Abstract
De novo methylation of CpG islands within the promoters of eukaryotic genes is often associated with their transcriptional repression, yet the methylation of CpG islands located downstream of promoters does not block transcription. We investigated the kinetics of mRNA induction, demethylation, and remethylation of the p16 promoter and second-exon CpG islands in T24 cells after 5-aza-2′-deoxycytidine (5-Aza-CdR) treatment to explore the relationship between CpG island methylation and gene transcription. The rates of remethylation of both CpG islands were associated with time but not with the rate of cell division, and remethylation of the p16 exon 2 CpG island occurred at a higher rate than that of the p16 promoter. We also examined the relationship between the remethylation of coding sequence CpG islands and gene transcription. The kinetics of remethylation of the p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 loci were examined following 5-Aza-CdR treatment because these genes contain exonic CpG islands which are hypermethylated in T24 cells. Remethylation occurred most rapidly in the p16, PAX-6, and c-ABL genes, shown to be transcribed prior to drug treatment. These regions also exhibited higher levels of remethylation in single-cell clones and subclones derived from 5-Aza-CdR-treated T24 cells. Our data suggest that de novo methylation is not restricted to the S phase of the cell cycle and that transcription through CpG islands does not inhibit their remethylation.
DNA methylation is essential for normal embryonic development, possibly due to its roles in transcriptional silencing (7, 32, 45, 50), X-chromosome inactivation (41, 46), and genomic imprinting (10, 31). Cytosine methylation normally occurs at CpG dinucleotides, which are represented at lower-than-expected frequencies in the eukaryotic genome, with the exception of regions known as CpG islands, which have the statistically expected frequency of CpGs (12). Analyses of the spatial relationship between CpG islands and eukaryotic genes have shown that CpG islands often reside within gene promoters and extend further downstream into transcribed regions (5); however, they can also occur in regions remote from the promoter (30). CpG islands normally remain unmethylated in the germ line and rarely become methylated in somatic cells (1); however, alterations in these methylation patterns are associated with many human cancers (2, 4, 23).
Numerous investigations suggest that hypermethylation of promoter CpG islands correlates with transcriptional inhibition (7, 13, 17–19, 39, 44, 47, 53). On the other hand, additional studies show that de novo methylation of CpG islands residing within transcribed regions is permissive for gene expression (22, 24, 54) and that methylation of exonic CpG islands does not inhibit transcriptional elongation in mammalian cells (14). Paradoxically, hypermethylation of promoter CpG islands is often associated with transcriptional silencing, whereas increased CpG island methylation downstream of transcription initiation correlates with gene expression (24).
Additional evidence from our laboratory suggests that gene transcription does not block the de novo methylation of CpG islands, though most studies in the field have concentrated on the effects of promoter methylation on gene silencing. First, genome-scanning techniques have led to the identification of CpG islands within transcribed regions of genes which are overexpressed and hypermethylated in tumor cells (33, 48); second, hypermethylation of p16 exon 2 has been observed in primary tumors and tumor cell lines which express the gene (13); and third, this report shows that remethylation of the p16 exon 2 CpG island after 5-aza-2′-deoxycytidine (5-Aza-CdR) treatment occurs more rapidly than that of the p16 promoter CpG island.
These observations led us to investigate the roles of cell division and gene transcription in DNA methylation to further clarify the association between gene transcription and the remethylation of CpG islands, including those within exonic sequences. First, we analyzed the kinetics of p16 activation and demethylation by 5-Aza-CdR in the T24 bladder carcinoma cell line and observed that demethylation of the p16 promoter CpG island was directly associated with transcription of the gene. Next, the rates of remethylation of the p16 gene following this transient demethylation after 5-Aza-CdR treatment were shown to depend on time but not on the rate of cell division, and it was shown that p16 exon 2 became remethylated at a higher rate than the promoter. We also explored how gene transcription might influence remethylation of the coding sequence CpG islands in PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3. Remethylation occurred most rapidly in the actively transcribed regions of p16 exon 2, PAX-6 exon 5, and c-ABL exon 11, whereas the MYF-3 gene, which is not transcribed in T24 cells, exhibited a lower rate of remethylation than the other loci examined. The transcribed CpG islands of p16 exon 2, PAX-6 exon 5, and c-ABL exon 11 also became remethylated to greater degrees in clones and subclones derived from T24 cells treated with 5-Aza-CdR, showing that the increased levels of methylation observed were due to de novo methylation of previously unmethylated sequences rather than to the selection of cells which had been unaffected by treatment with a demethylating agent. These results show that cell division does not increase the rate of remethylation and that the transcription of endogenous genes does not block remethylation of CpG islands.
MATERIALS AND METHODS
Cell lines.
The J82 and T24 bladder transitional cell carcinoma cell lines were obtained from the American Type Tissue Collection, Rockville, Md. T24 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 5% penicillin-streptomycin. J82 cells were cultured in minimal essential medium supplemented with 10% FCS, 5% penicillin-streptomycin, nonessential amino acids, and sodium pyruvate.
5-Aza-CdR treatments.
Cells were plated (3 × 105 cells per 100-mm dish) and treated 24 h later with 5-Aza-CdR (5 × 10−7 M). This dose was selected because it exhibited the greatest effect on DNA demethylation with minimal cytotoxicity. To increase the cell survival rate and facilitate the isolation of single-cell clones, 3 × 10−7 M 5-Aza-CdR was utilized. The medium was changed 24 h after drug treatment and every 3 days subsequently. RNA and DNA were isolated after specific time periods following treatment, as described previously (13).
Analysis of the relationship between cell division and the de novo methylation of CpG islands following 5-Aza-CdR treatment.
T24 cells originally maintained in DMEM containing 10% FCS and 5% penicillin-streptomycin were plated (3 × 105 per 100-mm dish) in DMEM containing 1% FCS and 5% penicillin-streptomycin. Cells were treated 24 h later with 5-Aza-CdR (5 × 10−7 M; Sigma Chemical Co., St. Louis, Mo.). The drug was removed and the medium was replaced 24 h after addition of the drug, with half the plates containing medium supplemented with 1% FCS and the remaining half containing medium supplemented with 10% FCS. The media were subsequently changed every 3 days. RNA and DNA were isolated after specific time periods following treatment, as described previously (13). The total cell number was assessed at each time point by using a Coulter Counter to determine the differences in doubling times between cells grown in medium supplemented with 1% FCS and those grown in 10% FCS.
Generation of T24 single-cell clones and subclones.
Cells were plated (105 per 75-cm2 flask) and treated 24 h later with 5-Aza-CdR (3 × 10−7 M). This lower dose was selected (instead of 5 × 10−7 M) to reduce the immediate cytotoxic effects of the drug and increase the cell survival rate, thus facilitating the isolation of individual cells. Clones 2, 3, 4, and 7 were isolated from single cells between 48 and 72 h after drug removal, as described previously (14). Conditioned medium was used for the initial culturing of single-cell clones. One of these clones (clone 4) was used for the repeated isolation of single-cell subclones (designated 4:1, 4:2, 4:5, and 4:9). RNA and DNA were extracted from each individual clone or subclone at least 20 cell population doublings after isolation.
Quantitation of methylation by Ms-SNuPE.
Methylation of PAX-6 exon 5, c-ABL exon 11, MYF-3 exon 3, p16 exon 2, and the p16 promoter was measured by the methylation-sensitive single nucleotide primer extension (Ms-SNuPE) assay as described previously (15). T24 cells were treated with 5-Aza-CdR (5 × 10−7 M), and DNA was harvested at various times after treatment (proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation). DNA was also isolated from T24 single-cell subclones. Genomic DNA was treated with sodium bisulfite (8, 11) to convert unmethylated cytosines to uracil, leaving 5-methylcytosine unchanged. The regions of interest were amplified with PCR primers specific for bisulfite-converted DNA (Table 1). The PCR conditions were as follows: for the p16 promoter, 95°C for 3 min, 37 cycles of 95°C for 50 s, 67°C for 50 s, and 72°C for 45 s, and 72°C for 2 min; for p16 exon 2, 95°C for 3 min, 38 cycles of 95°C for 50 s, 60°C for 50 s, and 72°C for 45 s, and 72°C for 2 min; for PAX-6 exon 5, 95°C for 3 min, 38 cycles of 95°C for 1 min, 50°C for 30 s, and 72°C for 1 min, and 72°C for 2 min; for c-ABL exon 11, 95°C for 3 min, 37 cycles of 95°C for 50 s, 47°C for 50 s, and 72°C for 45 s, and 72°C for 2 min; and for MYF-3 exon 3, 95°C for 3 min, 36 cycles of 95°C for 50 s, 49°C for 40 s, and 72°C for 45 s, and 72°C for 2 min. SNuPE was performed with primers internal to the region amplified, with each primer terminating immediately 5′ of the CpG site to be assayed. The methylation status of individual CpG sites within the amplified regions of interest was analyzed, and the average methylation value for two or three sites within each region was obtained. Ms-SNuPE sequences specific for each region are shown in Table 1. The conditions for primer extension were as follows: for the p16 promoter and exon 2, 95°C for 2 min, 50°C for 1 min, and 72°C for 1 min; for PAX-6 exon 5, 95°C for 2 min, 60°C for 1 min, and 72°C for 1 min; for c-ABL exon 11, 95°C for 2 min, 47°C for 2 min, and 72°C for 1 min; and for MYF-3 exon 3, 95°C for 2 min, 43°C for 1 min, and 72°C for 1 min.
TABLE 1.
Oligonucleotide sequences
| Assay and gene | Sequence | Direction |
|---|---|---|
| Bisulfite PCR | ||
| p16 promoter | 5′-GTAGGTGGGGAGGAGTTTAGTT-3′ | Sense |
| 5′-TCTAATAACCAACCAACCCCTCC-3′ | Antisense | |
| p16 exon 2 | 5′-TTGATTATTTTGTTTTTTTTGGTAGGTT-3′ | Sense |
| 5′-CAAATTCTCAAATCATCAATCCTCACC-3′ | Antisense | |
| PAX-6 exon 5 | 5′-GGGAGGATTATTTGTAG-3′ | Sense |
| 5′-CTTTCCTCAAATCACAAC-3′ | Antisense | |
| c-ABL exon 11 | 5′-GTTTTTGTAGGGAAGGTTGG-3′ | Sense |
| 5′-ACTAAACTAATAAAAATCCC-3′ | Antisense | |
| MYF-3 exon 3 | 5′-GAGTTTAGATTATTTGTTTAG-3′ | Sense |
| 5′-AAACATTTAAATTCAATCTTTTAAAC-3′ | Antisense | |
| Ms-SNuPE | ||
| p16 promoter | 5′-TTTGAGGGATAGGGT-3′ | |
| 5′-TTTTAGGGGTGTTATATT-3′ | ||
| 5′-TTTTTTTGTTTGGAAAGATAT-3′ | ||
| p16 exon 2 | 5′-GTTGGTGGTGTTGTAT-3′ | |
| 5′-AGGTTATGATGATGGGTAG-3′ | ||
| 5′-TATTAGAGGTAGTAATTATGTT-3′ | ||
| PAX-6 exon 5 | 5′-AGTTAGTTTTATAATTTTTTGT-3′ | |
| 5′-GAGGATTATTTGTAGAATT-3′ | ||
| 5′-GTTGATAAAGATATTAT-3′ | ||
| c-ABL exon 11 | 5′-GGAGGTAGTTTTGGG-3′ | |
| 5′-TTTGGTTGATGTTGTGAA-3′ | ||
| 5′-GTAGAGGGTTTTAAAAAGTT-3′ | ||
| MYF-3 exon 3 | 5′-TTTTGAGGGGGATGTGGT-3′ | |
| 5′-AGGGAGAGAGTAG-3′ | ||
| RT-PCR | ||
| p16 | 5′-AGCCTTCGGCTGACTGGCTGG-3′ | Sense |
| 5′-CTGCCCATCATCATGACCTGGA-3′ | Antisense | |
| PAX-6 | 5′-CTAATGGGCCAGTGAGGAG-3′ | Sense |
| 5′-TACTCACACATCCGTTGGACAC-3′ | Antisense | |
| c-ABL | 5′-GGCTGCCCAGAGAAGGTCTA-3′ | Sense |
| 5′-GAGCAATGGAGACACGGCAG-3′ | Antisense | |
| MYF-3 | 5′-TCCAAACCAGCGGTTGCCCAAG-3′ | Sense |
| 5′-TGGAGATGCGCTCCACGATGCT-3′ | Antisense |
RT-PCR.
Total RNA was isolated from 2 × 106 cells lysed in 2 ml of buffer containing guanidine isothiocyanate (4 M; Gibco BRL, Palo Alto, Calif.), N-lauryl sarcosine (0.5%), sodium citrate (25 mM; Fisher Scientific, Fair Lawn, N.J.), and 2-mercaptoethanol (0.1 M; Sigma Chemical Co.). RNA was precipitated (1 h, −20°C) in 50% isopropanol–50% lysis buffer following standard phenol-chloroform extraction of the cell lysate. After centrifugation (10 min, 10,000 × g), the supernatant was decanted and the RNA pellet was washed twice in 70% ethanol prepared with diethylpyrocarbonate-treated double-distilled water. The RNA pellet was dissolved in 100% diethylpyrocarbonate-treated water. Two micrograms of total RNA was reverse transcribed with random hexamers, deoxynucleoside triphosphates, and SuperScript II reverse transcriptase (Gibco BRL) in a 25-μl reaction mixture, as described previously (13). cDNA was amplified with primers specific for either PAX-6, c-ABL, MYF-3, p16, or GAPDH. PCR primer sequences and conditions for p16 and GAPDH were utilized as described previously (13). The primer sequences for PAX-6, c-ABL, and MYF-3 expression analysis are listed in Table 1. The reverse transcription (RT)-PCR conditions were as follows: for PAX-6, 95°C for 3 min, 30 cycles of 95°C for 1 min, 60°C for 30 s, and 72°C for 1 min, and 72°C for 2 min; for c-ABL, 95°C for 3 min, 28 cycles of 95°C for 50 s, 60°C for 50 s, and 72°C for 45 s, and 72°C for 2 min; and for MYF-3, 95°C for 3 min, 27 cycles of 95°C for 1 min, 58°C for 45 s, and 72°C for 45 s, and 72°C for 2 min. PCRs were performed with cDNA template concentrations equivalent to 100 ng of RNA. All reactions were analyzed in the linear range of amplification. PCR products were resolved on 2% agarose gels and subsequently transferred to nylon membranes (Zetaprobe; Bio-Rad, Richmond, Calif.) under alkaline conditions. All blots were hybridized with digoxigenin-labeled oligonucleotide probes (Genius; Boehringer, Mannheim, Germany).
Determination of cytotoxicity.
T24 cells (200 per 60-mm dish) were plated in triplicate sets for a colony formation assay. Cells were treated with 10−7, 5 × 10−7, or 10−6 M 5-Aza-CdR 24 h later. Once cell colonies were visible (after 7 days), cells were fixed in 100% methanol and stained with 10% Giemsa stain. The cell survival percentage was assessed by dividing the mean colony number on the 5-Aza-CdR-treated plates by the mean colony number on the untreated plates and multiplying the quotient by 100. This number was subtracted from 100 to determine the percent toxicity.
RESULTS
Kinetics of p16 mRNA induction and p16 promoter demethylation by 5-Aza-CdR in T24 cells.
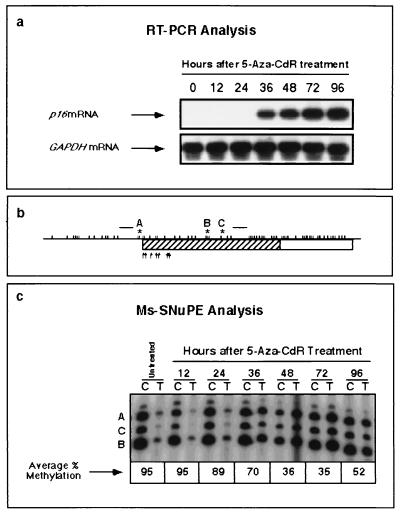
Members of our group have previously shown that activation of the p16 gene by 5-Aza-CdR in the T24 bladder carcinoma cell line is associated with significant demethylation of the p16 promoter CpG island (3, 14). The kinetics of p16 mRNA induction and p16 promoter demethylation were studied with precision by the quantitative Ms-SNuPE assay (15) to determine the relationship between these two processes (Fig. 1a). Preliminary experiments (data not shown) demonstrated that the concentration of 5-Aza-CdR most effective at inducing p16 expression and at reducing p16 methylation with minimal toxicity was 5 × 10−7 M. p16 expression was detected beginning 36 h after treatment of T24 cells with 5-Aza-CdR. The kinetics of demethylation within the proposed critical region of the p16 promoter (14) were investigated in parallel by the quantitative Ms-SNuPE technique (Fig. 1b and c). Reduced methylation was first apparent after 36 h (Fig. 1c). Maximal demethylation was detected between 48 and 72 h, with average methylation values of 36 and 35%, respectively, and the average methylation value increased to 52% by 96 h after treatment (Fig. 1c). These results support recent studies of the roles of critical CpG sites in the p16 promoter, whose hypermethylation is associated with transcriptional silencing (14), and they indicate that demethylation of this region by 5-Aza-CdR may play a direct role in drug-mediated mRNA p16 induction. The data are also important for our future quantitative studies, which will describe the kinetics of remethylation following maximal demethylation after 72 h.
FIG. 1.
Kinetics of p16 mRNA induction and promoter demethylation by 5-Aza-CdR. T24 cells were exposed to 5-Aza-CdR (5 × 10−7 M) for 24 h. DNA and RNA were isolated at 12-h intervals after drug addition. (a) Levels of p16 mRNA expression at each time point were determined by RT-PCR analysis. Relative levels of GAPDH mRNA expression were measured to control for relative cDNA input. PCR products were resolved on 2% agarose, transferred to a nylon membrane, and hybridized with an internal oligonucleotide probe specific for the cDNA sequence of either p16 or GAPDH. (b) Schematic map of the p16 5′ CpG island. A 149-bp region (hatching) was amplified by PCR with primers specific for bisulfite-converted DNA (raised horizontal bars), and three CpG sites were analyzed by the Ms-SNuPE technique (sites A, B, and C). Each tick mark represents an individual CpG dinucleotide, and the arrows show putative transcription initiation sites. (c) Demethylation of the p16 promoter by 5-Aza-CdR at each time point, quantitated by Ms-SNuPE. Ms-SNuPE reaction mixtures were resolved on a 16% denaturing polyacrylamide gel and subsequently exposed by autoradiography, using purified bisulfite PCR products as the templates for primer extension. The presence of a band indicates primer extension at a given CpG site. A band in the “C” lane indicates the detection of DNA molecules which are methylated, and a band in the “T” lane indicates the detection of unmethylated molecules.
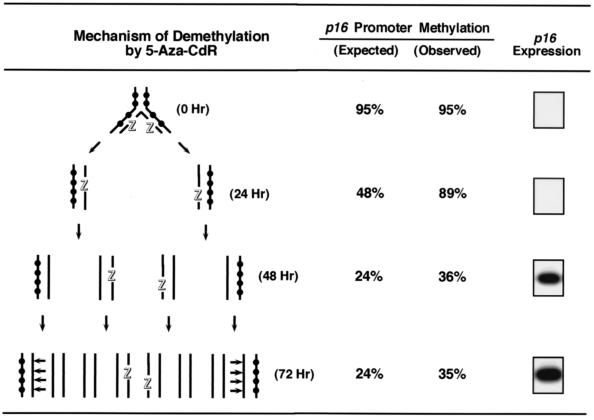
Model for the mechanism of p16 promoter demethylation by 5-Aza-CdR.
Figure 2 illustrates the kinetics of demethylation expected to result from the incorporation of 5-Aza-CdR into replicating, hypermethylated DNA. Hemimethylated duplex DNAs prepared from cells treated with 5-Aza-CdR present favorable substrates for DNA methyltransferase (27, 40, 51), and the dose used (5 × 10−7 M) has been shown to inhibit DNA methyltransferase almost completely and to deplete cells of active enzyme upon incorporation into a CpG site opposite a methylated CpG (26, 51). T24 cells would enter one or two S phases during the 24-h treatment with 5-Aza-CdR because the drug has no immediate effect on the cells’ division, and members of our group confirmed a doubling time of 21 h in treated and untreated cells, as previously described (3). The “active” inhibitor of DNA methyltransferase is present in hemimethylated DNA up to 48 h after treatment, and maintenance methylase activity is expected to methylate its hemimethylated substrate after 72 h, assuming that recovery synthesis of the DNA methyltransferase has occurred (51). Consistent with this model, it has been demonstrated by bisulfite sequencing that 5-Aza-CdR incorporation into replicating DNA results in the formation of individual DNA molecules in the p16 promoter containing patches of demethylation spanning 200 to 400 bp (14).
FIG. 2.
Model for the mechanism of DNA demethylation by 5-Aza-CdR. Unsynchronized T24 cells in the log phase of growth have a doubling time of 21 h (3) and were treated with 5-Aza-CdR (5 × 10−7 M) for 24 h. DNA and RNA were isolated at 24-h intervals, p16 promoter methylation levels were quantitated by Ms-SNuPE, and the presence of p16 mRNA was determined by RT-PCR. Cell numbers were assessed at each time point to determine the cell population doubling time. Strand breakage of 5-Aza-CdR-containing DNA after bisulfite treatment is likely because of the lability of the drug under alkaline conditions, which explains why the observed and expected values for methylation differ. Methylated CpG sites are indicated by black circles, DNA strands containing incorporated 5-Aza-CdR are each indicated by an outlined letter “Z,” and horizontal arrows show sites of hemimethylated DNA where maintenance methylation is expected to occur.
The methylation values quantitated for the p16 promoter after 5-Aza-CdR treatment were higher than expected. One explanation is that the Ms-SNuPE technique entails sodium bisulfite treatments under alkaline conditions, which would facilitate the rapid hydrolysis and breakage of 5-Aza-CdR-containing DNA strands due to the analog’s lability in alkaline solutions (9). The p16 promoter methylation values were consistent with a model for the activity of 5-Aza-CdR which assumes that the 5-Aza-CdR-containing DNA molecules, fragmented upon exposure to sodium bisulfite, should not contribute to the measured methylation levels. This technical bias against measuring the methylation status of analog-containing molecules becomes less significant during subsequent rounds of DNA replication after drug removal.
Figure 2 further elucidates the association between drug-induced p16 promoter demethylation and the reactivation of p16 mRNA expression. The data are consistent with previous analyses of clones of T24 cells treated with 5-Aza-CdR, where efficient p16 mRNA expression was observed in clones with extensive demethylation of the p16 promoter (14). They also suggest that p16 transcription was not initiated from a hemimethylated promoter template, because no p16 mRNA was detected 24 h after treatment. It is likely that the sequence must become demethylated on both DNA strands to facilitate p16 transcription; however, additional experiments must be performed to address this issue.
Role of cell division in remethylation of p16.
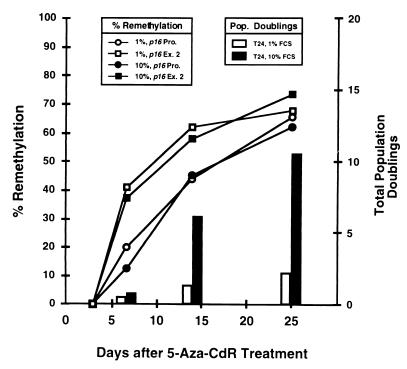
p16 promoter methylation is restored to original levels during passage in culture following 5-Aza-CdR treatment, with complete remethylation after 21 population doublings associated with decreased p16 mRNA expression and restoration of the doubling time to the same as that of untreated cells (3). Thus, we investigated whether remethylation of the promoter and exon 2 CpG islands followed similar kinetics and whether their rates of remethylation would be influenced by the rate of cell division. Figure 3 illustrates the relationship between time and the remethylation of the two CpG islands of p16 in T24 cells after 5-Aza-CdR treatment. Interestingly, cells grown in 1 and 10% FCS exhibited nearly identical kinetics of p16 remethylation, although their growth characteristics differed significantly. The CpG island in exon 2 also remethylated at a higher rate than the promoter under both growth conditions, and the rate of cell division after drug treatment had no effect on the rate of remethylation of either CpG island. For example, the two islands had become similarly remethylated 25 days after treatment, even though cells grown in 1% FCS had undergone only 2.5 population doublings, versus 10 population doublings for cells cultured in 10% FCS. The CpG island in exon 2 also became completely remethylated over a 2-week period if treated cells were arrested in G1 by culturing them in medium containing 0.1% serum immediately after treatment. This resulted in an almost complete loss of histone H4 mRNA, whose expression is restricted to S phase (data not shown). These results suggest that the remethylation of CpG islands after drug-induced demethylation is a time-dependent but not a cell division-dependent process and can occur in the G1 phase of the cell cycle.
FIG. 3.
Investigation of the association between the rate of cell division and the rate of remethylation. T24 cells treated with 5-Aza-CdR (5 × 10−7 M) were maintained in medium supplemented with either 1 or 10% FCS. DNA was isolated at specific times after 5-Aza-CdR treatment and subsequently treated with sodium bisulfite (8, 11). Methylation of the p16 promoter (p16 Pro.) and exon 2 (p16 Ex. 2) CpG islands was determined by Ms-SNuPE (15) before and after 5-Aza-CdR treatment to ascertain whether remethylation of these regions was associated with the rate of cell division. Remethylation was determined as the degree of recovery (compared to original levels) following maximal demethylation at 72 h. T24 cells grown in medium supplemented with 10% FCS were analyzed as controls. If de novo methylation of p16 is linked to the rate of cell division, then cells maintained in 10% FCS would be expected to remethylate p16 more rapidly than cells dividing more slowly in 1% FCS.
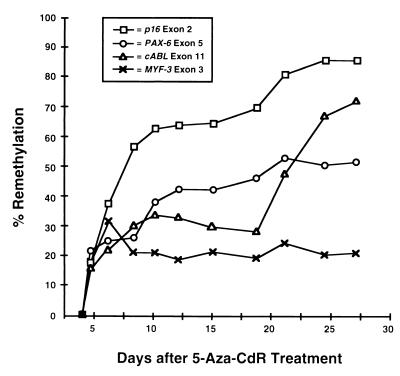
Kinetics of p16, PAX-6, c-ABL, and MYF-3 remethylation in T24 cells after 5-Aza-CdR treatment.
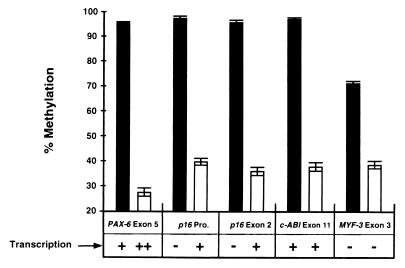
We further investigated the effects of gene transcription on the remethylation of CpG islands downstream from the region of transcription initiation to possibly explain why the p16 promoter became remethylated more slowly than p16 exon 2 (Fig. 3). The remethylation kinetics of the p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 CpG islands were examined because these islands are hypermethylated in T24 cells, they reside within coding sequences, and they become significantly demethylated by 5-Aza-CdR in vitro (Fig. 4). Levels of demethylation were measured beginning 72 h after drug addition because maximal demethylation has been observed in most loci examined at this time point. The levels of p16, PAX-6, c-ABL, and MYF-3 transcription were determined by RT-PCR in the same cells. Transcription of p16, PAX-6, and c-ABL was detected in T24 cells both before and after 5-Aza-CdR treatment; however, MYF-3 was not transcribed in either case. The remethylation kinetics of the p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 CpG islands were analyzed by Ms-SNuPE in T24 cells treated with 5-Aza-CdR (Fig. 5). All the loci examined showed significant remethylation between 3 and 7 days after 5-Aza-CdR treatment; however, the methylation levels of the p16 exon 2, PAX-6 exon 5, and c-ABL exon 11 CpG islands continued to increase for up to 27 days, whereas the methylation levels of MYF-3 exon 3 remained constant. One explanation for this observation is that the absence of MYF-3 transcription somehow prevents remethylation of this gene. Alternatively, these data are consistent with the hypothesis that transcription does not block remethylation of endogenous genes because the loci which were transcribed demonstrated higher rates of remethylation than the MYF-3 gene, which was not transcribed (Fig. 5).
FIG. 4.
Effects of 5-Aza-CdR on the demethylation and transcription of the p16 promoter and exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 in T24 cells. Average methylation values for specific sites within the p16, PAX-6, c-ABL, and MYF-3 exonic CpG islands were measured in T24 cells prior to drug treatment (black bars) and 72 h after treatment with 5 × 10−7 M 5-Aza-CdR (white bars). Error bars indicate the ranges of values obtained. Relative transcription levels of each gene were also estimated by comparison of band intensities (−, +, or ++) before and after drug treatment by RT-PCR analysis (data not shown). The CpG and GC contents of these regions were also analyzed to determine if these exonic sequences fulfilled the criteria of CpG islands in which a DNA sequence of ≥200 bp must have a GC content of ≥0.50 and an observed/expected CpG ratio of ≥0.60 (12). Fragments of 800 bp from each gene, all of which fulfilled the established criteria for CpG islands, were analyzed. In untreated T24 cells, transcription through the p16 promoter and exon 2 CpG islands is not initiated from the p16 promoter but is initiated from the upstream p14 promoter (38). Pro., promoter.
FIG. 5.
Remethylation kinetics of CpG islands in T24 cells after 5-Aza-CdR treatment. T24 cells were treated with 5-Aza-CdR (5 × 10−7 M), and DNA was harvested every 1 to 3 days for up to 27 days. The population doubling time increased approximately 1.8-fold after 5-Aza-CdR treatment, as previously described (3). Only eight cell population doublings were attained between days 3 and 27 because cells transiently entered lag phase each time they were split and reseeded. Methylation of p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 was quantitated at each time point by the Ms-SNuPE technique, and the degree of remethylation at each locus was determined as the degree of methylation compared to the original level in untreated cells at 72 h. Methylation averages from three independent experiments are shown.
Relationship between gene transcription and the remethylation of downstream CpG islands in clones and subclones derived from T24 cells.
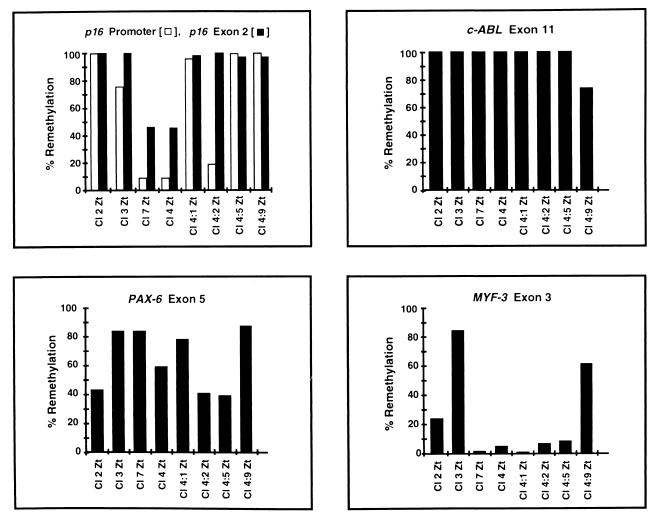
Methylation levels within p16, PAX-6, c-ABL, and MYF-3 were quantitated in clones derived from single T24 cells after 5-Aza-CdR treatment (clones 2, 3, 4, and 7) to determine whether the remethylation observed (Fig. 5) resulted from de novo methylation or from the selection of cells in which CpG islands had not become demethylated by the drug (Fig. 6). Four additional subclones derived from clone 4 were also analyzed (subclones 4:1, 4:2, 4:5, and 4:9). The p16 promoter and exon 2 regions showed complete remethylation in clone 2, which does not express p16 (14). Remethylation of both CpG islands was also more apparent in clone 2 (Fig. 6) than in clones 3, 7, and 4, which expressed p16 at increasing levels (14). It is possible that clone 2 resulted from the selection of cells in which p16 never became demethylated by 5-Aza-CdR. Clones 7 and 4 showed less remethylation of exon 2 than clone 3. This does not disprove the hypothesis that transcription does not block de novo methylation, however, because the p16 gene sequence is transcribed as part of the p14 gene transcript initiating from the upstream p14 promoter (38) in all eight clones and subclones (reference 15 and data not shown). Another explanation is that p16 exon 2 did not become demethylated by 5-Aza-CdR in clone 3. The subclones derived from clone 4 (4:1, 4:2, 4:5, and 4:9) showed complete remethylation of p16 exon 2, while evidence for de novo methylation of the p16 promoter was demonstrated in subclones 4:1, 4:5, and 4:9. These clones and subclones were derived from individual parental cells, and the methylation results clearly demonstrated that de novo methylation rather than selection occurred in these cells. Whether this methylation is due to spreading of methylation from a few sites left unaffected by 5-Aza-CdR or is the same process as that responsible for de novo methylation of completely unmodified sequences is not clear.
FIG. 6.
Remethylation of CpG islands after 5-Aza-CdR treatment in T24 clones and subclones. Clones 2, 3, 4, and 7 were isolated following treatment of parent T24 cultures with 5-Aza-CdR (3 × 10−7 M). This lower dose was utilized (instead of 5 × 10−7 M) to increase the cell survival rate and to facilitate the isolation of the clones. Clone 4 was used for the repeated isolation of single-cell subclones. DNA was isolated, and methylation of the p16 promoter, p16 exon 2, PAX-6 exon 5, c-ABL exon 11, and MYF-3 exon 3 CpG islands was quantitated by Ms-SNuPE. Clones 2, 3, 4, and 7 completed approximately 20 cell population doublings at the time DNA was harvested, whereas subclones derived from clone 4 (4:1, 4:2, 4:5, and 4:9) completed approximately 40 population doublings (data not shown). The degree of remethylation at each locus was determined as the degree of methylation recovery (compared to original levels) following maximal demethylation at 72 h. Methylation averages from three independent experiments are shown.
All clones and subclones expressed c-ABL and PAX-6 (data not shown). Complete remethylation of c-ABL exon 11 was observed in all but subclone 4:9, and remethylation of PAX-6 exon 5 varied significantly (from 40 to 88%) among all eight clones and subclones (Fig. 6). The MYF-3 exon 3 CpG island, which was not transcribed in any of the clones or subclones (data not shown), demonstrated significant remethylation only in clone 3 and subclone 4:9. Increased methyltransferase expression in these two cases may be responsible for this clonal variation in methylation levels. Alternatively, transcription may facilitate but not be required for de novo methylation. Altogether, these results have revealed (i) that de novo methylation of CpG islands occurs in T24 cells after 5-Aza-CdR treatment, (ii) that the observed methylation patterns show clonal variability, and (iii) that gene transcription may be associated with the remethylation of CpG islands within the transcribed regions of genes.
DISCUSSION
CpG islands frequently reside within promoter regions and extend downstream into the transcribed regions of genes (5, 30), and it has been widely documented that hypermethylation of promoter sequence CpG islands causes transcriptional repression (7, 13, 17–19, 39, 44, 47, 53). We investigated the roles of cell division and gene transcription in the remethylation of CpG islands within the p16 promoter, p16 exon 2, and the coding sequences of several additional genes. Our results show that the rate of DNA remethylation is not associated with the rate of cell division and that hypermethylation of CpG islands downstream of promoter sequences does not block transcription initiation and elongation.
Quantitative analyses showed that the kinetics of demethylation of the p16 promoter were directly associated with the activation of p16 mRNA, consistent with previous investigations of critical CpGs in the p16 promoter, where hypermethylation is associated with transcriptional silencing (14). Demethylation of the promoter may directly mediate p16 mRNA induction by 5-Aza-CdR; however, factors associated with demethylation could also include chromatin decondensation, changes in protein-DNA interactions, or activation in trans. The results also demonstrate the utility of the Ms-SNuPE technique (15), with which minimal CpG demethylation within the p16 promoter could be reliably measured. Thus, the sensitivity of this assay allowed us to investigate remethylation of p16 as a function of time in more detail. The fact that the kinetics of remethylation of the p16 gene were not influenced by the number of cell divisions after treatment was interesting in view of the fact that levels of the DNA methyltransferase I (Dnmt 1) mRNA are known to vary in the cycle and to be increased in S-phase cells (49). It is therefore possible that the remethylation observed may be catalyzed by one of the newly isolated putative methyltransferase enzymes Dnmt 3a and 3b (43), which may not show such cell cycle regulation.
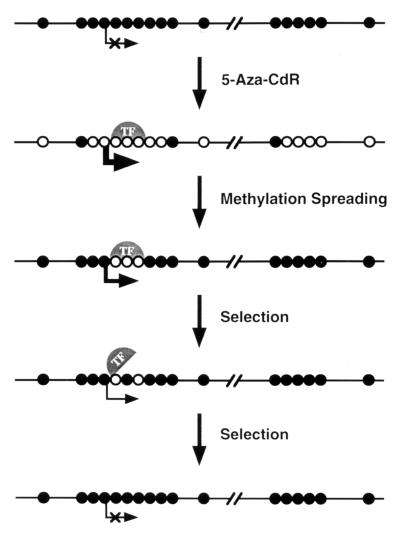
Our results also showed that the p16 exon 2 CpG island became remethylated more rapidly than the p16 promoter CpG island after drug treatment. One explanation for this observation is that protein-DNA interactions within the p16 promoter but not exon 2 interfered with remethylation of this region subsequent to drug treatment (see Fig. 7). This prediction is consistent with studies by Macleod et al. (35, 36) and Brandeis et al. (6), who showed that mutagenesis of Sp1 sites within the CpG islands of the mouse and hamster aprt promoters, respectively, resulted in the de novo methylation of these sequences. Sp1 elements were therefore implicated in the prevention of methylation spreading, and Macleod et al. (35, 36) have proposed that the presence of a functional promoter at the 5′ end of a CpG island maintains its methylation-free status. Alternatively, Brandeis et al. (6) suggested that protein-occupied Sp1 sites in the hamster aprt promoter prevent methylation spreading by “protecting” CpGs from methylation. With regard to this model, it is possible that transcription factors associated with regions of the p16 promoter protect it from methylation following demethylation by 5-Aza-CdR. This is indirectly supported by analyses of p16 promoter methylation in single-cell T24 clones, which revealed a localized patch of demethylation induced by 5-Aza-CdR within a region containing putative transcription initiation sites (14). Additional experiments must be performed to identify factors which possibly bind to this region, block methylation and prevent further spreading of the patch.
FIG. 7.
Model for the remethylation of promoter and coding sequence CpG islands of the p16 gene after 5-Aza-CdR treatment. Promoter and coding sequence CpG islands of growth-regulatory genes such as p16 become remethylated at different rates after 5-Aza-CdR treatment. Following 5-Aza-CdR-mediated demethylation and transcriptional activation, remethylation may first appear in a CpG island downstream of the promoter, whereas transcription factors (TF) associated with a demethylated promoter CpG island protect it from remethylation. Patches of demethylation have been observed by bisulfite genomic sequencing of the p16 promoter in single-cell clones after 5-Aza-CdR treatment, providing support for this interpretation (14). Cells which acquire promoter methylation in one or more growth-regulatory genes may subsequently exhibit selective growth advantages due to the obstruction of transcription factor binding, leading to gene silencing. The protection of demethylated promoter sequences by transcription factors may explain how exonic CpG islands can become remethylated more rapidly than promoter islands after 5-Aza-CdR treatment, as DNA-binding proteins may protect promoter sequences from methylation. Methylated CpG sites are depicted as filled circles; unmethylated sites are shown as open circles.
The model in Fig. 7 illustrates how promoter and coding sequence CpG islands of a growth-regulatory gene (like p16) may become de novo methylated at different rates after 5-Aza-CdR treatment in vitro. Following 5-Aza-CdR-mediated demethylation and transcriptional activation, remethylation may first appear in CpG islands located downstream of promoter sequences because transcription factors bound to a demethylated promoter protect it from methylation (6, 25). Cells which do acquire promoter methylation may subsequently be selected by the acquisition of a selective growth advantage due to methylation-coupled “resilencing” of growth-regulatory genes (3, 23).
The relative sizes and CpG densities of the p16 promoter and exon 2 CpG islands may also explain why transcription was associated with the increased rate of remethylation of exon 2. These characteristics of CpG islands have been proposed to influence transcriptional repression more than their relative positions within genes (20, 21). Based on this theory, hypermethylation of a smaller CpG island with fewer CpGs should have a lesser effect on transcriptional repression than a larger island with a higher CpG density. However, the higher rate of remethylation of p16 exon 2 associated with gene expression was probably not due to a significantly smaller size and/or fewer CpGs in p16 exon 2, because the promoter and exon 2 CpG islands have similar CpG densities, with the p16 exon 2 island spanning a region approximately 20% smaller than the promoter CpG island (data not shown).
Recent studies by Wutz et al. (52) have demonstrated that methylation of the intronic CpG island of the maternal copy of the mouse insulin-like growth factor 2 receptor gene (Igf2r) was associated with its expression. This region, which serves as a promoter for transcription in the opposite direction, was unmethylated in the paternal copy, correlating with methylation of the upstream CpG island of Igf2r. Additionally, CpG sites in the hypoxanthine phosphoribosyltransferase gene on the active X chromosome are more methylated than those on the inactive allele (34). These observations, in addition to our studies of p16 exon 2, PAX-6 exon 5, and c-ABL exon 11, are consistent with a role for transcription in the de novo methylation of CpG islands downstream of gene promoters. Nevertheless, transcription is probably not always required for methylation, because CpG islands within the coding sequences of transcribed genes are often unmethylated in eukaryotic cells (24). Transcription-coupled mechanisms which may facilitate methylation of CpG islands include helical unwinding and/or DNA strand separation by presenting DNA substrates more accessible and favorable for de novo methylation. Additional molecular processes or conditions which may further influence de novo methylation include the following activities: protein-DNA interactions (6, 35, 36), chromatin decondensation and structural changes associated with replication (1, 25, 29), histone deacetylation (28, 42), or the proximity of Alu sequences to certain CpG islands (16, 37). Our studies raise the intriguing question of whether there is a causal link between transcription and the de novo methylation of CpG islands; thus, the association between these processes requires further examination.
ACKNOWLEDGMENTS
We thank Gangning Liang for his assistance with the figures and TuDung Nguyen for detailing the primer sequences and reaction conditions for PCR-based amplification of the c-ABL gene.
This work was supported by USPHS grant R37 CA49758 from the National Cancer Institute.
REFERENCES
- 1.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 2.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–146. [PubMed] [Google Scholar]
- 3.Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 4.Bender C M, Zingg J M, Jones P A. DNA methylation as a target for drug design. Pharmacol Res. 1998;15:175–187. doi: 10.1023/a:1011946030404. [DOI] [PubMed] [Google Scholar]
- 5.Bird A P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 6.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 7.Cedar H, Stein R, Gruenbaum Y, Naveh-Many T, Sciaky-Gallili N, Razin A. Effect of DNA methylation on gene expression. Cold Spring Harbor Symp Quant Biol. 1983;47:605–609. doi: 10.1101/sqb.1983.047.01.071. [DOI] [PubMed] [Google Scholar]
- 8.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantinides P G, Jones P A, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 11.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 14.Gonzalgo M L, Hayashida T, Bender C M, Pao M M, Tsai Y C, Gonzales F A, Nguyen H D, Nguyen T T, Jones P A. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- 15.Gonzalgo M L, Jones P A. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graff J R, Herman J G, Myohanen S, Baylin S B, Vertino P M. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 17.Herman J G, Jen J, Merlo A, Baylin S B. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 18.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 20.Hsieh C-L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh C-L. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol Cell Biol. 1997;17:5897–5904. doi: 10.1128/mcb.17.10.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa J P, Vertino P M, Boehm C D, Newsham I F, Baylin S B. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci USA. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones P A. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 24.Jones P A. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 25.Jones P A, Laird P W. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 26.Jones P A, Taylor S M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 27.Jones P A, Taylor S M. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981;9:2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 29.Kass S U, Goddard J P, Adams R L. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 31.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 32.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 33.Liang G, Salem C E, Yu M C, Nguyen H D, Gonzales F A, Nguyen T-D T, Nichols P W, Jones P A. DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics. 1998;53:260–268. doi: 10.1006/geno.1998.5502. [DOI] [PubMed] [Google Scholar]
- 34.Lock L F, Melton D W, Caskey C T, Martin G R. Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol Cell Biol. 1986;6:914–924. doi: 10.1128/mcb.6.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macleod D, Ali R R, Bird A. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: implications for the origin of CpG islands. Mol Cell Biol. 1998;18:4433–4443. doi: 10.1128/mcb.18.8.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 37.Magewu A N, Jones P A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. A novel p16INK4A transcript. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 39.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 40.Michalowsky L A, Jones P A. Differential nuclear protein binding to 5-azacytosine-containing DNA as a potential mechanism for 5-aza-2′-deoxycytidine resistance. Mol Cell Biol. 1987;7:3076–3083. doi: 10.1128/mcb.7.9.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohandas T, Sparkes R S, Shapiro L J. Reactivation of an inactive X human chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 42.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 43.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 44.Ottaviano Y L, Issa J P, Parl F F, Smith H S, Baylin S B, Davidson N E. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- 45.Razin A. DNA methylation patterns: formation and biological functions. In: Razin A, Cedar H, Riggs A D, editors. DNA methylation: biochemistry and biological significance. New York, N.Y: Springer-Verlag Press; 1984. pp. 127–146. [Google Scholar]
- 46.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 47.Sakai T, Toguchida J, Ohtani N, Yandell D W, Rapaport J M, Dryja T P. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- 48.Salem, C. E., I. D. C. Markl, C. M. Bender, P. A. Jones, and G. Liang. PAX6 expression and methylation in human tumor cells. Submitted for publication. [DOI] [PubMed]
- 49.Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991;266:10027–10030. [PubMed] [Google Scholar]
- 50.Tate P H, Bird A P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 51.Taylor S M, Jones P A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982;162:679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- 52.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Shields T, Crenshaw T, Hao Y, Moulton T, Tycko B. Imprinting of human H19: allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am J Hum Genet. 1993;53:113–124. [PMC free article] [PubMed] [Google Scholar]
- 54.Zion M, Ben-Yehuda D, Avraham A, Cohen O, Wetzler M, Melloul D, Ben-Neriah Y. Progressive de novo DNA methylation at the bcr-abl locus in the course of chronic myelogenous leukemia. Proc Natl Acad Sci USA. 1994;91:10722–10726. doi: 10.1073/pnas.91.22.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]