Abstract
Huntington’s disease (HD) is caused by expansion of polyglutamine repeats in the protein huntingtin, which affects the corpus striatum of the brain. The polyglutamine repeats in mutant huntingtin cause its aggregation and elicit toxicity by affecting several cellular processes, which include dysregulated organellar stress responses. The Golgi apparatus not only plays key roles in the transport, processing, and targeting of proteins, but also functions as a sensor of stress, signaling through the Golgi stress response. Unlike the endoplasmic reticulum (ER) stress response, the Golgi stress response is relatively unexplored. This review focuses on the molecular mechanisms underlying the Golgi stress response and its intersection with cysteine metabolism in HD.
Keywords: Golgi apparatus, Huntington’s disease, cysteine, transsulfuration, Golgi stress response, integrated stress response
1. Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease which profoundly affects the corpus striatum of the brain; it results from expansion of polyglutamine repeats in the protein huntingtin [1]. Mutant huntingtin (mHtt) aggregates and affects cellular processes in multiple ways [2]. mHtt affects basic neuronal processes such as transcription, translation, nuclear-cytoplasmic transport, redox homeostasis, mitochondrial function and amino acid metabolism in addition to a myriad of physiological processes [3,4,5,6,7].
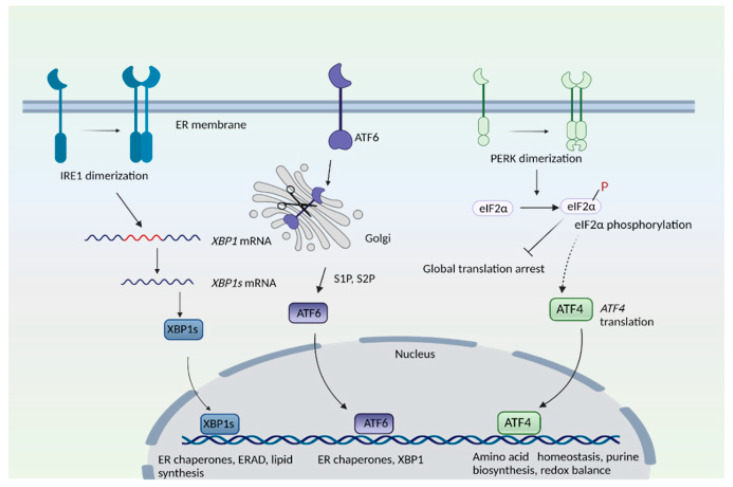
HD has also been linked to impaired stress responses involving redox homeostasis and endoplasmic stress response [5,7]. In addition to essential functions in cellular function, organelles serve important roles as sensors of stress and as hubs for signaling pathways. For instance, the endoplasmic reticulum (ER) plays central roles in protein folding, post-translational modifications, quality control of proteins and Ca2+ handling, among many other functions [8,9,10,11]. During ER stress—a state of functional imbalance—adaptive and restorative programs such as the unfolded protein response (UPR) and ER-associated protein degradation (ERAD), or autophagy, come into play [12,13]. One stimulus that triggers the ER stress response is the accumulation of unfolded or misfolded proteins in the ER lumen. Three arms exist in the ER stress response: the protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1) pathways, where each of the sensor proteins is a membrane protein (Figure 1). In the PERK arm, during stress, PERK dissociates from the chaperone protein, binding immunoglobulin protein/glucose-regulated protein 78 (BiP/GRP78), and undergoes dimerization and phosphorylation. PERK, (a component of the integrated stress response) then phosphorylates the eukaryotic translation initiation factor 2 subunit −α (eIF2α), which results in global translational arrest. Under these conditions only certain mRNAs such as those encoding activating transcription factor 4 (ATF4) are translated, in order to maintain functions important for cell survival. ATF4 regulates amino acid homeostasis, purine metabolism, response to oxidative stress, autophagy and apoptosis.
Figure 1.
The endoplasmic reticulum (ER) stress response. The mammalian ER stress response consists of three arms: the inositol-requiring enzyme 1 (IRE1), protein kinase R (PKR)-like ER kinase (PERK), and activating transcription factor 6 (ATF6) pathways. IRE1 senses ER stress, which leads to its dimerization and to the activation of its endonuclease role, that is, to splice a specific intron from the mRNA of X-box binding protein 1, XBP1 to create XBP1s. T XBP1s protein translocates to the nucleus and transactivates its target genes. In the ATF6 arm, binding immunoglobulin protein/glucose-regulated protein 78 (BiP/GRP78) dissociates from ATF6 when unfolded proteins accumulate in the ER. ATF6 translocates to the Golgi complex, where it undergoes proteolytic cleavage by site 1 and site 2 proteases (S1P and S2P). The N-terminal cytosolic fragment of ATF6 migrates to the nucleus and induces expression of target genes. In the PERK arm, dissociation of BiP causes its dimerization and autophosphorylation. PERK then phosphorylates eukaryotic initiation factor 2α (eIF2α), which results in global translational arrest. Under these conditions, only certain mRNAs such as ATF4 are translated, in order to maintain cellular functions during stress.
In the inositol-requiring enzyme (IRE) branch, the ER-resident IRE1 senses unfolded proteins or lipid disequilibrium and undergoes dimerization and autophosphorylation, activating IRE1′s cytosolic endonuclease domain, which then splices a specific intron from the mRNA of X-box binding protein 1u, XBP1u to create XBP1s. The XBP1s protein translocates to the nucleus and transactivates genes involved in protein degradation, protein folding, and lipid metabolism [14,15]. The third arm of the UPR consists of ATF6, an ER transmembrane protein that translocates to the Golgi when activated. During ER stress, when unfolded proteins accumulate, BiP/GRP78 dissociates from ATF6 to cause translocation of ATF6 into the Golgi. In the Golgi, site 1 protease (S1P) and site 2 protease (S2P) cleave ATF6 [16]. The N-terminal region of ATF6 functions as a transcription factor and stimulates expression of target genes, such as protein disulfide isomerase (PDI), XBP1, and C/EBP Homologous Protein (CHOP) [16,17]. When proteins cannot be repaired or folded back into their functional configurations, they are targeted for degradation by ERAD [18]. When the repair capacity of ERAD is crossed, portions of the ER can be specifically targeted for degradation through autophagy (ER-phagy) [19]. Recently, we elucidated the involvement of signaling pathways modulated by the Golgi apparatus in HD [20,21]. This review focuses on the involvement of the Golgi and stress signaling coordinated by this organelle in neurodegenerative states.
2. The Golgi Apparatus
2.1. Organization of the Golgi Apparatus
The Italian anatomist Camillo Golgi was the first to describe the Golgi apparatus in 1898 [22]. He developed the staining protocol for the Golgi, termed the ‘Black Reaction’ (La reazione nera) or Golgi’s staining, which accelerated the study of the brain by facilitating the microscopic visualization of the complexity of the human nervous system [23,24,25]. The Golgi apparatus both processes and sorts lipids and proteins through the secretory pathway. The Golgi complex is organized as a stack of cisternae, with the cis-face receiving cargo from the ER and the trans-face or trans Golgi network (TGN) sorting cargo for export to their respective destinations [26]. The stacks are interconnected by tubular membranes into a continuous structure named the Golgi ribbon, which is a feature of the organelle in mammals. Within cells the Golgi is positioned close to the centrosomes, the primary microtubule-organizing center (MTOC) in dividing cells; vital for the maintenance of cell polarity, this center in turn modulates cell migration and neurite outgrowth [27]. The Golgi undergoes changes in morphology during various cellular processes such as cell cycle progression and stress responses [28]. The Golgi is highly dynamic, and undergoes disassembly during mitosis in early prophase and reassembly in telophase [29,30]. During mitosis in mammals, the Golgi ribbon is disassembled and partitioned into daughter cells. This disassembly also regulates mitotic progression [31].
2.2. The Golgi Stress Response
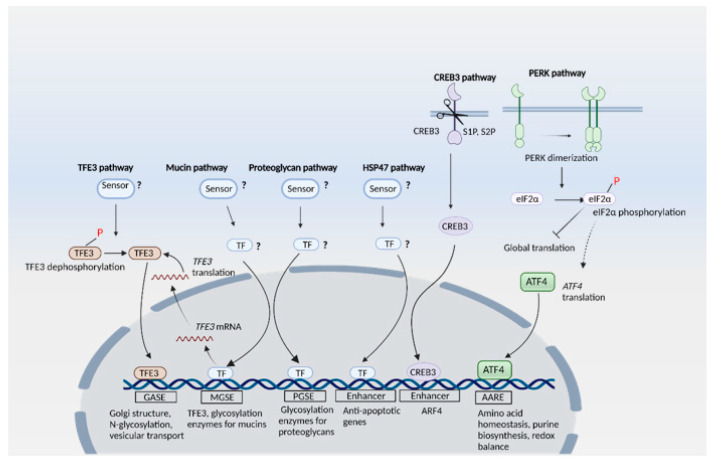
The Golgi plays central roles in glycosylation of proteins and harbors glycosyltransferases, glycosidases, and nucleotide sugar transporters, which orchestrate addition of various sugars that result in a mature glycan [32]. The cargo may also undergo several post-translational modifications including acetylation and phosphorylation, sulfation, methylation or proteolytic cleavage [33]. When the capacity of the Golgi is exceeded, it causes Golgi stress in a manner analogous to ER stress. In order to counter Golgi stress, cells mount signaling cascades which constitute the Golgi stress response [34]. Although the Golgi stress response is not as extensively characterized as ER stress, mounting evidence suggests the involvement of signaling cascades. Several signaling cascades have been reported with sensor proteins and effector proteins, which together elicit the Golgi stress response (Figure 2). These include the TFE3, proteoglycan, CREB3, PERK and HSP47 pathways [21,35,36].
Figure 2.
The Golgi stress response. Pathways involving the transcription factor E3 (TFE3), mucin, proteoglycan, heat shock protein 47 (HSP47), cAMP responsive element binding protein 3 (CREB3) and the protein kinase R (PKR)-like ER kinase (PERK) have been identified. The sensors for TFE3, mucin, proteoglycan and HSP47 have not yet been identified. The transcription factors for the mucin, proteoglycan and HSP47 pathways are also not well characterized. In the TFE3 pathway, dephosphorylation of TFE3 causes its nuclear translocation, where it activates transcription of its target genes by interacting with the Golgi apparatus stress response element (GASE) enhancer element. The mucin pathway, which is activated in response to insufficiency of mucin glycosylation, displays crosstalk with the TFE3 pathway. The mucin-type Golgi stress response element (MGSE) is present on the promoter of the TFE3 gene as well. The proteoglycan pathway involves the ER-localized CREB3 (which functions as a sensor for Golgi stress) translocating from the ER to the Golgi to be cleaved by S1P and S2P proteases. The cytosolic region of the truncated CREB3 migrates to the nucleus and activates transcription of ARF4, leading to apoptosis. In the HSP47 pathway, expression of HSP47 (an ER chaperone involved in collagen folding and maturation) inhibits Golgi stress-induced apoptosis. The PERK pathway acts via the eIF2α/ATF4 axis, however, BiP/GRP78 is not induced when this pathway is activated by Golgi stress.
2.2.1. The TFE3 Pathway
The TFE3 pathway involves the activation of proteins responsible for the maintenance of Golgi functions. TFE3 targets include the Golgi structural proteins (GCP60, Giantin, and GM130) which maintain the structural integrity of the Golgi. GM130 (Golgin95) and Golgin-160 are cis-Golgi-localized harboring coiled-coils, and mediate stacking of Golgi cisternae and vesicular transport by serving as a vesicle tethering factor [37]. GM130 anchors adjacent stacks by interacting with Golgi reassembly and stacking protein of 65 kD (GRASP65) and the p115 protein through its C-terminal and N-domains, respectively [37]. GCP60 (also called acyl-CoA binding domain containing 3, or ACBD3) is a Golgi structural protein associated with Golgi integral membrane protein (Giantin). Overexpressing a dominant negative mutant of GCP60 triggers disassembly of the Golgi and blockage of protein transport from the ER to the Golgi [38]. Other targets of TFE3 include: N-glycosylation enzymes, such as ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (SIAT4A/ST3GAL1), SIAT10, fucosyltransferase 1 (FUT1), Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 (B3GAT2) and UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 (UAP1L1); proteins involved in vesicular transport, such as Syntaxin 3A (STX3A), WD-repeat protein Interacting with phosphoinositide (WIPI)-1alpha (WIPI49/WIP1α) and RAB20 (Ras-related protein Rab-20); and Golgi proteases. Analysis of the promoters of ACBD3 and SIAT4A has revealed the presence of an enhancer element, termed the Golgi apparatus stress response element (GASE) [39]. Two transcription factors, TFE3 and MLX, were reported to bind GASE, which has a consensus sequence of ACGTGGC. Increasing the expression of TFE3 while increasing expression of MLX decreased transcription of genes with the GASE sequence. It has also been reported that Golgi stressors cause dephosphorylation of TFE3 at Ser108 and its nuclear translocation [40]. Similarly, MLX also translocates to the nucleus in response to Golgi stress [41]. Similar to ER stress, Golgi stress may trigger proteosomal degradation, which acts to restore Golgi homeostasis and organelle autoregulation. When Golgi stress persists, Golgi-apparatus related degradation (GARD) may ensue, analogous to ERAD associated with the ER. Golgi stress induces proteasomal degradation of GM130, which causes Golgi dispersal [42]. Other mechanisms proposed for regulation of Golgi tethering factors and morphology include caspase 3 mediated cleavage of GRASP-65 [43].
2.2.2. The Proteoglycan Pathway
The proteoglycan (PG) pathway involves upregulation of glycosylation enzymes when PG glycosylation capacity in the Golgi is suboptimal. Genes encoding these glycosylation enzymes harbor the enhancer elements PGSE-A and PGSE-B (with consensus sequences CCGGGGCGGGGCG and TTTTACAATTGGTC, respectively) in their promoters.
2.2.3. The CREB3 Pathway
In the CREB3 (ATF3) pathway, the ER-resident membrane protein CREB3 senses Golgi stress and translocates from the ER to the Golgi to be cleaved by S1P and S2P proteases. The truncated CREB3 derived from the cytosolic region moves to the nucleus and activates transcription of ARF4 to induce apoptosis [36].
2.2.4. The HSP47 Pathway
The HSP47 pathway, on the other hand, acts to prevent apoptosis induced by Golgi stress by stimulating the expression of the ER-resident chaperone HSP47 to suppress Golgi stress-induced apoptosis [44].
2.2.5. The Mucin Arm
The mucin arm, first proposed in 2019, is activated in response to inadequate mucin-type glycosylation in the Golgi. Mucins are high molecular weight, heavily glycosylated proteins produced by epithelial cells, which often form gel-like structures and are components of mucous. Expression of glycosylation enzymes for mucins such as GALNT5, GALNT8, and GALNT18 has been observed in this case. The mucin-type Golgi stress intersects with the TFE3 pathway as well, by inducing the expression and activation of TFE3. An enhancer element regulating transcriptional induction of TFE3 upon mucin-type Golgi stress was identified and designated as the mucin-type Golgi stress response element, with a consensus sequence ACTTCC (N9) TCCCCA [45].
2.2.6. The PERK Pathway
The PERK pathway, which is also activated during ER stress, has additionally been identified as a pathway activated by the Golgi stressor monensin [21]. However, unlike ER stress, the ER-resident chaperone BiP/GRP78 is not induced during Golgi stress, suggesting that Golgi stress response is a distinct type of stress despite the phosphorylation of PERK in both types of stress response.
3. Golgi Stress Response and Redox Imbalance in Neurodegeneration: Focus on Huntington’s Disease
Accumulating evidence reveals that abnormalities in the structure and function of Golgi apparatus occur in neurodegenerative diseases including Alzheimer’s disease (AD), Amyotrophic lateral sclerosis (ALS), Parkinson’s disease (PD), Huntington’s disease (HD) and Creutzfeldt–Jakob disease [20,46,47,48,49,50,51]. The Golgi has also been reported to be fragmented during viral infection [52]. Depletion of the golgin GM130 has been reported to cause Golgi disruption, Purkinje neuron loss, and ataxia in mice [53]. Golgi fragmentation in dopaminergic neurons in the substantia nigra has been observed in Parkinson’s disease patients [54]. Early studies revealed that the Golgi apparatus may be fragmented in a population of neurons without neurofibrillary tangles (NFTs) [55]. In JNPL3 transgenic mice—which express the P301L mutant of tau, a component of NFTs and paired helical filaments (PHFs)—the Golgi complex was fragmented; however, mitochondria or other membranous organelles appeared normal, indicating that Golgi fragmentation is one of the earliest events that occur during pathogenesis, a finding which has been suggested by other laboratories as well [56,57,58]. Structural deformities in the Golgi complex were also linked to accumulation of phospho-tau in the P301S mouse model of AD [59]. Aging is a major risk factor for neurodegeneration; not surprisingly, increased Golgi fragmentation in neurons was observed with aging. GRASP65 and Golgin-84 were also diminished in the aging mouse brain [60].
In HD, we showed that elevated levels of ACBD3 occurred in cell culture and mouse models as well as human HD [20]. ACBD3 bound to the striatal protein ras homolog enriched in striatum (Rhes), which binds to mutant huntingtin (mHtt) and mediates cell death in HD [61]. More recently, we identified another arm of the Golgi stress signaling pathway in a striatal progenitor cell line model of HD. HD is a neurodegenerative disorder triggered by expansion of CAG repeats (encoding polyglutamine repeats) in the gene encoding huntingtin and which primarily affects the corpus striatum of the brain, manifesting as abnormal involuntary movements along with motor and cognitive deficits [1,2]. mHtt affects multiple cellular processes such as DNA replication and repair, transcription, translation, nucleocytoplasmic trafficking, mitochondrial function and proteostasis, to name a few [4,5,6,62,63,64,65,66].
3.1. Redox Imbalance and Cysteine Metabolism in HD
A hallmark of HD is increased oxidative stress. Oxidative stress occurs when the balance between prooxidant and antioxidant pathways tilts in favor of the former. However, oxidative stress has more recently been defined as a disruption of redox signaling pathways [67]. Elevated oxidative stress is at the heart of several neurodegenerative diseases, as well as other conditions [68,69]. Decreased levels and/or dysregulated metabolism of the antioxidants such as ascorbate (vitamin C), glutathione (GSH) and cysteine, and coenzyme Q10 (CoQ10) have been observed in HD and contribute to disease pathology [70,71,72]. Both biosynthesis and the uptake of cysteine or its oxidized form cystine are compromised in HD, causing elevated oxidative stress [5,73,74]. The activity of the neuronal cysteine transporter EAAT3/EAAC1 is reduced in HD due to decreased trafficking to the cell membrane [75]. Decreased expression of cystathionine γ-lyase (CSE) (the biosynthetic enzyme for cysteine) also occurs in HD as mHtt sequesters specificity protein 1 (SP1), the transcription factor for CSE during basal conditions [71,76,77]. CSE is also regulated by activating transcription factor 4 (ATF4) in response to stress. In HD-affected cells the induction of ATF4 is suboptimal, leading to decreased CSE expression and cysteine biosynthesis [5]. Cysteine is utilized in the biosynthesis of sulfur-containing molecules such as coenzyme A, taurine, lanthionine, homolanthionine, and cystamine [78]. It is also the substrate for production of the gaseous signaling molecule hydrogen sulfide (H2S) [79,80]. H2S signals by sulfhydration or persulfidation, a posttranslational modification which occurs on the –SH group of reactive cysteine residues, leading to formation of –SSH or persulfide groups [81]. Sulfhydration modulates the function of several proteins and signaling cascades, including response to inflammation, mitochondrial bioenergetics and stress responses [82,83]. Both cysteine metabolism and sulfhydration are altered in HD, which contributes to increased protein oxidation [5,71,84].
3.2. Golgi Stress Response and Links to Redox Homeostasis
ATF4, a master regulator of amino acid homeostasis and stress responses, is a central player in the integrated stress response [85,86,87]. It also regulates purine biosynthesis and regulates mTOR function [88]. Furthermore, ATF4 modulates the switch from synthesis of fetal hemoglobin to adult hemoglobin by stimulating transcription of BCL11A, a repressor of γ-globin synthesis [89]. ATF4 harbors a basic leucine-zipper (bZIP) domain and can either form homodimers or heterodimerize with other members of the bZIP family (FOS/JUN, ATF and CCAAT enhancer-binding protein (C/EBP) bZIP transcription factors) to control transcription [90,91]. Nuclear factor erythroid 2-related factor 2 (Nrf2), the master regulator of redox regulation, also forms heterodimers with ATF4 and stimulates the transcription of cytoprotective genes during oxidative stress [92]. ATF4 may have dual functions, modulating either cell survival or cell death, and excessive stimulation of ATF4 signaling may cause cell death [93]. The pro-survival or apoptotic function of ATF4 has been attributed in part to the identity of its heterodimerization partner and the physiologic context; the mechanisms are still being elucidated [94].
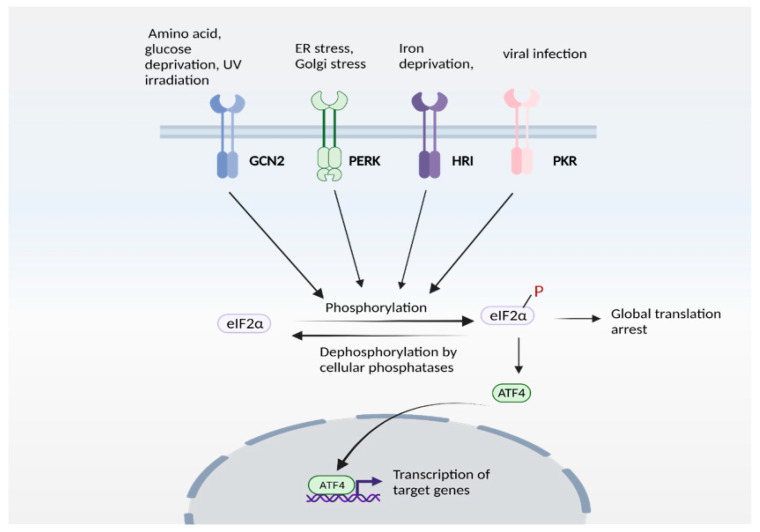
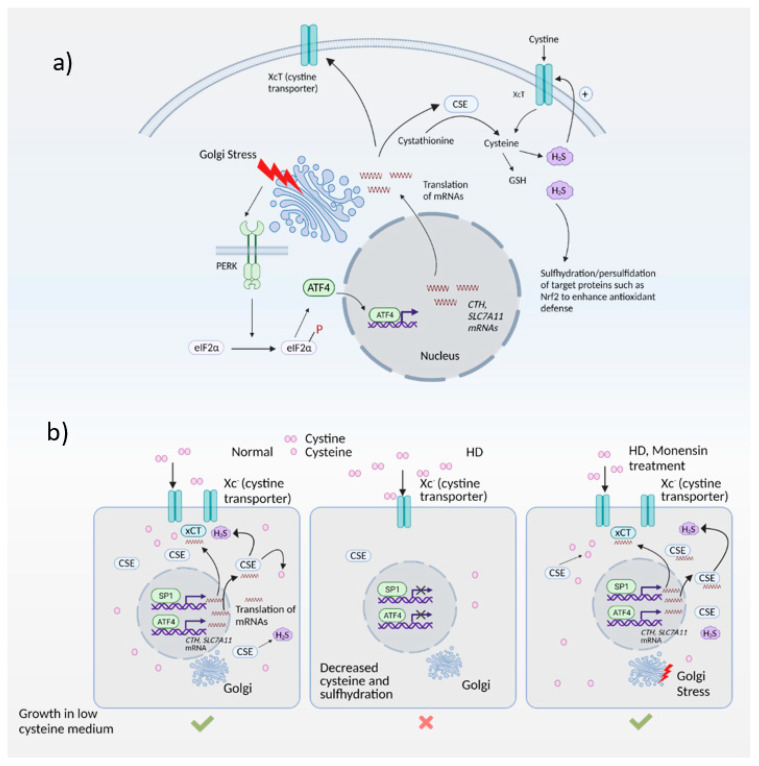
Expression of ATF4 is regulated at the transcriptional as well as the translational level, and is a vital part of the integrated stress response [95,96]. The integrated stress response (Figure 3) is engaged in response to stress stimuli, including but not limited to amino acid and nutrient deprivation, ER stress, mitochondrial stress, iron dysregulation and viral infection. Four kinases, namely general control non-derepressible 2 (GCN2), PKR-like ER kinase (PERK) 19, double-stranded RNA-dependent protein kinase (PKR), and heme-regulated eIF2α kinase (HRI) sense the stress and phosphorylate the eukaryotic initiation 2 α (eIF2α), which abrogates its catalytic activity resulting in global translational arrest [85,97,98,99,100,101]. Under these conditions, only mRNAs responsible for maintenance of cell survival and essential signaling (such as ATF4) are translated. Recently, we showed that the Golgi stressors, monensin and nigericin, activated the integrated stress response by eliciting phosphorylation of PERK, resulting in translation of ATF4 and expression of its targets [21] (Figure 4). Among these targets were enzymes involved in cysteine biosynthesis and uptake, cystathionine γ-lyase (CSE) and SLC7A11, a subunit of the XcT transporter, which imports cystine, the oxidized form of cysteine (Figure 4a).
Figure 3.
The integrated stress response (ISR). ISR is activated by several stress stimuli. In mammals, these are sensed by four kinases: general control non-depressible 2 (GCN2), protein kinase R protein kinase R (PKR)-like ER kinase (PERK), Heme-regulated eIF2α kinase (HRI) and double stranded RNA dependent protein kinase (PKR), each of which respond to a set of stimuli. These kinases undergo autophosphorylation and phosphorylate the eukaryotic initiation factor 2α (eIF2α), which inhibits its catalytic activity and its function of delivering the initiator tRNA to the ribosome, thereby arresting global translation. Under these conditions, only certain mRNAs (such as that encoding ATF4) are translated. ATF4 functions either as a homodimer or heterodimer to transactivate its target genes. The pathway is referred to as ISR because signaling mediated by diverse stress stimuli converge upon a common pathway (eIF2α/ATF4 axis).
Figure 4.
The Golgi stress response and its intersection with redox homeostasis in Huntington’s disease (HD). (a) Golgi stress response in normal cells. Golgi stress activates PERK, which phosphorylates eIF2α to inhibit general protein synthesis. Only mRNAs such as ATF4 are translated. ATF4 regulates amino acid homeostasis and one of the genes induced by ATF4 is CTH (which encodes the biosynthetic enzyme for cysteine, also called CSE). CSE utilizes cysteine to produce the gaseous signaling molecule hydrogen sulfide (H2S). H2S signals by a post-translational modification termed sulfhydration/persulfidation and modulates the activity of target proteins. H2S stimulates cystine uptake by the cystine transporter, leading to increased cysteine levels in cells. ATF4 also regulates expression of SLC7A11 (xCT), a subunit of the cystine transporter, by activating its transcription through heterodimerization with Nrf2, a master regulator of redox homeostasis. (b) Harnessing the Golgi stress response to elicit cytoprotection in HD. Normal cells express CSE and ATF4 during basal conditions and during stress to produce cysteine. Cysteine is also imported into cells via the cystine transporter, Xc-. In HD, both basal expression of CSE (regulated by specificity protein1, SP1) as well as stress-induced expression of CSE and the xCT subunit of the cystine transporter by ATF4 are compromised, causing a cysteine deficit which leads to decreased H2S levels and sulfhydration. When cells are treated with monensin, a Golgi stressor, CSE is induced via the PERK/ATF4 pathway to increase cysteine and H2S levels and mediate cytoprotection.
In the striatal progenitor cell culture models STHdhQ7/Q7 (Q7) and STHdh1117/Q111 (Q111), treatment with monensin rescued cell death associated with cysteine deprivation. Monensin increased the expression of ATF4 in the Q111 cells, which have compromised stress responses. We had shown previously that Q111 cells had decreased expression of CSE and thus could not grow in the absence of cysteine [5,71]. These cells were also compromised in their ability to upregulate ATF4, the transcription factor responsible for induction of CSE during cysteine deprivation. Treatment with monensin rescued growth in cysteine-free media and decreased oxidative stress in a manner dependent on PERK (Figure 4b). Monensin failed to upregulate ATF4 and CSE expression in cells deleted for PERK. Thus, Golgi stress engages the PERK pathway in HD cells. We further showed that mild Golgi stress can be harnessed to elicit cytoprotective effects [21].
4. Conclusions
Cells are outfitted with an array of defense mechanisms to counter stress. When exposed to stress stimuli, adaptive and cytoprotective pathways are engaged to restore balance. When the damage induced by stress cannot be resolved, apoptosis ensues. It is becoming increasingly clear that exposure to low-grade stress may precondition cells in a hermetic manner. Thus, mild stress such as low-grade Golgi stress, which does not cause toxicity, can up-regulate defense mechanisms to precondition cells to withstand future insults. Novel therapeutics which target the points of intersection between stress and adaptive responses may be beneficial in a wide variety of diseases.
Acknowledgments
Biorender is acknowledged for the use of their software for preparation of figures.
Abbreviations
| ACBD3 | Acyl CoA-Binding Domain Containing 3 |
| ATF4 | Activating Transcription Factor 4 |
| BGAT2 | Galactosyl Galactosyl Xylosyl Protein 3-β Glucuronosyltransferase 2 |
| BiP | Binding Immunoglobulin Protein |
| CHOP | C/EBP Homologous Protein |
| CoQ10 | Coenzyme Q10 |
| CREB | cAMP Response Element Binding Protein 3 |
| CSE | Cystathionine γ-lyase |
| GCN2 | General Control non-Derepressible 2 |
| eIF2α | Eukaryotic Translation Initiation Factor 2 Subunit-α |
| EAAT3/EAAC1 | Excitatory Amino Acid Transporter 3 |
| ER | Endoplasmic Reticulum |
| ERAD | ER-Associated Degradation |
| FUT1 | Fucosyltransferase 1 |
| GARD | Golgi-Apparatus Related Degradation |
| GASE | Golgi Apparatus Stress Response Element |
| GRASP65 | Golgi Reassembly and Stacking Protein of 65 kD |
| HD | Huntington’s Disease |
| HRI | Heme-Regulated eIF2α Kinase |
| IRE1 | Inositol-Requiring Enzyme 1 |
| MTOC | Microtubule Organizing Center |
| NFTs | Neurofibrillary Tangles |
| Nrf2 | Nuclear Factor Erythroid 2-Related Factor 2 |
| PDI | Protein Disulfide Isomerase |
| PERK | Protein Kinase R Protein Kinase R (PKR)-Like ER Kinase |
| PG | Proteoglycan |
| PHF | Paired Helical Filament |
| Rab20 | Ras-Related Protein Rab-20 |
| Rhes | Ras Homolog Enriched in Striatum |
| RIDD | Regulated IRE1-Dependent Decay |
| S1P | Site 1 Protease |
| S2P | Site 2 Protease |
| SIAT4A/ST3GAL1ST3 | β-Galactoside Alpha-2,3-Sialyltransferase 1 |
| SP1 | Specificity Protein 1 |
| STX3 | Syntaxin 3A |
| TFE3 | Transcription Factor E3 |
| TGN | Trans Golgi Network |
| UAP1L1 | UDP-N-Acetylhexosamine Pyrophosphorylase-Like Protein 1 |
| UPR | Unfolded Protein Response |
| WIPI49/WIP1α | WD-Repeat Protein Interacting with PhosphoInosides 1α |
| XBP1 | X-box Binding Protein 1 |
Funding
This work was funded by the American Heart Association (AHA)/Paul Allen Frontiers Group; grant number 19PABH134580006 and National Institutes of Health, NIA grant 1R21AG073684—01 to B.D.P.
Conflicts of Interest
The author declares no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.O’Donovan M.C. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R., et al. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed I., Sbodio J.I., Harraz M.M., Tyagi R., Grima J.C., Albacarys L.K., Hubbi M., Xu R., Kim S., Paul B.D., et al. Huntington’s disease: Neural dysfunction linked to inositol polyphosphate multikinase. Proc. Natl. Acad. Sci. USA. 2015;112:9751–9756. doi: 10.1073/pnas.1511810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grima J.C., Daigle J.G., Arbez N., Cunningham K., Zhang K., Ochaba J., Geater C., Morozko E., Stocksdale J., Glatzer J.C., et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron. 2017;94:93–107. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sbodio J.I., Snyder S.H., Paul B.D. Transcriptional control of amino acid homeostasis is disrupted in Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2016;113:8843–8848. doi: 10.1073/pnas.1608264113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eshraghi M., Karunadharma P.P., Blin J., Shahani N., Ricci E.P., Michel A., Urban N.T., Galli N., Sharma M., Ramírez-Jarquín U.N., et al. Mutant Huntingtin stalls ribosomes and represses protein synthesis in a cellular model of Huntington disease. Nat. Commun. 2021;12:1–20. doi: 10.1038/s41467-021-21637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul B.D., Snyder S.H. Impaired Redox Signaling in Huntington’s Disease: Therapeutic Implications. Front. Mol. Neurosci. 2019;12:68. doi: 10.3389/fnmol.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao S.S., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox. Signal. 2014;21:396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid D.W., Nicchitta C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015;16:221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz D.S., Blower M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roscoe J.M., Sevier C.S. Pathways for Sensing and Responding to Hydrogen Peroxide at the Endoplasmic Reticulum. Cells. 2020;9:2314. doi: 10.3390/cells9102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemmer I.L., Willemsen N., Hilal N., Bartelt A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol. Metab. 2021;47:101169. doi: 10.1016/j.molmet.2021.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter P., Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 15.Frakes A.E., Dillin A. The UPR(ER): Sensor and Coordinator of Organismal Homeostasis. Mol. Cell. 2017;66:761–771. doi: 10.1016/j.molcel.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R., Brown M.S., Goldstein J.L. ER Stress Induces Cleavage of Membrane-Bound ATF6 by the Same Proteases that Process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 17.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemberg M.K., Strisovsky K. Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol. Cell. 2021;81:2507–2519. doi: 10.1016/j.molcel.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson S. Emerging Principles of Selective ER Autophagy. J. Mol. Biol. 2020;432:185–205. doi: 10.1016/j.jmb.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sbodio J.I., Paul B.D., Machamer C.E., Snyder S.H. Golgi Protein ACBD3 Mediates Neurotoxicity Associated with Huntington’s Disease. Cell Rep. 2013;4:890–897. doi: 10.1016/j.celrep.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sbodio J.I., Snyder S.H., Paul B.D. Golgi stress response reprograms cysteine metabolism to confer cytoprotection in Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2018;115:780–785. doi: 10.1073/pnas.1717877115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golgi C., Lipsky N.G. On the structure of nerve cells. J. Microsc. 1989;155:3–7. doi: 10.1111/j.1365-2818.1989.tb04294.x. [DOI] [PubMed] [Google Scholar]
- 23.Friend D.S., Murray M.J. Osmium Impregnation of the Golgi Apparatus. Am. J. Anat. 1965;117:135–149. doi: 10.1002/aja.1001170109. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S.K. Camillo Golgi (1843–1926): Scientist extraordinaire and pioneer figure of modern neurology. Anat. Cell Biol. 2020;53:385–392. doi: 10.5115/acb.20.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodstad H. The neuron theory. Stereotact. Funct. Neurosurg. 2001;77:20–24. doi: 10.1159/000064596. [DOI] [PubMed] [Google Scholar]
- 26.Lowe M. Structural organization of the Golgi apparatus. Curr. Opin. Cell Biol. 2011;23:85–93. doi: 10.1016/j.ceb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Wei J.H., Seemann J. Unraveling the Golgi ribbon. Traffic. 2010;11:1391–1400. doi: 10.1111/j.1600-0854.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altan-Bonnet N., Sougrat R., Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Robbins E., Gonatas N.K. The Ultrastructure of a Mammalian Cell during the Mitotic Cycle. J. Cell Biol. 1964;21:429–463. doi: 10.1083/jcb.21.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayala I., Colanzi A. Mitotic inheritance of the Golgi complex and its role in cell division. Biol. Cell. 2017;109:364–374. doi: 10.1111/boc.201700032. [DOI] [PubMed] [Google Scholar]
- 31.Guizzunti G., Seemann J. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc. Natl. Acad. Sci. USA. 2016;113:E6590–E6599. doi: 10.1073/pnas.1610844113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanley P. Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 2011;3:a005199. doi: 10.1101/cshperspect.a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potelle S., Klein A., Foulquier F. Golgi post-translational modifications and associated diseases. J. Inherit. Metab. Dis. 2015;38:741–751. doi: 10.1007/s10545-015-9851-7. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki K., Yoshida H. Organelle autoregulation-stress responses in the ER, Golgi, mitochondria and lysosome. J. Biochem. 2015;157:185–195. doi: 10.1093/jb/mvv010. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki K., Yoshida H. Golgi stress response and organelle zones. FEBS Lett. 2019;593:2330–2340. doi: 10.1002/1873-3468.13554. [DOI] [PubMed] [Google Scholar]
- 36.Reiling J.H., Olive A., Sanyal S., Carette J., Brummelkamp T.R., Ploegh H.L., Starnbach M.N., Sabatini D.M. A CREB3–ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 2013;15:1473–1485. doi: 10.1038/ncb2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura N. Emerging New Roles of GM130, a cis-Golgi Matrix Protein, in Higher Order Cell Functions. J. Pharmacol. Sci. 2010;112:255–264. doi: 10.1254/jphs.09R03CR. [DOI] [PubMed] [Google Scholar]
- 38.Sohda M., Misumi Y., Yamamoto A., Yano A., Nakamura N., Ikehara Y. Identification and Characterization of a Novel Golgi Protein, GCP60, That Interacts with the Integral Membrane Protein Giantin. J. Biol. Chem. 2001;276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- 39.Oku M., Tanakura S., Uemura A., Sohda M., Misumi Y., Taniguchi M., Wakabayashi S., Yoshida H. Novel Cis-acting Element GASE Regulates Transcriptional Induction by the Golgi Stress Response. Cell Struct. Funct. 2011;36:1–12. doi: 10.1247/csf.10014. [DOI] [PubMed] [Google Scholar]
- 40.Taniguchi M., Nadanaka S., Tanakura S., Sawaguchi S., Midori S., Kawai Y., Yamaguchi S., Shimada Y., Nakamura Y., Matsumura Y., et al. TFE3 is a bHLH-ZIP-type transcription factor that regulates the mammalian Golgi stress response. Cell Struct. Funct. 2015;40:13–30. doi: 10.1247/csf.14015. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi M., Sasaki-Osugi K., Oku M., Sawaguchi S., Tanakura S., Kawai Y., Wakabayashi S., Yoshida H. MLX Is a Transcriptional Repressor of the Mammalian Golgi Stress Response. Cell Struct. Funct. 2016;41:93–104. doi: 10.1247/csf.16005. [DOI] [PubMed] [Google Scholar]
- 42.Eisenberg-Lerner A., Benyair R., Hizkiahou N., Nudel N., Maor R., Kramer M.P., Shmueli M.D., Zigdon I., Lev M.C., Ulman A., et al. Golgi organization is regulated by proteasomal degradation. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-14038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane J., Lucocq J., Pryde J., Barr F., Woodman P.G., Allan V., Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyata S., Mizuno T., Koyama Y., Katayama T., Tohyama M. The Endoplasmic Reticulum-Resident Chaperone Heat Shock Protein 47 Protects the Golgi Apparatus from the Effects of O-Glycosylation Inhibition. PLoS ONE. 2013;8:e69732. doi: 10.1371/journal.pone.0069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamaludin M.I., Wakabayashi S., Taniguchi M., Sasaki K., Komori R., Kawamura H., Takase H., Sakamoto M., Yoshida H. MGSE Regulates Crosstalk from the Mucin Pathway to the TFE3 Pathway of the Golgi Stress Response. Cell Struct. Funct. 2019;44:137–151. doi: 10.1247/csf.19009. [DOI] [PubMed] [Google Scholar]
- 46.Gonatas N.K., Stieber A., Gonatas J.O. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J. Neurol. Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Mourelatos Z., Gonatas N.K., Stieber A., Gurney M.E., Canto M.C.D. The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu, Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc. Natl. Acad. Sci. USA. 1996;93:5472–5477. doi: 10.1073/pnas.93.11.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi G., Bekier M.E., 2nd, Wang Y. Golgi fragmentation in Alzheimer’s disease. Front. Neurosci. 2015;9:340. doi: 10.3389/fnins.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rendón W.O., Martínez-Alonso E., Tomás M., Martínez-Martínez N., Martínez-Menárguez J.A. Golgi fragmentation is Rab and SNARE dependent in cellular models of Parkinson’s disease. Histochem. Cell Biol. 2012;139:671–684. doi: 10.1007/s00418-012-1059-4. [DOI] [PubMed] [Google Scholar]
- 50.Strehlow A.N., Li J., Myers R.M. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum. Mol. Genet. 2006;16:391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- 51.Sakurai A., Okamoto K., Fujita Y., Nakazato Y., Wakabayashi K., Takahashi H., Gonatas N.K. Fragmentation of the Golgi apparatus of the ballooned neurons in patients with corticobasal degeneration and Creutzfeldt-Jakob disease. Acta Neuropathol. 2000;100:270–274. doi: 10.1007/s004010000182. [DOI] [PubMed] [Google Scholar]
- 52.Campadelli G., Brandimarti R., Di Lazzaro C., Ward P.L., Roizman B., Torrisi M.R. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus. Proc. Natl. Acad. Sci. USA. 1993;90:2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C., Mei M., Li Q., Roboti P., Pang Q., Ying Z., Gao F., Lowe M., Bao S. Loss of the golgin GM130 causes Golgi disruption, Purkinje neuron loss, and ataxia in mice. Proc. Natl. Acad. Sci. USA. 2017;114:346–351. doi: 10.1073/pnas.1608576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomas M., Martinez-Alonso E., Martinez-Martinez N., Cara-Esteban M., Martinez-Menarguez J.A. Fragmentation of the Golgi complex of dopaminergic neurons in human substantia nigra: New cytopathological findings in Parkinson’s disease. Histol. Histopathol. 2021;36:47–60. doi: 10.14670/HH-18-270. [DOI] [PubMed] [Google Scholar]
- 55.Stieber A., Mourelatos Z., Gonatas N.K. In Alzheimer’s disease the Golgi apparatus of a population of neurons without neurofibrillary tangles is fragmented and atrophic. Am. J. Pathol. 1996;148:415–426. [PMC free article] [PubMed] [Google Scholar]
- 56.Liazoghli D., Perreault S., Micheva K.D., Desjardins M., Leclerc N. Fragmentation of the Golgi Apparatus Induced by the Overexpression of Wild-Type and Mutant Human Tau Forms in Neurons. Am. J. Pathol. 2005;166:1499–1514. doi: 10.1016/S0002-9440(10)62366-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis J., McGowan E., Rockwood J., Melrose H., Nacharaju P., Van Slegtenhorst M., Gwinn K., Murphy M.P., Baker M., Yu X., et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 2000;25:402–405. doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- 58.Nakagomi S., Barsoum M.J., Bossy-Wetzel E., Sütterlin C., Malhotra V., Lipton S.A. A Golgi fragmentation pathway in neurodegeneration. Neurobiol. Dis. 2008;29:221–231. doi: 10.1016/j.nbd.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Antón-Fernández A., Merchán-Rubira J., Avila J., Hernández F., DeFelipe J., Muñoz A. Phospho-Tau Accumulation and Structural Alterations of the Golgi Apparatus of Cortical Pyramidal Neurons in the P301S Tauopathy Mouse Model. J. Alzheimer’s Dis. 2017;60:651–661. doi: 10.3233/JAD-170332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Q., Wang L., Guan Y., Xu H., Niu Y., Han L., Wei Y.-P., Lin L., Chu J., Wang Q., et al. Golgin-84-associated Golgi fragmentation triggers tau hyperphosphorylation by activation of cyclin-dependent kinase-5 and extracellular signal-regulated kinase. Neurobiol. Aging. 2014;35:1352–1363. doi: 10.1016/j.neurobiolaging.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 61.Subramaniam S., Sixt K.M., Barrow R., Snyder S.H. Rhes, a Striatal Specific Protein, Mediates Mutant-Huntingtin Cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu X.H., Mattis V.B., Wang N., Al-Ramahi I., van den Berg N., Fratantoni S.A., Waldvogel H., Greiner E., Osmand A., Elzein K., et al. Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington’s disease. Sci. Transl. Med. 2014;6:268ra178. doi: 10.1126/scitranslmed.3010523. [DOI] [PubMed] [Google Scholar]
- 63.Iyer R.R., Pluciennik A. DNA Mismatch Repair and its Role in Huntington’s Disease. J. Huntingtons Dis. 2021;10:75–94. doi: 10.3233/JHD-200438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luthi-Carter R., Hanson S.A., Strand A.D., Bergstrom D.A., Chun W., Peters N.L., Woods A.M., Chan E.Y., Kooperberg C., Krainc D., et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: Parallel changes in muscle and brain. Hum. Mol. Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 65.Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K., Pletnikova O., Borchelt D.R., Ross C.A., Margolis R.L., et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soares T.R., Reis S.D., Pinho B.R., Duchen M.R., Oliveira J.M.A. Targeting the proteostasis network in Huntington’s disease. Ageing Res. Rev. 2019;49:92–103. doi: 10.1016/j.arr.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones D.P. Redefining Oxidative Stress. Antioxid. Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 68.Sbodio J.I., Snyder S.H., Paul B.D. Redox Mechanisms in Neurodegeneration: From Disease Outcomes to Therapeutic Op-portunities. Antioxid. Redox Signal. 2019;30:1450–1499. doi: 10.1089/ars.2017.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paul B.D., Lemle M.D., Komaroff A.L., Snyder S.H. Redox imbalance links COVID-19 and myalgic encephalomyelitis/chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA. 2021;118:e2024358118. doi: 10.1073/pnas.2024358118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrich J., Saft C., Gerlach M., Schneider B., Arz A., Kühn W., Müller T. Focus on Extrapyramidal Dysfunction. Volume 68. Springer; Vienna, Austria: 2004. Coenzyme Q10 serum levels in Huntington’s disease; pp. 111–116. [DOI] [PubMed] [Google Scholar]
- 71.Paul B.D., Sbodio J.I., Xu R., Vandiver M.S., Cha J.Y., Snowman A.M., Snyder S.H. Cystathionine gamma-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature. 2014;509:96–100. doi: 10.1038/nature13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Acuña A.I., Esparza M., Kramm C., Beltrán F.A., Parra A.V., Cepeda C., Toro C.A., Vidal R.L., Hetz C., Concha I.I., et al. A failure in energy metabolism and antioxidant uptake precede symptoms of Hun-tington’s disease in mice. Nat. Commun. 2013;4:2917. doi: 10.1038/ncomms3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang C., Rodriguez C., Spaulding J., Aw T.Y., Feng J. Age-dependent and tissue-related glutathione redox status in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2012;28:655–666. doi: 10.3233/JAD-2011-111244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frederick N.M., Bertho J., Patel K.K., Petr G.T., Bakradze E., Smith S.B., Rosenberg P.A. Dysregulation of system xc(-) expression induced by mutant huntingtin in a striatal neuronal cell line and in R6/2 mice. Neurochem. Int. 2014;76:59–69. doi: 10.1016/j.neuint.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X., Valencia A., Sapp E., Masso N., Alexander J., Reeves P., Kegel K.B., Aronin N., DiFiglia M. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J. Neurosci. 2010;30:4552–4561. doi: 10.1523/JNEUROSCI.5865-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 77.Paul B.D., Snyder S.H. Neurodegeneration in Huntington’s disease involves loss of cystathionine gamma-lyase. Cell Cycle. 2014;13:2491–2493. doi: 10.4161/15384101.2014.950538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paul B.D., Sbodio J.I., Snyder S.H. Cysteine Metabolism in Neuronal Redox Homeostasis. Trends Pharmacol. Sci. 2018;39:513–524. doi: 10.1016/j.tips.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paul B.D., Snyder S.H. H2S: A Novel Gasotransmitter that Signals by Sulfhydration. Trends Biochem. Sci. 2015;40:687–700. doi: 10.1016/j.tibs.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sen N., Paul B.D., Gadalla M.M., Mustafa A.K., Sen T., Xu R., Kim S., Snyder S.H. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol. Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K., Barrow R.K., Yang G., Wang R., Snyder S.H. H2S Signals Through Protein S-Sulfhydration. Sci. Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paul B.D., Snyder S.H. Gasotransmitter hydrogen sulfide signaling in neuronal health and disease. Biochem. Pharmacol. 2018;149:101–109. doi: 10.1016/j.bcp.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paul B.D., Snyder S.H., Kashfi K. Effects of hydrogen sulfide on mitochondrial function and cellular bioenergetics. Redox Biol. 2021;38:101772. doi: 10.1016/j.redox.2020.101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zivanovic J., Kouroussis E., Kohl J.B., Adhikari B., Bursac B., Schott-Roux S., Petrovic D., Miljkovic J.L., Thomas-Lopez D., Jung Y., et al. Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration. Cell Metab. 2019;30:1152–1170.e13. doi: 10.1016/j.cmet.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harding H., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R.S., et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 86.Kilberg M.S., Shan J., Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rzymski T., Milani M., Pike L., Buffa F., Mellor H.R., Winchester L., Pires I., Hammond E., Ragoussis I., Harris A.L. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29:4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 88.Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang P., Peslak S.A., Lan X., Khandros E., Yano J.A., Sharma M., Keller C.A., Giardine B.M., Qin K., Abdulmalik O., et al. HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. Blood. 2020;135:2121–2132. doi: 10.1182/blood.2020005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huggins C.J., Mayekar M.K., Martin N., Saylor K.L., Gonit M., Jailwala P., Kasoji M., Haines D.C., Quiñones O.A., Johnson P.F. C/EBPgamma Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF. Mol. Cell Biol. 2015;36:693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He C.H., Gong P., Hu B., Stewart D., Choi M.E., Choi A.M.K., Alam J. Identification of Activating Transcription Factor 4 (ATF4) as an Nrf2-interacting Protein. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 93.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M., Geschwind D.H., Ratan R.R. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wortel I.M.N., van der Meer L.T., Kilberg M.S., van Leeuwen F.N. Surviving Stress: Modulation of ATF4-Mediated Stress Re-sponses in Normal and Malignant Cells. Trends Endocrinol. Metab. 2017;28:794–806. doi: 10.1016/j.tem.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dey S., Baird T., Zhou D., Palam L.R., Spandau D.F., Wek R.C. Both Transcriptional Regulation and Translational Control of ATF4 Are Central to the Integrated Stress Response. J. Biol. Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berlanga J.J., Herrero S., de Haro C. Characterization of the hemin-sensitive eukaryotic initiation factor 2alpha kinase from mouse nonerythroid cells. J. Biol. Chem. 1998;273:32340–32346. doi: 10.1074/jbc.273.48.32340. [DOI] [PubMed] [Google Scholar]
- 98.Chen J.J., Throop M.S., Gehrke L., Kuo I., Pal J., Brodsky M., London I.M. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: Homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc. Natl. Acad. Sci. USA. 1991;88:7729–7733. doi: 10.1073/pnas.88.17.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi Y., Vattem K.M., Sood R., An J., Liang J., Stramm L., Wek R.C. Identification and Characterization of Pancreatic Eukaryotic Initiation Factor 2 α-Subunit Kinase, PEK, Involved in Translational Control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/MCB.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meurs E., Chong K., Galabru J., Thomas N.B., Kerr I.M., Williams B., Hovanessian A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-N. [DOI] [PubMed] [Google Scholar]
- 101.Donnelly N., Gorman A., Gupta S., Samali A. The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]