Abstract
Simple Summary
The MYC oncoprotein is deregulated in up to 70% of human tumors and has a crucial role in the initiation, progression, and maintenance of the tumorigenic process. The MYC modulator MNT belongs to the MXD family of MYC antagonists and transcriptional repressors. It differs from the other MXD family members due to its essential role in the cell and its diverse and complex functions that can either facilitate or impair MYC-driven tumorigenesis. As MYC is a difficult therapeutic target, increasing our knowledge of other proteins in the MYC network will provide the basis for alternative strategies to impair MYC activity in cancer.
Abstract
MNT is a crucial modulator of MYC, controls several cellular functions, and is activated in most human cancers. It is the largest, most divergent, and most ubiquitously expressed protein of the MXD family. MNT was first described as a MYC antagonist and tumor suppressor. Indeed, 10% of human tumors present deletions of one MNT allele. However, some reports show that MNT functions in cooperation with MYC by maintaining cell proliferation, promoting tumor cell survival, and supporting MYC-driven tumorigenesis in cellular and animal models. Although MAX was originally considered MNT’s obligate partner, our recent findings demonstrate that MNT also works independently. MNT forms homodimers and interacts with proteins both outside and inside of the proximal MYC network. These complexes are involved in a wide array of cellular processes, from transcriptional repression via SIN3 to the modulation of metabolism through MLX as well as immunity and apoptosis via REL. In this review, we discuss the present knowledge of MNT with a special focus on its interactome, which sheds light on the complex and essential role of MNT in cell biology.
Keywords: MNT, MYC, MAX, REL, transcriptional regulation, proliferation, cancer
1. Introduction
The discovery of MNT (MAX’s Next Tango) was reported by two independent laboratories in 1997. One group found MNT through a two-hybrid screening in yeasts using a murine cDNA library with MAX as the bait [1], while the other group isolated MNT (originally called ROX) in a search for transcribed sequences of the human chromosome 17p13.3 [2].
The MNT transcription factor belongs to the MXD family (originally called MAD), which shares a basic helix–loop–helix–leucine zipper (bHLHLZ) domain as well as a SIN3 interaction domain (SID). The bHLHLZ is necessary for forming dimers with MAX and for binding to DNA on E-boxes (Enhancer-boxes). The SID establishes the interaction with SIN3 proteins, facilitating transcriptional repression (see below) [3,4]. Notably, the SID repressor domain linked to Cas9 has been described as an efficient tool in CRISPR-mediated repression of targeted genes [5]. The canonical MXD proteins are MXD1, MXI1, MXD3, and MXD4. There is yet another member, MGA [6], that shows little similarity with the other MXDs, as it lacks the SID but binds to MAX through its bHLHLZ domain. MGA is a very large protein (3065 amino acids) and besides the bHLHLZ, it contains a T-box domain, which is described in Brachyury proteins and is a characteristic of the TBX (T-Box) transcription factor family. Its structure allows the regulation of both MAX-network and T-domain target genes, either as an activator or repressor [6,7,8]. Inactivation of MGA is frequent in human cancer, and a recent study has demonstrated that MGA functions as a tumor suppressor in murine models of lung carcinoma and in colorectal cancer organoids, partly due to the de-repression of MYC target genes [9].
MNT is a unique member of the MXD family due to several features; it is (i) the largest protein of the family; (ii) the most divergent gene in its sequences; (iii) ubiquitously expressed in human tissues; (iv) essential for mouse development; (v) expressed in both quiescent and proliferating cells; (vi) altered in human cancer; and (vii) is frequently involved in tumorigenesis (Table 1). It also plays an important role in modeling the oncogenic activities of MYC, as an antagonist in some models or as a cooperator in others (reviewed in [10]).
Table 1.
Main differences between the human MNT, other human MXD proteins, MGA, and MYC. The sizes of the proteins are the largest canonical forms, according to the UniProt database. Aa, amino acids; ND, not done; DEL, deletions; AMP, amplifications; MUT, mutations; CTCL, Cutaneous T-cell lymphoma; MB, medulloblastoma; SS, Sezary Syndrome; ALL, acute lymphoblastic leukemia.
| MNT | MXD1 | MXI1 (MXD2) | MXD3 | MXD4 | MYC | References | |
|---|---|---|---|---|---|---|---|
| Protein size | 582 aa | 221 aa | 228 aa | 206 aa | 209 aa | 439 aa | Uniprot database |
| P-rich sequences | Yes | No | No | No | No | Yes | [2] |
| Phenotype of KO mice | Perinatally lethal/Craniofacial abnormalities | Viable/Increased immature granulocyte progenitors | Viable/Hyperplasia | Viable/Enhanced sensitivity to apoptotic stimuli | ND | Embryonic lethal E9.5–E10.5 | [11,12,13,14] |
| Proximal MYC Network interactors | MNT, MAX, MLX | MAX, MLX | MAX | MAX | MAX, MLX | MAX | Reviewed in [15] |
| Expression in cells | Quiescent and proliferating | Quiescent | Quiescent and proliferating * | Proliferating (S-phase) | Quiescent | Proliferating | Reviewed in [15] |
| Pan-cancer copy number alterations (%) | DEL 10 AMP 3 MUT 1 |
DEL 2 AMP 6 MUT < 0.5 |
DEL 8 AMP 4 MUT < 0.5 |
DEL 7 AMP 8 MUT < 0.5 |
DEL 6 AMP 5 MUT < 0.5 |
DEL 2 AMP 21 MUT 1 |
[16] |
| Involvement in human cancer | DEL in CTCL-SS and ALL. Reduced expression in MB | None or weak | None or weak | None or weak | None or weak | Strongly, 70% of tumors have deregulation | [17,18,19,20,21,22] |
* central nervous system, epidermis, and the myeloid lineage.
MNT is evolutionarily conserved, an argument for its relevant role in cell biology. It is the sole MXD protein identified in the fly Drosophila melanogaster [23]. Flies express a “minimal” network composed of dMax, a single MYC protein (dMyc), and a sole MXD protein (dMnt). In general, MNT and MXD1 are present along the phylogenetic tree, whereas the other MXD are only present in vertebrates (reviewed in [15]). Interestingly, dMnt is recruited to a higher number of Drosphila genes than dMyc is [24]. MNT orthologues are found in primitive pluricellular organisms from the Cnidaria (Hydra spp., Nemastotella spp.) and Placozoa (Trichoplax spp.) phylum [25]. In contrast, the nematodes C. elegans, C. briggsae, and Brugia malayi lack MNT or MYC orthologs. However, these species, together with the trematode Schistosoma mansoni, present orthologs for MAX, MLX, MXD, and MLXIP [15,25,26].
In contrast to the other members of the MXD family, MNT is an essential protein during development, as Mnt knockout mice die soon after birth [11,27]. Mice with null homozygous deletions for Mxd1, Mxd2, or Mxd3 show different defects [12,13,28] but survive to adult ages. Mnt-deficient embryos exhibit small size throughout development and, interestingly, show reduced levels of c-Myc and N-Myc [11]. Mnt-overexpressing embryos also have developmental defects, are of a smaller size, and have decreased cellularity [1]. These data suggest that a tight control of MNT levels is necessary for the correct development.
2. MNT Structure and Regulation
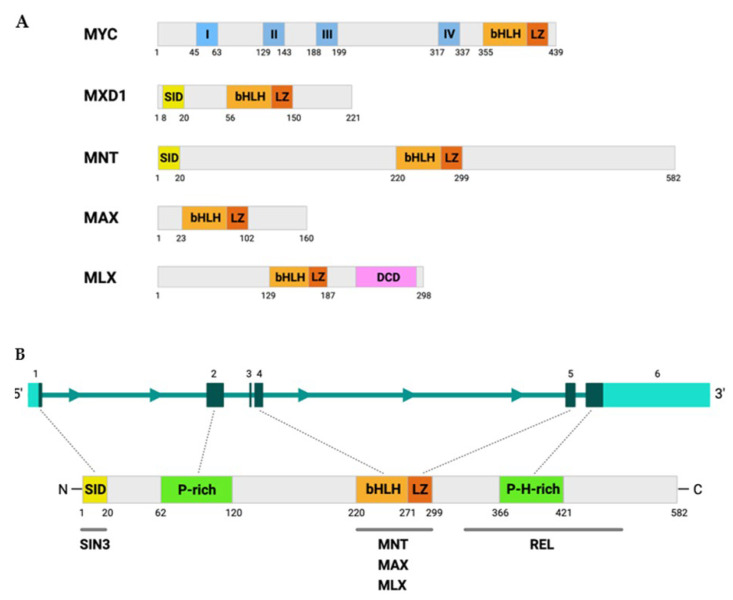
The MNT gene is localized at human chromosome 17p13.3 and mouse chromosome 11B5. The human gene is 1.7 kb long and contains six exons according to the NCBI Gene browser (GRCh38/hg38). Human MNT encodes a 582-amino acid protein that contains (i) a coiled-coil α-helix structure for interaction with SIN3 (SID); (ii) a highly proline-rich region, similar to the activation domains of other transcription factors like MYC; (iii) a bHLHLZ domain, for homodimerization, interaction with MAX and MLX, and DNA-binding at E-boxes, and (iv) a proline and histidine-rich region at its C-terminal domain [1,2]. Proline-rich regions are usually involved in protein–protein interactions [29] and are typical of transcriptional activation domains and are absent in the other MXD proteins. The SID is necessary for the repressive function of MNT, as its deletion turns the protein into a transcriptional activator [1]. We recently described that the C-terminal domain of MNT (amino acids 306–537) is necessary for its interaction with REL [30], a member of the NF-κB family. The MNT, MYC, MXD1, MAX, and MLX proteins are schematized in Figure 1A. The MNT gene, protein, and its main interactors are shown in Figure 1B.
Figure 1.
MNT and MYC protein structure. (A) Comparison between MYC, MXD1, MNT, MAX, and MLX protein sizes and domains. The canonical isoforms are shown; specifically, the p22 isoform for MAX and the gamma isoform for MLX. MYC boxes are represented in blue with the numbers I–IV. (B) The MNT gene structure is schematized at the top, with boxes representing the six exons, and a scheme of the MNT protein at the bottom. The MNT exons that encode the most important regions are indicated as boxes, and the dotted lines connect the exons with the corresponding protein domains. The main MNT-interacting proteins are shown at the bottom. SID: SIN3 interacting domain; P-rich: proline-rich sequence; bHLH: basic helix–loop–helix; LZ: leucine zipper; P-H-rich: proline and histidine-rich region; DCD: dimerization and cytoplasmic localization domain. Figures were created with BioRender.com (Toronto, ON, Canada).
The basic region of MNT differs from the other bHLHLZ proteins in the proximal MYC network, and MNT–MAX dimers show a higher preference for non-canonical CACGCG E-boxes though they can also bind to the canonical CACGTG [2]. This difference in the basic regions of the DNA explains why MNT has a subset of unique target genes apart from the ones shared with MYC and the other MXDs.
MNT is a mainly nuclear protein with a half-life of 30–60 min, and it appears as a doublet of 72–74 kDa in Western blots [1,2]. The 72 kDa form is the form detected in growth-arrested cells. In contrast, MNT is detected at 74 kDa in serum-stimulated cells and corresponds to the hyperphosphorylated protein. This phosphorylation is conducted by MKK/ERK kinases and impairs the interaction with SIN3B and, consequently, the repressive function of MNT [31]. So far, no other post-translational modifications have been described for MNT except for ubiquitination (described below) [30].
3. MNT Expression
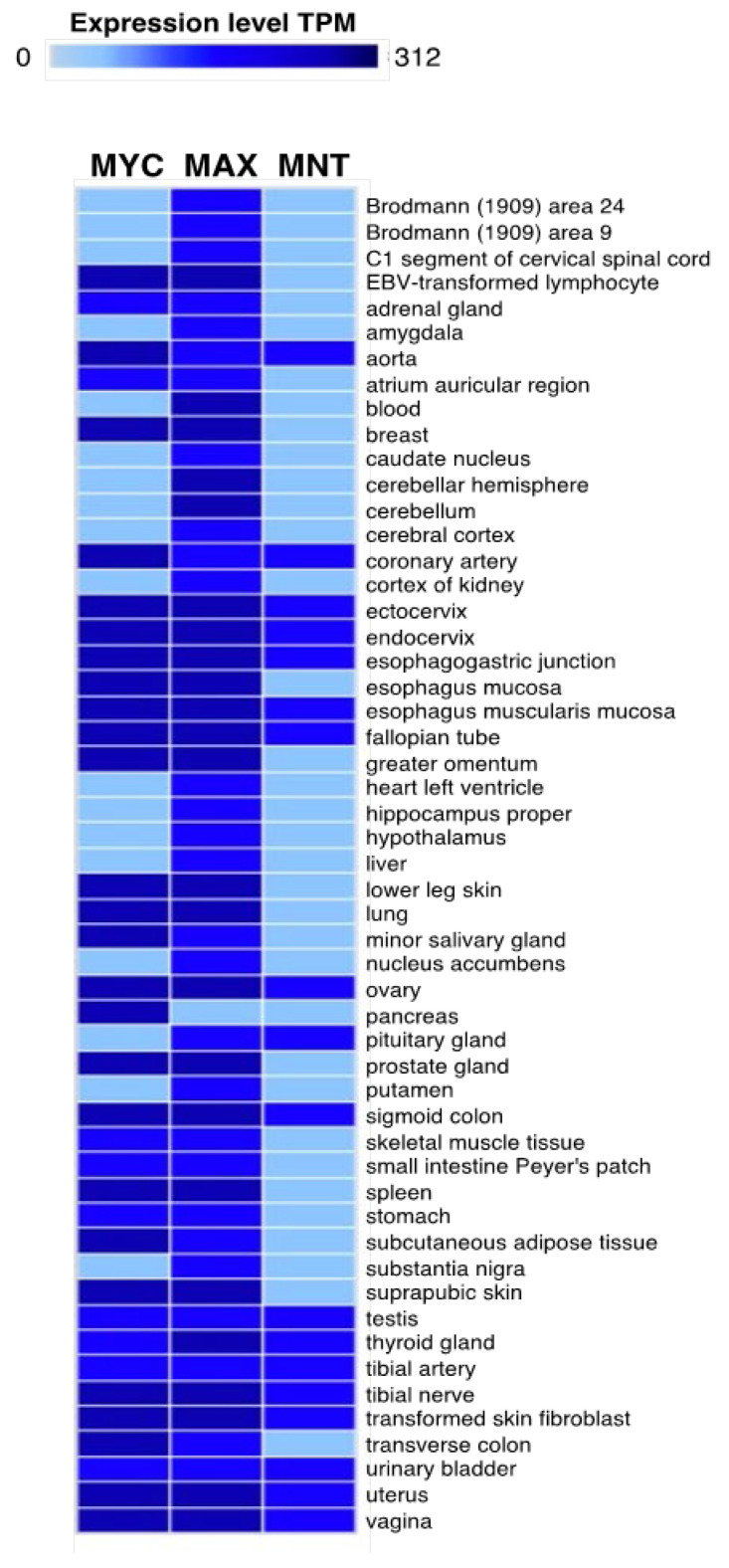
MNT mRNA is expressed in most human tissues according to a massive RNA-sequencing study [32]. MNT mRNA levels were highest in the bone marrow, brain, endometrium, spleen, and testis, while the lowest levels were in the heart, liver, pancreas, and salivary glands. The GTEX database gives a more restricted pattern of MNT mRNA expression in human tissues, peaking in the esophagus, intestine, and pancreas (Figure 2).
Figure 2.
Expression of MYC, MAX, and MNT in human tissues. Data obtained using the GTEX Portal (gtexportal.org). TPM, transcripts per million.
Mnt is broadly expressed during mouse development. Its mRNA has already been detected in early conceptus and in multiple tissues during development (informatics.jax.org/marker/MGI:109150) until stage E10.5. At later stages, Mnt expression is the highest in neural structures [1].
During Xenopus laevis development, the MNT orthologue XMnt is expressed in early embryos at the neurula stage and in migrating cranial neural crest cells [33]. The expression of Mnt in the neural tissues of mice and Xenopus seems consistent with the cleft palates and craniofacial deformities observed in Mnt-deficient mice [11,34]. In Xenopus, XMnt, XMad1, and XMad3 exhibit distinct areas of expression during embryogenesis, suggesting non-redundant functions [33].
Both quiescent cells and fibroblasts express MNT throughout the cell cycle. MNT levels do not increase upon serum stimulation of quiescent cells [31]. This is in contrast with the expression pattern of MYC (expressed mainly in proliferating cells) and most other MXD proteins, such as MXD1, MXI1, and MXD4 (mainly expressed in quiescent cells). Thus, MNT–MAX dimers coexist with MYC–MAX in proliferating cells [1]. MNT represses its own expression in a MAX-dependent manner by direct binding to its promoter. E-box 2, which is mapped at −788 bp from the transcription start site, is critical for this autoregulation. A decrease or absence of MAX results in high levels of MNT, which are distributed among cytoplasm and nucleus. MNT also binds weakly to its own promoter in the absence of MAX [35]. The presence of E-boxes in the MNT promoter indicates that other bHLHLZ proteins could also control its expression.
MNT protein levels are also regulated by the E6-associated protein (E6AP), an E3 ubiquitin ligase that induces its ubiquitination and degradation by the proteasome. This was observed in myeloid differentiation experiments, in which all-trans retinoic acid, vitamin D3, or phorbol 12-myristate 13-acetate downregulated E6AP and consequently reduced the E6AP-mediated degradation of MNT [30].
4. MNT Interactions in the Proximal MYC Network
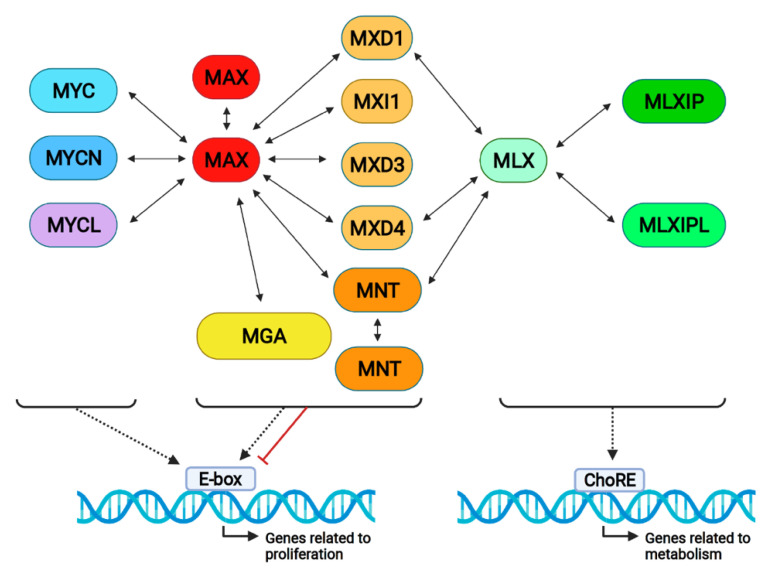
The proximal MYC network is composed of thirteen proteins distributed into three protein families: MYC, MXD, and MLXIP. The MYC family includes c-MYC (also called MYC), MYCN (or N-MYC), and MYCL (or L-MYC). MAX connects MYC with the MXD proteins while MLX connects the MXD with the MLXIP (also known as MONDO) proteins. A scheme of this network is shown in Figure 3.
Figure 3.
The proximal MYC network and its interactions. MYC proteins form dimers with MAX and bind DNA on E-boxes, generally activating transcription. All MXD proteins bind to MAX and MXD1, and MXD4 and MNT also bind to MLX, activating or repressing gene transcription. MLX forms complexes with MLXIP (MONDOA) and MLXIPL (MONDOB), binding on ChoRE (carbohydrate response elements) and regulating metabolic genes. Notably, MAX and MNT form homodimers, as represented in the Figure.
In contrast to the MYC and the MXD proteins, MNT can form homodimers. This was initially discovered in yeast two-hybrid assays and with recombinant proteins [1,2]. More recently, the formation of MNT dimers has been shown in rat and human cell lines. Yet, MNT–MAX complexes are formed preferentially to MNT–MNT dimers [35]. Electrophoretic mobility shift assay (EMSA) assays with in vitro synthesized proteins also showed that MNT–MNT can bind to E-boxes but with a lower affinity compared to MNT–MAX [2]. Importantly, it has been shown that MNT homodimers not only bind DNA, but they are also able to regulate gene transcription [35].
In summary, MNT can form heterodimers with MAX [1,2], heterodimers with MLX [36], or homodimers [35] through the bHLHLZ domain (Figure 3). Thus, MNT serves as a bridge between MYC–MAX (the proliferative arm of the network) and MLX–MLXIP/MLXIPL (the metabolic arm of the network) [37].
5. MNT Interactions Outside the Proximal MYC Network
5.1. SIN3 Proteins
SIN3A and SIN3B are transcriptional corepressors that recruit histone deacetylases and other chromatin modifying enzymes, inhibiting transcription upon binding to E-boxes. MNT represses transcription through direct interaction with the SIN3 proteins through the SID, a domain in the N-termini that binds to the SIN3 s paired amphipathic helix (PAH2) domain of SIN3B [3,22]. SIN3A and SIN3B, in turn, recruit histone deacetylases through their histone interaction domain (HID) or co-factors including N-CoR, SDS3, SAP30, SAP18, RBP1, or ING1/2 that work by bridging and stabilizing the complex and/or by enhancing the chromatin remodeling activities [4,38]. In the case of MXD1, this is mediated by the recruitment of HDAC1/3 by SIN3 [39]. The ability of MNT to block MYC-dependent transformation and cell cycle progression is lost in SID mutants, indicating a close link between biological function and repression [1,40].
5.2. REL
Recently, we identified the NF-κB family protein REL (also known as c-REL) as a MNT interacting partner through the proteomic analysis of MNT co-immunoprecipitating proteins in a MAX-independent setting. MNT acts as a repressor of the NF-κB pathway by two suggested mechanisms: (i) the retention of REL in the cytoplasm in REL–MNT complexes and (ii) the repression of REL target genes [41].
Once a stimulus is detected by the cell, the NF-κB pathway is activated and IκBα is degraded. However, NF-κB also induces a negative feedback loop, leading to the transcription of NFKBIA/IκBα. The newly synthesized IκBα enters the nucleus and shuttles the NF-κB dimers back to the cytoplasm to terminate transcription [42,43,44]. MNT–REL can bind and repress the NFKBIA/IκBα gene at least in some cell types. Therefore, MNT might be acting to limit NF-κB activity in the absence of specific pathway activators. In addition, the requirement of MNT for cell proliferation is dependent on REL in two cellular models. This indicates that the pro-survival role of MNT can be mediated through REL. Interestingly, MNT regulates the NF-κB pathway independently of MAX [41].
5.3. MATα-1
Methionine adenosyltransferase a1 (MATα-1) forms a complex with MNT–MAX in the normal liver and bile duct epithelial cells that binds E-boxes. In cholestasis and cholangiocarcinoma cells, MATα-1-MNT interaction drops, and there is a switch towards MYC and MAF protein complexes [45]. The switch from MNT–MAX to MYC–MAX during cholestasis has been previously described, and it is responsible for p53 and cyclin D1 up-regulation and apoptosis [46].
5.4. Other Proteins
Recent studies have identified at least four other proteins as MNT interactors [41]: CCDC6, AMPD2, QSER1, and TPP2. CCDC6 is involved in DNA damage response [47]; AMPD2 participates in purine metabolism as an AMP deaminase [48]; QSER1 has recently been described as a demethylation regulator [49]; and TPP2 is a peptidase with roles in antigen processing, lipid metabolism and CCK8 hormone regulation [50].
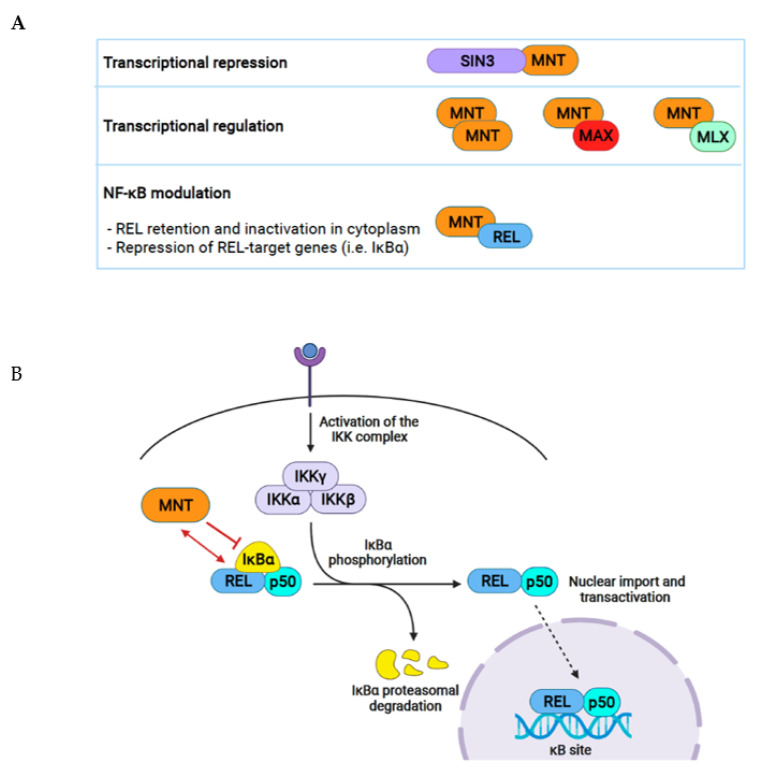
The main MNT functional interactions are represented in Figure 4A and include NF-κB modulation by MNT in Figure 4B.
Figure 4.
MNT functional interactions. (A) MNT represses transcription through SIN3, which recruits a HDAC complex and turns DNA into a closed conformation. MNT homodimers and MNT–MAX and MNT–MLX heterodimers regulate transcription by binding DNA at E-boxes. MNT–REL interaction results in NF-κB regulation by retaining REL dimers inactive in the cytoplasm and by repressing REL target genes that would usually be activated by REL. (B) Once the cell receives an NF-κB activation signal, the IKK complex is activated and is able to phosphorylate IκBα, which is bound to REL-p50 dimers in the cytoplasm. This phosphorylation targets IκBα for ubiquitination and posterior proteasomal degradation. Finally, the REL-p50 dimers are free to translocate into the nucleus and regulate their target genes by binding to κB sites on the DNA. MNT forms complexes with REL and inhibits its functions.
6. MNT Alterations in Cancer
MNT has been classified both as an oncogene or as a tumor suppressor gene based on its diverse activities in the cells. The data arguing that it behaves as tumor suppressor gene can be summarized as follows: (a) the tissue-specific loss of Mnt in mouse models results in thymic lymphoma [51] or mammary tumors [27,52] (discussed below in point 7.1); (b) MEFs deficient in Mnt proliferate faster than wild-type cells and prematurely enter into the S phase [27] (discussed below in point 7.1); (c) in a pan-cancer study of The Cancer Genome Atlas (TCGA) data, heterozygous MNT deletion was found in 10% of the tumors (overall frequency) and in more than 20% of liver hepatocellular carcinomas, lung adenocarcinomas, sarcomas, and uterine carcinosarcomas [16].
The MNT locus at chromosome 17p13.3 is a hot spot for the loss of heterozygosity (LOH) in tumors such as sporadic breast cancer, medulloblastomas, or chronic lymphocytic leukemia (CLL) [21,53,54,55] as well as in ovarian cancer [56], astrocytomas [57], bladder cancer [58], or osteosarcoma [59]. However, no inactivating mutations in MNT have been found in these tumors. MNT deletions were also found in acute lymphoblastic leukemia [20], and a decrease in its levels in medulloblastomas, which was caused by the haploinsufficiency of chromosome 17p13.3 [21]. In the Sézary Syndrome, a malignant variant of cutaneous T-cell lymphoma, more than 60% of the patients show a loss of one or both MNT alleles [17,18], thus being the tumor with the highest frequency of MNT deletions. In agreement, the down-regulation of MNT expression has also been reported in Sézary Syndrome [19].
The analysis of the genomic data available in TCGA shows that 0.6% of tumors carry MNT mutations, with missense mutations being the most common. Most of these missense mutations are observed in colorectal, endometrial, melanoma, esophagic, and head and neck cancer. The majority of mutations map in the C-terminal half of the protein, i.e., downstream of the bHLHLZ domain.
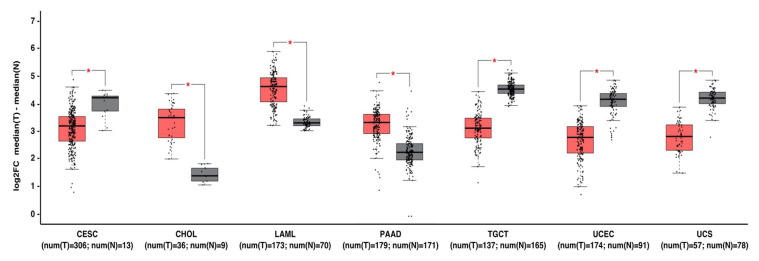
Analysis of MNT mRNA expression in tumors shows that it is down-regulated in tumor samples compared to their normal tissue counterparts, including testicular germ cell tumors, uterine corpus endometrial carcinoma, and uterine carcinosarcoma. However, the opposite occurs in other tumor types, including cervical carcinoma, cholangiocarcinoma, acute myeloid leukemia, and pancreatic adenocarcinoma (Figure 5). These differences likely reflect the complex biological activities of MNT, which depend on the cellular context and the repertoire of proteins to interact with.
Figure 5.
MNT can act as a tumor suppressor gene or oncogene depending on the tumor type. Gene expression profiling interactive analysis (GEPIA, http://gepia.cancer-pku.cn/, accessed on 7 July 2021) was performed to validate MNT mRNA expression in selected cancer type samples (in red) vs. normal samples (in grey). The expression data were first log2(TPM+1) transformed for differential analysis, and the log2FC was defined as median (tumor)–median (normal). Data are represented as mean ± SD (* p < 0.01). Abbreviations: CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; LAML, acute myeloid leukemia; PAAD, pancreatic adenocarcinoma; TGCT, testicular germ cell tumors; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma.
7. Functions of MNT in the Cell
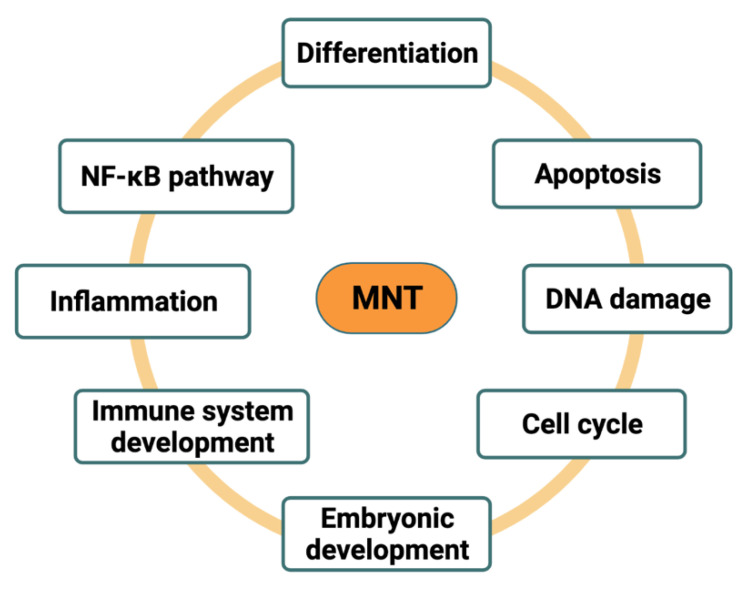
MNT plays a pivotal role in controlling cell proliferation, differentiation, and cellular transformation. The cellular processes that MNT regulates are represented in Figure 6 and are described below. Its functions can result in either antagonism or cooperation with MYC.
Figure 6.
Cellular functions of MNT. MNT is involved in several cellular functions, including differentiation, apoptosis, DNA damage, cell cycle, embryonic development, immune system development, inflammation, and NF-κB pathway regulation. Depending on the model, MNT will either antagonize or cooperate with MYC.
7.1. MNT as MYC Antagonist
The expression of MNT is ubiquitous, and its levels are constant throughout the cell cycle. Thus, MNT–MAX dimers coexist with MYC–MAX dimers along all phases of the cell. The antagonism between MNT and MYC is achieved at three different levels: (i) competition for forming complexes with MAX, as MNT and MYC show similar affinities towards MAX; (ii) binding to the E-Boxes of their shared target genes; and (iii) the transcriptional repression of genes that are normally activated by MYC–MAX [1,24]. In fact, it has been hypothesized that a major role of MYC is to overcome MNT transcriptional repression rather than its transactivation capacity (reviewed in [22]).
MNT–MYC counteraction was initially studied in wild-type mouse embryonic fibroblasts (wt MEFs) versus Mnt knockout fibroblasts (Mnt-/- MEFs). Mnt-deficient MEFs proliferated faster and entered the S phase prematurely, which was accompanied by an increase in Cdk4 and Ccne1 (Cyclin E) and a decrease in Myc. Mnt-/- MEFs have higher apoptotic rates and can efficiently escape senescence compared to wt MEFs. Notably, Mnt-/- MEFs can be transformed by oncogenic RAS alone, mimicking cells with Myc overexpression. The deletion of Myc in MEFs causes a proliferation arrest that can be partially rescued by the simultaneous deletion of Mnt [27,60,61,62].
In serum-stimulated quiescent cells, Myc is induced at the G0 to G1 transition, whereas Mnt levels remain constant. During this transition, there is a switch from Mnt–Max to Myc–Max dimers, which activate cell cycle progression genes, such as Cdk4, Ccnd2 (Cyclin D2), ODC, or E2F2. Both Mnt overexpression or Myc loss blocks cell cycle entry, suggesting that the ratio of Mnt versus Myc levels could determine the quiescent or proliferative state of the cell [62].
Additional data supporting a MYC–MNT antagonism derive from studies using mice with the conditional deletion of Mnt in mammary tissue. This revealed an important role of MNT in mammary epithelium development and tumorigenesis. These mice carry a floxed Mnt gene and a Cre transgene under the control of the mouse mammary tumor virus (MMTV), which drives Cre expression in mammary epithelium. The conditional deletion of Mnt in this model led to the formation of an adenocarcinoma with a tumor latency of 6–20 months, similar to the results of Myc overexpression in mammary epithelium. Indeed, the mRNA expression patterns of mammary tumors resulting from Mnt deletion or Myc overexpression were very similar [27,52].
The conditional deletion of MNT in T-cells caused an increase in both proliferation and apoptosis. This was accompanied by tumor formation with a long latency, the disruption of T-cell development, and the enlargement of the secondary lymphoid organs. There was a modest increase in the expression of Cdk4 and the cyclins D2, E1, A, and B1 and a slight downregulation of Bcl-2 and Bcl-xL. The polarized differentiation of CD4+ T cells into Th1 (T-helper cell type 1) caused inflammation and, consequently, predisposition to T-cell lymphoma [51,63].
Studies in Drosophila melanogaster also show similar MYC–MNT antagonism to the one found in mouse models and in humans. Indeed, dMnt and dMyc have opposing activities in cell growth in vivo [23,24]. There is an important overlap between the dMyc, dMnt, and dMax DNA binding regions, revealing shared target genes. For instance, dMnt antagonized the growth stimulatory effects of dMyc by downregulating pre-rRNA synthesis [64,65]. In this model, dMnt overexpression rescued the viability and cell growth defects caused by dMyc deletion.
However, both in mice [52] and in Drosophila [24], the overlap between Myc- and Mnt-regulated genes is only partial. This has been attributed to at least two facts: (i) the sequence of the bHLH domains and thus the affinities for DNA are not identical, and (ii) whereas Myc can only bind to Max, Mnt may form at least three different complexes, as discussed in Section 4.
Another proof of MNT and MYC antagonism is the response to hypoxia, a common feature in solid tumors. During hypoxia there is an increase in HIF-1α and HIF-2α, which, in turn, induce the expression of the microRNA miR-210. Since miR-210 downregulates MNT, its levels are lower in hypoxia. Furthermore, MNT downregulation has caused a switch to MYC–MAX dimers and the activation of MYC-target genes in several models. Consequently, tumoral cells can override cell cycle arrest and apoptosis by releasing MNT–MYC antagonism [46,66,67,68]. Hypoxia has an important impact on the cells, which is in part because of the impairment of the circadian rhythm [69]. Apart from the roles of HIF-1α and mTOR in the regulation of the transcriptional clock program, the disturbed MYC/MAX/MNT balance can lead to changes in the expression of circadian rhythm genes. In fact, MNT–MAX were recently described to bind to the PER2, CRY1, and CRY2 promoters [70].
MNT–MYC counteraction was also shown in a myeloid differentiation model described by Kapoor and colleagues [30]. Treating HL60 cells with differentiating agents, including all-trans retinoic acid, led to a decrease in E6AP and subsequently to a loss of E6AP-mediated degradation of MNT. In this model, increased levels of MNT antagonized MYC and induced cell cycle arrest followed by myeloid differentiation.
Niu et al. [71] described that MYC induced miR-378a-3p, which, in turn, down-regulated MNT in Burkitt’s lymphoma. MNT reduction releases MNT–MYC antagonism and enables the MYC-driven transformation process.
Altogether, these data support the role of MNT as a MYC antagonist and show how MNT restricts the pro-proliferative activities of MYC.
7.2. MNT as MYC Cooperator
In contrast to the data presented above, MNT can also act as an oncogene cooperating with Myc as: (a) T-cell-specific homozygous Mnt deletion prevented thymic lymphoma development in mice overexpressing the Myc protein in T cells; (b) Mnt heterozygosity slowed Myc-driven tumorigenesis; (c) Mnt deletion in homozygosis impairs lymphomagenesis in Eμ-Myc mice.
The pro-tumorigenic role of MNT is supported by its pro-survival functions, which seem necessary for sustaining MYC activities. In the study of Link et al., Mnt was necessary for Myc-driven thymic lymphomagenesis in mouse models [72]. Mnt knockout together with Myc overexpression in the thymocytes resulted in higher apoptotic levels than in cells with Mnt knockout or with Myc overexpression alone. This was not related to increased p53 but rather to enhanced levels of reactive oxygen species (ROS). Mnt deficient thymocytes were highly sensitive to the inhibition of several ROS detoxification systems. This effect on survival became worse when MYC levels increased, as shown by using models from no MYC to endogenous levels and further to ectopic high MYC expression [73], which is in line with the fact that that MYC-driven proliferation and apoptosis are triggered by certain MYC thresholds. Consequently, MYC levels influence MYC outputs; thus, they determine the differential need for MNT [73].
The pro-survival functions of MNT and its counteracting role on MYC were also described in the response of T-cells to OX40, a T-cell costimulatory molecule from the tumor-necrosis receptor ligand family [74]. This ligand produces an increase in Myc but also in Mxd4 and Mnt. Myc drives T-cell proliferation while Mxd4 and Mnt evade cell death inflicted by Myc excess.
Another study showed that Mnt heterozygosity slows down Myc-driven B and T-lymphomagenesis in two Myc transgenic models, vavP-MYC10 mice, and Eμ-myc mice [75]. However, the exact mechanism was not defined, as the authors did not find changes in the preleukaemic populations, cell size, cell cycle, apoptosis, or senescence of the murine lymphoma cells. In a second study [76], Mnt was deleted in homozygosis using two Mnt conditional knockdown models: the Mntfl/fl;Eμ-Myc/Rag1Cre and Mntfl/fl;Eμ-Myc/CreERT2 mice, to avoid the early embryonic lethality of homozygous Mnt deletion. In the Mntfl/fl;Eμ-Myc/Rag1Cre model, the mice carried floxed Mnt alleles and the Rag1Cre35 transgene, which allowed the expression of the Cre recombinase in lymphoid progenitor cells only. In the Mntfl/fl;Eμ-Myc/CreERT2 model, the CreERT2 transgene encoded a Cre recombinase fused to the modified hormone-binding domain of the estrogen receptor in the Rosa26 locus. Thus, Cre recombinase expression can be induced by 4-hydroxy(4OH)-tamoxifen. Both the Mntfl/fl;Eμ-Myc/Rag1Cre and Mntfl/fl;Eμ-Myc/CreERT2 models showed that Mnt loss impairs the characteristic Myc-driven lymphomagenesis of Eμ-Myc mice, mainly affecting the premalignant early B lineage cell proliferation. Mnt deletion also impaired normal lymphopoiesis in wild-type mice, suggesting that MNT has an important role in B-cell development. MNT anti-apoptotic functions must be key to compensate for increases in MYC levels during lymphopoiesis. In MYC-driven B lymphomagenesis, the IL-7 receptor signaling up-regulates both Mnt and Myc, and Mnt suppresses apoptosis by repressing Bim thus facilitating MYC activity.
8. MAX-Independent Roles of MNT
MAX is an essential protein that supports the functions of the proximal MYC network, as it is a shared partner between the MYC and MXD proteins. Little is known about the function of these proteins in the absence of MAX. However, MAX is deleted in some cancers of neuroendocrine origin, such as pheochromocytomas, paragangliomas, gastrointestinal stromal tumors, and small cell lung cancer [77,78,79,80,81]. This raises the question of how MYC and MXD proteins can conduct their functions without MAX.
We studied the MAX-independent functions of MNT using UR61 cells, which derive from the rat pheochromocytoma PC12 cell line. PC12 cells express a truncated Max protein that lacks helix 2, the leucine zipper, and the C-terminal region. Thus, Max in PC12 cannot homo- or heterodimerize, nor repress transcription [82]. In this model, we observed that Mnt expression is higher in the absence of Max and that it distributes in both the nucleus and the cytoplasm, in contrast to MAX-expressing cells, in which MNT is mostly nuclear [1,2]. This is due to the negative regulation of Mnt–Max dimers over the Mnt promoter. Thus, in the absence of Max, Mnt cannot autoregulate its expression and the excess of Mnt spreads towards the cytoplasm. Max re-expression led to a decrease of Mnt levels, as Mnt–Max dimers repress the Mnt promoter. Interestingly, Mnt was required for sustaining cell proliferation even in the absence of Max and regulated gene expression, especially DNA damage and cell cycle genes, in a Max-independent way. We confirmed Mnt–Mlx heterodimerization and Mnt homodimerization in UR61 cells. The latter had only been shown in yeast two-hybrid assays and in vitro using recombinant proteins [1,2]. Chromatin immunoprecipitation (ChIP)-qPCR and ChIP-seq revealed that Mnt binds to the DNA on E-boxes in the absence of Max. We also observed Mnt binding to fork head factors sites, in accordance with a study on the coordinated regulation of MNT and FOXO of some cell cycle control genes [83]. Some of the Mnt direct target genes that we detected were BIRC5/Survivin, CDK1, BRCA1, ERCC6, and FBXO32, the latter of which was previously described [83]. Despite this, we believe that in the absence of MAX, MNT binding to the DNA is weaker, as the overall results of our ChIP-seq in Max-deficient cells was low compared to other available ChIP-seq data for MNT in MAX-expressing cells (ENCODE project) [35]. This is in line with results in wild-type and knockout Max B-cells, where the genomic occupancy of Mnt decreases in the absence of Max, although it does not disappear, as is the case for Myc [84].
As described above, MNT can form dimers with MLX, which, in turn, interact with MLXIP and MLXIPL. These complexes play an important role in the regulation of the genes involved in glycolytic and lipogenic metabolism by binding to ChoRE elements, which are composed of two E-boxes separated by five nucleotides (Figure 3) [85]. Thus, it is conceivable that MNT impinges on metabolism by modifying the levels of MLX available for binding to MLXIP or MLXIPL.
9. Concluding Remarks and Therapeutical Perspectives
MYC has been one main focus of cancer research since it was identified in 1977–1979 as the oncogene captured by an avian oncogenic retrovirus [86,87,88] and was later found deregulated in 70% of human tumors [89]. However, in recent years, the interest in understanding the entire proximal MYC network has grown. The studies mentioned in this review place MNT in a key position as a MYC modulator and demonstrate its diverse and essential roles in cell biology (summarized in Figure 6).
The functions of MNT are generally connected to MYC. Yet, studies in MYC knockout cells raised the question of whether MYC is a MNT antagonist rather than MNT a MYC antagonist [22]. Mnt knockdown in Myc-deficient rat fibroblasts showed that Mnt also conducts pro-survival functions in the absence of Myc [61]. Thus, the most important role of MYC might be to relieve MNT-mediated repression rather than activating transcription. In this scenario, MNT functions would remain unaffected by the presence or absence of MYC, while excessive MYC levels would impact MNT by impairing its transcriptional repressing activity.
The dual role of MNT in cancer, either as a MYC antagonist and or as a MYC cooperator, offers the possibility of using MNT as a therapeutical target. For instance, in MYC-driven lymphomas, tumor cells depend on MNT to survive. Thus, drugs that inhibit MNT activities (e.g., impairing critical interactions) would increase the efficacy of current therapies by enhancing apoptosis [76]. One possibility would be to test the MYC:MAX inhibitors Omomyc, 10058-F4, 10074-G5, Mad, or MYCMI-6, 11, and 14 on MNT. Some of these, such as Omomyc or Mad peptides, preferentially bind to MAX and not to MYC. Hence, they might be used as MNT inhibitors by impairing MNT–MAX interaction. However, this possibility has not been formally addressed yet [89,90,91,92,93]. In addition, as MNT downregulation stimulates NF-κB signaling through the interaction with REL (Figure 4B), NF-κB inhibitors could be used in tumors with MNT loss, such as in CTCL (Table 1). Further research will uncover new roles of MNT in cell biology and will provide strategies for its use as a clinical target.
Author Contributions
Writing, figures, and tables, J.L.-P. and J.L. review and expertise, M.A.-H. All authors have read and agreed to the published version of the manuscript.
Funding
J.L.-P. was supported by a postdoctoral scholarship from the Radiumhemmet Research Funds, Stockholm. M.A.-H. was supported by grants from the Swedish Cancer Society, the Swedish Childhood Cancer Fund, the Swedish Research Council, Radiumhemmet Research Funds, and Karolinska Institutet, and J.L. was supported by grant SAF2017-88026-R from Agencia Estatal de Investigación, from the Spanish Government.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hurlin P.J., Quéva C., Eisenman R.N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 2.Meroni G., Reymond A., Alcalay M., Borsani G., Tanigami A., Tonlorenzi R., Lo Nigro C., Messali S., Zollo M., Ledbetter D.H., et al. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer D.E., Lawrence Q.A., Eisenman R.N. Mad-max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 4.Grzenda A., Lomberk G., Zhang J.S., Urrutia R. Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta Gene Regul. Mech. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raffeiner P., Hart J.R., García-Caballero D., Bar-Peled L., Weinberg M.S., Vogt P.K. An MXD1-derived repressor peptide identifies noncoding mediators of MYC-driven cell proliferation. Proc. Natl. Acad. Sci. USA. 2020;117:6571–6579. doi: 10.1073/pnas.1921786117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurlin P.J., Steingrimsson E., Copeland N.G., Jenkins N.A., Eisenman R.N. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA- binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rikin A., Evans T. The Tbx/bHlH Transcription Factor mga regulates gata4 and Organogenesis. Dev. Dyn. 2010;239:535–547. doi: 10.1002/dvdy.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa H., Ishiguro K., Gaubatz S., Livingston D.M., Nakatani Y. A Complex with Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 9.Mathsyaraja H., Catchpole J., Eastwood E., Babaeva E., Geuenich M., Cheng P.F., Freie B., Ayers J., Yu M., Wu N., et al. Loss of MGA mediated Polycomb repression promotes tumor progression and invasiveness. Elife. 2021;28:e64212. doi: 10.7554/eLife.64212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G., Hurlin P.J. MNT and Emerging Concepts of MNT-MYC Antagonism. Genes. 2017;8:83. doi: 10.3390/genes8020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyo-oka K., Hirotsune S., Gambello M.J., Zhou Z., Olson L., Rosenfeld M.G., Eisenman R., Hurlin P., Wynshaw-Boris A. Loss of the Max-interacting protein Mnt in mice results in decreased viability, defective embryonic growth and craniofacial defects: Relevance to Miller-Dieker syndrome. Hum. Mol. Genet. 2004;13:1057–1067. doi: 10.1093/hmg/ddh116. [DOI] [PubMed] [Google Scholar]
- 12.Foley K.P., McArthur G.A., Quéva C., Hurlin P.J., Soriano P., Eisenman R.N. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreiber-Agus N., Meng Y., Hoang T., Hou H., Chen K., Greenberg R., Cordon-Cardo C., Lee H.-W., DePinho R.A. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 14.Queva C., McArthur G.A., Iritani B.M., Eisenman R.N. Targeted Deletion of the S-Phase-Specific Myc Antagonist Mad3 Sensitizes Neuronal and Lymphoid Cells to radiation-induced apoptosis. Mol. Cell. Biol. 2001;21:703–712. doi: 10.1128/MCB.21.3.703-712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diolaiti D., McFerrin L., Carroll P.A., Eisenman R.N. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta. 2015;1849:484–500. doi: 10.1016/j.bbagrm.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaub F.X., Dhankani V., Berger A.C., Trivedi M., Richardson A.B., Shaw R., Zhao W., Zhang X., Ventura A., Liu Y., et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300.e2. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boonk S.E., Zoutman W.H., Marie-Cardine A., van der Fits L., Out-Luiting J.J., Mitchell T.J., Tosi I., Morris S.L., Moriarty B., Booken N., et al. Evaluation of Immunophenotypic and Molecular Biomarkers for Sézary Syndrome Using Standard Operating Procedures: A Multicenter Study of 59 Patients. J. Investig. Dermatol. 2016;136:1364–1372. doi: 10.1016/j.jid.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 18.Van Doorn R., Dijkman R., Vermeer M.H., Out-Luiting J.J., Van Der Raaij-Helmer E.M.H., Willemze R., Tensen C.P. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sézary syndrome identified by gene expression analysis. Cancer Res. 2004;64:5578–5586. doi: 10.1158/0008-5472.CAN-04-1253. [DOI] [PubMed] [Google Scholar]
- 19.Vermeer M.H., Van Doorn R., Dijkman R., Mao X., Whittaker S., Van Voorst Vader P.C., Gerritsen M.J.P., Geerts M.L., Gellrich S., Söderberg O., et al. Novel and highly recurrent chromosomal alterations in Sézary Syndrome. Cancer Res. 2008;68:2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 20.Guo X., Pan L., Zhang X., Suo X., Niu Z., Zhang J., Wang F., Dong Z., Da W., Ohno R. Expression and mutation analysis of genes that encode the Myc antagonists Mad1, Mxi1 and Rox in acute leukaemia. Leuk. Lymphoma. 2007;48:1200–1207. doi: 10.1080/10428190701342018. [DOI] [PubMed] [Google Scholar]
- 21.Cvekl A., Zavadil J., Birshtein B.K., Grotzer M.A., Cvekl A. Analysis of transcripts from 17p13.3 in medulloblastoma suggests ROX/MNT as a potential tumour suppressor gene. Eur. J. Cancer. 2004;40:2525–2532. doi: 10.1016/j.ejca.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Wahlström T., Henriksson M. Mnt Takes Control as Key Regulator of the Myc/Max/Mxd Network. Adv. Cancer Res. 2007;97:61–80. doi: 10.1016/S0065-230X(06)97003-1. [DOI] [PubMed] [Google Scholar]
- 23.Loo L.W.M., Secombe J., Little J.T., Leni-Sue Carlos L.-S., Yost C., Cheng P.-F., Flynn E.M., Edgar B.A., Eisenman R.N. The Transcriptional Repressor dMnt Is a Regulator of Growth in Drosophila melanogaster. Mol. Cell. Biol. 2005;25:7078–7091. doi: 10.1128/MCB.25.16.7078-7091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orian A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McFerrin L.G., Atchley W.R. Evolution of the Max and Mlx networks in animals. Genome Biol. Evol. 2011;3:915–937. doi: 10.1093/gbe/evr082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D.W., Llop J.R., Farrell S.F., Yuan J., Stolzenburg L.R., Samuelson A.V. The Caenorhabditis elegans Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals. PLoS Genet. 2014;10:e1004278. doi: 10.1371/journal.pgen.1004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurlin P.J., Zhou Z., Toyo-oka K., Ota S., Walker W.L., Hirotsune S., Wynshaw-boris A. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J. 2003;22:4584–4596. doi: 10.1093/emboj/cdg442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A., Depinho R.A. An Amino-Terminal Domain of Mxi1 Mediates Anti-Myc Oncogenic Activity and Interacts with a Homolog of the Yeast Transcriptional Repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 29.Kay B.K., Williamson M.P., Sudol M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. doi: 10.1096/fasebj.14.2.231. [DOI] [PubMed] [Google Scholar]
- 30.Kapoor I., Kanaujiya J., Kumar Y., Thota J.R., Bhatt M.L.B., Chattopadhyay N., Sanyal S., Trivedi A.K. Proteomic discovery of MNT as a novel interacting partner of E3 ubiquitin ligase E6AP and a key mediator of myeloid differentiation. Oncotarget. 2016;7:7640–7656. doi: 10.18632/oncotarget.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popov N., Wahlström T., Hurlin P.J., Henriksson M. Mnt transcriptional repressor is functionally regulated during cell cycle progression. Oncogene. 2005;24:8326–8337. doi: 10.1038/sj.onc.1208961. [DOI] [PubMed] [Google Scholar]
- 32.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juergens K., Rust B., Pieler T., Henningfeld K.A. Isolation and comparative expression analysis of the Myc-regulatory proteins Mad1, Mad3, and Mnt during Xenopus development. Dev. Dyn. 2005;233:1554–1559. doi: 10.1002/dvdy.20470. [DOI] [PubMed] [Google Scholar]
- 34.Sadaghiani B., Thiébaud C.H. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- 35.Lafita-Navarro M.C., Liaño-Pons J., Quintanilla A., Varela I., Blanco R., Ourique F., Bretones G., Aresti J., Molina E., Carroll P., et al. The MNT transcription factor autoregulates its expression and supports proliferation in MYC-associated factor X (MAX)-deficient cells. J. Biol. Chem. 2020;295:2001–2017. doi: 10.1074/jbc.RA119.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meroni G., Cairo S., Merla G., Messali S., Brent R., Ballabio A., Reymond A. Mlx, a new Max-like bHLHZip family member: The center stage of a novel transcription factors regulatory pathway? Oncogene. 2000;19:3266–3277. doi: 10.1038/sj.onc.1203634. [DOI] [PubMed] [Google Scholar]
- 37.Conacci-Sorrell M., McFerrin L., Eisenman R.N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzel T., Lavinsky R.M., Mullen T., Soderstrom M., Lahertyll C.D., Torchia J., Yang W., Branl G., Ngo S.D., Davie J.R., et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 39.Laherty C.D., Yang W., Sun J., Davie J.R., Seto E., Eisenman R.N. Histone Deacetylases associated with the mSin3 Corepressor mediate Mad Transcriptional Repression. Cell. 1997;89:349–356. doi: 10.1016/S0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 40.Koskinen P.J., Ayer D.E., Eisenman R.N. Ras Cotransformation by Mad Is Mediated by Multiple Interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 41.Liaño-Pons J., Lafita-Navarro M.C., García-Gaipo L., Colomer C., Rodríguez J., von Kriegsheim A., Hurlin P.J., Delgado M.D., Bigas A., Espinosa M.L., et al. A novel role of MNT as a negative regulator of REL and the NF-κB pathway. Oncogenesis. 2021;10:1–14. doi: 10.1038/s41389-020-00298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayden M.S., Ghosh S. Shared Principles in NF-κB Signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Kanarek N., London N., Schueler-Furman O., Ben-Neriah Y. Ubiquitination and Degradation of the Inhibitors of NF-kB. Cold Spring Harb. Perspect. Biol. 2010;2:1–16. doi: 10.1101/cshperspect.a000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun S.C., Ganchi P.A., Ballard D.W., Greene W.C. NF-kappa B controls expression of inhibitor I kappa B alpha: Evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 45.Yang H., Liu T., Wang J., Li T.W.H., Fan W., Peng H., Krishnan A., Gores G.J., Mato J.M., Lu S.C. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans‡. Hepatology. 2016;64:439–455. doi: 10.1002/hep.28541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H., Li T.W.H., Ko K.S., Xia M., Lu S.C. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49:860–870. doi: 10.1002/hep.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerrato A., Merolla F., Morra F., Celetti A. CCDC6: The identity of a protein known to be partner in fusion. Int. J. Cancer. 2018;142:1300–1308. doi: 10.1002/ijc.31106. [DOI] [PubMed] [Google Scholar]
- 48.Van den Bergh F., Sabina R.L. Characterization of human AMP deaminase 2 (AMPD2) gene expression reveals alternative transcripts encoding variable N-terminal extensions of isoform L. Biochem. J. 1995;312:401–410. doi: 10.1042/bj3120401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon G., Pan H., Yang D., Rosen B.P., Jashari T., Verma N., Pulecio J., Caspi I., Lee K., Stransky S., et al. QSER1 protects DNA methylation valleys from de novo methylation. Science. 2021;372:eabd0875. doi: 10.1126/science.abd0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockel B., Kopec K.O., Lupas A.N., Baumeister W. Structure and function of tripeptidyl peptidase II, a giant cytosolic protease. Biochim. Biophys. Acta-Proteins Proteom. 2012;1824:237–245. doi: 10.1016/j.bbapap.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Dezfouli S., Bakke A., Huang J., Wynshaw-Boris A., Hurlin P.J. Inflammatory Disease and Lymphomagenesis Caused by Deletion of the Myc Antagonist Mnt in T Cells. Mol. Cell. Biol. 2006;26:2080–2092. doi: 10.1128/MCB.26.6.2080-2092.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyo-oka K., Bowen T.J., Hirotsune S., Li Z., Jain S., Ota S., Lozach L.E., Bassett I.G., Lozach J., Rosenfeld M.G., et al. Mnt-Deficient Mammary Glands Exhibit Impaired Involution and Tumors with Characteristics of Myc Overexpression. Cancer Res. 2006;66:5565–6894. doi: 10.1158/0008-5472.CAN-05-2683. [DOI] [PubMed] [Google Scholar]
- 53.Edelmann J., Holzmann K., Miller F., Winkler D., Buhler A., Zenz T., Bullinger L., Kuhn M.W.M., Gerhardinger A., Bloehdorn J., et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;6:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- 54.Lo Nigro C., Venesio T., Reymond A., Meroni G., Alberici P., Cainarca S., Enrico F., Stack M., Ledbetter D.H., Liscia D.S., et al. The Human ROX Gene: Genomic Structure and Mutation Analysis in Human Breast Tumors. Genomics. 1998;282:275–282. doi: 10.1006/geno.1998.5241. [DOI] [PubMed] [Google Scholar]
- 55.Sommer A., Waha A., Tonn J., Sörensen N., Hurlin P.J., Eisenman R.N., Lüscher B., Pietsch T. Analysis of the MAX-binding protein MNT in human medulloblastomas. Int. J. Cancer. 1999;82:810–816. doi: 10.1002/(SICI)1097-0215(19990909)82:6<810::AID-IJC7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 56.Phillips N., Ziegler M., Saha B., Xynos F. Allelic loss on chromosome 17 in human ovarian cancer. Int. J. Cancer. 1993;54:85–91. doi: 10.1128/MCB.26.6.2080-2092.2006. [DOI] [PubMed] [Google Scholar]
- 57.Saxena A., Clark W.C., Robertson J.T., Ikejiri B., Oldfield E.H., Ali I.U. Erratum: Evidence for the involvement of a potential second tumor suppressor gene on chromosome 17 distinct from p53 in malignant astrocytomas (Cancer Research (December 1, 1992) (6716–6721)) Cancer Res. 1993;53:1472. [PubMed] [Google Scholar]
- 58.Williamson M.P., Elder P.A., Knowles M.A. The spectrum of TP53 mutations in bladder carcinoma. Genes Chromosom. Cancer. 1994;9:108–118. doi: 10.1002/gcc.2870090206. [DOI] [PubMed] [Google Scholar]
- 59.Andreassen Å., Oyjord T., Hovig E., Holm R., Florenes V.A., Nesland J.M., Myklebost O., Hoie J., Bruland S., Borresen A.-L., et al. p53 Abnormalities in Different Subtypes of Human Sarcomas. Cancer Res. 1993;53:468–471. [PubMed] [Google Scholar]
- 60.Hurlin P.J., Zhou Z.Q., Toyo-Oka K., Ota S., Walker W.L., Hirotsune S., Wynshaw-Boris A. Evidence of Mnt-Myc antagonism revealed by Mnt gene deletion. Cell Cycle. 2004;3:97–99. doi: 10.4161/cc.3.2.638. [DOI] [PubMed] [Google Scholar]
- 61.Nilsson J.A., Maclean K.H., Keller U.B., Pendeville H., Baudino T.A., Cleveland J.L. Mnt loss triggers Myc transcription targets, proliferation, apoptosis, and transformation. Mol. Cell. Biol. 2004;24:1560–1569. doi: 10.1128/MCB.24.4.1560-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker W., Zhou Z.Q., Ota S., Wynshaw-Boris A., Hurlin P.J. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J. Cell Biol. 2005;169:405–413. doi: 10.1083/jcb.200411013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Link J.M., Hurlin P.J. The activities of MYC, MNT and the MAX-interactome in lymphocyte proliferation and oncogenesis. Biochim. Biophys. Acta-Gene Regul. Mech. 2015;1849:554–562. doi: 10.1016/j.bbagrm.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Pierce S.B., Yost C., Anderson S.A.R., Flynn E.M., Delrow J., Eisenman R.N. Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 2008;315:303–316. doi: 10.1016/j.ydbio.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pierce S.B., Yost C., Britton J.S., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131:2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- 66.Dang C.V., Kim J., Gao P., Yustein J. The Interplay Between MYC and HIF in the Warburg Effect. Nat. Rev. Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 67.Yang W., Wei J., Guo T., Shen Y., Liu F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Exp. Cell Res. 2014;326:22–35. doi: 10.1016/j.yexcr.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z., Sun H., Dai H., Walsh R.M., Imakura M., Schelter J., Burchard J., Dai X., Chang A.N., Diaz R.L., et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 69.Yu C., Yang S.L., Fang X., Jiang J.X., Sun C.Y., Huang T. Hypoxia disrupts the expression levels of circadian rhythm genes in hepatocellular carcinoma. Mol. Med. Rep. 2015;11:4002–4008. doi: 10.3892/mmr.2015.3199. [DOI] [PubMed] [Google Scholar]
- 70.Blaževitš O., Bolshette N., Vecchio D., Guijarro A., Croci O., Campaner S., Grimaldi B. MYC-associated factor max is a regulator of the circadian clock. Int. J. Mol. Sci. 2020;21:2294. doi: 10.3390/ijms21072294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niu F., Dzikiewicz-Krawczyk A., Koerts J., de Jong D., Wijenberg L., Hernandez M.F., Slezak-Prochazka I., Winkle M., Kooistra W., van der Sluis T., et al. Mir-378a-3p is critical for burkitt lymphoma cell growth. Cancers. 2020;12:3546. doi: 10.3390/cancers12123546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Link J.M., Ota S., Zhou Z., Daniel C.J., Sears R.C., Hurlin P.J. A critical role for Mnt in Myc-driven T-cell proliferation and oncogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:19685–19690. doi: 10.1073/pnas.1206406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murphy D.J., Junttila M.R., Pouyet L., Karnezis A., Shchors K., Bui D.A., Brown-Swigart L., Johnson L., Evan G.I. Distinct Thresholds Govern Myc’s Biological Output In Vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vasilevsky N.A., Ruby C.E., Hurlin P.J., Weinberg A.D. OX40 engagement stabilizes Mxd4 and Mnt protein levels in antigen-stimulated T cells leading to an increase in cell survival. Eur. J. Immunol. 2011;41:1024–1034. doi: 10.1002/eji.201040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell K.J., Vandenberg C.J., Anstee N.S., Hurlin P.J., Cory S. Mnt modulates Myc-driven lymphomagenesis. Cell Death Differ. 2017;24:2117–2126. doi: 10.1038/cdd.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen H.V., Vandenberg C.J., Ng A.P., Robati M.R., Anstee N.S., Rimes J., Hawkins E.D., Cory S. Development and survival of MYC-driven lymphomas require the MYC antagonist MNT to curb MYC-induced apoptosis. Blood. 2020;135:1019–1031. doi: 10.1182/blood.2019003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burnichon N., Cascoń A., Schiavi F., Morales N.P., Comino-Méndez I., Abermil N., Inglada-Pérez L., De Cubas A.A., Amar L., Barontini M., et al. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin. Cancer Res. 2012;18:2828–2837. doi: 10.1158/1078-0432.CCR-12-0160. [DOI] [PubMed] [Google Scholar]
- 78.Comino-Méndez I., Gracia-Aznárez F.J., Schiavi F., Landa I., Leandro-García L.J., Letón R., Honrado E., Ramos-Medina R., Caronia D., Pita G., et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 79.Pantaleo M.A., Urbini M., Indio V., Ravegnini G., Nannini M., De Luca M., Tarantino G., Angelini S., Gronchi A., Vincenzi B., et al. Genome-Wide Analysis Identifies MEN1 and MAX Mutations and a Neuroendocrine-Like Molecular Heterogeneity in Quadruple WT GIST. Mol. Cancer Res. 2017;15:553–562. doi: 10.1158/1541-7786.MCR-16-0376. [DOI] [PubMed] [Google Scholar]
- 80.Robledo M. MAX and MYC: A Heritable Breakup. Cancer Res. 2012;72:1–7. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 81.Romero O.A., Torres-Diz M., Pros E., Savola S., Gomez A., Moran S., Saez C., Iwakawa R., Villanueva A., Montuenga L.M., et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2014;4:292–303. doi: 10.1158/2159-8290.CD-13-0799. [DOI] [PubMed] [Google Scholar]
- 82.Hopewell R., Ziff E.B. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol. Cell. Biol. 1995;15:3470–3478. doi: 10.1128/MCB.15.7.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terragni J., Nayak G., Banerjee S., Medrano J., Graham J.R., Brennan J.F., Sepulveda S., Cooper G.M. The E-box Binding Factors Max/Mnt, MITF, and USF1 Act Coordinately with FoxO to Regulate Expression of Proapoptotic and Cell Cycle Control Genes by Phosphatidylinositol 3-Kinase/ Akt/Glycogen Synthase Kinase 3 Signaling. J. Biol. Chem. 2011;286:36215–36227. doi: 10.1074/jbc.M111.246116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mathsyaraja H., Freie B., Cheng P.F., Babaeva E., Catchpole J.T., Janssens D., Henikoff S., Eisenman R.N. Max deletion destabilizes MYC protein and abrogates Eµ-Myc lymphomagenesis. Genes Dev. 2019:1252–1264. doi: 10.1101/gad.325878.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carroll P.A., Diolaiti D., McFerrin L., Gu H., Djukovic D., Du J., Cheng P.F., Anderson S., Ulrich M., Hurley J.B., et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer Cell. 2015;27:271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheiness D., Fanshier L., Bishop J.M. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J. Virol. 1978;28:600–610. doi: 10.1128/jvi.28.2.600-610.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sheiness D., Bishop J.M. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J. Virol. 1979;31:514–521. doi: 10.1128/jvi.31.2.514-521.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duesberg P.H., Bister K., Vogt P.K. The RNA of avian acute leukemia virus MC29. Proc. Natl. Acad. Sci. USA. 1977;74:4320–4324. doi: 10.1073/pnas.74.10.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Massó-Vallés D., Soucek L. Blocking Myc to Treat Cancer: Reflecting on Two Decades of Omomyc. Cells. 2020;9:883. doi: 10.3390/cells9040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vita M., Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 91.Castell A., Yan Q., Fawkner K., Hydbring P., Zhang F., Verschut V., Franco M., Zakaria S.M., Bazzar W., Goodwin J., et al. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci. Rep. 2018;8:10064. doi: 10.1038/s41598-018-28107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Demma M.J., Hohn M.J., Sun A., Mapelli C., Hall B., Walji A., O’Neil J. Inhibition of Myc transcriptional activity by a mini-protein based upon Mxd1. FEBS Lett. 2020;594:1467–1476. doi: 10.1002/1873-3468.13759. [DOI] [PubMed] [Google Scholar]
- 93.Demma M.J., Mapelli C., Sun A., Bodea S., Ruprecht B., Javaid S., Wiswell D., Muise E., Chen S., Zelina J., et al. Omomyc Reveals New Mechanisms To Inhibit the MYC Oncogene. Mol. Cell. Biol. 2019;39:1–27. doi: 10.1128/MCB.00248-19. [DOI] [PMC free article] [PubMed] [Google Scholar]