Abstract
Centrosomes were first described by Edouard Van Beneden and named and linked to chromosome segregation by Theodor Boveri around 1870. In the 1960–1980s, electron microscopy studies have revealed the remarkable ultrastructure of a centriole -- a nine-fold symmetrical microtubular assembly that resides within a centrosome and organizes it. Less than two decades ago, proteomics and genomic screens conducted in multiple species identified hundreds of centriole and centrosome core proteins and revealed the evolutionarily conserved nature of the centriole assembly pathway. And now, super resolution microscopy approaches and improvements in cryo-tomography are bringing an unparalleled nanoscale-detailed picture of the centriole and centrosome architecture. In this chapter, we summarize the current knowledge about the architecture of human centrioles. We discuss the structured organization of centrosome components in interphase, focusing on localization/function relationship. We discuss the process of centrosome maturation and mitotic spindle pole assembly in centriolar and acentriolar cells, emphasizing recent literature.
Keywords: centrosome, centriole, pericentriolar material, centrosome maturation
1. Introduction
Centrosomes are multifunctional membrane-less organelles. In the core of a centrosome is a nine-fold symmetrical cylindrical microtubule (MT)-based structure called a centriole (Figure 1), which organizes the second major part of the centrosome, the pericentriolar material (PCM). A typical vertebrate somatic cell has only two centrosomes (Figure 2A), often localized adjacent to each other and situated in the physical center of the cell near the nucleus [1] (hence the name ‘central body’). But even when adjacent, two centrosomes retain their individuality and functional and structural differences (for the review detailing structural and functional differences between two resident centrosomes please see [2]). In interphase, centrosomes nucleate, anchor, and regulate nucleation of interphase microtubules (MTs), participate in cellular signaling, and influence tissue architecture and cell motility [3–9]. Centrosomes can move close to the cell membrane, where the older centriole converts into a basal body and assembles a primary or a motile cilium (Figure 1A). Primary cilia transmit signals between the cell environment and cell interior and are critically important for tissue homeostasis and development [10–13]. Motile cilia move sperm [14, 15], in differentiated epithelial multiciliated cells promote the flow of fluids critical for mucus clearance, left-right patterning during development, or transport the egg cell from the ovary to the uterus [11, 16–18]. Finally, centrosomes facilitate formation of mitotic spindles. Before mitosis, centrosomes undergo a process called centrosome maturation, in which they expand in size, increase MT nucleation and form the poles of mitotic spindles (Figure 2A and 3B).
Figure 1. Architecture of a human centrosome.

(A) Electron micrographs of centrioles from RPE-1 cells. Left: Longitudinally sectioned centriole (basal body) associated with a cilium. Right: Cross-sectioned centriole at its proximal end. Nine microtubule triplets surrounded by electron-denser pericentriolar material are prominent. (B) A scheme depicts structural features of a centrosome containing a fully developed (mature) centriole. Centriole walls are composed of nine sets of triplet microtubules (A, B, and C), which are arranged in a nine-fold symmetry. On the proximal end, A and C microtubules are connected via a linker (in red). The inner scaffold (in magenta) is proposed to confer stability to the centriole. Putative scaffold components are Poc1B, FAM161A, POC5, Centrin-2, and WDR90. The inner scaffold is connected to microtubule walls and is connected to microtubules via specific densities (in magenta) localized at the A and B tubule junction. Centriole is polarized from proximal to distal end. The proximal end is surrounded by the Pericentriolar material (PCM, yellow), which harbors components for microtubule nucleation and anchoring, such as ɣ-TuRC. On the distal end, centriole has a “cap” of proteins such as CP110 and Cep290, which is involved in regulation of centriole length. The distal end also harbors two sets of appendages. Subdistal appendages are robust and anchor microtubules. Distal appendages are required for ciliogenesis (see Figure 2A) functions). The order of subdistal and distal appendage components illustrates their approximate mutual localization on appendages.
Figure 2. Centriole duplication cycle and procentriole structure.
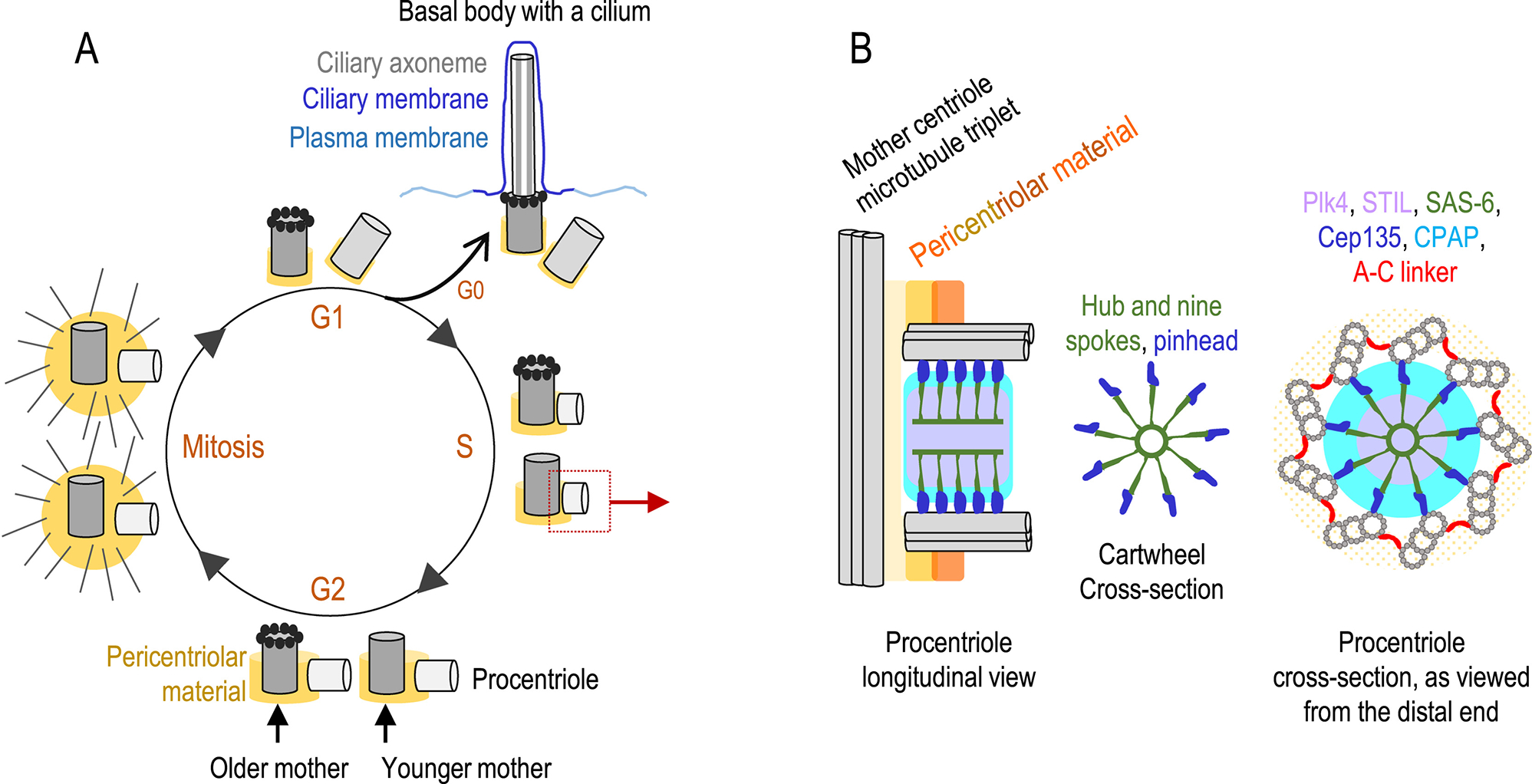
(A) Centriole cycle. After mitosis, somatic vertebrate cells have two centrioles. One centriole is older, and harbors appendages (black ring on the distal end). The older centriole can, usually after exiting the cell cycle, convert to a basal body and form a primary cilium, a sensory organelle. In cycling cells, at the beginning of S phase, both resident (mother) centrioles duplicate by forming one procentriole at a right angle, adjacent to their proximal end. Procentrioles continue to elongate through S, G2 and mitosis, and gradually distance from the mother centriole, but remain in the same complex (they are engaged). Before mitosis, centrosomes undergo maturation, in which they expand PCM and increase MT-nucleation production. In mitosis, centrosomes organize mitotic spindle poles. After mitosis, each daughter cell inherits one mother centriole and one procentriole (now called a daughter centriole). Mother and daughter centriole separate (disengage) and each forms a centrosome. (B) Procentriole architecture. Procentriole assembly begins with the assembly of a nine-fold symmetrical structure called a cartwheel, which helps establish procentriole symmetry. The cartwheel has three prominent elements: a hub, nine spokes, and nine pinheads. Plk4 kinase initiates centriole assembly by phosphorylating STIL, which promotes the recruitment of SAS-6 to the site of procentrioles and its oligomerization results in cartwheel formation. Other proteins like Cep135 and CPAP are also incorporated to the centriole. Cep135 is a putative pinhead component and helps the assembly of procentriole microtubules. Cartwheel is, in vertebrate cells, removed from procentrioles in mitosis.
Figure 3. Organization of centrosomal components in interphase and mitosis.
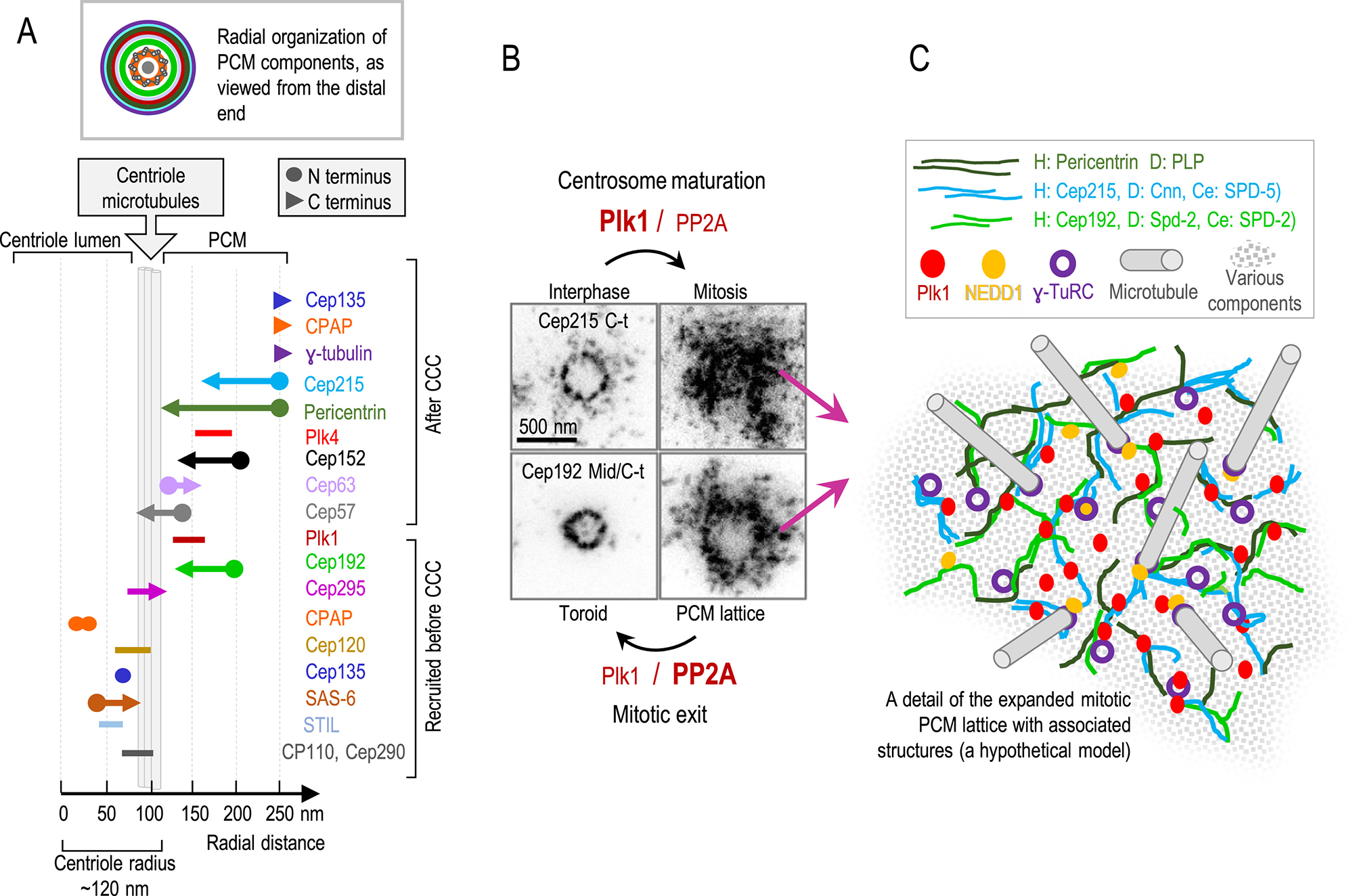
(A) Radial distribution of centrosomal components from the centriole center. Centrosome components are recruited gradually and hierarchically during centrosome formation which is reflected in their radial distribution from the centriole center. STIL, SAS-6, Cep135, Cep120 and CPAP, are recruited early and localize closer to the centriole lumen. Cep295 recruits in S phase and recruits Cep192. Plk1 drives centriole-to centrosome conversion (CCC), a process which allows procentrioles to accumulate PCM and gain the ability to duplicate and nucleate microtubules. Cep57, Cep63, and Cep152 recruit centriole initiating kinase Plk4. Pericentrin/Cep215/Cep192, build the PCM which harbors components needed for microtubule nucleation. The roles of CPAP and Cep135 in the PCM are less understood. The scheme was generated based on imaging data from [103–105, 108, 111, 114, 119, 120]. (B) Stimulated emission depletion microscopy images of two PCM components, Cep192 and Cep215, from HeLa cells labeled using Bethyl anti rabbit A302–324A-M and IHC-00063, respectively. Both proteins are localized in toroids in interphase. Increased phosphorylation by Plk1 before mitosis promotes PCM expansion called centrosome maturation. Toroidal organization of Cep192 and Cep215 is replaced with a lattice-like arrangement. At the end of mitosis, drop in Plk1 levels and dephosphorylation of PCM by PP2A promote PCM disassembly and return it to an interphase state. (C) A hypothetical model of PCM lattice in mitosis. Cep192, Cep215 and pericentrin form a porous lattice populated by functional components such as γ-tubulin recruiting factor NEDD1, γ-TuRC and Plk1. H: human; D: Drosophila; Ce: C. elegans.
In humans, defects in centriole structure, centrosome protein function or centrosome number cause mitotic and chromosome segregation errors, aneuploidy, cell cycle arrest [19–22], defects in cell polarity and tissue architecture [4, 23, 24], and negatively affect cilia formation and function. Expectedly, the list of human disorders linked to centrosome defects is large and growing. Centrosome structural and numerical defects can cause cellular transformation [25, 26], are prevalent in various types of cancer [27], and are the cause of hereditary diseases such as microcephaly and ciliopathies [28–30]. The effects of centrosome aberrations on human health have been extensively reviewed in excellent recent reviews [4, 7, 11, 16, 31–37].
2. Centrosome organization
Centrioles and basal bodies (CBBs) are present in all major eukaryotic groups, while they are absent in yeast and higher plants, likely due to loss during evolution [38–40]. CBBs are structurally conserved across different species in the sense that they are stable nine-fold symmetrical cylinders with the cylinder walls made of MTs (Figure 1) [41–43]. In turn, basic organizational concepts are also conserved across centrosomes, although ultrastructural analyses by electron microscopy and cryo-tomography show that CBBs from different species carry many unique features [41, 44]. One notable difference is in MTs that make up a centriole’s wall, which can be organized as nine single, double, or triple MTs, depending on the species. Another notable difference is centriole length, which can be as small as 100 nm (e.g. in C. elegans) and as large as 2000 nm (e.g. Drosophila early spermatocytes) or even 4000 nm as in the flagellate Trichonympha [45]. The third prominent ultrastructural difference is the presence of a cartwheel. The cartwheel is a nine-fold symmetrical structure, which forms during centriole early development and helps establish the nine-fold symmetry of the centriole (Figure 2B) [46–48]. The cartwheel remains a permanent fixture within the centrioles of some species (Chlamydomonas [49], Drosophila [50], and Paramecium [51]) for example, while it degrades during centriole maturation in vertebrate centrioles [42]. Additionally, more subtle species-specific ultrastructural differences have been detected even between centrioles built of nine MT triplets [52–54]. Nevertheless, a common model of the canonical centriole built of nine MT triplets can still be drawn.
2.1. Architecture of human centrioles
In this chapter, we will discuss the organization of human centrioles from somatic cycling cells, as representatives of vertebrate centrioles. Human somatic centrioles measure ~240 nm in outer diameter and 330–600 nm in length [55]. They exhibit proximal-distal polarity, which can be determined by the radial angle of MT triplets, which always turn clockwise if centrioles are viewed from the distal end [56] (Figure 1B). The inmost MT in the triplet -- A tubule -- contains 13 α/β-tubulin protofilaments, while the middle B and the outmost C tubules have 10 [52, 57, 58]. Toward the centriole distal end, in human centrioles, MT triplets turn into doublets by discontinuing the C tubule. On the proximal end, the A and the C tubules of adjacent MT triplets are connected via a linker of unknown composition [42, 43, 45, 57, 59, 60]. Central and distal parts of the centriole MT wall are underlined by an inner electron dense scaffold [42, 61, 62]. Recent cryo-tomography reconstruction of centrioles from Paramecium and Chlamydomonas revealed that the inner scaffold is sheet-like with underlying helical patterns which differ amongst species [61]. In human centrioles, a short stem connects the scaffold with the junction between A and B tubules (Figure 1B). In Paramecium and Chlamydomonas, MTs and the scaffold are connected with the stem and additional elements [61]. The scaffold is thought to provide support for centriole MTs. Based on their approximate colocalization, centriolar proteins Poc1B, FAM161A, POC5, Centrin-2 and WDR90 are suggested scaffold candidates [58, 61].
On their distal end, centrioles carry two types of attachments called subdistal and distal appendages (SDAs and DAs, respectively, Figure 1). By classical electron microscopy, subdistal appendages are bulky, usually cone-shaped densities. They vary in shape and number, even within the same centrosome or between cells of the same population [2, 63, 64]. SDAs emanate from centriole MTs and on their ends anchor cytosolic MTs, which helps regulate centrosome positioning [65–68]. SDA components are organized orderly with ODF2 closest to the centriole MTs, followed by CEP128 and Centriolin with CCDC120, Ninein and CEP170 localized toward the periphery [69, 70]. SDAs are removed from centrioles during G2, they are not detectable in mitosis, and they are rebuilt in G1. The physiological significance for this remodeling is unclear.
Just above SDAs, ~100 nm from the distal centriole end, emanate nine DAs, that appear as ~150 nm long protrusions [42, 71]. Looking from the side, DAs are angled ~34° from centriole longitudinal axis (Figure 1). Each DA is attached to two adjacent MT triplets (or doublets when the outer MT is lost). The DA base further continues into a stem, which, in turn, ends with a fibrous and more electron dense head. DAs are required for the attachment of ciliary vesicles to the centriole during ciliogenesis and their subsequent fusion with the cytoplasmic membrane (Figure 1A), and they are thus, essential for ciliogenesis [72–76]. The components of DAs, similarly to SDAs, follow hierarchical organization [73]. CCDC41/Cep83 localizes to the site of the DA base and is needed for the recruitment of SCLT1. SCLT1 recruits more distal components FBF1 and Cep164. The C-terminal portion of Cep164 colocalizes with the head densities, while its N terminus stretches between and beyond DA heads [71]. Additional outer DA proteins such as TTBK2 (a kinase required for ciliogenesis [77]), ANKRD26 [71], and PIDDosome component PIDD1 also dynamically co-localize with Cep164 at outer portions of DAs [78, 79]. ANKRD26, which localizes to DAs downstream from SCLT1 but independently of Cep164, initiates a centriole-driven signal to limit cell proliferation following centrosome amplification [78, 79]. ANKRD26 was found to recruit PIDD1 to the DAs, which is critical for activating the PIDDosome in response to supernumerary centrosomes and initiating p53-dependent cell cycle arrest [79]. Thus, in addition to their more notable roles in ciliogenesis, DAs appear to serve as a platform linking the centrosome with the cell cycle.
DAs dismantle during late G2 and during mitosis, so that the outer DA components such as Cep164, TTBK2, ANKRD26, and FBF1 are displaced from DAs, while the inner components such as Cep83 and SCLT1 remain associated with the centriole in preserved nine-fold arrangement. Such mitotic DA remodeling is regulated by mitotic kinase Nek2 and to some extent Plk1 [71, 80]. Inhibition of these two kinases perturbed displacement of DAs components and reabsorption of primary cilia markers before mitosis, resulting in asymmetric inheritance of the DAs and ciliary signaling components [80].
2.2. A quick guide through the centriole duplication cycle
Unlike cytosolic MTs which are highly dynamic, centrioles of somatic cells are permanent structures, which segregate to daughter cells during cell division and continue to organize centrosomes through generations of cells. To maintain the required centriole number, two resident centrioles inherited after cell division duplicate precisely once in each cell cycle (Figure 2A). Duplication begins in early S phase when one new centriole (procentriole) initiates orthogonally near the proximal region of the resident (mother) centriole. New centrioles form ~50 nm from the proximal end of mother centriole wall (Figure 2A and B). In most species including humans, initiation of a new centriole starts with the formation of a cartwheel, a nine-fold symmetrical structure which is thought to provide symmetry and stability to the procentriole (Figure 2B, [48, 81]). In the center of the cartwheel is a ~22 nm wide central hub and nine spokes that radially emanate from it. [42, 43, 45]. The hub and the spokes are composed of SAS-6 which can homodimerize and assemble a higher order structure. SAS-6 dimers then interact in such a way that the N termini form the cartwheel hub, while the C termini extend outwards, forming nine spokes [82]. Such nine-fold symmetrical SAS-6 assembly units are additionally laterally stacked and make the proximal procentriole core (Figure 2B) [52, 61, 83]. The spokes and the A tubules are further connected through a pinhead, likely containing Cep135/Bld10 [84–86]. The cartwheel is thought to provide symmetry to centrioles and is a scaffold for the formation of procentriole MTs, which subsequently assemble around it and elongate [41, 81, 87]. In vertebrates, the cartwheel dismantles from procentrioles in mitosis [42], although it is a permanent structure in centrioles of Drosophila [50] and Paramecium [51], among other organisms. The mechanisms of cartwheel removal from procentrioles in vertebrates are not clear, but degradation of key cartwheel proteins STIL and SAS-6 in mitosis and G1 phase by proteasome [88, 89] could be the contributing factor.
Cartwheel formation is triggered upon S phase entry by interaction of the kinase Plk4 (SAK in Drosophila) [90, 91] with cartwheel component STIL [92, 93]. During cartwheel initiation, Plk4, which in G1 is distributed around the mother centriole, concentrates, together with STIL, to the site of the future procentriole [94, 95]. Phosphorylated STIL is thought to, in turn, recruit SAS-6 to the Plk4/STIL focus [96–98], promoting its self-oligomerization into a nine-fold cartwheel scaffold that is capable of lateral stacking and interacting with other centriolar components [47, 82, 99]. Plk4/STIL complex is, at the same time, inhibitory for the initiation of additional procentrioles around the same mother centriole. This fascinating mechanism of internal centrosome regulation has yet to be understood mechanistically (for detailed discussions about centriole initiation mechanisms please see [93, 95, 100–102]). It is also currently unclear how other components necessary for centriole initiation, such as CPAP (Sas-4 in Drosophila) and Cep135 get integrated to the Plk4/STIL/SAS-6 complex.
Following the assembly and elongation of MTs around the cartwheel (A tubules form first and C tubules last), dozens of centriolar components gradually accumulate in the lumen and around the procentriole MT wall, contributing to its structural stability and allowing a gradual assembly of PCM components (at this stage a new centriole is also referred to as a daughter centriole). Through mitosis, a mother and its daughter centriole remain embedded in the same PCM cloud and segregate together to the new daughter cell (Figure 2A). After mitotic exit, the daughter centriole becomes autonomous, gradually accumulates PCM components and gains the ability to duplicate and nucleate MTs. Before mitosis, in somatic cells, both centrosomes have comparable capacity for MT nucleation, a critical feature for successful participation in mitosis (the human centrosome assembly process is reviewed in [2]).
2.3. Organization of pericentriolar material.
Proximal ends of centrioles organize PCM, creating a milieu for the assembly of new centrioles, and MT nucleation and anchoring (Figure 1 and Figure 2B). While centrioles are easily distinguishable by electron microscopy, the PCM, is unremarkable. By light microscopy, until recently, most PCM components had appeared as spotty signals that, to some extent, colocalize with each other. However, a recent development of super resolution microscopy has brought a revitalization in centrosome research. It has allowed charting of centrosome components in nano scale resolution, revealing a remarkable order of the centrosomal components during interphase. In combination with in vitro analyses of PCM components, it provided new insights into the (seemingly) disordered nature of centrosomes in mitosis.
2.3.1. Interphase pericentriolar material: a marvel of hierarchical organization
Mapping N-terminally or C-terminally fused fluorescent tags and centrosome protein domains by antibodies, has revealed that many centrosome components extend radially from the centriole center in an N-C terminus specific fashion, localizing in toroids of distinct sizes within and outside MT walls (Figure 3A, [103–108]. Because of the largely reproducible radial distance of centrosome components from the centriole center, several scenarios were put forward to categorize components of Drosophila and human centrosomes into concentric zones [103, 108, 109]. But systematizing centrosome components to the zones is challenging, as it cannot be universally applied to all types of centrosomes, to all cell cycle phases, and to all centrosomal components. For instance, in humans, ɣ-tubulin, CPAP, and Cep135 are localized both in the centriole lumen and PCM [103, 110] (Figure 3A). Some radially extended and large proteins such as Cep152 and pericentrin stretch over several proposed concentric zones [103–105]. Cep57, for instance, has one of its termini within and another outside the centriole wall [111] and would, thus, fall somewhere in between the zones. Radial distribution of centrosomal components largely recapitulates the order by which these components are recruited during centriole and centrosome biogenesis. Proteins localized within and closer to the centriole MT wall mostly associate with the centriole at early assembly stages and often assist in the incorporation of more radially extended proteins. In continuation, we will summarize the current information about the organization of centrosome components in human centrosomes in interphase, not adhering to any proposed classification but pointing to their localization/function relationship.
But first, it is important to digress and describe a process termed centriole-to-centrosome conversion (CCC) [112]. Procentrioles are initially bare and cannot duplicate or nucleate MTs. In order to accumulate PCM on the outer MT wall, which is needed to gain these functions, procentrioles must undergo CCC, which, in cycling cells, occurs around the time of their first mitosis and the ensuing G1. During that time, centrioles gradually assemble more and new PCM components. For CCC to occur, the centriole needs to be properly structured and should undergo a “priming” process by Plk1 kinase (Polo in Drosophila and PLK1 in C. elegans), which takes place during G2 and mitosis [113]. Which molecular events underly such “priming” is still an open question. How are centrosome components, especially PCM components, loaded and retained on the centrosomes is also only partially understood, but recent research has provided us with many new details.
In humans, CCC requires a scaffold protein Cep295 [112, 114, 115]. Depletion of Cep295 resulted in a failure of nascent centrioles to stabilize after mitosis and accumulate PCM components including Cep192, Cep152, ɣ-tubulin, and pericentrin [112, 114, 115]. Cep295 was proposed to promote CCC by directly binding and loading the PCM component Cep192 to procentrioles [114]. However, Cep295/Cep192 interaction may not be sufficient for CCC, because similar defects in the PCM recruitment were observed in the absence of other centriole scaffold proteins such as POC1B and Cep44, which reportedly act downstream from Cep295 [115]. Atorino and colleagues have found that depletion of either Cep295, Cep44 or POC1B leads to defects in structuring of centriole MT triplets and to reduced glutamylation of centriole MTs [115, 116]. Thus, Cep295, together with other centriole inner scaffold proteins might support CCC also by assuring centriole MT integrity. Cep295 localizes around the proximal part of the centriole with the C terminus adjacent to the outer MT wall (Figure 3A) [114]. However, although present on centrioles before and after CCC, Cep295 does not seem to be required for Cep192 maintenance on mother centrioles. This is consistent with the observation that, unlike Cep295 which is restricted only to the proximal end, Cep192 localizes along the entire centriole wall (Fig. 3B, [114, 117]). The hierarchy of Cep295 recruitment with respect to cartwheel proteins is not entirely clear, as some authors place Cep295 upstream of the cartwheel initiation complex [114], and others downstream [112, 116, 118]. Ana1, which is a Cep295 counterpart in Drosophila, is loaded to procentrioles downstream of cartwheel protein Cep135 [119].
Regardless of the upstream mechanisms leading to Cep192 recruitment, Cep192 seems to be critical for the subsequent CCC, and is also one of the first PCM components to organize around growing centrioles. It starts to accumulate sometime in late S/G2, and its radial distance increases as procentriole grows. After CCC, the C and N termini of Cep192 are detected at a similar radial distance of ~200 nm by structured illumination microscopy by some authors [104], which would imply that Cep192 is not radially extended within the PCM. However, other authors place Cep192 C terminus at a radial distance of ~120–140 nm [103, 120], which is also consistent with our analysis (Figure 3B). Cep192 seems to be a predominant ɣ-tubulin ring complex (γ-TuRC) binding factor on the human centrosome [9, 121]. γ-TuRC is a multiprotein complex which, in association with various adaptor and activator proteins drives the nucleation of MTs ([122–125], reviewed in [126, 127]). Although other PCM components such as Cep215 (also called CDK5RAP2) and pericentrin bind γ-TuRC [121, 128, 129], depletion of Cep192 reduces the amount of centrosome ɣ-tubulin and centrosome MT nucleation by 50% and reduces the centrosomal levels of Cep215 by ~20%. In controlling MT nucleation on the centrosome, Cep192 acts somewhat antagonistically to pericentrin [121]. In the absence of Cep192, cells maintain normal MT-nucleating capacity by increasing the production of MTs on other sites, such as the Golgi [9, 121].
Within the centrosome, pericentrin shows an extended configuration with its C terminal PACT (pericentrin-AKAP450 centrosomal targeting) domain [130] localized in the centriole vicinity and its N terminal domain facing outwards, at a radial distance of ~250 nm (Figure 3A). Extended radial localization is also characteristic of Cep215, which has its N terminus at ~250 nm and the C terminus closer to the centriole center. Through N-terminal Centrosomin motif 1 (CM1), Cep215 can, in addition to binding, also activate γ-TuRC complex to nucleate MTs [131, 132]. Consistently, γ-tubulin can be detected as a toroid at a radial distance of 200–250 nm, which is similar to the position of the Cep215 CM1 domain. Considering its relatively small size, γ-tubulin would not be expected to show an extended configuration. γ-tubulin is also present in the lumen of mother centrioles, although its function there has not been determined yet [103].
In its second role in interphase, Cep192 functionally cooperates with Cep152 in loading of centriole initiating kinase Plk4 to the centrosome [94, 117]. Per suggested mechanism, both, Cep192 and Cep152, act as independent scaffolds forming distinct complexes with Plk4 on their N termini [94]. Cep192 is proposed to bind Plk4 first. Then, Plk4 is switched to the Cep152 scaffold via Cep152 N terminus, where it remains [94]. However, centrioles still initiate in the absence of Cep192 [94, 117, 121, 129], so clarifying the exact requirement of Cep192 for centriole initiation would be warranted. Cep192 removal also reduces (but does not eliminate) Cep152 associated with centrosomes [117]. It is unclear how removal of Cep192 reduces centrosomal Cep152 levels, since for centrosomal loading, Cep152 was shown to require Cep63 and Cep57 [111, 133] (and Cep57 paralog Cep57/1 [134]). Cep57, which localizes at the centriole wall, is proposed to bind centriole MTs via its C terminus and with its N terminus to interact with the N terminus of Cep63. The C terminus of Cep63, in turn, interacts with the C terminus of Cep152, tethering it around the centriole [111].
After CCC, in addition to the centriole lumen, PCM also contains CPAP and Cep135, although their N-C orientation has not been determined yet. CPAP was shown to interact with Cep152, pericentrin and Cep192, which could, potentially, all mediate its PCM localization [135–137]. CPAP/Sas-4 N terminus has also been suggested to tether cytoplasmic PCM components for their recruitment to centrosomes [138]. However, following work demonstrated that PCM components do not recruit to the centrosomes as a part of the Sas-4 complex [139], in agreement with their monomeric nature in the cytoplasm in C. elegans [140]. In human and Drosophila cells, CPAP also recruits Plk1 to the MTs of nascent centrioles before CCC [141, 142]. Plk1, a kinase that drives CCC [113], is usually not detectable on procentrioles in their early stages. On mature centrioles, it is localized on centriole outer MT wall.
It is apparent from many published studies that there is a dependency between various PCM components for their centrosomal localization during interphase. It would be critically important to dissect how removal of individual PCM components changes the sub-centrosomal localization pattern of other components and combine this knowledge with biochemical data to gain a better insight into a functional /architectural relationship between centrosome components.
Here we described the interphase organization of key PCM components of a human centrosome. However, different mechanistic schemes operate in building of interphase centrosomes from other species. For instance, Cep63 is not found in either Drosophila or worms. The Cep152 homolog Asterless exists in Drosophila (but is absent from worms) and is recruited hierarchically via Ana1 and Cep135. Cep295/Ana1 is not conserved in worms but in C. elegans, recruitment of the Cep192 homolog SPD-2 occurs via its functional homolog SAS-7, which, like Cep295/Ana1 localizes near MT wall [143]. The pericentrin homolog PLP (pericentrin-like protein) is present in Drosophila but again, has not been identified in worms. Likewise, a Cep44 homolog has not been found in Drosophila, pointing again to mechanistic and structural differences between centrosomes from different species.
2.3.2. Mitotic centrosomes: when toroids turn into lattices
In contrast to highly structured interphase PCM, a predominant feature of mitotic PCM is an extended and amorphous-looking appearance [104, 105, 144] (Figure 3B and C). Mitotic centrosomes are formed in a process called centrosome maturation [145], which starts in G2 and is characterized by a gradual expansion of the PCM volume due to increased incorporation of PCM components and various MT-nucleating and MT-associating proteins, such as the ɣ-TuRC to the expanded PCM matrix [146]. During mitosis, some centrosomal proteins, such as centriole structural elements, Cep295, Cep63, and Cep152, retain their original interphase radial organization and remain at similar levels as in interphase. By contrast, PCM components which make up expanded PCM, such as ɣ-tubulin, Cep192, pericentrin, and Cep215, lose radial arrangement and appear amorphic and unstructured (Figure 3B).
The exact structural organization of mitotic PCM is not known. The most frequent perception is based on the ultrastructural analysis of isolated centrosomes from surf clam oocytes after their exposure to potassium iodide [147]. The salt treatment stripped away many PCM components from oocyte centrosomes, including MT-nucleating factors such as ɣ-tubulin, and revealed the presence of a three-dimensional complex PCM lattice composed of 12–15 nm wide filaments. In addition, if mixed with the oocyte extract, the stripped lattice re-gained ɣ-tubulin and the ability to nucleate MTs. These experiments provided the first clues that mitotic PCM is built of a porous scaffold into which components from the cytoplasm can be incorporated (Figure 3C).
In human cells, three PCM components expand and possibly build the mitotic PCM lattice; Cep192, pericentrin and Cep215 (centrosomin (Cnn) in Drosophila) [132, 144, 148–153]. In C. elegans, mitotic PCM expansion relies on SPD-2 and SPD-5 (which are functional analogs of Cep192 and Cep215/Cnn, respectively), while pericentrin is absent [153, 154]. It is also of note that, unlike in human cells, PLP does not expand in Drosophila and remains localized close to the centriole during mitosis [108]. Regardless of these differences, the principles underlying the assembly of the expanded PCM lattice are currently thought to be generally conserved. PCM proteins which make up the PCM lattice are rich in coiled coil domains (α-helical protein motifs known to facilitate protein–protein interactions [155]). Cep215 family of proteins also contains centrosomin motif 2 (CM2) in the C terminus, which mediates multimerization, in addition to the CM1 motif in the N terminus that can bind to and activate microtubule nucleation complexes [131, 156, 157].
Centrosome maturation in all studied species requires the activity of mitotic kinase Plk1 (Polo in Drosophila, PLK-1 in C. elegans) [158–160]. Plk1 phosphorylates multiple PCM proteins [149, 161–163], and promotes their centrosomal recruitment [148, 149, 151, 159, 162, 164–167]. Plk1 activity also drives the recruitment and anchoring of the γ-TuRC and its adaptors MZT1 and NEDD1 to the PCM lattice, stimulating microtubule-nucleating activity at the centrosome [167–169]. Another important centrosome-localized mitotic kinase is Aurora A, which phosphorylates and activates Plk1 promoting its localization to the centrosome [170]. Aurora A promotes the recruitment of the MT-nucleating complexes and MT-associated proteins to the expanded PCM, contributing to PCM maturation and functionality [171]. Inhibition of Plk1 during mitosis results in a rapid reduction of PCM components, while inhibition of Aurora A results in spindle collapse but with preserved PCM [164, 171].
The formation of the mitotic PCM lattice is initiated by docking of the active Plk1 to the centrosome [161], presumably through binding to Cep192/Spd-2 [163, 172]. Plk1, in turn, phosphorylates multiple PCM components, including Cep192, Cep215 and pericentrin, promoting their three-dimensional oligomerization. Important insight into the initial molecular events underlying the formation of the mitotic PCM lattice is based on work in Drosophila [151] and C. elegans [160, 162, 173] (reviewed in detail in [174]). In Drosophila, Polo first phosphorylates the phospho-regulated multimerization (PReM) domain within the central part of Cnn. PReM phosphorylation, promotes interaction between the adjacent leucin zipper domain with the CM2 motif from another Cnn molecule [150, 152, 175]. In vitro, interaction of these two domains resulted in the formation of micron-scale stable assemblies, prompting the idea that similar oligomerization processes underly the formation of the mitotic PCM lattice in vivo. Similarly, purified SPD-5, promoted by Plk1 and SPD-2, readily assembled into a PCM-like lattice and even self-assembled when a crowding agent was used to approximate cellular conditions. It has yet to be demonstrated whether human proteins follow a similar oligomerization principle. In human cells, the C-terminal CM2 motif of Cep215 is important for its centrosomal accumulation and its interaction with pericentrin [176, 177]. Overexpression of Cep215, or pericentrin, resulted in enlargement of interphase PCM around centrioles, which contained components and functions normally present on interphase centrosomes [104, 132, 178]. In addition, overexpression of Cep215 resulted in cytosolic aggregates containing ɣ-tubulin and pericentrin and showing MT- nucleating ability.
In human mitotic centrosomes, Cep192, Cep215 and pericentrin are all found in the expanded mitotic PCM lattice (see Figure 3B for Cep192 and Cep215). Yet, multiple studies have demonstrated that only Cep192 is critical for the formation of bipolar spindle poles in somatic cultured cells [129, 161, 179, 180]. Removal of Cep192 resulted in the loss of nearly all centrosome-associated γ-tubulin, and a substantial decrease in pericentrin and Cep215 levels in mitosis [179], mislocalization of Aurora A from mitotic centrosomes [161], and severe spindle defects. By contrast, the loss of pericentrin or Cep215 seems to be better tolerated and individual or joint pericentrin and Cep215 removal reduced, but did not eliminate, centrosome-associated levels of Cep192 and γ-tubulin in mitosis, and centrioles could still form smaller but functional spindle poles [129, 132, 148, 179, 180]. Nevertheless, the percentage of cells showing mitotic errors was slightly elevated in cells depleted for either pericentrin or Cep215 [180]. In cells lacking both, pericentrin and Cep215, Cep192 was present on mitotic cells only in a thin layer adjacent to the centriole wall [179]; thus, functional spindles formed without a typical expanded PCM lattice. In addition, Cep215 and pericentrin partially co-depend for localization to mitotic centrosomes [148], and removal of either protein, although largely tolerated, still has consequences. For instance, astral MTs were lost from spindle poles after Cep215 depletion [132], mitotic spindles tend to be shorter and misoriented after pericentrin perturbation [179, 181], and mitotic errors after depletion of PCM components have been widely reported (reviewed in [182]). The presence of multiple PCM lattice elements could be variably important in different cellular contexts. For instance, the astral MTs that emanate from mitotic centrosomes and interact with the cell cortex are critically important for spindle positioning, directionality of cell division, and tissue patterning [183]. Thus, it is possible that multiple PCM components are available to ensure the robustness in spindle formation and chromosome segregation across different cell types. Finally, pericentrin and Cep215 are critical in the formation of acentriolar microtubule organizing centers (discussed in Section 3).
2.3.3. PCM during mitotic exit: what goes up must come down
During mitotic exit, mitotic centrosomes swiftly disassemble their PCM lattice and reverse to their interphase state but how this process is regulated is just starting to emerge. Prophase and early mitosis are characterized by an increase in phosphorylation of PCM components and various mitotic substrates, in large part through an increased activity of Plk1 [148, 149, 158, 184]. During transition to anaphase, Plk1 activity decreases, while phosphatase activity increases [185, 186]. Available data from C. elegans embryos [187–189] argues that the size of mitotic PCM, its density and mechanical properties are balanced by the opposing actions of Plk1 and phosphatase PP2A [190]. While Plk1 increases phosphorylation and promotes oligomerization of the PCM components and PCM expansion, PP2A, active on the centrosomes throughout mitosis, dephosphorylates PCM components driving PCM toward disassembly (Figure 3B). Thus, an increased Plk1 activity before and during the first part of mitosis would tilt the balance toward PCM assembly. A sharp decline in the Plk1 activity in anaphase would change the balance toward dephosphorylation of centrosomal components, favoring PCM disassembly. Indeed, PCM disassembly was found to be coincidental with dephosphorylation of SPD-5 and depletion of PP2A slowed down PCM disassembly [187, 189]. Time-lapse analysis of centrosome dynamics in C. elegans embryos also demonstrated that, after anaphase onset, Plk1 and SPD-2 are lost first, followed by γ-tubulin and SPD-5. This pattern coincided with changes in PCM appearance. The loss of Plk1 and SPD-2 was followed by the deformation of PCM, followed by the “rupture” phase in which outer PCM components formed small packets containing SPD-5, which were removed from the centrosome [188, 189]. Elimination of cortical MT-dependent pulling forces abolished SPD-5 rupture and packet formation, which instead gradually dissolved from centrosomes [189]. Consistently, the strength and ductility of the PCM also abruptly changes at anaphase onset [188]. Both, resistance to deformation and resistance to fracture diminishes sharply in anaphase coincidently with the loss of Plk1 and SPD-2 from the centrosomes. Since both inhibition of Plk1 and defects in SPD-2 decreased PCM resistance to stress and fracture, these data strongly indicate that Plk1 and SPD-2 provide strength and resistance to PCM before anaphase [188]. Collectively, Plk1 inactivation, phosphatase activity and MT-pulling forces disassemble PCM during mitotic exit. Future research should clarify whether this principle can apply to vertebrate centrosome.
3. Division without centrioles
Centrioles and centriole-assembled centrosomes are not strictly required for cell proliferation. Cell divisions and even morphogenesis can occur in the complete absence of centrioles, and some animal cells inherently don’t have them [191–193]. There are several classic examples. Vertebrate oocytes naturally lack centrioles [194]. Centrioles are also absent from the earliest mitotic divisions in mice embryos and are only formed after the 64-cell stage [195]. In the planarian S. mediterranea, cells without centrioles proliferate to form the entire body [191, 196].
Acentriolar dividing cells use diverse schemes to form bipolar spindles and segregate chromosomes. During spindle formation in mouse oocytes, numerous small acentriolar MTOCs first form throughout the ooplasm in prophase [197]. After nuclear membrane breakdown, they congress and form a multipolar spindle around chromosomes. Congressed MTOCs then progressively cluster into two dominant poles, which ultimately form an elongated bipolar spindle that segregates chromosomes. Such acentriolar MTOCs contain pericentrin, Cep215, and γ-tubulin and are organized by dynein and nuclear mitotic apparatus (NuMA) protein [198, 199], which is known to interact with MTs and organize them in asters [200, 201]. In this system, pericentrin and Cep215 are indispensable for the formation of MTOCs. By contrast, in human oocytes, spindle formation is, instead of acentriolar MTOCs, mediated by chromosomes and the small GTPase Ran [202]. MTs are initially formed in the middle of aggregated chromosomes. As the MT aster grows, chromosomes get distributed along the aster surface with kinetochores facing inwards. This is followed by the extension of the aster and formation of unstable bipolar spindle, where chromosomes reach alignment only ~16 h after nuclear envelope breakdown. This long process of spindle assembly, characterized by intrinsic spindle instability, could be the reason for the very high rate of aneuploidy specific to human eggs (reviewed in [203]).
In early divisions of acentriolar mouse embryos, mitotic spindles form from randomly distributed acentriolar MTOCs containing pericentrin which cluster into a bipolar spindle through a multipolar intermediary. Dynein is essential for the maturation of acentriolar MTOCs and the spindle, whereas kinesin-5 is required for spindle bipolarization [204]. Until late blastocyst stage the process of spindle assembly gradually transitions from meiotic to mitotic mode.
The requirement for centrioles has also been studied in vertebrate and Drosophila cells after centrosome removal by micromanipulations, after centriole loss due to the inhibition or depletion of centriole initiation factors [22, 180, 205–207], and in a mutant Drosophila acentriolar cell line 1182–4, which constitutively lacks centrioles [193, 208]. These analyses showed that in the absence of centrioles, somatic cells can continuously divide employing acentriolar spindle assembly pathways that normally operate in centriolar cells and contribute to spindle assembly [205, 209–211]. However, acentriolar cells frequently suffer from prolonged spindle assembly, chromosome mis-segregation, misoriented divisions, DNA damage, cell cycle arrest, and apoptosis [20, 22, 206, 208, 212–215]. Centrioles appear critical during early stages of spindle formation, and early acentriolar mitotic spindles are initially disorganized, with unfocused MTs, and multipolar prior to bipolarization [22]. The presence of multipolar spindles is known to promote merotelic chromosome attachments and lagging chromosomes [216–218], which may result in aneuploidy, DNA breaks, and proliferative defects [19–21, 215, 219]. Finally, both centriole removal from immortalized human RPE-1 cells and prolonged mitosis can trigger a USP28-53BP1-TP53-dependent cell cycle arrest, which is independent of DNA damage or chromosome segregation errors [19–21], to suppress the growth of impaired cells.
Recent studies show that acentriolar human somatic cells in culture can employ diverse pathways for spindle formation [180]. For instance, spindle formation in acentriolar RPE-1 and HeLa cells is mediated by MT asters containing Cep192, Cep215 and pericentrin, are all indispensable for the assembly of these asters. In these cells, aster assembly also requires the Cep215 CM2 oligomerization motif, Plk1, and microtubules, while the Cep215 CM1 motif is used to dock γ-tubulin complexes [180]. NuMA, and dynein are also needed to form asters (in HeLa) [207]. In contrast, acentriolar DLD1 and U2OS cells formed error-prone spindles that contained none or very little of Cep192, Cep215 and pericentrin, although these proteins were abundantly present in the cytoplasm and on centriole-containing spindle poles [180]. Altogether, these exciting new results highlight inherent differences in the mechanisms that generate mitotic spindle poles in the absence of centrioles. More research will need to be done to understand the origins of these differences.
4. Perspectives
Recent years have provided new insights into the processes underlying centriole and centrosome structuring and centrosome maturation, in large part thanks to the great work on fly and worm centrosomes. But there are numerous questions that must be answered, especially for human centrosomes, which are less understood and structurally and biochemically more complex. It will be important to understand which principles underlying centrosome organization and formation can be applied across species and which are unique only to the human centrosome. For instance, data shows that some of components used to stabilize human centrioles are unique to the vertebrate centriole, yet centrioles across species are stable, indicating redundant mechanisms for centriole stabilization. Similarly, the assembly of functional mitotic PCM lattice can be achieved using a smaller or larger number of lattice components. This, which begs the question as to whether the mitotic PCM lattice of a fly, worm, and human centrosome are organized as in the surf clam, or if different patterns are used in different species? It will be important to understand the co-dependency and functional redundancy observed between components of a human centrosome and how that operates in centrosomes of different tissues. It is unclear how centrosome components transition from the centriole-bound toroidal to the lattice-like configuration in mitosis. Does the mitotic lattice form de novo and how much of their original interphase organization remains preserved during mitosis? Answering these questions will be a fascinating task but thanks to the development of precise genetic tools and ever more sophisticated imaging tools allowing nanoscale view into centrosome architecture and dynamics, many outstanding questions may be unraveled faster than we can currently imagine.
Table 1.
List of used abbreviations.
| Ana1: Anastral spindle 1 |
| ANKRD26: Ankyrin repeat domain-containing protein 26 |
| CBB: Centrioles and basal bodies |
| CCC: Centriole to centrosome conversion |
| CCDC120: Coiled-coil domain-containing protein 120 |
| CCDC41: Coiled-coil domain-containing protein 41 |
| CEP: Centrosomal protein |
| CM1: Centrosomin motif-1 |
| CM2: Centrosomin motif-2 |
| Cnn: Centrosomin |
| CPAP: Centrosomal P4.1-associated protein |
| DA: Distal Appendages |
| DLD-1: Human Colon Adenocarcinoma epithelial cells |
| FAM161A: FAM161 centrosomal protein A |
| FBF1: Fas-binding factor 1 |
| HeLa: Human Cervical Adenocarcinoma epithelial cells |
| MT: Microtubules |
| MTOCs: Microtubule organizing centers |
| MZT1: Mitotic-spindle organizing protein 1 |
| NEDD1: Neural precursor cell expressed developmentally down-regulated protein 1 |
| Nek2: NimA-related protein kinase 2 |
| NuMA: Nuclear mitotic apparatus protein |
| ODF2: Outer dense fiber protein 2 |
| PACT domain: Pericentrin-AKAP450 Centrosomal Targeting domain |
| PCM: Pericentriolar material |
| PIDD1: p53-induced death domain-containing protein 1 |
| Plk1: Polo-like kinase 1 |
| Plk4: Polo-like kinase 4 |
| PLP: Pericentrin-like protein |
| POC1B: Proteome of the centriole 1 centriolar protein B |
| POC5: Proteome of the centriole 5 centriolar protein |
| PP2A: Protein phosphatase 2A |
| PReM: Phospho-regulated multimerization domain |
| RPE-1: Human retinal pigment epithelial-1 cells |
| SAK: Snk/Plk-akin kinase |
| Sas-4: Spindle assembly abnormal protein 4 |
| SAS-6: Spindle assembly abnormal protein 6 homolog |
| SAS-7: Spindle assembly abnormal protein 7 |
| SCLT1: Sodium channel and clathrin linker 1 |
| SDA: Subdistal appendages |
| Spd-2, SPD-2: Spindle-defective protein 2 |
| SPD-5: Spindle-defective protein 5 |
| STIL: SCL-interrupting locus protein |
| TTBK2: Tau-tubulin kinase 2 |
| U2OS: Human Bone Osteosarcoma epithelial cells |
| WDR90: WD repeat-containing protein 90 |
| γ-TuRC: γ-Tubulin Ring Complex |
Acknowledgements
Many thanks to Dr. Catherine Sullenberger, Dr. Yien Che Tsai, and other members of the Loncarek lab for reading of the manuscript and critical input, and Dr. Dong Kong and Dr. Delgermaa Luvsanjav for electron micrographs.
Funding source
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute to JL.
Footnotes
Declaration of interest:
The authors declare no competing financial or other interests.
Extended list of abbreviations including the full names of centrosomal proteins is now provided as a separate table (Table 1)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jimenez AJ, et al. , Acto-myosin network geometry defines centrosome position. Curr Biol, 2021. 31(6): p. 1206–1220.e5. [DOI] [PubMed] [Google Scholar]
- 2.Sullenberger C, et al. , With Age Comes Maturity: Biochemical and Structural Transformation of a Human Centriole in the Making. Cells, 2020. 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arquint C, Gabryjonczyk AM, and Nigg EA, Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg EA, Schnerch D, and Ganier O, Impact of Centrosome Aberrations on Chromosome Segregation and Tissue Architecture in Cancer. Cold Spring Harb Symp Quant Biol, 2017. 82: p. 137–144. [DOI] [PubMed] [Google Scholar]
- 5.Vertii A, Hehnly H, and Doxsey S, The Centrosome, a Multitalented Renaissance Organelle. Cold Spring Harb Perspect Biol, 2016. 8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J and Akhmanova A, Microtubule-Organizing Centers. Annu Rev Cell Dev Biol, 2017. 33: p. 51–75. [DOI] [PubMed] [Google Scholar]
- 7.Nigg EA and Holland AJ, Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol, 2018. 19(5): p. 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roman AC, et al. , Centriole Positioning: Not Just a Little Dot in the Cell. Results Probl Cell Differ, 2019. 67: p. 201–221. [DOI] [PubMed] [Google Scholar]
- 9.Gavilan MP, et al. , The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep, 2018. 19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anvarian Z, et al. , Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol, 2019. 15(4): p. 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchison HM and Valente EM, Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol, 2017. 241(2): p. 294–309. [DOI] [PubMed] [Google Scholar]
- 12.Nachury MV, How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura Y, et al. , Primary Cilia as Signaling Hubs in Health and Disease. Adv Sci (Weinh), 2019. 6(1): p. 1801138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avidor-Reiss T, Carr A, and Fishman EL, The sperm centrioles. Mol Cell Endocrinol, 2020. 518: p. 110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindemann CB and Lesich KA, Functional anatomy of the mammalian sperm flagellum. Cytoskeleton (Hoboken), 2016. 73(11): p. 652–669. [DOI] [PubMed] [Google Scholar]
- 16.Legendre M, Zaragosi LE, and Mitchison HM, Motile cilia and airway disease. Semin Cell Dev Biol, 2020. [DOI] [PubMed] [Google Scholar]
- 17.Lee L and Ostrowski LE, Motile cilia genetics and cell biology: big results from little mice. Cell Mol Life Sci, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons RA, Saridogan E, and Djahanbakhch O, The reproductive significance of human Fallopian tube cilia. Hum Reprod Update, 2006. 12(4): p. 363–72. [DOI] [PubMed] [Google Scholar]
- 19.Lambrus BG, et al. , A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol, 2016. 214(2): p. 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meitinger F, et al. , 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol, 2016. 214(2): p. 155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong CS, et al. , 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sir JH, et al. , Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol, 2013. 203(5): p. 747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganier O, et al. , Structural centrosome aberrations promote non-cell-autonomous invasiveness. Embo j, 2018. 37(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godinho SA, et al. , Oncogene-like induction of cellular invasion from centrosome amplification. Nature, 2014. 510(7503): p. 167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine MS, et al. , Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev Cell, 2017. 40(3): p. 313–322 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basto R, et al. , Centrosome amplification can initiate tumorigenesis in flies. Cell, 2008. 133(6): p. 1032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Assoro AB, Lingle WL, and Salisbury JL, Centrosome amplification and the development of cancer. Oncogene, 2002. 21(40): p. 6146–53. [DOI] [PubMed] [Google Scholar]
- 28.Marthiens V, et al. , Centrosome amplification causes microcephaly. Nat Cell Biol, 2013. 15(7): p. 731–40. [DOI] [PubMed] [Google Scholar]
- 29.Phan TP, et al. , Centrosome defects cause microcephaly by activating the 53BP1-USP28-TP53 mitotic surveillance pathway. Embo j, 2021. 40(1): p. e106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond J, et al. , A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet, 2005. 37(4): p. 353–5. [DOI] [PubMed] [Google Scholar]
- 31.Chavali PL, Putz M, and Gergely F, Small organelle, big responsibility: the role of centrosomes in development and disease. Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nigg EA, Cajanek L, and Arquint C, The centrosome duplication cycle in health and disease. FEBS Lett, 2014. 588(15): p. 2366–72. [DOI] [PubMed] [Google Scholar]
- 33.LoMastro GM and Holland AJ, The Emerging Link between Centrosome Aberrations and Metastasis. Developmental Cell, 2019. 49(3): p. 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goundiam O and Basto R, Centrosomes in disease: how the same music can sound so different? Curr Opin Struct Biol, 2020. 66: p. 74–82. [DOI] [PubMed] [Google Scholar]
- 35.Marthiens V and Basto R, Centrosomes: The good and the bad for brain development. Biology of the Cell, 2020. 112(6): p. 153–172. [DOI] [PubMed] [Google Scholar]
- 36.Monteiro P and Godinho SA, Structural Centrosomal Abnormalities Push Cells toward Invasion. Dev Cell, 2018. 45(3): p. 286–288. [DOI] [PubMed] [Google Scholar]
- 37.Megraw TL, Sharkey JT, and Nowakowski RS, Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol, 2011. 21(8): p. 470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho-Santos Z, et al. , Stepwise evolution of the centriole-assembly pathway. J Cell Sci, 2010. 123(Pt 9): p. 1414–26. [DOI] [PubMed] [Google Scholar]
- 39.Azimzadeh J, Exploring the evolutionary history of centrosomes. Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho-Santos Z, et al. , Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol, 2011. 194(2): p. 165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta A and Kitagawa D, Ultrastructural diversity between centrioles of eukaryotes. J Biochem, 2018. 164(1): p. 1–8. [DOI] [PubMed] [Google Scholar]
- 42.Vorobjev IA and Chentsov Yu S, Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol, 1982. 93(3): p. 938–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson RG and Brenner RM, The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol, 1971. 50(1): p. 10–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jana SC, Centrosome structure and biogenesis: Variations on a theme? Semin Cell Dev Biol, 2021. [DOI] [PubMed] [Google Scholar]
- 45.Gibbons IR and Grimstone AV, On flagellar structure in certain flagellates. J Biophys Biochem Cytol, 1960. 7: p. 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acs P, et al. , A novel form of ciliopathy underlies hyperphagia and obesity in Ankrd26 knockout mice. Brain structure & function, 2015. 220(3): p. 1511–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakazawa Y, et al. , SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol, 2007. 17(24): p. 2169–74. [DOI] [PubMed] [Google Scholar]
- 48.Hirono M, Cartwheel assembly. Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Toole ET, Giddings TH, and Dutcher SK, Understanding Microtubule Organizing Centers by Comparing Mutant and Wild-Type Structures with Electron Tomography, in Methods in Cell Biology. 2007, Academic Press. p. 125–143. [DOI] [PubMed] [Google Scholar]
- 50.Callaini G, Whitfield WG, and Riparbelli MG, Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp Cell Res, 1997. 234(1): p. 183–90. [DOI] [PubMed] [Google Scholar]
- 51.Dippell RV, The development of basal bodies in paramecium. Proc Natl Acad Sci U S A, 1968. 61(2): p. 461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nazarov S, et al. , Novel features of centriole polarity and cartwheel stacking revealed by cryo-tomography. Embo j, 2020. 39(22): p. e106249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klena N, et al. , Architecture of the centriole cartwheel-containing region revealed by cryo-electron tomography. The EMBO Journal, 2020. 39(22): p. e106246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li S, et al. , Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. Embo j, 2012. 31(3): p. 552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong D, et al. , Prolonged mitosis results in structurally aberrant and over-elongated centrioles. J Cell Biol, 2020. 219(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uzbekov R and Prigent C, Clockwise or anticlockwise? Turning the centriole triplets in the right direction! FEBS Lett, 2007. 581(7): p. 1251–4. [DOI] [PubMed] [Google Scholar]
- 57.Li S, et al. , Electron cryo-tomography provides insight into procentriole architecture and assembly mechanism. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steib E, et al. , WDR90 is a centriolar microtubule wall protein important for centriole architecture integrity. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guichard P, et al. , Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr Biol, 2013. 23(17): p. 1620–8. [DOI] [PubMed] [Google Scholar]
- 60.Greenan GA, et al. , Insights into centriole geometry revealed by cryotomography of doublet and triplet centrioles. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Guennec M, et al. , A helical inner scaffold provides a structural basis for centriole cohesion. Sci Adv, 2020. 6(7): p. eaaz4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rattner JB and Phillips SG, Independence of centriole formation and DNA synthesis. J Cell Biol, 1973. 57(2): p. 359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uzbekov R and Alieva I, Who are you, subdistal appendages of centriole? Open Biol, 2018. 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paintrand M, et al. , Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol, 1992. 108(2): p. 107–28. [DOI] [PubMed] [Google Scholar]
- 65.Pizon V, et al. , hVFL3/CCDC61 is a component of mother centriole subdistal appendages required for centrosome cohesion and positioning. Biol Cell, 2020. 112(1): p. 22–37. [DOI] [PubMed] [Google Scholar]
- 66.Piel M, et al. , The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol, 2000. 149(2): p. 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mogensen MM, et al. , Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci, 2000. 113 (Pt 17): p. 3013–23. [DOI] [PubMed] [Google Scholar]
- 68.Delgehyr N, Sillibourne J, and Bornens M, Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci, 2005. 118(Pt 8): p. 1565–75. [DOI] [PubMed] [Google Scholar]
- 69.Huang N, et al. , Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat Commun, 2017. 8: p. 15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chong WM, et al. , Super-resolution microscopy reveals coupling between mammalian centriole subdistal appendages and distal appendages. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowler M, et al. , High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat Commun, 2019. 10(1): p. 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joo K, et al. , CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci U S A, 2013. 110(15): p. 5987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanos BE, et al. , Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes & Development, 2013. 27(2): p. 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graser S, et al. , Cep164, a novel centriole appendage protein required for primary cilium formation. The Journal of Cell Biology, 2007. 179(2): p. 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sillibourne JE, et al. , Primary ciliogenesis requires the distal appendage component Cep123. Biol Open, 2013. 2(6): p. 535–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye X, et al. , C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci U S A, 2014. 111(6): p. 2164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goetz SC, Liem KF, and Anderson KV, The Spinocerebellar Ataxia-associated Gene Tau Tubulin Kinase 2 (TTBK2) Controls the Initiation of Ciliogenesis. Cell, 2012. 151(4): p. 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans LT, et al. , ANKRD26 recruits PIDD1 to centriolar distal appendages to activate the PIDDosome following centrosome amplification. The EMBO Journal, 2020n/a(n/a): p. e105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burigotto M, et al. , Centriolar distal appendages activate the centrosome-PIDDosome-p53 signalling axis via ANKRD26. Embo j, 2021. 40(4): p. e104844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viol L, et al. , Nek2 kinase displaces distal appendages from the mother centriole prior to mitosis. J Cell Biol, 2020. 219(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guichard P, Hamel V, and Gonczy P, The Rise of the Cartwheel: Seeding the Centriole Organelle. Bioessays, 2018. 40(4): p. e1700241. [DOI] [PubMed] [Google Scholar]
- 82.Kitagawa D, et al. , Structural basis of the 9-fold symmetry of centrioles. Cell, 2011. 144(3): p. 364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guichard P, et al. , Cartwheel architecture of Trichonympha basal body. Science, 2012. 337(6094): p. 553. [DOI] [PubMed] [Google Scholar]
- 84.Hiraki M, et al. , Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol, 2007. 17(20): p. 1778–83. [DOI] [PubMed] [Google Scholar]
- 85.Matsuura K, et al. , Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol, 2004. 165(5): p. 663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohta T, et al. , Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol, 2002. 156(1): p. 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banterle N and Gönczy P, Centriole Biogenesis: From Identifying the Characters to Understanding the Plot. Annu Rev Cell Dev Biol, 2017. 33: p. 23–49. [DOI] [PubMed] [Google Scholar]
- 88.Arquint C and Nigg EA, STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr Biol, 2014. 24(4): p. 351–60. [DOI] [PubMed] [Google Scholar]
- 89.Strnad P, et al. , Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell, 2007. 13(2): p. 203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Habedanck R, et al. , The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol, 2005. 7(11): p. 1140–6. [DOI] [PubMed] [Google Scholar]
- 91.Bettencourt-Dias M, et al. , SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol, 2005. 15(24): p. 2199–207. [DOI] [PubMed] [Google Scholar]
- 92.Vulprecht J, et al. , STIL is required for centriole duplication in human cells. J Cell Sci, 2012. 125(Pt 5): p. 1353–62. [DOI] [PubMed] [Google Scholar]
- 93.Arquint C and Nigg EA, The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc Trans, 2016. 44(5): p. 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim TS, et al. , Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc Natl Acad Sci U S A, 2013. 110(50): p. E4849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamamoto S and Kitagawa D, Self-organization of Plk4 regulates symmetry breaking in centriole duplication. Nat Commun, 2019. 10(1): p. 1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moyer TC, et al. , Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol, 2015. 209(6): p. 863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohta M, et al. , Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun, 2014. 5: p. 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dzhindzhev NS, et al. , Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr Biol, 2014. 24(21): p. 2526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cottee MA, et al. , The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. Elife, 2015. 4: p. e07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takao D, Yamamoto S, and Kitagawa D, A theory of centriole duplication based on self-organized spatial pattern formation. J Cell Biol, 2019. 218(11): p. 3537–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leda M, Holland AJ, and Goryachev AB, Autoamplification and Competition Drive Symmetry Breaking: Initiation of Centriole Duplication by the PLK4-STIL Network. iScience, 2018. 8: p. 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gonczy P and Hatzopoulos GN, Centriole assembly at a glance. J Cell Sci, 2019. 132(4). [DOI] [PubMed] [Google Scholar]
- 103.Sonnen KF, et al. , 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open, 2012. 1(10): p. 965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lawo S, et al. , Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol, 2012. 14(11): p. 1148–58. [DOI] [PubMed] [Google Scholar]
- 105.Mennella V, et al. , Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol, 2012. 14(11): p. 1159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei Z, et al. , Requirement of the Cep57-Cep63 interaction for proper Cep152 recruitment and centriole duplication. Mol Cell Biol, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim TS, et al. , Molecular architecture of a cylindrical self-assembly at human centrosomes. Nat Commun, 2019. 10(1): p. 1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu J and Glover DM, Structured illumination of the interface between centriole and peri-centriolar material. Open Biol, 2012. 2(8): p. 120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Varadarajan R and Rusan NM, Bridging centrioles and PCM in proper space and time. Essays Biochem, 2018. 62(6): p. 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dahl KD, et al. , A Short CEP135 Splice Isoform Controls Centriole Duplication. Curr Biol, 2015. 25(19): p. 2591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei Z, et al. , Requirement of the Cep57-Cep63 Interaction for Proper Cep152 Recruitment and Centriole Duplication. Mol Cell Biol, 2020. 40(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Izquierdo D, et al. , Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep, 2014. 8(4): p. 957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang WJ, et al. , The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol, 2011. 193(4): p. 727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsuchiya Y, et al. , Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat Commun, 2016. 7: p. 12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Atorino ES, et al. , CEP44 ensures the formation of bona fide centriole wall, a requirement for the centriole-to-centrosome conversion. Nat Commun, 2020. 11(1): p. 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang C-W, et al. , CEP295 interacts with microtubules and is required for centriole elongation. Journal of Cell Science, 2016. 129(13): p. 2501–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sonnen KF, et al. , Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci, 2013. 126(Pt 14): p. 3223–33. [DOI] [PubMed] [Google Scholar]
- 118.Chen HY, et al. , Human microcephaly protein RTTN interacts with STIL and is required to build full-length centrioles. Nat Commun, 2017. 8(1): p. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fu J, et al. , Conserved molecular interactions in centriole-to-centrosome conversion. Nat Cell Biol, 2016. 18(1): p. 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watanabe K, et al. , The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat Commun, 2019. 10(1): p. 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O’Rourke BP, et al. , Cep192 controls the balance of centrosome and non-centrosomal microtubules during interphase. PLoS One, 2014. 9(6): p. e101001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng Y, et al. , Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature, 1995. 378(6557): p. 578–83. [DOI] [PubMed] [Google Scholar]
- 123.Consolati T, et al. , Microtubule Nucleation Properties of Single Human γTuRCs Explained by Their Cryo-EM Structure. Dev Cell, 2020. 53(5): p. 603–617.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wieczorek M, et al. , Asymmetric Molecular Architecture of the Human γ-Tubulin Ring Complex. Cell, 2020. 180(1): p. 165–175.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu P, et al. , Insights into the assembly and activation of the microtubule nucleator γ-TuRC. Nature, 2020. 578(7795): p. 467–471. [DOI] [PubMed] [Google Scholar]
- 126.Tovey CA and Conduit PT, Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem, 2018. 62(6): p. 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu P, et al. , Microtubule nucleation: The waltz between γ-tubulin ring complex and associated proteins. Curr Opin Cell Biol, 2020. 68: p. 124–131. [DOI] [PubMed] [Google Scholar]
- 128.Zhu F, et al. , The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol, 2008. 18(2): p. 136–41. [DOI] [PubMed] [Google Scholar]
- 129.Gomez-Ferreria MA, et al. , Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol, 2007. 17(22): p. 1960–6. [DOI] [PubMed] [Google Scholar]
- 130.Gillingham AK and Munro S, The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep, 2000. 1(6): p. 524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Choi YK, et al. , CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol, 2010. 191(6): p. 1089–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fong KW, et al. , CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell, 2008. 19(1): p. 115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lukinavicius G, et al. , Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr Biol, 2013. 23(3): p. 265–70. [DOI] [PubMed] [Google Scholar]
- 134.Zhao H, et al. , Cep57 and Cep57l1 function redundantly to recruit the Cep63-Cep152 complex for centriole biogenesis. J Cell Sci, 2020. 133(13). [DOI] [PubMed] [Google Scholar]
- 135.Firat-Karalar EN, et al. , Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol, 2014. 24(6): p. 664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng X, et al. , Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc Natl Acad Sci U S A, 2014. 111(3): p. E354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cizmecioglu O, et al. , Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol, 2010. 191(4): p. 731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gopalakrishnan J, et al. , Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun, 2011. 2: p. 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Conduit PT, et al. , Re-examining the role of Drosophila Sas-4 in centrosome assembly using two-colour-3D-SIM FRAP. Elife, 2015. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wueseke O, et al. , The Caenorhabditis elegans pericentriolar material components SPD-2 and SPD-5 are monomeric in the cytoplasm before incorporation into the PCM matrix. Mol Biol Cell, 2014. 25(19): p. 2984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shukla A, et al. , Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat Commun, 2015. 6: p. 8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Novak ZA, et al. , Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev Cell, 2016. 37(6): p. 545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sugioka K, et al. , Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dictenberg JB, et al. , Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol, 1998. 141(1): p. 163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Palazzo RE, et al. , Centrosome maturation. Curr Top Dev Biol, 2000. 49: p. 449–70. [DOI] [PubMed] [Google Scholar]
- 146.Kollman JM, et al. , Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol, 2011. 12(11): p. 709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schnackenberg BJ, et al. , The disassembly and reassembly of functional centrosomes in vitro. Proc Natl Acad Sci U S A, 1998. 95(16): p. 9295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haren L, Stearns T, and Luders J, Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One, 2009. 4(6): p. e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee K and Rhee K, PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol, 2011. 195(7): p. 1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Conduit PT, et al. , Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol, 2010. 20(24): p. 2178–86. [DOI] [PubMed] [Google Scholar]
- 151.Conduit PT, et al. , A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife, 2014. 3: p. e03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Feng Z, et al. , Structural Basis for Mitotic Centrosome Assembly in Flies. Cell, 2017. 169(6): p. 1078–1089.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hamill DR, et al. , Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell, 2002. 3(5): p. 673–84. [DOI] [PubMed] [Google Scholar]
- 154.Kemp CA, et al. , Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell, 2004. 6(4): p. 511–23. [DOI] [PubMed] [Google Scholar]
- 155.Lupas AN and Bassler J, Coiled Coils - A Model System for the 21st Century. Trends Biochem Sci, 2017. 42(2): p. 130–140. [DOI] [PubMed] [Google Scholar]
- 156.Samejima I, et al. , Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J Cell Sci, 2008. 121(Pt 23): p. 3971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang J and Megraw TL, Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol Biol Cell, 2007. 18(10): p. 4037–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Golsteyn RM, et al. , Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol, 1995. 129(6): p. 1617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lane HA and Nigg EA, Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol, 1996. 135(6 Pt 2): p. 1701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wueseke O, et al. , Polo-like kinase phosphorylation determines Caenorhabditis elegans centrosome size and density by biasing SPD-5 toward an assembly-competent conformation. Biol Open, 2016. 5(10): p. 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Joukov V, Walter JC, and De Nicolo A, The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol Cell, 2014. 55(4): p. 578–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Woodruff JB, et al. , Regulated assembly of a supramolecular centrosome scaffold in vitro. Science, 2015. 348(6236): p. 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Meng L, et al. , Bimodal Interaction of Mammalian Polo-Like Kinase 1 and a Centrosomal Scaffold, Cep192, in the Regulation of Bipolar Spindle Formation. Mol Cell Biol, 2015. 35(15): p. 2626–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cabral G, et al. , Differential Requirements for Centrioles in Mitotic Centrosome Growth and Maintenance. Dev Cell, 2019. 50(3): p. 355–366.e6. [DOI] [PubMed] [Google Scholar]
- 165.Woodruff JB, Wueseke O, and Hyman AA, Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci, 2014. 369(1650). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mahen R, et al. , Continuous polo-like kinase 1 activity regulates diffusion to maintain centrosome self-organization during mitosis. Proc Natl Acad Sci U S A, 2011. 108(22): p. 9310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ohta M, et al. , Polo-like kinase 1 independently controls microtubule-nucleating capacity and size of the centrosome. J Cell Biol, 2021. 220(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lin TC, et al. , MOZART1 and γ-tubulin complex receptors are both required to turn γ-TuSC into an active microtubule nucleation template. J Cell Biol, 2016. 215(6): p. 823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wieczorek M, et al. , MZT Proteins Form Multi-Faceted Structural Modules in the γ-Tubulin Ring Complex. Cell Rep, 2020. 31(13): p. 107791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Seki A, et al. , Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science, 2008. 320(5883): p. 1655–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hannak E, et al. , Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol, 2001. 155(7): p. 1109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alvarez-Rodrigo I, et al. , Evidence that a positive feedback loop drives centrosome maturation in fly embryos. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Woodruff JB, et al. , The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell, 2017. 169(6): p. 1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- 174.Gupta GD and Pelletier L, Centrosome Biology: Polymer-Based Centrosome Maturation. Curr Biol, 2017. 27(17): p. R836–r839. [DOI] [PubMed] [Google Scholar]
- 175.Citron YR, et al. , The centrosomin CM2 domain is a multi-functional binding domain with distinct cell cycle roles. PLoS One, 2018. 13(1): p. e0190530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim S and Rhee K, Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One, 2014. 9(1): p. e87016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang Z, et al. , Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem, 2010. 285(29): p. 22658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Loncarek J, et al. , Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol, 2008. 10(3): p. 322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chinen T, et al. , Centriole and PCM cooperatively recruit CEP192 to spindle poles to promote bipolar spindle assembly. J Cell Biol, 2021. 220(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Watanabe S, et al. , Centriole-independent mitotic spindle assembly relies on the PCNT-CDK5RAP2 pericentriolar matrix. J Cell Biol, 2020. 219(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen CT, et al. , A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr Biol, 2014. 24(19): p. 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Delaval B and Doxsey SJ, Pericentrin in cellular function and disease. J Cell Biol, 2010. 188(2): p. 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.di Pietro F, Echard A, and Morin X, Regulation of mitotic spindle orientation: an integrated view. EMBO Rep, 2016. 17(8): p. 1106–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Santamaria A, et al. , The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics, 2011. 10(1): p. M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wurzenberger C and Gerlich DW, Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol, 2011. 12(8): p. 469–82. [DOI] [PubMed] [Google Scholar]
- 186.Lindon C and Pines J, Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol, 2004. 164(2): p. 233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Enos SJ, et al. , Phosphatase PP2A and microtubule-mediated pulling forces disassemble centrosomes during mitotic exit. Biol Open, 2018. 7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mittasch M, et al. , Regulated changes in material properties underlie centrosome disassembly during mitotic exit. J Cell Biol, 2020. 219(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Magescas J, Zonka JC, and Feldman JL, A two-step mechanism for the inactivation of microtubule organizing center function at the centrosome. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Janssens V and Goris J, Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J, 2001. 353(Pt 3): p. 417–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bettencourt-Dias M, Q&A: Who needs a centrosome? BMC Biology, 2013. 11(1): p. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Debec A, Sullivan W, and Bettencourt-Dias M, Centrioles: active players or passengers during mitosis? Cellular and Molecular Life Sciences, 2010. 67(13): p. 2173–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Szöllösi A, et al. , A centriole-free Drosophila cell line. A high voltage EM study. Eur J Cell Biol, 1986. 40(1): p. 100–4. [PubMed] [Google Scholar]
- 194.Manandhar G, Schatten H, and Sutovsky P, Centrosome reduction during gametogenesis and its significance. Biol Reprod, 2005. 72(1): p. 2–13. [DOI] [PubMed] [Google Scholar]
- 195.Gueth-Hallonet C, et al. , gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci, 1993. 105 (Pt 1): p. 157–66. [DOI] [PubMed] [Google Scholar]
- 196.Azimzadeh J, et al. , Centrosome loss in the evolution of planarians. Science, 2012. 335(6067): p. 461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schuh M and Ellenberg J, Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell, 2007. 130(3): p. 484–98. [DOI] [PubMed] [Google Scholar]
- 198.Kolano A, et al. , Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci U S A, 2012. 109(27): p. E1858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang X, et al. , CEP215 and AURKA regulate spindle pole focusing and aMTOC organization in mouse oocytes. Reproduction, 2020. 159(3): p. 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gaglio T, Saredi A, and Compton DA, NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol, 1995. 131(3): p. 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Radulescu AE and Cleveland DW, NuMA after 30 years: the matrix revisited. Trends Cell Biol, 2010. 20(4): p. 214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Holubcová Z, et al. , Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science, 2015. 348(6239): p. 1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thomas C, Cavazza T, and Schuh M, Aneuploidy in human eggs: contributions of the meiotic spindle. Biochem Soc Trans, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Courtois A, et al. , The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol, 2012. 198(3): p. 357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Khodjakov A, et al. , Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol, 2000. 10(2): p. 59–67. [DOI] [PubMed] [Google Scholar]
- 206.Basto R, et al. , Flies without centrioles. Cell, 2006. 125(7): p. 1375–86. [DOI] [PubMed] [Google Scholar]
- 207.Chinen T, et al. , NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. Embo j, 2020. 39(2): p. e102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Debec A, et al. , Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J Cell Sci, 1995. 108 (Pt 7): p. 2645–53. [DOI] [PubMed] [Google Scholar]
- 209.O’Connell CB and Khodjakov AL, Cooperative mechanisms of mitotic spindle formation. J Cell Sci, 2007. 120(Pt 10): p. 1717–22. [DOI] [PubMed] [Google Scholar]
- 210.Moutinho-Pereira S, et al. , Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc Natl Acad Sci U S A, 2013. 110(49): p. 19808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lüders J and Stearns T, Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol, 2007. 8(2): p. 161–7. [DOI] [PubMed] [Google Scholar]
- 212.Poulton JS, Cuningham JC, and Peifer M, Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev Cell, 2014. 30(6): p. 731–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wong YL, et al. , Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science, 2015. 348(6239): p. 1155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bazzi H and Anderson KV, Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci U S A, 2014. 111(15): p. E1491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Uetake Y and Sluder G, Activation of the apoptotic pathway during prolonged prometaphase blocks daughter cell proliferation. Mol Biol Cell, 2018. 29(22): p. 2632–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ganem NJ, Godinho SA, and Pellman D, A mechanism linking extra centrosomes to chromosomal instability. Nature, 2009. 460(7252): p. 278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cimini D, et al. , Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol, 2001. 153(3): p. 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Salmon ED, et al. , Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci, 2005. 360(1455): p. 553–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Uetake Y and Sluder G, Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol, 2010. 20(18): p. 1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


