Abstract
Extrachromosomal circular DNA molecules of chromosomal origin have been detected in many organisms and are thought to reflect genomic plasticity in eukaryotic cells. Here we report a developmentally regulated formation of extrachromosomal circular DNA that occurs de novo in preblastula Xenopus embryos. This specific DNA population is not detected in the male or female germ cells and is dramatically reduced in later developmental stages and in adult tissues. The activity responsible for the de novo production of extrachromosomal circles is maternally inherited, is stored in the unfertilized egg, and requires genomic DNA as a template. The formation of circular molecules does not require genomic DNA replication but both processes can occur simultaneously in the early development. The production of extrachromosomal circular DNA does not proceed at random since multimers of the tandemly repeated sequence satellite 1 were over-represented in the circle population, while other sequences (such as ribosomal DNA and JCC31 repeated sequence) were not detected. This phenomenon reveals an unexpected plasticity of the embryonic genome which is restricted to the early developmental stage.
Plasticity of the eukaryotic genome, characterized by rearrangements, transposition, translocation, or amplification, is observed during evolution as well as in the development of specific organisms. A massive occurrence of such phenomena leads to genomic instability, which is a hallmark of neoplastic processes in mammals (54). The production of small extrachromosomal circular DNA molecules, also named small polydispersed circular DNA, is one indication of genome plasticity (18). These molecules, consisting mainly of repetitive sequences, are found in the tissues and cells of many organisms and are thought to emerge from the chromosomes but by a mechanism not yet determined. Elevated levels of extrachromosomal circular DNA have been detected in response to carcinogen treatment of human and rodent cells (12, 14, 53), and they were proposed to play a role in gene amplification (57). In addition, an increased amounts of circular molecules have been observed in patients suffering from genetic diseases which are characterized by genomic instability and premature aging, such as Fanconi’s anemia (14, 39) and Werner syndrome (30). Interestingly, it has recently been reported that extrachromosomal circles of ribosomal DNA (rDNA) accumulate in aged yeast cells and in mutants of Sgs 1, the yeast homolog of the human Werner syndrome gene (50). Circular DNA is also detected during the rearrangement of the T-cell receptor (17, 42) and immunoglobulin class switch, which leads to antibody diversity (36, 56). Extrachromosomal circular molecules have been observed in Drosophila and mouse embryos (44, 52, 59), but their specificity to embryonic stages or their developmental significance remain obscure.
Although circular DNA has been observed in many eukaryotes for more than two decades, the study of circular DNA has often been limited due to the lack of convenient techniques and physiological model systems. We have combined a well-characterized system for embryonic development, Xenopus laevis, and a useful assay for the detection and characterization of circular DNA, two-dimensional (2D) gel electrophoresis (12), to investigate these phenomena during embryogenesis.
Here we describe a developmentally regulated formation of circular DNA that occurs at a high rate during early embryogenesis of X. laevis. A specific pattern of multimeric circles consisting of the tandemly repeated sequence satellite 1 was identified in the circular molecules population. A cell-free system which mimics the in vivo phenomena was used, and we found that circular DNA is formed de novo by an activity which is stored in the egg and which is independent of DNA replication. The possible significance of circular DNA during development is discussed.
MATERIALS AND METHODS
Embryos and egg extracts.
Preparation of embryos and egg extracts was performed as previously described (15, 25).
Injection of plasmids into fertilized eggs.
Twenty nanoliters of a solution containing 20 pg of each plasmid (2.7, 5, and 9.4 kb) were injected into fertilized eggs as previously described (46). Embryos were developed until the 512-cell stage (pre-MBT embryos), and the DNA was purified.
Preparation of DNA.
Total genomic DNA was prepared by rapidly homogenizing the eggs, embryos, or liver tissues in 5 volumes of 30 mM EDTA–1% sodium dodecyl sulfate (SDS)–0.5% Triton X-100–0.3 M NaCl, followed by incubation at 37°C for 16 h with 0.1 mg of proteinase K per ml. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and digested with 0.2 mg of RNase A per ml for 2 h at 37°C. After ethanol precipitation and resuspension in 10 mM Tris–1 mM EDTA (pH 8), the DNA samples were electrophoresed.
When cell-free systems were used, reactions were stopped by the addition of 5 volumes of 30 mM EDTA–1% SDS–0.5% Triton X-100–0.3 M NaCl, and total DNA was prepared as described above.
Low-molecular-weight DNA was prepared by a neutral lysis technique, according to the method of Hirt (23). Embryos were homogenized in the presence of at least 10 volumes of 10 mM Tris-HCl (pH 7.9)–10 mM EDTA–0.6% SDS, and NaCl was added to a final concentration of 1 M. The homogenate was incubated 16 h at 4°C, and subsequent steps were performed as described earlier (23).
Neutral-neutral 2D gel electrophoresis.
Separation of DNA in neutral-neutral 2D gels was performed according to the method of Brewer and Fangman (6), as described previously (14).
Blotting and hybridization.
Southern blot analyses were done with Hybond N+ nylon membranes (Amersham), and probes were labelled by using the Ready-to-Go Kit (Pharmacia). Radiolabelled DNA was detected by autoradiography (Hyperfilm MP; Amersham) and with a PhosphorImager (Molecular Dynamics) for quantification by using the ImageQuant 1.1 software.
RESULTS
Detection of extrachromosomal circular molecules in Xenopus embryos.
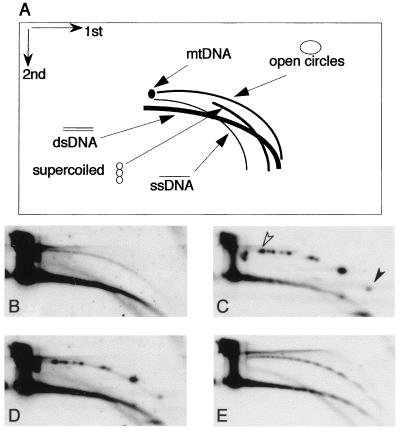
In the 2D gel electrophoresis used in this study, a population of molecules sharing the same structure but of heterogeneous molecular mass generates a continuous arc, and thus typical arcs of supercoiled molecules, open circles, and linear molecules can be distinguished after hybridization with total DNA or with specific probes (Fig. 1A). Single-stranded DNA and mitochondrial DNA can be identified in the same gel, and the structural identity of the DNA in each arc has been previously determined by electron microscopy and biochemical means (12, 14). After 2D gel analysis of a low-molecular-weight DNA fraction from Xenopus embryos at the cleavage stage, before mid-blastula transition (pre-MBT), we detected a continuous arc of circles, homologous to total Xenopus DNA, as well as arcs of double- and single-stranded linear molecules (Fig. 1B).
FIG. 1.
Detection of extrachromosomal circular DNA in early development by neutral-neutral 2D gel analysis. (A) Diagram of 2D gel electrophoretic patterns of genomic DNA generated by populations of linear and circular molecules heterogeneous in size (adapted from previous studies (12, 14). Each arc consists of molecules sharing the same structure but differing in mass. (B to E) A DNA sample enriched for low-molecular-weight DNA was isolated from Xenopus embryos at the early blastula stage (2,000-cell stage), mixed with plasmid-derived open circle size markers (see text), and separated on a 2D gel. The blot was first hybridized with a sperm DNA probe to detect total genomic sequences (B) and then with a plasmid probe to identify the open circles (C). The plasmids range from 2.7 kb (solid arrowhead) to 11.2 kb (open arrowhead). (D) Comigration of the nonlinear genomic DNA arc with the markers by superposition of panels B and C. (E) Rehybridization with a Xenopus satellite 1 probe (the insert of pE190 [31]) shows ladders of circular and linear multimers of the satellite 1 unit (multimers of 740 to 750 bp). The sizes of the linear and circular multimers were determined by circular (in panel C) and linear (not shown) size markers.
The identity of the DNA which migrated with the nonlinear DNA arc was validated by mixing the embryonic DNA with open-circle markers consisting of plasmid molecules of various lengths which were relaxed by use of DNA topoisomerase I. Upon sequential hybridization with the two probes (Xenopus DNA and then plasmid DNA), comigration of the open-circle markers with the extrachromosomal circle arc was observed (Fig. 1B–D). Hence, the nonlinear DNA arc is identified as a heterogeneous population of open circles consisting of genomic sequences, with molecular masses of several hundreds of base pairs up to 12 kbp.
Extrachromosomal circular molecules homologous to total DNA were not detected in DNA isolated from unfertilized eggs, indicating the absence of maternal stocks of such molecules (data not shown; see also Fig. 7C).
FIG. 7.
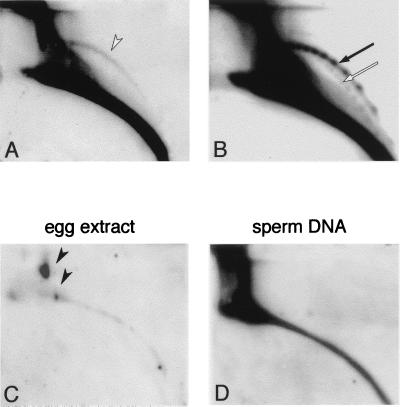
De novo formation of circular DNA in a cell-free system. 2D gel analysis of DNA purified from 2 × 105 demembranated sperm nuclei after incubation in 200 μl of egg extract for 2 h at 23°C as described previously (15) (A and B), from 200 μl of low-speed activated egg extract which was incubated without addition of sperm nuclei (C), and from 2 × 105 demembranated sperm nuclei (D). For panels A, C, and D, blots were hybridized with a sperm DNA probe, and the arc of open circles is indicated in panel A by an open arrowhead. The vast majority of the DNA in the egg extract is circular and linear mitochondrial DNA (4 ng per egg [21], ca. 800 ng in this sample). It is slightly labelled due to nonspecific hybridization with the sperm DNA probe (solid arrowheads in panel C). Rehybridization of blots from panels C and D with the satellite 1 probe did not reveal circular DNA (data not shown), while rehybridization of the blot from panel A with satellite 1 probe revealed ladders of circular multimers in an arc of open circles (solid arrow) and a faint arc of supercoiled circles (open arrow), as well as a massive linear-DNA arc (B). The structural identity of the arc DNA and the size of the multimers was determined by their comigration with supercoiled and relaxed plasmid markers, which were mixed with the DNA sample and visualized after hybridization with a plasmid probe (data not shown).
As repetitive DNA may be involved in genomic plasticity (55), we tested for the presence of the Xenopus satellite 1 DNA sequence, a tandemly repeated element of 750 bp representing 1 to 2% of the Xenopus genome (1, 31, 37) in the extrachromosomal DNA population. We detected extrachromosomal satellite 1 DNA in two series of spots defining two arcs (Fig. 1E) when intact DNA was analyzed. Using circular and linear size markers, we determined that these ladders consist of circular and linear multimers of the 750-bp unit of satellite 1. The arc of circular DNA was sensitive to restriction enzymes which cut inside the satellite 1 unit (e.g., HindIII, HinfI, and BglII; also data not shown) but was resistant to those that cut outside the unit (e.g., AlwNI, EcoRI, and XhoI) (data not shown; see also Fig. 8C). We conclude that multimers of satellite 1 DNA are present in the extrachromosomal circular DNA. Transcription followed by reverse transcription, a mechanism involved in the formation of some classes of transposons, is unlikely to be responsible for the production of the extrachromosomal satellite 1 DNA. Satellite 1 DNA is transcribed only after the midblastula stage (35), and its transcript, which is 180 bp long (1), would not give multimers of 750 bp and would not be sensitive to BglII, whose site is external to the transcription unit.
FIG. 8.
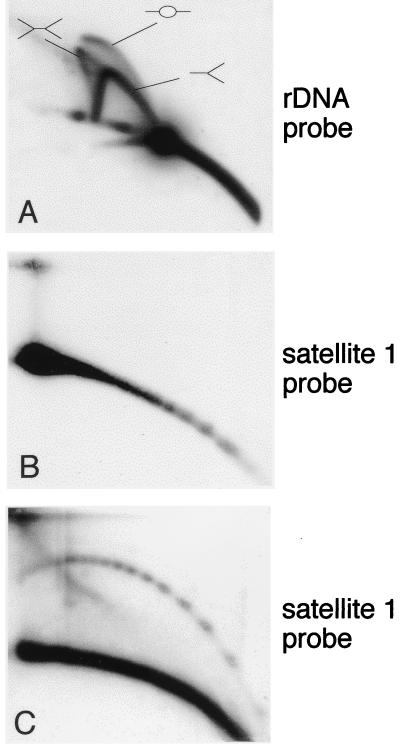
Formation of circular DNA in egg extracts competent for DNA replication. DNA purified after in vitro incubation of sperm nuclei in egg extract was cleaved with EcoRI and then analyzed by 2D electrophoresis. (A) Hybridization with a 5-kb coding sequence of the rDNA gene cluster (probe B [25]) reveals the replication intermediates of this fragment in a typical triple-arc pattern, representing initiation (–○–), termination (>–<), and elongation (>–) events as expected in this system (26). Rehybridization with the satellite 1 probe reveals linear multimers of satellite 1 unit after a short exposure (B) and circular multimers with a longer exposure (C), similar to the patterns observed with early embryo DNA (e.g., Fig. 1E and 2C).
The broad size range of multimers in the satellite 1 extrachromosomal population suggests that their formation involves intramolecular homologous recombination, an activity previously detected in activated Xenopus eggs, and has been suggested to serve for embryonic functions (32).
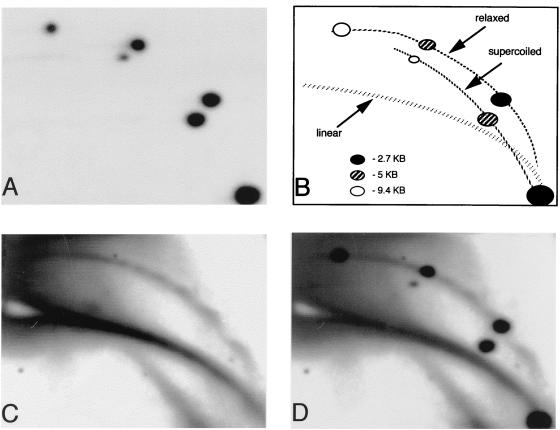
We did not detect supercoiled DNA in our DNA preparations from embryos, although the same purification method successfully recovered from developing embryos supercoiled plasmids which were injected into fertilized eggs (Fig. 2). The DNA content of the injected embryos was analyzed after eight cell divisions (512-cell stage). Supercoiled and relaxed forms of all injected plasmids were observed, while linear sequences were not detected (Fig. 2A). This result shows that supercoiled DNA can persist in the embryos and that it can be successfully purified with genomic DNA. It further indicates that the injected molecules were not subjected to nuclease activity or other mechanical breakage. Rehybridization of the same blot with satellite 1 probe revealed linear DNA and open circles (Fig. 2C) that comigrate with the open circle form of the injected plasmids (Fig. 2D).
FIG. 2.
Supercoiled plasmids can be recovered from embryos while endogenous circles are relaxed. Supercoiled plasmids (2.7, 5, and 9.4 kbp) were injected into fertilized eggs before the first cellular division. Total DNA was purified from the embryos at the pre-MBT stage (512 cells/embryo) and separated on a 2D gel. (A) Hybridization with plasmid probe revealed the supercoiled and the relaxed forms of each plasmid but no linear DNA. (B) Scheme indicating the identity of the plasmid spots found in panel A and the positions of the linear and circular DNA. (C) Rehybridization with satellite 1 probe revealed the position of the genomic circular DNA which contained only open circles. (D) Superposition of panels A and C.
We cannot eliminate the possibility that supercoiled molecules appear only in a transient manner in the cells and are immediately metabolized into other forms (e.g., linearization or integration) and therefore are not accumulated as standard extrachromosomal episomes.
It should be noted that throughout this study, normal development of the embryos was verified prior to DNA extraction and, thus, circular DNA is unlikely to result from necrosis. As an additional control, DNA was purified from abnormally developing embryos at the pre-MBT stage and was found to contain a reduced amount of extrachromosomal circular satellite 1 DNA molecules compared to the normally developing counterparts (data not shown). Apoptosis is also unlikely to be responsible for our observations since (i) apoptosis is not detected in Xenopus embryos before the gastrula stage (22, 49, 51) and (ii) apoptosis would generate DNA fragments which are multimers of 180 bp and not sequence specific.
Sequence content of circular DNA is not random.
Hybridization of the same blot to different probes revealed that satellite 1 is over-represented in the population of circular DNA relative to its proportion in the genome, since the ratio of circular to linear satellite 1 sequences is at least threefold higher than the ratio of circular to linear total genomic DNA (compare Fig. 1B and E, 3B and C, 6A and C, and 7A and B).
We have also identified 5S rDNA sequences in the extrachromosomal DNA molecules, at a level similar to satellite 1 DNA (Fig. 3A and C). The 5S rDNA genes are organized in tandem repeats close to the telomeres of most of the chromosomes. Their size (ca. 690 bp) and frequency are similar to those of satellite 1. However, the repeating units are very heterogeneous in size (10), and even adjacent repeats can be heterogeneous (9). The circular form of 5S rDNA sequences are observed in a continuous arc (Fig. 3A), a finding in agreement with this size heterogeneity. The circles containing 5S rDNA were resistant to cleavage with EcoRI, which does not have a site in the rDNA sequence, and sensitive to cleavage with DraI, which has two sites (data not shown).
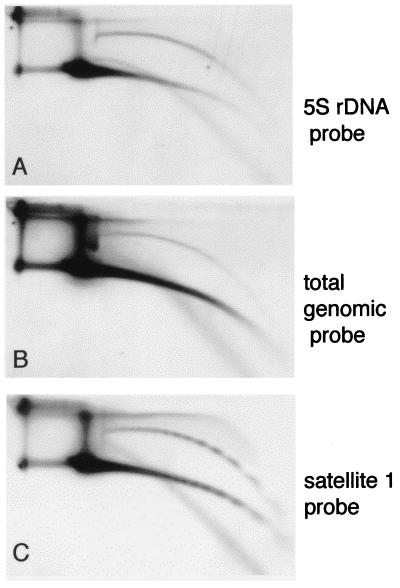
FIG. 3.
5S rDNA in circular DNA from early embryos. A DNA sample enriched for low-molecular-weight DNA was isolated from Xenopus embryos at the early blastula stage and separated on a 2D gel. The blot was hybridized with 5S rDNA probe (pXbs115/77) (3) (A), with total Xenopus genomic DNA (B), and with satellite 1 (C).
With total cellular DNA extracted from pre-MBT embryos, circular DNA migrated with the same pattern as with low-molecular-weight DNA, namely, extrachromosomal circles separated from the bulk of chromosomal DNA (Fig. 2C, 4A and C, and 5A).
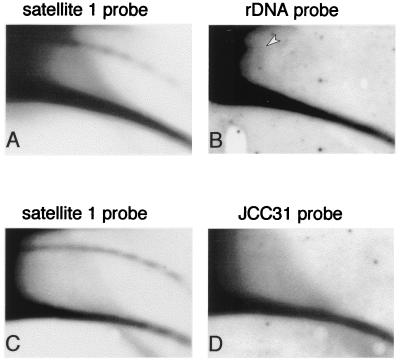
FIG. 4.
Sequence content of circular DNA. (A and C) Total genomic DNA from early embryos was separated on 2D gel and hybridized with satellite 1. One blot was then hybridized with a 5-kb coding sequence of the rDNA gene cluster (probe B [25]) (B), and the second was hybridized with a 500-bp dispersed repetitive sequence (JCC31 [11]) (D). To compare the relative amounts of circular DNA of each probe and to avoid the effect of different copy numbers, the exposure time was adapted to obtain similar intensity of the linear DNA arc. The position of maternally amplified circular rDNA is marked by an arrowhead.
High-molecular-weight extrachromosomal rDNA is maternally amplified during early oogenesis by a rolling circle mechanism and is accumulated in the egg (7). Electron microscopy studies have shown that most of the molecules are linear, and only a small fraction (2 to 5%) of the amplified DNA consisted of circular molecules representing multimers of the rDNA unit (24, 48), which varies between 11 and 15 kbp (47, 58). When analyzed on 2D gel electrophoresis, the vast majority of rDNA present in the egg migrated with the arc of linear DNA, but a low amount of circular rDNA corresponding to one unit could be identified (data not shown and Fig. 4B). Molecules larger than one unit are beyond the resolution limits of the technique. The population of circular extrachromosomal rDNA molecules decreased in embryos, in agreement with the degradation of the amplified rDNA population during early development (8). An arc of small circular molecules homologous to the rDNA gene cluster but smaller than the size of one unit was never detected either in eggs or embryos (Fig. 4B), implying that an abundant sequence is not prone to form extrachromosomal circles by stochastic events of breakage and ligation.
In addition, a repetitive sequence of 500 bp (pJCC31) which is dispersed in the Xenopus genome (11) is not detected in the population of circular DNA (Fig. 4D), further demonstrating that the formation of extrachromosomal circles does not occur at random.
Quantification of the gels of Fig. 4A and C and Fig. 5A and gels from additional experiments revealed that ca. 1% of the cellular satellite 1 DNA sequences is present as circular molecules during early development. As it was not possible to quantify the linear satellite 1 multimers comigrating with linear chromosomal DNA, this is probably an underestimation of the extrachromosomal satellite 1. Interestingly, in HeLa cells a similar fraction (1%) of the genomic alphoid satellite Sau3A element is represented as extrachromosomal multimers of the basic 170-bp unit (27, 28).
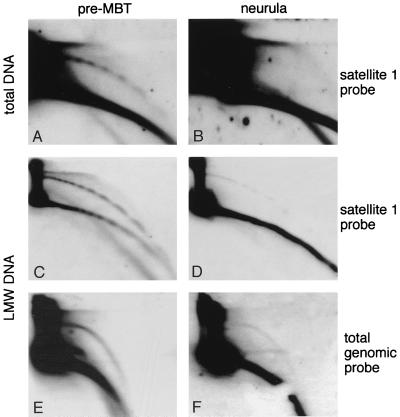
FIG. 5.
Circular DNA is produced during the early stages of development. (A and B) Total genomic DNA hybridized with a satellite 1 probe: A, 590 ng of early blastula embryos (pre-MBT, 1,000-cell stage); B, 1.4 μg of late-neurula-stage embryos (stage 23, 90,000 cells/embryo). The quantity of DNA was estimated from the number and the stage of the embryos and was verified directly on the gel by hybridization with a genomic DNA probe and comparison to a genomic standard calibrated spectrophotometrically at 260 nm and loaded on the same gel (not shown). (C to F) Low-molecular-weight (LMW) DNA was isolated from early blastula embryos (pre-MBT [C and E]) and from stage 13 embryos (neurula [D and F]), and similar amounts of DNA were compared on the same 2D gel. The blots were hybridized with a satellite 1 probe (C and D) and with a sperm DNA probe (total genomic probe [E to F]). The pre-MBT panels (C and E) represent low-molecular-weight DNA from 32 embryos of the 1,000- to 2,000-cell stage (i.e., maximum of 60,000 cells) and the neurula panels (D and F) represent low-molecular-weight DNA from five embryos at the 80,000-cell stage (i.e., 400,000 cells).
The production of extrachromosomal DNA circles is developmentally regulated.
To determine whether the accumulation of circular DNA was stage specific, we compared on the same blot samples of DNA taken from pre-MBT embryos and from late-neurula-stage embryos. While circular DNA of satellite 1 could be easily identified in the early embryos (Fig. 5A), we could not detect circular molecules in neurula embryos when using 2.4-fold more DNA (Fig. 5B). To further increase the sensitivity of the assay, the analysis was repeated with the low-molecular-weight fraction of embryonic DNA. In Fig. 5C and D, similar amounts of low-molecular-weight DNA purified from pre-MBT and neurula embryos were loaded on the same gel. Hybridization to satellite 1 probe shows a reduced amount (seven- to eightfold) of circular molecules in the DNA sample extracted from neurula embryos. As the neurula DNA was extracted from 6-fold-more cells, our data indicate a 50-fold decrease in the amount of extrachromosomal circles per cell at the neurula stage compared to pre-MBT stage. In addition, the linear multimers of satellite 1 are no longer detected in the low-molecular-weight neurula DNA, even with shorter exposures (data not shown). Hybridization of the low-molecular-weight DNA samples with a total genomic DNA probe further indicates that the decrease in the amount of circular DNA is not restricted to satellite 1 DNA (Fig. 5E and F).
The earliest developmental stage that could be tested by the 2D gel analysis was that of the embryos of the 32- to 64-cell stage. Circular DNA can be detected at this stage, and its proportion relative to total genomic satellite 1 content was similar to that observed in 1,000- to 2,000-cell-stage embryos.
Circular molecules were not detected in adult liver tissue (Fig. 6) or in Xenopus somatic A6 cells in culture (data not shown). We conclude that the formation of extrachromosomal circular DNA is a developmentally regulated phenomenon, one specific to early development.
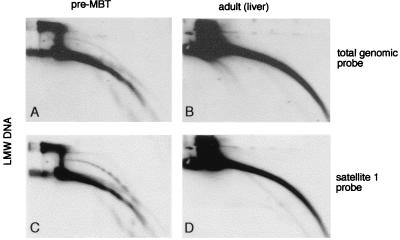
FIG. 6.
Circular DNA is not detected in adult liver tissue. Low-molecular-weight (LMW) DNA was isolated from early blastula embryos (pre-MBT [A and C]) and from adult liver (B and D) and compared on the same 2D gel. Hybridizations with a sperm DNA probe (total genomic probe [A and B]) and with a satellite 1 probe (C and D) show that, in adult liver cells, circular DNA is not detectable, even when loading an amount of low-molecular-weight DNA which was three times greater than that of the early embryo.
Extrachromosomal circles are generated de novo by an activity present in the egg.
To further investigate the mechanism that produces extrachromosomal circles, we used cell-free systems derived from Xenopus eggs (2, 34). After incubation of sperm nuclei in egg extract, extrachromosomal circles were detected both by the total genomic probe (Fig. 7A) and the satellite 1 probe (Fig. 7B). In some cases, a small amount of supercoiled molecules was also detected, which might result from a less-efficient processing mechanism of the circular DNA in vitro. Hence, the in vitro system reproduced the phenomena observed in vivo.
Egg extracts incubated in vitro without additional DNA template did not produce circular DNA homologous to total genomic sequences (Fig. 7C) or to satellite 1. We conclude that the circles are not produced from a preexisting free cytoplasmic egg DNA.
Total DNA purified from sperm nuclei did not contain detectable extrachromosomal circles after hybridization with total sperm DNA probe or with satellite 1 probe (Fig. 7D and data not shown). This result confirms that the formation of extrachromosomal circles occurs de novo due to an activity already present in the egg and that the template for this activity is nuclear genomic DNA.
Circular DNA is produced by egg extract which reproduces the early cell cycles in vitro.
Demembranated sperm nuclei are replicated in Xenopus activated egg extracts that mimic the early cell cycles, including genomic DNA replication (2, 41). DNA isolated from sperm nuclei that were incubated in egg extract was digested with EcoRI to produce a specific rDNA fragment and then subjected to 2D gel electrophoresis. The satellite 1 repeated sequence does not contain an EcoRI site and therefore remains intact in these conditions.
DNA replication in early embryos and in egg extracts is characterized by three types of replication intermediates while testing a single DNA fragment (26). These are bubbles for initiations in the fragment, Y shape for forks that pass through, and H shapes for termination events. After hybridization with an rDNA probe, all of the expected chromosomal rDNA replication intermediates were identified, including replication bubbles (Fig. 8A). Rehybridization of the same gel with the satellite 1 probe revealed both the linear and circular extrachromosomal multimers (Fig. 8B and C, respectively). These data show that the egg extracts competent for DNA replication can also form circular extrachromosomal DNA molecules. The presence of intact replication intermediates further rules out the possibility that circle production refers to abnormal processes of defective extracts. The amount of circles detected was proportional to the concentration of sperm nuclei in the extract (data not shown). This observation suggests that the activity that produces circles is present in excess in the egg. The formation of extrachromosomal DNA in extracts that reproduce the normal cell cycle shows that the extracts are competent for both processes but does not necessarily indicate that circle formation requires DNA replication.
The formation of extrachromosomal circular DNA does not require chromosomal replication.
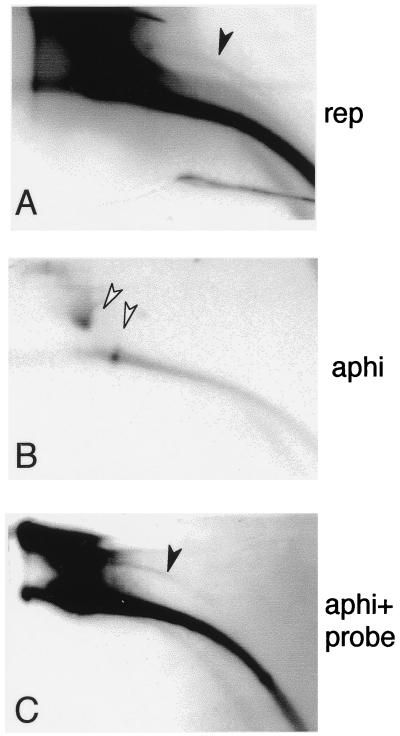
To determine whether DNA replication was involved in the production of the extrachromosomal circular DNA, newly replicated DNA was radiolabelled by adding [α-32P]dATP and sperm nuclei simultaneously to the egg extracts. 2D gel analysis revealed a distinct arc of labelled extrachromosomal circles (Fig. 9A). No labelled arcs were detected in the absence of sperm nuclei (data not shown). However, the labelled extrachromosomal DNA circles could have arisen from replicated DNA in this experiment. To test whether DNA replication could be uncoupled from the production of extrachromosomal circular molecules, DNA replication was blocked by aphidicolin, which was added at the beginning of the reaction. Extrachromosomal circles were not detected by direct labelling (Fig. 9B) but were revealed by hybridization with a total genomic DNA probe (Fig. 9C). These data demonstrate that the formation of extrachromosomal circles can be uncoupled from genomic DNA replication since it is resistant to aphidicolin.
FIG. 9.
The production of circular DNA does not require DNA replication. To label the newly replicated DNA, 105 sperm nuclei were incubated in 100 μl of activated egg extract as described above in the presence of 5 μl of [α-32P]dCTP (100 μCi). Aphidicolin (50 μg/ml) was added in panels B and C (aphi). Samples were taken after 95 min of incubation, and the DNA was separated on 2D gels and transferred to a nylon membrane. An arc of circular DNA (solid arrowhead) was detected among the labelled newly replicated DNA (A), while labelled circular molecules were not detected in the presence of aphidicolin (B). The weak labelling of the large amount of mitochondrial DNA present in the egg (21) in the presence of aphidicolin (panel B, open arrowheads) may be due to its replication by DNA polymerase-γ (16), which is aphidicolin resistant. (C) Hybridization of the blot from panel B with a total Xenopus genomic DNA probe revealed circular molecules (solid arrowhead).
Circular DNA was already detected at 30 min of incubation of sperm nuclei in the egg extract. At that time, DNA synthesis had not yet started, as determined by incorporation of [α-32P]dATP, which was added to a sample of the tested reaction mixture at time zero (data not shown). This result is in accordance with the appearance of circular DNA in the presence of aphidicolin. The formation of circles before the onset of DNA synthesis or in the presence of aphidicolin may suggest an excision by recombination. We cannot yet determine whether the circular molecules can be further replicated as extrachromosomal DNA in a DNA polymerase α-dependent manner or are excised from newly replicated chromosomal molecules, generating the labelled circular DNA.
DISCUSSION
In this study we report a phenomenon of genomic plasticity identified by the formation of extrachromosomal circles from a chromosomal origin during early embryonic development in X. laevis. We show that this activity is developmentally regulated and is particularly pronounced in the early embryos.
Several features strongly argue against an artificial or a random production of extrachromosomal circles. (i) The circular DNA population was detected after several different procedures for DNA isolation which involve phenol detergent and high salt extraction. Random breakage, if it occurs during extraction, is unlikely to be followed by ligation under these conditions. (ii) Nuclease activity in the embryos or during DNA preparation is unlikely since supercoiled plasmids that were injected into fertilized eggs were successfully recovered from embryos after eight cell divisions and using our standard extraction procedures (Fig. 2). Furthermore, replication intermediates remained intact (Fig. 6A), as well as most of mitochondrial DNA (17 kbp, circular), which is very abundant in the eggs and in early embryos (Fig. 7C and 9B). (iii) Circles are not likely be formed artificially by reannealing of single-stranded DNA during DNA purification since we did not observe a correlation between the presence of single-stranded DNA and the formation of extrachromosomal circles. In addition, pretreatment of the input DNA used in vitro with nuclease S1 does not affect circle formation (13). Still, we cannot exclude the possibility that the mechanism by which circular DNA is formed includes single-stranded intermediates as in the single-strand annealing mechanism (32, 33, 45). (iv) Apoptosis or necrosis cannot be responsible for circle formation since apoptosis is not detected in early embryos (22, 49, 51) and normally developing embryos were used in this study. When abnormal embryos were deliberately selected, their DNA contained a reduced amount of circular DNA. (v) The formation of circular DNA can be reproduced in egg extracts which mimic the cell cycle in vitro. (vi) The under-representation of the repetitive pJCC31 sequence in the circular molecule population compared to a random genomic sequence rules out the possibility of accidental breakage-ligation in vivo. The maternally amplified rDNA did not form circles smaller than the size of its repeating unit, thus further supporting the conclusion that random breakage-ligation of abundant sequences is not likely. (vii) Furthermore, the over-representation of satellite 1 sequence and the formation of sharply defined circular multimers of satellite 1 suggest the presence of a controlled mechanism which may involve intramolecular homologous recombination.
Circular DNA in mammalian cells is correlated with genomic instability and aging (18) and was proposed to appear as an early intermediate of gene amplification during malignant processes (57). We found that they are also produced in early Xenopus embryos. Hence, the formation of circular DNA may be a marker of genome plasticity important for early development which is downregulated throughout the normal life of the organism.
Although the biological significance of the production of extrachromosomal circular DNA is difficult to address, we showed here that this phenomenon exists in normal Xenopus embryos, is developmentally regulated, affects the cellular genome, and does not proceed at random. A genetic rearrangement of some discrete sequences may be one explanation for this phenomenon during early development. Genetic rearrangements in developmental processes have been observed in unicellular organisms and specifically in the early development of some nematodes and hagfish (43). For example, a dramatic exception to the rule of DNA constancy is the developmentally controlled chromatin diminution first reported by T. Boveri in Parascaris, where specific chromosome fragmentation and telomere addition occurs in the presomatic blastomeres during the first divisions of the embryo (4, 40). In the presumptive somatic cells, 85% of the genomic DNA, including most repetitive DNA sequences, is eliminated (38). In hagfish (cyclostomata, vertebrata), highly repetitive DNA elements are restricted to the germ line and are deleted during early embryonic development (19, 29). However, significant changes in the amount of genomic satellite 1 sequences were not detected upon comparison of sperm DNA and somatic tissues (spleen and erythrocytes) of the same individual (13). Therefore, a massive elimination of satellite 1 elements in somatic tissues, as described in parascaris and in hagfish, is unlikely in X. laevis. An additional support to the idea that a loss of a large set of sequences during development is not likely in X. laevis arises from the experiments involving nuclear transplantation of somatic nuclei into enucleated eggs which resulted in normal fertile animals (20).
In higher vertebrates, large global programmed rearrangements of the genome have not yet been observed, but localized gene rearrangements have been demonstrated during the differentiation of the immune system cells (5). Small extrachromosomal circles are produced by intramolecular DNA deletions during the rearrangement of the T-cell receptor α and β genes in mouse thymocytes (17, 42) and during immunoglobulin class switch recombination (36, 56).
The presence of large amounts of specific repetitive sequences in the extrachromosomal population might represent by-products of cellular differentiation pathways or, alternatively, might indicate an active role for these sequences in developmental processes such as remodeling of the organization of the genome during embryogenesis. Extrachromosomal circles may also reflect elimination of sequences necessary for germ line functions, including fertilization and chromosome pairing, but unnecessary for further development. In any case, the production of extrachromosomal circles during Xenopus early embryogenesis is a novel example of genomic plasticity of a vertebrate genome during development. It occurs at a stage when there is no transcription in the embryo but when determination of the cell lineage is already taking place (16).
We believe that the physiological experimental system presented here will facilitate further study of genomic plasticity during early vertebrate embryogenesis and help with our understanding of the evolution and function of repetitive sequences in the genome.
ACKNOWLEDGMENTS
We thank P. Brooks for a critical reading of the manuscript.
This study was supported by grants from the CNRS, the Association pour la Recherche sur le Cancer, the Ligue Nationale contre le Cancer, and the Fondation pour la Recherche Médicale. S.C. was supported by postdoctoral fellowships from the European Molecular Biology Organization and the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Ackerman E J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2:1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blow J J, Laskey R A. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen D F, Sakonju S, Brown D D. A control region in the center of the 5S RNA gene directs specific initiation of transcription. II. The 3′ border of the region. Cell. 1980;19:27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- 4.Boveri T. Ueber Differenzierung der Zellkerne während der Furchung des Eies von Ascaris megalocephala. Anat Anz. 1887;2:688–693. [Google Scholar]
- 5.Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978;15:1–14. doi: 10.1016/0092-8674(78)90078-8. [DOI] [PubMed] [Google Scholar]
- 6.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 7.Brown D D, Dawid I B. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- 8.Busby S J, Reeder R H. Fate of amplified nucleoli in Xenopus laevis embryos. Dev Biol. 1982;91:458–467. doi: 10.1016/0012-1606(82)90052-5. [DOI] [PubMed] [Google Scholar]
- 9.Carroll D, Brown D D. Adjacent repeating units of Xenopus laevis 5S DNA can be heterogeneous in length. Cell. 1976;7:477–486. doi: 10.1016/0092-8674(76)90199-9. [DOI] [PubMed] [Google Scholar]
- 10.Carroll D, Brown D D. Repeating units of Xenopus laevis oocyte-type 5S DNA are heterogeneous in length. Cell. 1976;7:467–475. doi: 10.1016/0092-8674(76)90198-7. [DOI] [PubMed] [Google Scholar]
- 11.Chambers J C, Watanabe S, Taylor J H. Dissection of a replication origin of Xenopus DNA. Proc Natl Acad Sci USA. 1982;79:5572–5576. doi: 10.1073/pnas.79.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S, Lavi S. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol Cell Biol. 1996;16:2002–2014. doi: 10.1128/mcb.16.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, S., and M. Mechali. Unpublished data.
- 14.Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14:977–985. doi: 10.1038/sj.onc.1200917. [DOI] [PubMed] [Google Scholar]
- 15.Coue M, Kearsey S E, Mechali M. Chromatin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 16.Dunon-Bluteau D, Cordonnier A, Brun G. DNA synthesis in a mitochondrial lysate of Xenopus laevis oocytes. H strand replication in vitro. J Mol Biol. 1987;197:175–185. doi: 10.1016/0022-2836(87)90116-1. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto S, Yamagishi H. Isolation of an excision product of T-cell receptor alpha-chain gene rearrangements. Nature. 1987;327:242–243. doi: 10.1038/327242a0. . (Erratum, 327:439.) [DOI] [PubMed] [Google Scholar]
- 18.Gaubatz J W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res. 1990;237:271–292. doi: 10.1016/0921-8734(90)90009-g. [DOI] [PubMed] [Google Scholar]
- 19.Goto Y, Kubota S, Kohno S. Highly repetitive DNA sequences that are restricted to the germ line in the hagfish Eptatretus cirrhatus: a mosaic of eliminated elements. Chromosoma. 1998;107:17–32. doi: 10.1007/s004120050278. [DOI] [PubMed] [Google Scholar]
- 20.Gurdon J B. Nuclear transplantation in eggs and oocytes. J Cell Sci Suppl. 1986;4:287–318. doi: 10.1242/jcs.1986.supplement_4.17. [DOI] [PubMed] [Google Scholar]
- 21.Gurdon J B, Wickens M P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- 22.Hensey C, Gautier J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech Dev. 1997;69:183–195. doi: 10.1016/s0925-4773(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 23.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 24.Hourcade D, Dressler D, Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proc Natl Acad Sci USA. 1973;70:2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 26.Hyrien O, Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyama R, Matsui H, Oishi M. A repetitive DNA family (Sau3A family) in human chromosomes: extrachromosomal DNA and DNA polymorphism. Proc Natl Acad Sci USA. 1986;83:4665–4669. doi: 10.1073/pnas.83.13.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyama R, Matsui H, Okumura K, Oishi M. A group of repetitive human DNA families that is characterized by extrachromosomal oligomers and restriction-fragment length polymorphism. J Mol Biol. 1987;193:591–597. doi: 10.1016/0022-2836(87)90342-1. [DOI] [PubMed] [Google Scholar]
- 29.Kubota S, Ishibashi T, Kohno S. A germline restricted, highly repetitive DNA sequence in Paramyxine atami: an interspecifically conserved, but somatically eliminated, element. Mol Gen Genet. 1997;256:252–256. doi: 10.1007/s004380050567. [DOI] [PubMed] [Google Scholar]
- 30.Kunisada T, Yamagishi H, Ogita Z, Kirakawa T, Mitsui Y. Appearance of extrachromosomal circular DNAs during in vivo and in vitro ageing of mammalian cells. Mech Ageing Dev. 1985;29:89–99. doi: 10.1016/0047-6374(85)90050-8. [DOI] [PubMed] [Google Scholar]
- 31.Lam B S, Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat) J Mol Biol. 1983;165:567–585. doi: 10.1016/s0022-2836(83)80267-8. [DOI] [PubMed] [Google Scholar]
- 32.Lehman C W, Clemens M, Worthylake D K, Trautman J K, Carroll D. Homologous and illegitimate recombination in developing Xenopus oocytes and eggs. Mol Cell Biol. 1993;13:6897–6906. doi: 10.1128/mcb.13.11.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin F L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohka M J, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 35.Lund E, Dahlberg J E. Control of 4-8S RNA transcription at the midblastula transition in Xenopus laevis embryos. Genes Dev. 1992;6:1097–1106. doi: 10.1101/gad.6.6.1097. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka M, Yoshida K, Maeda T, Usuda S, Sakano H. Switch circular DNA formed in cytokine-treated mouse splenocytes: evidence for intramolecular DNA deletion in immunoglobulin class switching. Cell. 1990;62:135–142. doi: 10.1016/0092-8674(90)90247-c. [DOI] [PubMed] [Google Scholar]
- 37.Meyerhof W, Tappeser B, Korge E, Knochel W. Satellite DNA from Xenopus laevis: comparative analysis of 745 and 1037 base pair Hind III tandem repeats. Nucleic Acids Res. 1983;11:6997–7009. doi: 10.1093/nar/11.20.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moritz K B, Roth G E. Complexity of germline and somatic DNA in Ascaris. Nature. 1976;259:55–57. doi: 10.1038/259055a0. [DOI] [PubMed] [Google Scholar]
- 39.Motejlek K, Schindler D, Assum G, Krone W. Increased amount and contour length distribution of small polydisperse circular DNA (spcDNA) in Fanconi anemia. Mutat Res. 1993;293:205–214. doi: 10.1016/0921-8777(93)90071-n. [DOI] [PubMed] [Google Scholar]
- 40.Muller F, Bernard V, Tobler H. Chromatin diminution in nematodes. Bioessays. 1996;18:133–138. doi: 10.1002/bies.950180209. [DOI] [PubMed] [Google Scholar]
- 41.Murray A. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 42.Okazaki K, Davis D D, Sakano H. T cell receptor beta gene sequences in the circular DNA of thymocyte nuclei: direct evidence for intramolecular DNA deletion in V-D-J joining. Cell. 1987;49:477–485. doi: 10.1016/0092-8674(87)90450-8. [DOI] [PubMed] [Google Scholar]
- 43.Plasterk R H. Genetic switches: mechanism and function. Trends Genet. 1992;8:403–406. doi: 10.1016/0168-9525(92)90320-4. [DOI] [PubMed] [Google Scholar]
- 44.Pont G, Degroote F, Picard G. Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J Mol Biol. 1987;195:447–451. doi: 10.1016/0022-2836(87)90665-6. [DOI] [PubMed] [Google Scholar]
- 45.Pont-Kingdon G, Dawson R J, Carroll D. Intermediates in extrachromosomal homologous recombination in Xenopus laevis oocytes: characterization by electron microscopy. EMBO J. 1993;12:23–34. doi: 10.1002/j.1460-2075.1993.tb05628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prioleau M N, Huet J, Sentenac A, Mechali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 47.Reeder R H. rRNA synthesis in the nucleolus. Trends Genet. 1990;6:390–395. doi: 10.1016/0168-9525(90)90298-k. [DOI] [PubMed] [Google Scholar]
- 48.Rochaix J D, Bird A, Barkken A. Ribosomal RNA gene amplification by rolling circles. J Mol Biol. 1974;87:473–487. doi: 10.1016/0022-2836(74)90098-9. [DOI] [PubMed] [Google Scholar]
- 49.Sible J C, Anderson J A, Lewellyn A L, Maller J L. Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Dev Biol. 1997;189:335–346. doi: 10.1006/dbio.1997.8683. [DOI] [PubMed] [Google Scholar]
- 50.Sinclair D A, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 51.Stack J H, Newport J W. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development. 1997;124:3185–3195. doi: 10.1242/dev.124.16.3185. [DOI] [PubMed] [Google Scholar]
- 52.Stanfield S, Helinski D R. Small circular DNA in Drosophila melanogaster. Cell. 1976;9:333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- 53.Sunnerhagen P, Sjoberg R M, Bjursell G. Increase of extrachromosomal circular DNA in mouse 3T6 cells on perturbation of DNA synthesis: implications for gene amplification. Somatic Cell Mol Genet. 1989;15:61–70. doi: 10.1007/BF01534670. [DOI] [PubMed] [Google Scholar]
- 54.Tlsty T D, Briot A, Gualberto A, Hall I, Hess S, Hixon M, Kuppuswamy D, Romanov S, Sage M, White A. Genomic instability and cancer. Mutat Res. 1995;337:1–7. doi: 10.1016/0921-8777(95)00016-d. [DOI] [PubMed] [Google Scholar]
- 55.Vogt P. Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code.”. Hum Genet. 1990;84:301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- 56.von Schwedler U, Jack H M, Wabl M. Circular DNA is a product of the immunoglobulin class switch rearrangement. Nature. 1990;345:452–456. doi: 10.1038/345452a0. [DOI] [PubMed] [Google Scholar]
- 57.Wahl G M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 58.Wellauer P K, Reeder R H, Dawid I B, Brown D D. Arrangement of length heterogeneity in repeating units of amplified and chromosomal ribosomal DNA from Xenopus laevis. J Mol Biol. 1976;105:487–505. doi: 10.1016/0022-2836(76)90230-8. [DOI] [PubMed] [Google Scholar]
- 59.Yamagishi H, Kunisada T, Iwakura Y, Nishimune Y, Ogiso Y, Matsushiro A. Emergence of the extrachromosomal circular DNA complexes as one of the earliest signals of cellular differentiation in the early development of mouse embryo. Dev Growth Differ. 1983;25:563–569. doi: 10.1111/j.1440-169X.1983.00563.x. [DOI] [PubMed] [Google Scholar]