 93.3% (615/659) | 99.9% (669/670) | 99.8% (824/826) | 95.4% (683/716) | 88.8% (404/455) | 98.3% (353/359) |
| Brevistachys variabilis CBS 141,057 † | 79.9% (222/278) | 77.2% (319/413) | 93.5% (772/826) | 72.0% (321/446) | 92.6% (137/148) | 80.8% (298/369) |
| Capitofimbria compacta CBS 111,739 *,†,‡ | 77.7% (313/402) | 75.8% (503/664) | 98.6% (815/827) | 79.6% (577/725) | 97.3% (143/147) | 82.7% (296/358) |
| Dimorphiseta terrestris CBS 127,345 *,† | 75.0% (437/583) | 81.9% (550/672) | 98.6% (815/827) | 85.6% (612/715) | 94.4% (135/143) | 88.6% (327/369) |
| Grandibotrys pseudotheobromae CBS 136,170 *,† | -- | 74.7% (519/695) | 96.7% (771/797) | 72.9% (510/700) | 91.6% (131/143) | 81.7% (294/360) |
| Gregatothecium humicola CBS 205.96 *,† | 77.8% (312/401) | 80.6% (555/689) | 99.2% (820/827) | 82.7% (596/721) | 98.6% (139/141) | 84.1% (301/358) |
| Inaequalispora prestonii CBS 175.73 *,†,‡ | 73.4% (315/429) | 79.0% (527/667) | 97.8% (809/827) | 79.6% (575/722) | 91.6% (130/142) | 80.3% (236/294) |
| Kastanostachys aterrima CBS 101,310 *,† | -- | -- | 95.5% (790/827) | 72.6% (525/723) | -- | -- |
| Myrothecium inundatum CBS 275.48 * | 74.7% (331/443) | 80.7% (453/673) | 98.6% (815/827) | -- | 96.5% (136/141) | 84.1% (310/358) |
| Myrothecium simplex CBS 582.93 * | 75.5% (320/424) | 81.0% (545/673) | 98.6% (815/827) | -- | 97.2% (137/141) | 83.8% (300/358) |
| Neomyrothecium humicola CBS 310.96 *,† | 76.3% (318/417) | 82.7% (548/663) | 99.2% (820/827) | 83.4% (601/721) | 98.6% (139/141) | -- |
| Paramyrothecium cupuliforme CBS 127,789 *,†,‡ | 78.7% (329/418) | 81.0% (554/684) | 99.2% (820/827) | 81.2% (521/642) | 91.5% (129/141) | 85.0% (306/360) |
| P. roridum NRRL 2183 †,‡ | 78.5% (328/418) | 94.1% (642/682) | 98.9% (818/827) | 82.0% (591/721) | 89.3% (134/150) | 84.9% (304/358) |
| P. viridisporim CBS 873.85 * | 78.7% (329/418) | 81.1% (553/682) | 98.9% (818/827) | 82.4% (594/721) | 89.8% (132/147) | 83.8% (300/358) |
| Parvothecium terrestre CBS 198.89 *,† | 75.1% (320/426) | 79.9% (531/665) | 97.9%(810/827) | 80.6% (582/722) | 93.1% (134/144) | 82.4% (305/370) |
| Smaragdiniseta bisetosa CBS 459.82 *,†,‡ | 76.8% (447/582) | 82.7% (554/670) | 99.2% (819/826) | 83.8% (604/721) | 93.6% (132/141) | 87.1% (317/364) |
| Stachybotrys chartarum CBS 182.80 * | 74.6% (309/414) | 76.3% (509/667) | 96.3% (796/827) | 75.8% (547/722) | 92.9% (131/141) | 81.8% (296/362) |
| Striaticonidium cinctum CBS 932.69 *,†,‡ | 76.0% (319/420) | 74.9% (500/668) | 97.9% (809/826) | 78.9% (569/721) | -- | 82.3% (307/373) |
| Tangerinosporium thalitricola CBS 317.61 *,† | 73.3% (431/588) | 80.5% (542/673) | 97.9% (810/827) | -- | -- | 84.7% (305/360) |
| Xenomyrothecium tongaense CBS 598.80 *,†,‡ | 72.5% (427/589) | 79.3% (548/691) | 98.9% (818/827) | 82.1% (592/721) | 97.9% (140/143) | 86.6% (310/358) |
* Type specimen; † gen. nov. from Lombard et al. [16]; ‡ formerly placed into Myrothecium.
Figure 2.
Consensus six-gene concatenated neighbor-joining tree placing the bioherbicidal isolate IMI 368,023 (highlighted in red) within Albifimbria verrucaria.
4. Conclusions
The morphological and genetic observations here support the renaming of CABI-IMI 368,023 as Albifimbria verrucaria, consistent with the assignment of other isolates previously recognized as Myrotheicum verrcuaria.
Current Name:
Albifimbria verrucaria (Alb. and Schwein.) L. Lombard and Crous, in Lombard, Houbraken, Decock, Samson, Meijer, Réblová, Groenewald and Crous, Persoonia 36: 177 (2016).
Synonymy:
Peziza verrucaria Alb. and Schwein, Conso. Fungi. (Leipzig): 340 (1805) (also the Basionym).
Gliocladium fimbriatum J.C. Gilman and E.V. Abbott, Iowa St. Coll. J. Sci. 1: 304 (1927).
Metarhizium glutinosum S.A. Pope, Mycologia 36(4): 346 (1944).
Myrothecium verrucaria (Alb. and Schwein.) Ditmar, in Sturm, Deutschl. Fl., 3 Abt. (Pilze Deutschl.) 1(1): 7 (1813).
Acknowledgments
The authors would like to thank Nahreen Mirza for assistance in sequencing.
Author Contributions
Conceptualization, methodology and visualization, M.A.W. and S.P.B.; validation and formal analysis, S.P.B.; writing—original draft preparation, M.A.W.; funding acquisition, R.E.H. and S.P.B.; writing—review and editing M.A.W., R.E.H., C.D.B. and S.P.B. All authors have read and agreed to the published version of the manuscript.
Funding
Partial support for this work was provided the Department of Biological Sciences at University of Memphis.
Data Availability Statement
Gene sequences have been deposited with National Center for Biotechnology Information, GenBank. Accession numbers MZ673262–MZ673264.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tulloch M. The genus Myrothecium Tode ex Fr. Mycol. Pap. 1972;130:1–44. [Google Scholar]
- 2.Vyas P., Deshpande M.V. Chitinase Production by Myrothecium verrucaria and its Significance for Fungal Mycelia Degradation. J. Gen. Appl. Microbiol. 1989;35:343–350. doi: 10.2323/jgam.35.343. [DOI] [Google Scholar]
- 3.Sulistyaningdyah W.T., Ogawa J., Tanaka H., Maeda C., Shimizu S. Characterization of Alkaliphilic Laccase Activity in the Culture Supernatant of Myrothecium verrucaria 24G-4 in Comparison with Bilirubin Oxidase. FEMS Microbiol. Lett. 2004;230:209–214. doi: 10.1016/S0378-1097(03)00892-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhao D., Zhang X., Cui D., Zhao M. Characterisation of a Novel White Laccase from the Deuteromycete Fungus Myrothecium verrucaria NF-05 and its Decolourisation of Dyes. PLoS ONE. 2012;7:e38817. doi: 10.1371/journal.pone.0038817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proctor R.H., McCormick S.P., Kim H.-S., Cardoza R.E., Stanley A.M., Lindo L., Kelly A., Brown D.W., Lee T., Vaughn M.M., et al. Evolution of Structural Diversity of Trichothecenes, a Family of Toxins Produced by Plant Pathogenic and Entomopathogenic Fungi. PLoS Pathog. 2018;14:e1006946. doi: 10.1371/journal.ppat.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.B., Kim J.-C., Hong K.-S., Kim C.-J. Evaluation of a Fungal Strain, Myrothecium roridum F0252, as a Bioherbicide Agent. Plant Pathol, J. 2008;24:453–460. doi: 10.5423/PPJ.2008.24.4.453. [DOI] [Google Scholar]
- 7.Okunowo W.O., Osuntoki A.A., Adekunle A.A., Gbenle G.O. Occurrence and Effectiveness of an Indigenous Strain of Myrothecium roridum Tode: Fries as a Bioherbicide for Water Hyacinth (Eichhornia crassipes) in Nigeria Biocontr. Sci. Technol. 2013;12:1387–1401. [Google Scholar]
- 8.Clarke T.C., Shetty K.G., Jayachandran K., Norland M.R. Myrothecium verrucaria-A Potential Biological Control Agent for the Invasive ‘Old World Climbing Fern’ (Lygodium microphyllum) Bio. Control. 2007;52:399–411. doi: 10.1007/s10526-006-9035-3. [DOI] [Google Scholar]
- 9.Walker H.L., Tilley A.M. Evaluation of an Isolate of Myrothecium verrucaria from Sicklepod (Senna obtusfolia) as a Potential Mycoherbicide Agent. Biol. Control. 1997;10:104–112. doi: 10.1006/bcon.1997.0559. [DOI] [Google Scholar]
- 10.Boyette C.D., Reddy K.N., Hoagland R.E. Glyphosate and Bioherbicide Interaction for Controlling Kudzu (Pueraria lobata), Redvine (Brunnichia ovata), and Trumpetcreeper (Campsis radicans) Biocontr. Sci. Technol. 2006;16:1067–1077. doi: 10.1080/09583150600828742. [DOI] [Google Scholar]
- 11.Boyette C.D., Hoagland R.E., Stetina K.C. Biological Control of the Weed Hemp Sesbania (Sesbania exaltata) in Rice (Oryza sativa) by the Fungus Myrothecium verrucaria. Agronomy. 2014;4:74–89. doi: 10.3390/agronomy4010074. [DOI] [Google Scholar]
- 12.Hoagland R.E., McCallister T.S., Boyette C.D., Weaver M.A., Beecham R.V. Myrothecium verrucaria as a Bioherbicidal Agent Against Morning-glory (Ipomoea) Species. Allelopathy J. 2011;27:151–162. [Google Scholar]
- 13.Weaver M.A., Boyette C.D., Hoagland R.E. Rapid Kudzu Eradication and Switchgrass Establishment through Herbicide, Bioherbicide and Integrated Programs. Biocontr. Sci. Technol. 2016;26:640–650. doi: 10.1080/09583157.2016.1141175. [DOI] [Google Scholar]
- 14.Weaver M.A., Shearer J.F., Grodowitz M.J., Boyette C.D. Potential of Myrothecium Species as Bioherbicides for Giant Salvinia (Salvinia molesta) J. Aquat. Plant Manag. 2018;56:120–122. [Google Scholar]
- 15.Chen Y., Ran S.F., Dai D.Q., Wang Y., Hyde K.D., Wu Y.M., Jiang Y. Mycosphere Essays 2. Myrothecium. Mycosphere. 2016;7:64–80. doi: 10.5943/mycosphere/7/1/7. [DOI] [Google Scholar]
- 16.Lombard L., Houbraken J., Decock C., Samson R.A., Meijer M., Réblová M., Crous P.W. Generic Hyper-diversity in Stachybotriaceae. Persoonia. 2016;36:156–246. doi: 10.3767/003158516X691582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Gene sequences have been deposited with National Center for Biotechnology Information, GenBank. Accession numbers MZ673262–MZ673264.