Abstract
mRNAs are monitored for errors in gene expression by RNA surveillance, in which mRNAs that cannot be fully translated are degraded by the nonsense-mediated mRNA decay pathway (NMD). RNA surveillance ensures that potentially deleterious truncated proteins are seldom made. NMD pathways that promote surveillance have been found in a wide range of eukaryotes. In Saccharomyces cerevisiae, the proteins encoded by the UPF1, UPF2, and UPF3 genes catalyze steps in NMD and are required for RNA surveillance. In this report, we show that the Upf proteins are also required to control the total accumulation of a large number of mRNAs in addition to their role in RNA surveillance. High-density oligonucleotide arrays were used to monitor global changes in the yeast transcriptome caused by loss of UPF gene function. Null mutations in the UPF genes caused altered accumulation of hundreds of mRNAs. The majority were increased in abundance, but some were decreased. The same mRNAs were affected regardless of which of the three UPF gene was inactivated. The proteins encoded by UPF-dependent mRNAs were broadly distributed by function but were underrepresented in two MIPS (Munich Information Center for Protein Sequences) categories: protein synthesis and protein destination. In a UPF+ strain, the average level of expression of UPF-dependent mRNAs was threefold lower than the average level of expression of all mRNAs in the transcriptome, suggesting that highly abundant mRNAs were underrepresented. We suggest a model for how the abundance of hundreds of mRNAs might be controlled by the Upf proteins.
Nonsense and frameshift mutations cause premature termination of translation. In conjunction with this, they also trigger nonsense-mediated mRNA decay (NMD), which greatly accelerates the rate of degradation of the mRNA. By decreasing the half-life of the mRNA, nonsense mRNA accumulation is severely limited (15, 16). The phenomenon whereby mRNAs that would otherwise code for potentially deleterious protein fragments are degraded is called RNA surveillance (7, 26). Surveillance occurs in fungi (21), plants (29), nematodes (26), and vertebrates (22).
In Saccharomyces cerevisiae, three genes, UPF1, UPF2, and UPF3, are required for NMD (6, 11, 14–16). Sequence homologs of UPF1, an RNA helicase (8, 30), have been identified in Schizosaccharomyces pombe (4), Caenorhabditis elegans (26), Mus musculus (25), and Homo sapiens (1, 25). Comparison of the Upf1p-like proteins shows that they are related by a common central region containing conserved cysteine-rich and ATP-helicase domains flanked by divergent sequences at both ends. These results suggest that NMD pathways in eukaryotic organisms utilize at least one protein in common. This provides a measure of confidence that further studies of NMD in yeast will shed light on NMD in humans as well.
The biological purpose of RNA surveillance is to limit the accumulation of aberrant proteins that arise through errors in gene expression. Inefficient splicing of introns is one of the most frequent natural source of errors in gene expression, leading to the production of nonsense mRNAs that code for aberrant proteins. Expression of the CYH2 gene, which codes for a ribosomal protein in S. cerevisiae, is a good example where the consequences of inefficient splicing have been examined (12). The intron in CYH2 pre-mRNA, which contains stop codons, is inefficiently spliced. In wild-type cells, unspliced pre-mRNA is exported (19), translated up to the premature stop codon, and then rapidly degraded by the NMD pathway. When the NMD pathway is inactivated by a null mutation in any of the three UPF genes, the CYH2 pre-mRNA fails to be rapidly degraded and accumulates to a much higher level. The rapid decay of the pre-mRNA prevents the accumulation of a truncated protein that might assemble with ribosomal subunits and impair function.
Evidence that the Upf proteins in S. cerevisiae may serve a second purpose in addition to surveillance for errors in gene expression has been mounting. Several naturally occurring, intronless mRNAs whose normal level of accumulation depends on the presence of functional UPF genes have been identified. The accumulation of mRNAs coding for the transcriptional activator Ppr1p and several downstream target genes in the uracil biosynthetic pathway have been reported to be sensitive to inactivation of UPF1 (15, 24). Also, the mRNA encoding Ctf13p, a subunit of the kinetochore, depends on the presence of functional UPF genes (9). The mechanism through which the abundance of naturally occurring mRNAs are controlled by the Upf proteins is not clear.
Prior to this study, it was not known how many naturally occurring mRNAs might be affected by loss of UPF function. To assess the global effects of Upf proteins on gene expression, high-density oligonucleotide arrays (HDOA) representing over 6,000 open reading frames (ORFs) in S. cerevisiae were screened for their effects on mRNA accumulation with UPF1, UPF2, and UPF3 were individually inactivated or when all three genes were simultaneously inactivated. Our results indicate that the level of accumulation of hundreds of mRNAs is dependent on the presence of functional UPF genes.
MATERIALS AND METHODS
Construction of isogenic upf− strains.
To eliminate variation due to genetic background, we constructed a set of five isogenic strains for use in HDOA analysis that differ only at the three UPF loci. LRSy307 (MATa his3-11,15 ura3-52 trp1-Δ1 leu2 upf1-Δ1::URA3 upf2Δ1::HIS3 upf3-Δ1::TRP1) (3) was transformed with single-copy plasmids expressing pairwise combinations of the three wild-type UPF alleles. LRSy307 was transformed as follows: with pRS316 (CEN6 ARSH4 URA3) and pML2 (CEN6 ARSH4 LEU2 UPF2 UPF3) to generate a upf1− strain, with pRS316-UPF1 (CEN6 ARSH4 URA3 UPF1) and pLS74 (CEN6 ARSH4 LEU2 UPF3) to generate a upf2− strain, with pLS80 (CEN6 ARSH4 URA3 UPF1 UPF2) and pRS315 (CEN6 ARSH4 LEU2) to generate a upf3− strain, with pRS316 (CEN6 ARSH4 URA3) and pRS315 (CEN6 ARSH4 LEU2) to generate a upf1− upf2− upf3− strain, and with pRS316 (CEN6 ARSH4 URA3) and pML1 (CEN6 ARSH4 LEU2 UPF1 UPF2 UPF3) to generate a wild-type UPF1 UPF2 UPF3 strain. For convenience, we refer to the wild-type UPF1 UPF2 UPF3 genotype as UPF+ and the triple-mutation upf1− upf2− upf3− genotype as upf123−.
A second isogenic pair of strains, ML34 (MATα ura3-52::URA3) and ML51 (MATα ura3-52::URA3 upf1-Δ5), was constructed to monitor mRNA levels by HDOA analysis and Northern blotting in a strain background different from LRSy307. ML51 carries the upf1-Δ5 allele, which contains the same deletion as the upf1-Δ2 allele (15) except that it lacks the insertion of URA3 in the UPF1 coding region. To construct upf1-Δ5, a DNA fragment containing the upf1-Δ2 allele from pPL64 (16) but lacking the URA3 insertion was subcloned into the integrative plasmid pRS306 (28), resulting in pML3 (URA3 upf1-Δ5). pML3 was used to replace the wild-type UPF1 allele with upf1-Δ5 by two-step gene replacement (10) in strain ML27 (MATα ura3-52), resulting in strain ML49 (MATα ura3-52 upf1-Δ5). ML27 and ML49 were made prototrophic for uracil by integrating URA3 near the ura3-52 locus, to generate ML34 and ML51.
Additional strains used in HDOA analysis to assess potential strain-dependent changes in gene expression unrelated to the UPF genes included PLY107 (MATα his4-38 SUF1-1 ura3-52 leu2 trp1-Δ1 lys1-1), BSY1001 (MATα his4-38 SUF1-1 ura3-52 leu2 trp1-Δ1 lys1-1 upf3-Δ1) (14), YJB195 (MAT ade2-1 his3-11,-15 leu2-3,-112 trp1-1 ura3-1 can1-100,) and YJB1471 (MATa ade2-1 his3-11,-15 leu2-3,-112 trp1-1 ura3-1 can1-100 NMD2::HIS3 (17). NMD2 and UPF2 are synonymous (11).
Using quantitative Northern blotting, we confirmed that all strains displayed the expected NMD phenotype by assaying the accumulation of CYH2 pre-mRNA relative to mature CYH2 mRNA. The relative accumulation of CYH2 pre-mRNA serves to indicate whether the NMD pathway is functional because a stop codon in the intron targets the pre-mRNA for rapid decay (12). All of the upf− strains exhibited an average 5.5- ± 0.8-fold increase in the CYH2 pre-mRNA/CYH2 mRNA accumulation ratio, which is characteristic of an inactive NMD pathway.
Growth conditions.
LRSy307 transformants were grown at 30°C in synthetic complete medium (10) (Difco 0919-07 as base) with 2% dextrose supplemented with all amino acids except leucine and without the pyrimidine uracil. Strains ML34 and ML51 were grown in synthetic minimal medium with 2% dextrose without amino acids at 30°C. Overnight cultures grown in synthetic medium at 30°C were diluted 100-fold by resuspension in fresh synthetic medium and grown at 30°C to an optical density at 600 nm of 0.5 (mid-log phase), at which point total RNA was extracted as described below.
RNA methods.
Total yeast RNA was isolated by hot phenol extraction (15) of cells prepared by rapid centrifugation at 23°C, resuspension of the pellet in culture medium, and recentrifugation. Cell pellets were snap-frozen in ethanol mixed with dry ice. Northern blot analysis was performed as described previously (3) except that 15 μg of total RNA was denatured with glyoxal and C2H6SO. Either DNA or antisense RNA was used as the probe. To generate antisense RNA probes, templates for in vitro transcription were created by amplifying genomic DNA via PCR with a T7 polymerase site included in the 3′ oligonucleotide. Probes were labeled and used for hybridization as described previously (9). All experimental signals from Northern blot analysis were normalized against an ACT1-specific hybridization signal, which is unaffected by loss of UPF function (15). Signals were quantitated by using a Molecular Dynamics PhosphorImager (model 425) and ImageQuant software (version 3.3).
Hybridization and analysis of data from DNA microarrays.
The Affymetrix Ye6100 Set HDOA is divided into features that contain oligonucleotides intended to represent all genes coding for cellular mRNAs (31). Most of the ORFs are represented by 40 25-mers, including 20 that are perfectly complementary to the mRNA and 20 that contain a single-base-pair mismatch at the position 13. Together, the 20 probe pairs constitute a probe pair set. Probe pair sets corresponding to 6,218 unique ORFs are present on the HDOA. For 97% of these, it was possible to use stringent parameters to design complete probe pair sets (31). For the remaining 3%, two or three sets of less than ideal oligonucleotides were synthesized on the HDOA. These ORFs were represented by two or three different sets of probe pairs. We included all of these probe pair sets in our analyses to avoid making arbitrary choices regarding what data to include. Consequently, the HDOA contains 6,421 probe pair sets corresponding to 6,218 yeast ORFs, with approximately 3% of the ORFs represented by two or three different probe pair sets.
The preparation of poly(A)+ mRNAs, cDNAs, and cRNAs and the conditions for hybridization were as described by Wodicka et al. (31). cRNA probes were prepared by amplification using in vitro transcription in the presence of nucleotide triphosphates conjugated to biotin and were purified by using an RNeasy column (Qiagen, Santa Clarita, Calif.). The relative concentrations of individual cRNAs were previously shown to be proportional to the abundance of each poly(A) mRNA template (31). After hybridization, the arrays were washed with streptavadin-phycoerythrin. The fluorescent signal from each feature was quantitated with an Affymetrix confocal chip reader (31).
Fluorescent signals corresponding to hybridization intensities were analyzed with Affymetrix GeneChip software (version 3.0) using the following settings: difference threshold, 30; ratio threshold, 1.5; change threshold, 30; percent change threshold, 80. The parameters for the absolute decision matrix (analysis of a single HDOA) as designated by the software (detailed information on the definitions and uses of these parameters in calculations made by the software are available from Affymetrix) are Pos/Neg = (Min 3.0, Max 4.0), Pos/Total = (Min 0.33, Max 0.43), and Log Avg Ratio = (Min 0.9, Max 1.3). The parameters for the comparison decision matrix (comparison between two different HDOA) are Inc/Dec = (Min 3.0, Max 4.0), Inc/Total = (Min 0.33, Max 0.43), D Pos − D Neg Ratio = (Min 0.2, Max 0.3), and Log Avg Ratio Change = (Min 0.9, Max 1.3). In all of our analyses, we used the “normalize to all genes” function in GeneChip software.
To estimate the range of linearity, four different bacterial mRNAs were added to the hybridization cocktail at the following concentrations: BioB (1.5 pM), BioC (5 pM), BioD (25 pM), and Cre (100 pM). When plotted as a function of probe concentration, the fluorescence intensities associated with these transcripts were linear with respect to concentration over three log orders as described by Lockhart et al. (18).
To compare differences in the global expression levels of mRNAs from upf− and UPF+ strains, we used two outputs of the GeneChip software (version 3.0) as the basis of two different analytical methods, referred to as method 1 and method 2. Method 1 is based on one of 28 numerical assessments made by the GeneChip software. In this method, GeneChip subtracts the fluorescent signal for each mismatched oligonucleotide (MM) from the signal for each perfectly matched oligonucleotide (PM). The adjusted signals for each probe pair (PM-MM) are averaged across each probe pair set, excluding probe pairs that give signals 3 standard deviations (SD) or more from the mean. For incomplete probe pair sets consisting of fewer than 20 pairs (see above), GeneChip uses an algorithm to decide whether to exclude outliers when calculating an average for the entire probe pair set. The calculation used in method 1 yields an average adjusted primary signal (termed “average difference” in the software) for each probe pair set corresponding to each mRNA.
Fold changes in mRNA levels due to loss of UPF gene function can then be calculated by dividing the average adjusted primary signal in a mutant by the average adjusted primary signal in the wild type. Numerators and denominators used in this calculation were rounded to 20 for values less than 20. If both values were less than 20, then the fold change was not calculated (value = 1.0, indicating no change) and was not included when average fold changes across multiple trials were calculated. Average fold changes were calculated for each mRNA independently of the difference-call assigned to each mRNA (see below).
Method 2 utilizes a summary calculation (difference-call) made by GeneChip software version 3.0. For each mRNA, the software assigns a difference-call for each transcript as follows: increased, marginally increased, unchanged, marginally decreased, or decreased in a upf− strain compared to the isogenic UPF+ strain. The software utilizes various types of controls and numerical assessments to make adjustments in the data, each of which carries weight in making a difference-call.
RESULTS
Null mutations in UPF1, UPF2, and UPF3 have widespread effects on the transcriptome.
To visualize the effects of upf null mutations on the yeast transcriptome, we compared by HDOA analysis global steady-state mRNA levels in strains carrying upf null mutations with those in a UPF wild-type strain. To ensure that changes in the transcriptome could be attributed to loss of UPF function rather than to differences in genetic background, cRNA probes were prepared from five genetically defined, isogenic, haploid strains (Materials and Methods). Three of the strains carried single null mutations in UPF1, UPF2, or UPF3, the fourth strain carried all three UPF null mutations, and the fifth carried wild-type alleles for all three UPF genes. The wild-type UPF+ alleles in these strains were expressed from single-copy, centromeric plasmids.
Biotin-labeled cRNA probes were independently prepared four times, using poly(A)+ mRNAs and cDNAs as serial templates derived from each of the five strains (upf1−, upf2, upf3−, upf123−, and UPF123+) (Materials and Methods). After hybridization to the HDOAs, sets of data derived from digital images for each of the 20 trials (four trials per strain) were analyzed by using GeneChip software (see Materials and Methods). For each trial, the transcriptome of the UPF+ strain served as the baseline for comparison with the transcriptomes of the ufp1−, upf2−, upf3−, and upf123− strains. Using method 1 (Materials and Methods), all average primary signals for all mRNAs for all 16 trials (4 trials per upf− strain) were compared with the average primary signals for all four trials comprising the wild-type data set Probe pair sets producing the highest and lowest primary adjusted signals were not included in our calculations. We consistently detected >4,500 mRNAs, whereas a small subset of mRNAs were detected more sporadically in some but not all trials.
Of these mRNAs, 225 exhibited an average UPF-dependent fold increase of 2- to 11-fold. The standard deviation (n = 16) for the changes in abundance of all 225 mRNAs was less than or equal to 50% of the average Upf-dependent fold increase. These stringent criteria having been met, the results indicate that the minimal set of mRNAs affected by loss of UPF function is in the range of several hundred out of the >4,500 mRNAs detected.
Method 1 is simple to execute but fails to correct for sources of possible error that could cause an underestimate of the number of mRNA affected by loss of UPF gene function. Also, method 1 combines all upf− trials into one group and potentially ignores any differences in gene expression between the four upf− strains. For this reason, the data were analyzed more exhaustively by method 2 (Materials and Methods). GeneChip software version 3.0 assigns a difference-call of increase, marginal increase, no change, marginal decrease, or decrease (see Materials and Methods). To analyze difference-calls from four separate trials for each of the four upf− genotypes, we assigned numerical weights to the GeneChip difference-calls for each trial as follows: increased signal in single upf null mutant (+2), statistically marginal increase (+1), no change (0), statistically marginal decrease (−1), and decrease (−2). The numerical values of the difference-calls across all four trials for a single upf− strain and for a single mRNA species were summed and divided by the maximum potential score (+8) to generate an individual-knockout index (IKI) score ranging between −1 and +1. The IKI score for a given mRNA reflects the consistency of the change in abundance for each UPF-dependent mRNA in each upf− strain without regard to magnitude. IKI scores near −1 or +1 represent consistent UPF-dependent decreases or increases in the abundance of an mRNA in any of one of the upf− strains. IKI scores near zero signify that the abundance of an mRNA is not dependent on the loss of function of a specific UPF gene.
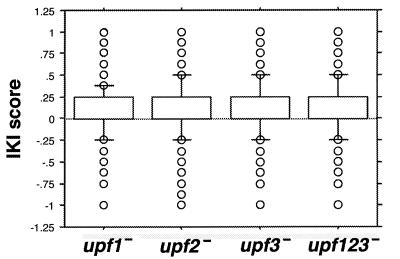
For each of the four upf− strains, we calculated the IKI scores for all 6,421 probe pair sets represented on the HDOA, which represents 6,218 unique ORFs (Materials and Methods; Table 1). When the four distributions of IKI scores corresponding to mRNAs levels in the upf1−, upf2−, upf3−, and upf123− strains were analyzed, we found that each distribution was skewed toward high scores approaching +1. This result would be expected if loss of upf gene function, which is known to block an mRNA decay pathway, primarily causes increased abundance of a substantial number of mRNAs. At the 95th percentile of each distribution, the top 5% (321 of 6,421 probe pair sets) had IKI scores of ≥0.50 (upf1−), ≥0.75 (upf2−), ≥0.63 (upf3−), and ≥0.75 (upf123−). In contrast, the bottom 5% of mRNAs in all four upf− strains had IKI scores of ≤−0.25. The difference between the IKI scores at the 5th and 95th percentiles demonstrates the positive skew in these distributions. This supports the result obtained with method 1 that hundreds of mRNAs increase in abundance in upf− strains. Box plots constructed for the IKI distributions revealed a close similarity between each distribution in each upf− strain (Fig. 1), suggesting that the upf− mutations may affect a similar subset of mRNAs regardless of which UPF gene is inactivated. In summary, genomewide screens failed to uncover major differences that would be expected to occur if a substantial number of mRNAs were differentially affected by mutations in one or two but not all three of the UPF genes.
TABLE 1.
Distribution of index scores
| Strain | Indexa | Median | Mean | SDb | Score at indicated percentile
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5th | 10th | 25th | 75th | 90th | 95th | |||||
| upf1− | IKI | 0.00 | 0.04 | 0.27 | −0.25 | −0.25 | 0.00 | 0.25 | 0.38 | 0.50 |
| upf2− | IKI | 0.00 | 0.07 | 0.27 | −0.25 | −0.25 | 0.00 | 0.25 | 0.50 | 0.75 |
| upf3− | IKI | 0.00 | 0.09 | 0.27 | −0.25 | −0.25 | 0.00 | 0.25 | 0.50 | 0.63 |
| upf123− | IKI | 0.00 | 0.07 | 0.28 | −0.25 | −0.25 | 0.00 | 0.25 | 0.50 | 0.75 |
| All strains | CKI | 0.00 | 0.07 | 0.24 | −0.19 | −0.13 | −0.06 | 0.13 | 0.38 | 0.59 |
IKI scores ranging from −1 to +1 monitor the consistency of changes in mRNA accumulation (see text); IKI scores are calculated from the average difference-calls across multiple trials of a single genotype. CKI scores are calculated manner similarly to IKI scores except that difference-calls are averaged across all trials for all genotypes, thus combining all data into a single quantitative measure of consistency. The IKI and CKI scores indicate whether the average call is for increased or decreased mRNA accumulation, but they are not related to the magnitudes of the changes.
Across four trials for each genotype.
FIG. 1.
Box plots representing the distribution of upf IKI scores. The box delineates the interquartile region between the 25th and 75th percentiles. Whiskers delineate the 10th and 90th percentiles. Values lying outside this region are displayed as circles. Each circle represents IKI scores for multiple mRNAs that happen to fall at the same position.
To further examine whether differences exist between sets of mRNAs affected by loss of function of UPF1, UPF2, or UPF3, the standard deviation for the set of IKI scores for each mRNA from upf1−, upf2−, and upf3− strains was calculated and rank ordered. We reasoned that if changes in abundance for a given mRNA were similar in all three strains, then the three associated IKI scores (one for each genotype) should be similar and consequently have a small standard deviation. Differential expression of an individual mRNA in one or two of the three strains in response to a particular upf− mutation would produce a larger standard deviation for a given set of IKI scores. Based on this, we examined 100 mRNAs that had the largest standard deviation in IKI scores (≥98.5 by percentile). Then, the average adjusted primary signal (method 1; see Materials and Methods) for each mRNA in each relevant trial was visually inspected. We found that only five mRNAs were consistently expressed at different levels depending on which UPF gene was inactivated. The differentially expressed mRNAs were YHR076W, YGR073C, UPF1 (YMR080C), UPF2 (YHR077C), and UPF3 (YGR072W).
To confirm this by another approach, we examined all RNAs represented in each trial, using an independently derived sorting algorithm that identifies subsets of mRNAs that are uniquely altered in only one or two of the three upf− strains. mRNAs were designated as altered within a single upf− strain if they were assigned a difference-call of “increase” or “decrease” in three of the four trials for that strain; otherwise, they were considered unchanged. mRNAs with difference-calls categorized as marginal were considered unchanged. This method produced a list of 104 mRNAs that changed in one or two of the three upf− strains without a corresponding change in the other upf− strains. The average adjusted primary signals for each of these mRNAs in each relevant trial were inspected visually. The same five mRNAs as described above were identified as the ones most likely to exhibit differential expression in different upf− strains.
HDOA data for the five mRNAs were analyzed in greater detail (Table 2). YHR076W mRNA, which is encoded by a gene adjacent to UPF2, was 3.5 (±0.4)- and 2.9 (±0.5)-fold more abundant in the upf1− and upf3− strains, respectively, than in the wild type but was not detected in upf2− and upf123− strains. YGR073C mRNA, which is encoded by a gene adjacent to UPF3, was increased 1.8 (±0.2)-fold in the upf2− strain but unchanged in all other upf− strains. We are not certain whether the observed patterns of differential expression are related to the locations of these genes near the upf2-Δ1::HIS3 and upf3-Δ1::TRP1 disruptions or to expression of wild-type UPF genes from centromeric plasmids in the strains, or to both factors.
TABLE 2.
Average fold increases of five differentially expressed mRNAs
| mRNA | Avg fold change in mRNA accumulation ISD
|
Method | |||
|---|---|---|---|---|---|
| upf1− | upf2− | upf3− | upf123− | ||
| YHR076W (near UPF2) | 3.5 ± 0.4 | Absent | 2.9 ± 0.5 | Absent | HDOA analysis |
| YGR073C (near UPF3) | 1.3 ± 0.2 | 1.8 ± 0.2 | 0.8 ± 0.0 | 0.9 ± 0.2 | HDOA analysis |
| YMR080C (UPF1) | Absent | 3.5 ± 2.5 | 2.1 ± 1.3 | Absent | HDOA analysis |
| YHR077C (UPF2) | 1.8 ± 0.5 | Absent | 1.3 ± 0.1 | Absent | HDOA analysis |
| YGR072W (UPF3) | 4.7 ± 1.3 | 5.6 ± 2.3 | Absentb | Absentb | HDOA analysis |
| YMR080C (UPF1) | Absent | 2.0 ± 0.3 | 1.8 ± 0.4 | Absent | Northern blotting |
| YGR072W (UPF3) | 5.7 ± 2.1 | 7.0 ± 1.8 | Absent | Absent | Northern blotting |
Calculated either from HDOA data (four trials for each upf− derivative of strain LRSy307 strain compared to four trials for the UPF+ derivative of LRSy307) or from Northern blotting (n = 4 for each genotype).
HDOA most likely detected hybridization to UPF3 sequences near the 3′ end of a UPF3-TRP1 fusion mRNA encoded by the UPF3::TRP1 allele, which does not produce a functional protein product (14). Native UPF3 mRNA is absent.
UPF1 mRNA was absent, as expected, in upf1− and upf123− strains but was two- to threefold more abundant in the upf2− and upf3− strains than in the UPF+ strain. UPF2 mRNA was absent as expected in upf2− and upf123− strains and unchanged in the upf3− strain but was increased 1.8 (±0.5)-fold in the upf1− strain. UPF3 mRNA was absent as expected in upf3− and upf123− strains but was increased an average of more than fivefold in upf1− and upf2− strains. We confirmed the increased levels of UPF1 and UPF3 mRNAs by Northern blotting of RNA from the LRSy307-based transformants that were used for HDOA analysis (Table 2).
To determine whether the differential expression of wild-type UPF1 and UPF3 genes might be related to their presence on plasmids in the LRSy307-based transformants, we used Northern blotting to assay additional sets of UPF+ and upf− strains that do not carry UPF genes on plasmids. The level of UPF1 mRNA was the same in strains PLy107 (UPF3+) and BSY1001 (upf3−) (1.2 ± 0.1-fold increase in upf3−, n = 2). Similarly, UPF3 mRNA levels were the same in strains ML34 (UPF1+) and ML51 (upf1−) (1.1 ± 0.6-fold increase in upf1−, n = 3).
When wild-type UPF genes were supplied on plasmids, UPF1 mRNA was overexpressed in a upf3− background and UPF3 mRNA was overexpressed in a upf1− background. No overexpression was observed when the wild-type alleles for these genes were located at their resident positions on chromosomes. This finding suggests that the overexpression from plasmids has no bearing on the expression of UPF genes in wild-type. Overexpression of UPF1 or UPF3 mRNA does not appear to cause any change in the global profile of mRNA accumulation that results from the disruption of a UPF gene because the same set of mRNAs was affected in the ufp123− triple mutant (data not shown). Barring the rare exceptions, our results suggest that loss of function of any one of the three UPF genes causes altered accumulation of the same subset of mRNAs.
Coincident changes in the abundance of mRNAs define the target size.
Since the patterns of mRNA accumulation were nearly identical in the upf1−, upf2−, upf3−, and upf123− strains, we developed a method to analyze the overall target size for UPF-dependent mRNAs by combining all upf− trials into a single set. This provided a sample based on 16 trials (4 independent trials in each of the four upf− genotypes: upf1−, upf2−, upf3−, and upf123−) normalized against four respective trials for the UPF+ strain. We used a scoring system similar to the IKI system to measure the consistency of the difference-calls associated with each mRNA. This index score, termed the combined-knockout index (CKI) score, measures the consistency of effects detected by HDOA analysis on a given mRNA in strains carrying a loss-of-function mutation of any or all of the UPF genes. CKI scores were calculated in a manner similar to IKI scores except that numerical values representing difference-calls were summed across 16 trials representing all four of the upf− strains rather than 4 trials from a single upf− strain.
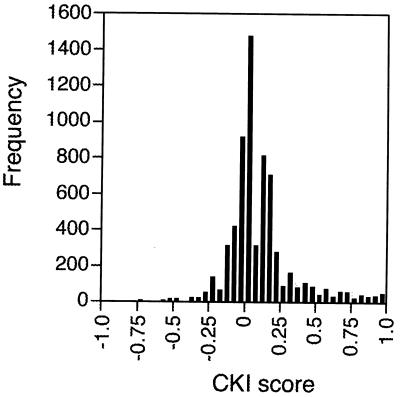
Like the IKI scores, the distribution of CKI scores was skewed toward high scores approaching +1. The standard deviation for CKI scores was smaller than for IKI scores due to the larger number of trials used to calculate the distribution (Table 1). Consequently, a tighter distribution was produced, which allowed for more accurate identification of mRNAs with outlying CKI scores. At the 95th percentile of the distribution, the top 5% (321 of 6,421 probe pair sets) had CKI scores of ≥0.59. The frequencies of CKI scores across for all probe pair sets are shown in a histogram (Fig. 2).
FIG. 2.
Distribution of all CKI scores across 6,421 mRNAs analyzed by HDOA. A score of −1.0 indicates a UPF-dependent decrease in every trial for every upf− strain; a score of +1.0 indicates a UPF-dependent increase in every trial for every upf− strain; a score of zero represents no average change.
The UPF-independent mRNAs are clustered around the median of 0.00 (Table 1). Of the 6,421 ORFs, 368 had CKI scores greater than 2 SD above the mean (+0.55); while 40 mRNAs had scores 2 SD below the mean (−0.41). Using the standard deviation associated with CKI score distribution, we divided the distribution into three sets of mRNAs: those that exhibit UPF-dependent increases (CKI ≥ +0.55), UPF-dependent decreases (CKI ≤ −0.41), and UPF-independent accumulation (−0.41 < CKI < +0.55).
The vast majority of mRNAs were not affected by loss of UPF function, as shown by the large accumulation of CKI scores near zero. However, there were a significant number of mRNAs affected by UPF loss of function. The distribution is skewed toward positive values, suggesting that loss of UPF function may cause a greater number of mRNAs to exhibit increased rather than decreased accumulation. To further define and empirically test the set of UPF-dependent mRNAs, Northern blot analysis was used to independently verify the results from HDOA analysis and to quantitate the changes in mRNA accumulation that result from inactivation of the NMD pathway. To accomplish this, we selected seven mRNAs with CKI scores near the score defined as the mean + 2 SD (+0.55). We selected mRNAs encoded by the genes GBP2 (YCL011C), UGA3 (YDL170W), ALR2 (YFL050C), YIL087C, MET14 (YKL001C), YLR130C, and PHO80 (YOL001W), which had CKI scores ranging from +0.44 and +0.59 (Table 3).
TABLE 3.
Quantitative comparison of changes in mRNA accumulation determined by HDOA analysis and Northern blotting
| ORF | Gene | CKI score | Avg fold change in mRNA accumulation ± SD
|
Ho: at 99% μ(A) = μ(N)c | ML34/ML51, Northern blottingd | |
|---|---|---|---|---|---|---|
| HDOAa | Northern blottingb | |||||
| YCL011C | GBP2 | 0.44 | 1.4 ± 0.2 | 2.5 ± 0.2 | Reject | 2.3 |
| YDL170W | UGA3 | 0.44 | 1.7 ± 0.5 | 1.6 ± 0.4 | Accept | 3.7 |
| YFL050C | ALR2 | 0.59 | 3.8 ± 3.0 | 4.2 ± 0.3 | Accept | 6.2 |
| YIL087C | 0.44 | 1.4 ± 0.2 | 1.8 ± 0.5 | Accept | 1.2 | |
| YKL001C | MET14 | 0.44 | 1.5 ± 0.3 | 1.6 ± 0.2 | Accept | 0.9 |
| YLR130C | 0.44 | 1.4 ± 0.4 | 1.8 ± 0.1 | Reject | 1.9 | |
| YOL001W | PHO80 | 0.47 | 2.1 ± 1.3 | 2.5 ± 0.4 | Accept | 2.7 |
| YCR020C | PET18 | 1.0 | 4.2 ± 0.8 | 7.1 ± 1.0 | Reject | 6.3 |
| YDR210W | 1.0 | 2.2 ± 0.3 | 5.1 ± 1.9 | Reject | 2.2 | |
| YEL073C | 1.0 | 11.0 ± 4.4 | 13.7 ± 5.5 | Accept | 6.3 | |
| YIL165C | 0.88 | 2.6 ± 0.6 | 2.7 ± 0.5 | Accept | 4.6 | |
| YLR014C | PPR1 | 0.19 | 2.0 ± 0.8 | 2.9 ± 0.5 | Accept | 3.4 |
| YLR363C | NMD4 | 0.84 | 3.4 ± 1.1 | 3.2 ± 0.5 | Accept | 3.4 |
| YML123C | PHO84 | −1.0 | −4.4 ± 1.4 | −3.3 ± 0.3 | Reject | |
| YNL046W | 1.0 | 2.8 ± 1.3 | 2.4 ± 0.4 | Accept | 2.8 | |
| YOL108C | INO4 | 0.9 | 2.2 ± 0.7 | 3.4 ± 1.9 | Accept | 2.3 |
| YOR303W | CPA1 | 0.81 | 1.8 ± 0.5 | 2.6 ± 0.2 | Reject | 2.2 |
Calculated from HDOA data, using four trials from each of four upf− derivatives of strain YRSy307 (upf1−, upf2−, upf3−, and upf123−) (n = 16 trials in all) compared with data from four trials for the UPF+ derivative of LRSy307.
mRNA accumulation was measured by quantitative Northern blot analysis. Values were calculated by comparing data from the upf123− and the UPF+ derivative of strain LRSy307. mRNAs for YCL011C, YDL170W, YFL050C, YIL165C, YKL001C, YLR130C, YLR363C, YNL046W, and YOR303W were tested at n = 4; all others were tested at n = 8.
Statistical similarities of the means associated with data from HDOA analysis and Northern blotting were compared by using two-tailed t test assuming unequal variances at α = 0.01. The null hypothesis (Ho) states that the means are equal [μ(A) = μ(N)], while the alternative hypothesis states that the means are not equal. Accepting the null hypothesis indicates that the mean calculations for both HDOA and Northern blot analyses are not statistically indistinguishable. Rejecting the null hypothesis indicates that the means are statistically different. μ(A), mean average fold change as measured by arrays: μ(N), mean average fold change as measured by Northern blotting.
mRNA accumulation was assayed by Northern blot analysis in strains ML34 (UPF1) and ML51 (upf1−) (see Materials and Methods; to test whether changes observed in strain LYRy307 were strain specific.
Using Northern blotting, we found that all seven mRNAs exhibited increased mRNA abundance ranging from fold increases of 1.6 (±0.2) for UGA3 to 4.2 (±0.3) for ALR2 (Table 3). When the results of Northern blotting were compared with the results from HDOA analysis by using a two-tailed t test (see footnote c to Table 3), no evidence of significant statistical differences was found for the average fold changes of five of the seven mRNAs. For the remaining two (GBP2 and YLR130C), the fold increases derived by Northern blotting were higher than those derived from HDOA analysis. To see if similar results would be obtained for a different set of strains, we also determined the fold increases for the seven mRNAs in another isogenic set of strains, ML34 (UPF+) and ML51 (upf1-Δ5) (Materials and Methods). The fold increases were similar except for MET14 mRNA (CKI = 0.44), which was not increased in ML51 (Table 3) but was increased in YJB1471 (upf2−) (discussed below).
To test the efficacy of HDOA in predicting the magnitude of changes in mRNA abundance, we examined 17 mRNAs (including the 7 discussed above) by comparing the changes predicted by HDOA analysis and Northern blotting (Table 3). Fifteen had CKI scores ranging from +0.44 to +1.0. PPR1 mRNA (CKI = 0.19) was included because of prior evidence that PPR1 mRNA abundance depends on a functional UPF1 gene (24). PHO84 mRNA (CKI = −1.0) was included to verify that a strongly negative CKI score corresponds to a decrease in abundance as determined Northern blotting.
Measured by both HDOA analysis and Northern blotting, the mean fold changes had a correlation coefficient of 0.95, indicating that the mean fold changes measured by HDOA analysis are similar to those measured by Northern blotting. We compared these means by using Student’s t tests to determine if the mean fold change as predicted by HDOA analysis was statistically different from that predicted by Northern blotting. Eleven of the 17 mRNAs produced mean fold changes that showed no evidence of a statistical difference between the two techniques; 6 of the 17 mRNAs displayed a statistical difference with Northern blotting typically reporting larger fold changes. However, the change predicted by HDOA analysis always showed the same positive or negative trend as did the change predicted by Northern blotting. When they occurred, deviations obtained when the two methods were used were less than a factor of 2 in magnitude.
To ensure that the UPF-dependent changes in expression were not unique to the LRSy307-based transformants, we used Northern blotting of RNA from additional sets of strains to examine the abundance of the 17 mRNAs discussed above. ML34 (UPF1+) and ML51 (upf1-Δ4) are isogenic derivatives of S288C differing only at the UPF1 locus (Materials and Methods). We detected 16 of the 17 mRNAs in ML34 and ML51. PHO84 could not be consistently detected in either ML34 or ML51. Of the 16 detectable mRNAs, 14 exhibited fold changes comparable to those observed in derivatives of strain LRSy307. The two mRNAs that exhibited anomalous behavior, MET14 and YIL087C, were detected as UPF-independent mRNAs in the isogenic strains ML34 and ML51. ML34 and ML51 were grown in a different synthetic medium than were the derivatives of strain LRSy307, which might account for differences in expression compared with expression in LRSy307 transformants. However, in another isogenic pair of strains, YJB1471 (upf2−) and YJB195 (UPF2+) (Materials and Methods), which were grown in the same synthetic medium as were strains ML34 and ML51, MET14 exhibited a 1.6 (±0.2)-fold increase and YIL087C exhibited a 1.9 (±0.7)-fold increase. Overall, the results indicate that most of the changes in mRNA levels identified in the LRSy307 transformants were also observed in other strains, but some of the mRNAs accumulated to different levels in different strains.
The Northern blotting experiments described above indicate that mRNAs with CKI scores above +0.44 are the best candidates for those exhibiting increased abundance when the UPF genes are inactivated. mRNAs with the highest CKI scores (≥+0.90) and the corresponding fold increases in abundance are shown in Table 4. CKI scores were ≥+0.44 for 539 of the 6,421 probe pair sets, including 529 unique mRNAs and 10 mRNAs tiled more than once on the HDOA (see Materials and Methods). Average fold changes for these mRNAs were increased as follows: 12 from 5- to 11-fold, 29 from 4- to 5-fold, 56 from 3- to 4-fold, 234 from 2- to 3-fold, 179 from 1.5- to 2-fold, and 28 from 1.2- to 1.5-fold. One mRNA was unchanged (CKI = 0.5).
TABLE 4.
Magnitude of changes for mRNAs with CKI scores of >+0.90
| ORF | Gene | CKI score | Avg fold change in mRNA accumulation ± SDa |
|---|---|---|---|
| YOL108C | INO4 | 0.91 | 2.2 ± 0.7 |
| YDL243c | 0.91 | 2.5 ± 0.6 | |
| YER186c | 0.91 | 2.4 ± 0.5 | |
| YEL049w | PAU2 | 0.91 | 2.6 ± 0.8 |
| YDR387C | 0.91 | 3.1 ± 0.8 | |
| YKR069W | 0.91 | 3.2 ± 0.5 | |
| YFR020W | 0.91 | 3.3 ± 0.9 | |
| YEL048c | 0.91 | 5.4 ± 2.9 | |
| YPL258C | 0.91 | 6.5 ± 3.0 | |
| YNL339C | 0.93 | 2.0 ± 0.3 | |
| YFL035C | 0.94 | 1.9 ± 0.4 | |
| YHL050C | 0.94 | 1.9 ± 0.4 | |
| YJL223C | 0.94 | 2.0 ± 0.3 | |
| YLR251W | 0.94 | 1.9 ± 0.4 | |
| YLL066c | 0.94 | 1.9 ± 0.3 | |
| YLR152c | 0.94 | 2.0 ± 0.4 | |
| YIR020C | 0.94 | 2.7 ± 0.7 | |
| YOR202W | HIS3 | 0.94 | 2.3 ± 0.4 |
| YHL047C | 0.94 | 2.6 ± 0.5 | |
| YHR199C | 0.94 | 2.5 ± 0.4 | |
| YKL224C | 0.94 | 2.6 ± 0.3 | |
| YGR040W | KSS1 | 0.94 | 2.7 ± 0.9 |
| YBR301w | 0.94 | 3.0 ± 0.4 | |
| YLR122c | 0.94 | 3.7 ± 1.9 | |
| YGL261C | 0.94 | 3.3 ± 0.9 | |
| YDL243c | 0.94 | 2.5 ± 0.6 | |
| YNR059W | 0.94 | 4.0 ± 2.7 | |
| YOL165C | 0.94 | 3.7 ± 1.3 | |
| YMR320W | 0.94 | 4.4 ± 1.7 | |
| YKR012C | 0.94 | 4.5 ± 1.9 | |
| YMR065W | 0.94 | 4.8 ± 2.3 | |
| YMR316C-b | 0.94 | 4.0 ± 1.1 | |
| YLL067c | 0.97 | 2.0 ± 0.5 | |
| YPL144W | 0.97 | 2.5 ± 0.5 | |
| YCR096c | A2 | 0.97 | 2.8 ± 0.5 |
| YFL057C | 0.97 | 2.9 ± 0.6 | |
| YNL270C | ALP1 | 0.97 | 3.3 ± 1.4 |
| YKL071W | 0.97 | 4.6 ± 1.9 | |
| YIR031C | DAL7 | 0.97 | 4.5 ± 1.0 |
| YER180C | ISC10 | 0.97 | 4.7 ± 1.4 |
| YFL020C | PAU5 | 0.97 | 3.7 ± 0.9 |
| YER076c | 0.97 | 3.9 ± 0.6 | |
| YER187w | 0.97 | 4.3 ± 1.3 | |
| YER188w | 0.97 | 4.5 ± 1.2 | |
| YFL061W | 0.97 | 4.2 ± 2.4 | |
| YLL060c | 0.97 | 4.9 ± 2.2 | |
| YLL064c | 1.0 | 2.2 ± 0.5 | |
| YCR039c | ALPHA2 | 1.0 | 2.6 ± 0.3 |
| YCL067c | ALPHA2 | 1.0 | 2.6 ± 0.3 |
| YKR061W | KTR2 | 1.0 | 2.5 ± 0.4 |
| YLR461W | PAU4 | 1.0 | 2.5 ± 1.0 |
| YDR210W | 1.0 | 2.1 ± 0.3 | |
| YBL113C | 1.0 | 2.2 ± 0.4 | |
| YLL025w | 1.0 | 2.8 ± 0.6 | |
| YML133C | 1.0 | 2.3 ± 0.7 | |
| YNL046W | 1.0 | 2.8 ± 1.3 | |
| YER092w | 1.0 | 2.5 ± 0.3 | |
| YER115C | spr6 | 1.0 | 2.7 ± 0.6 |
| YCR020C | PET18 | 1.0 | 4.2 ± 0.8 |
| YLR010c | 1.0 | 3.3 ± 0.7 | |
| YGR289C | AGT1 | 1.0 | 3.1 ± 0.7 |
| YLR460C | 1.0 | 3.1 ± 0.7 | |
| YBR008c | 1.0 | 3.9 ± 0.7 | |
| YHL046C | 1.0 | 3.3 ± 0.6 | |
| YGL193C | 1.0 | 4.0 ± 1.2 | |
| YCR102c | 1.0 | 3.9 ± 1.3 | |
| YAR023C | 1.0 | 4.0 ± 0.7 | |
| YGL263W | 1.0 | 4.2 ± 1.0 | |
| YEL028w | 1.0 | 4.2 ± 1.6 | |
| YER066w | 1.0 | 4.4 ± 1.6 | |
| YNL162W | RPL41A | 1.0 | 4.2 ± 1.1 |
| YNL335W | 1.0 | 4.9 ± 4.0 | |
| YFR026C | 1.0 | 5.1 ± 1.8 | |
| YOL162W | 1.0 | 6.7 ± 2.7 | |
| YGL262W | 1.0 | 8.2 ± 2.8 | |
| YEL073c | 1.0 | 11.0 ± 4.4 | |
| YIR043C | 1.0 | 9.4 ± 3.4 |
Averaged across all four trials for all four upf− strains (a total of 16 trials).
By comparison with the minimal set of 225 NMD-sensitive mRNAs defined earlier by method 1 (see above and Materials and Methods), the potential number of natural targets that increase in abundance when the UPF genes are inactivated is somewhere between 3 and 9% of the 6,218 unique mRNAs. We did not perform Northern blotting experiments on a sample of the mRNAs with CKI scores of between 0 and +0.44, but we presume that each mRNA in this range is less likely to be affected. However, an increase in the PPR1 mRNA level (CKI = 0.19) which was confirmed by Northern blotting indicates that some of the mRNAs in the 0 to +0.44 range could also exhibit increased abundance when the UPF genes are inactivated.
Using similar logic, we examined mRNAs with CKI scores of ≤−0.41, which fall 2 SD or more below the mean. Of the 40 mRNAs identified (Table 5), 1, PHO84 mRNA (CKI = −1.0), was decreased in abundance 4.4 (±1.4)-fold according to HDOA analysis. Northern blotting indicated that PHO84 mRNA was decreased 3.3 (±0.3)-fold in abundance. This result shows that inactivation of the NMD pathway can lead to reduced mRNA abundance. Six of the 40 mRNAs were decreased 2-fold or more in abundance; one of these six, YOR387C mRNA, was decreased 7.1-fold (Table 5). Overall, these results indicate that the number of mRNAs in upf− strains that increase in abundance outnumber those that decrease in abundance at least 10-fold. Data are available for all mRNAs on line (23b, 25a).
TABLE 5.
Minimal set of mRNAs with CKI scores that reflect a decrease in abundance when the NMD pathway is inactivated
| ORF | Gene | CKI score | Avg fold change in mRNA accumulation ± SDa |
|---|---|---|---|
| YDR281C | −0.53 | −2.0 ± 0.7 | |
| YGL258W | −0.56 | −3.4 ± 2.5 | |
| YCR097wa | Mata1 | −0.66 | −3.5 ± 1.3 |
| YOR387C | −0.87 | −7.1 ± 6.9 | |
| YML123C | PHO84 | −1.0 | −4.4 ± 1.4 |
| YHR136C | −1.0 | −6.9 ± 4.0 |
Averaged across all four trials for all four upf− strains (a total of 16 trials). The change for PHO84 was confirmed by Northern blotting (Table 8).
Distribution of UPF-dependent mRNAs by function and expression level.
mRNAs that change in abundance when the NMD pathway is inactivated were sorted according to function by using the categories described in the MIPS (Munich Information Center for Protein Sequences) database (23a). Table 6 shows the numbers and relative percentages of mRNAs in each functional category assigned by MIPS for all mRNAs and for the 529 unique UPF-dependent mRNAs that exhibit consistent, increased accumulation ranging from 1.2- to 11-fold with CKI scores of ≥0.44. A substantial number of mRNAs have no known function and are therefore are listed as “unclassified” in Table 6. For 13 of the 15 functional categories, the mRNAs are distributed similarly for both sets. mRNAs coding for products that function in protein synthesis were vastly underrepresented among the UPF-dependent mRNAs (5.5% for all mRNAs, compared to 0.9% for UPF-dependent mRNAs). A much more modest decrease in the frequency of representation was also observed for the “protein destination” category (8.2% for all mRNAs, compared to 3.7% for UPF-dependent mRNAs).
TABLE 6.
Functional distribution of UPF-dependent mRNAs
| MIPS category | No. (%)
|
|
|---|---|---|
| Transcriptome (6,233 ORFs)a | UPF-dependent (529 mRNAs)b | |
| Metabolism | ||
| Energy | 236 (3.7) | 23 (4.3) |
| Growth, division, DNA synthesis | 753 (12.1) | 44 (8.3) |
| Transcription | 722 (11.6) | 39 (7.3) |
| Protein synthesis | 343 (5.5) | 5 (0.9) |
| Protein destination | 512 (8.2) | 20 (3.8) |
| Transport facilitation | 300 (4.8) | 32 (6.0) |
| Intracellular transport | 414 (6.6) | 20 (3.8) |
| Cellular biogenesis | 170 (2.7) | 12 (2.3) |
| Signal transduction | 119 (1.9) | 4 (0.8) |
| Rescue, defense, death, aging | 311 (5.3) | 40 (7.6) |
| Ionic homeostasis | 114 (1.8) | 5 (0.9) |
| Cellular organization | 2,105 (33.8) | 107 (20.2) |
| Unclassified | 2,605 (41.8) | 260 (49.1) |
| Intron containingc | 248/6,218 (4.0) | 21/529 (3.6) |
Based on functional assignments made by MIPS (as of the 16 July 1998 MIPS update). To calculate the percentages, the denominator of 6,233 was used, which represents the number of unique ORFs in the MIPS database. The percentages do not sum to 100% because a substantial number of mRNAs have been assigned to more than one category.
Based on functional assignments of the UPF-dependent mRNAs that increase in abundance (529 unique mRNAs with CKI scores of ≥0.44). To calculate the percentages, the denominator of 6,218 was used, which represents the number of unique ORFs represented on the HDOA. The percentages do not sum to 100% because a substantial number of mRNAs are listed in more than one category.
Percentage of intron-containing mRNAs among 6,218 ORFs uniquely tiled on the HDOA and percentage of intron-containing mRNA among the 529 unique UPF-dependent mRNAs with CKI scores of ≥0.44.
Introns and exon-exon junctions can play a significant role in the NMD pathway in higher eukaryotes (23). Consequently, we compared the frequencies of intron-containing mRNAs in the set of 529 unique UPF-dependent mRNAs with CKI scores of ≥0.44 and the larger set of 6,218 unique mRNAs tiled on the HDOA. Intron-containing mRNAs were distributed similarly in both sets (Table 6, last row), suggesting that the presence of an intron is probably unrelated to UPF-mediated control of mRNA abundance in S. cerevisiae.
We assessed whether the average abundance for UPF-dependent mRNAs with CKI scores of ≥0.44 was higher or lower than the average abundance for all mRNAs (Table 7). To accomplish this, the average expression levels across the four UPF+ trials were calculated by using the average adjusted primary signal for each mRNA (method 1; see Materials and Methods). The mean values were 188 fluorescent units for UPF-dependent mRNAs with CKI scores of ≥0.44 and 546 fluorescent units for all mRNAs. The threefold difference is statistically significant as measured by Student’s t test assuming equal variance at 95% confidence. To confirm this, we examined a set of 529 mRNAs selected at random among all mRNAs and compared the mean value in fluorescent units for this set with mean value in fluorescent units for the set of 529 UPF-dependent mRNAs. For all three sets of data, the medians and interquartile regions were similar. However, the expression levels diverged at the 90th and 95th percentiles of the distribution. While there are some statistical caveats to conclusions based on comparing different probe pair sets (31), our results suggest that UPF-dependent mRNAs are underrepresented among mRNAs expressed in the upper quartile of relative expression levels. It therefore appears that the inactivation of UPF genes disproportionately affects the accumulation of mRNAs that are normally present at lower than average abundance in wild-type strains.
TABLE 7.
Average relative abundance of UPF-dependent mRNAs
| mRNAs | Median | Mean | SD | Avg relative abundance at indicated percentile
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 5th | 10th | 25th | 75th | 90th | 95th | ||||
| Alla | 86 | 546 | 1,550 | −2 | 8 | 31 | 291 | 1,133 | 2,790 |
| Randomb | 97 | 559 | 1,530 | 1 | 8 | 37 | 303 | 1,186 | 3,114 |
| UPF dependentc | 88 | 188 | 432 | 7 | 15 | 37 | 215 | 515 | 758 |
Distribution of mRNAs ranked according to relative level of accumulation for all ORFs tiled on the HDOA. The range for numbers listed by percentile is −50 (indicating an accumulation of near zero) to >10,000 (highest level of accumulation).
Sample distribution of 529 mRNAs selected at random.
Distribution of 529 mRNAs with CKI scores of ≥0.44.
Regulatory cascades among the UPF-dependent mRNAs.
Numerous subsets consisting of coregulated mRNAs were evident among the UPF-dependent mRNAs. We examined two such mRNA subsets in further detail. One coregulated subset consists of PPR1, which encodes a positive transcriptional activator, and downstream targets of Ppr1p-mediated transcriptional activation, including the URA1, URA3, URA4, and URA10 genes (Table 8) (20, 27). It was previously reported that PPR1 mRNA accumulation increases threefold when UPF1 is inactivated (24). According to HDOA data, the accumulation of PPR1 mRNA increased 2.0 (±0.8)-fold when NMD was inactivated. PPR1 mRNA was not included in the list of UPF-dependent mRNAs with CKI scores of ≥0.44. The CKI score was only +0.19 due to inconsistencies in the difference-calls across trials resulting from low signal intensities. However, we confirmed by quantitative Northern blotting that the accumulation of PPR1 mRNA increased 2.9 (±0.5)-fold when NMD was inactivated. PPR1 mRNA also exhibited increased accumulation in the upf− strain ML51 (Table 3).
TABLE 8.
Effects of NMD on regulatory cascades in metabolic pathways
| mRNA | CKI score | Avg fold change in mRNA accumulation ± SD
|
|
|---|---|---|---|
| HDOA | Northern blotting | ||
| PPR1 | +0.19 | 2.0 ± 0.8 | 2.9 ± 0.5 |
| URA1 | +0.69 | 1.5 ± 0.3 | NDa |
| URA10 | +0.87 | 2.1 ± 0.7 | ND |
| URA4 | +0.09 | 1.3 ± 0.2 | 1.3 ± 0.0 |
| PHO84 | −1.0 | −4.4 ± 1.4 | −3.3 ± 0.3 |
| PHO5 | −0.44 | −1.9 ± 1.1 | ND |
| PHO86 | −0.25 | −1.3 ± 0.4 | ND |
| PHO8 | −0.28 | −1.3 ± 0.3 | ND |
| PHO80 | +0.47 | 2.1 ± 1.3 | 2.5 ± 0.4 |
ND, not determined.
Since Ppr1p is a transcriptional activator, increased accumulation of the mRNA and the corresponding gene product should cause increased transcription of downstream target genes. This should lead to increased accumulation of the corresponding mRNAs. To test this, we examined the accumulation of the downstream targets URA1, URA4, and URA10. We could not obtain a meaningful assessment of URA3 mRNA accumulation because the URA3 gene was not at its usual chromosomal location but was instead contained on the single-copy plasmids used in the LRSy307-derived strains (Materials and Methods). We found by HDOA analysis that the URA1 (CKI = +0.69) and URA10 (CKI = +0.87) mRNAs exhibited increased accumulation (Table 8). The CKI score for URA4 mRNA was only +0.19. The accumulation of URA4 mRNA did not appear to be increased in response to an increase in PPR1 mRNA accumulation (1.3 ± 0.2 based on HDOA analysis and 1.3 ± 0.0 based on Northern blotting) although it is reportedly regulated by PPR1. By HDOA analysis, the accumulation of the URA2 and URA5 mRNAs were unchanged. These mRNAs are not regulated by PPR1 (20, 27) and would therefore not be expected to change.
We identified another subset of UPF-dependent mRNAs that code for proteins involved in phosphate utilization, including PHO5, which codes for the major secreted acid phosphatase (2), PHO84 and PHO86, which code for inorganic phosphate transporters (5), PHO8, which codes for an alkaline phosphatase (13), and PHO80, which codes for a cyclin-dependent protein kinase that inhibits transcription of PHO5 (6) (Table 8). According to HDOA analysis, PHO84, PHO5, PHO86, and PHO8 were all decreased in abundance whereas PHO80 was increased in abundance in upf− derivatives of strain LRSy307 and in the upf− strain ML51 (Table 3). All other mRNAs known to code for proteins involved in phosphate utilization, were unchanged in abundance.
DISCUSSION
The goal of this study was to establish the extent to which the Upf proteins affect the expression of the >6,000 genes that comprise the transcriptome of S. cerevisiae. To address this question, we probed HDOA with cRNAs corresponding to all polyadenylated mRNAs in strains carrying functional disruptions of the UPF genes. The fluorescent signals were analyzed by using two outputs of GeneChip 3.0 software as the basis of two different analytical methods (see Materials and Methods).
We identified a minimal set of 225 mRNAs that exhibited an average UPF-dependent fold increase of 2- to 11-fold with a standard deviation ≤50% of the average UPF-dependent fold increase. To mine the data further, we devised the IKI to compare changes in different upf− strains. By analyzing the distributions of IKI scores, which range from −1 to +1, we found that 99.9% of the observed changes in mRNA abundance were common to upf1−, upf2−, upf3−, and upf123− strains. There were only five exceptions: UPF1, UPF2, UPF3, YHR076W, and YGR073C.
The UPF genes exhibited a complex pattern of overexpression in the LRSy307 series of strains (Table 2). The anomalous behavior of the UPF genes might be explained in part by the fact that the genes were expressed from plasmids rather than from their normal chromosomal loci. In strains carrying UPF genes at their normal chromosomal loci, the UPF genes were expressed at normal levels and in a UPF-independent manner. Given this, we do not currently attach any physiological significance to the UPF-dependent overexpression of UPF genes from plasmids. However, we considered whether the overexpression could influence the number of mRNAs affected by loss of UPF function or the magnitudes of the effects. By comparing data from the strains that carry the single null alleles (upf1−, upf2−, or upf3−) with data from the triple-null strain (upf123−), we concluded that the overexpression of UPF genes had no effect on the number of UPF-dependent mRNAs or on the magnitude of the observed changes.
YHR076W is located immediately adjacent to UPF2. mRNA accumulation was increased about threefold in the upf1− and upf3− strains but was absent in the upf2− and upf123− strains. YGR073C is located immediately adjacent to UPF3. This mRNA was only marginally increased and only in the upf1− and upf2− strains. Possibly some of these changes can be related to the positions of these genes near the insertions in the upf2Δ1::HIS3 and upf3-Δ1::TRP1 null alleles. Although insertions could perturb local rates of transcription through local changes in chromatin structure, it is not clear why these mRNAs are differentially expressed depending on which of the UPF genes are being expressed from plasmids.
Barring the exceptions noted above, our results indicate that the same mRNAs respond to loss of UPF function regardless of which of the UPF genes is disrupted. This finding served as the basis for pooling all trials for all upf− strains to calculate a CKI index score for each mRNA. The CKI score measures the consistency (but not the magnitude) of the UPF-dependent effect on a given mRNA. This approach had the advantage of producing a tighter distribution with a much smaller standard deviation than any of the IKI distributions due to the increased number of trials used to calculate the CKI scores (16 in all).
Like the IKI scores, the CKI scores range from −1 to +1 and provide a measure of the consistency of difference-calls across all trials. CKI scores of +0.55 were 2 SD or more above the mean score. To determine whether mRNAs with scores near +0.55 were altered in abundance, we analyzed by Northern blotting seven mRNAs with scores ranging from +0.44 to +0.59. Northern blotting showed these mRNAs to be increased in abundance, indicating that mRNAs with CKI scores in the range of +0.44 are candidates for natural targets of the UPF genes. Overall, UPF-dependent changes in mRNA accumulation were as high as 11.0-fold (YEL073C). Thirteen mRNAs exhibited a greater than fivefold average increase in abundance. The average increase among 529 mRNAs with CKI scores of ≥+0.44 was 2.4-fold.
Although most of the observed changes in the transcriptome were in the direction of increased accumulation when UPF genes were inactivated, a smaller number of mRNAs decreased in abundance. Forty mRNAs had CKI scores ≤2 SD below the mean CKI score. Six of these had CKI scores ranging from −0.53 to −1.0 with two- to sevenfold downward changes in abundance. The change for one of these, PHO84 mRNA, was confirmed by Northern blotting. Overall, we detected about 10 times more mRNAs that increased in abundance as decreased in abundance.
We tested the efficacy of HDOA analysis in predicting the magnitude of changes in mRNA abundance by measuring the abundance of 17 UPF-dependent mRNAs by Northern blotting. The mean fold changes measured by HDOA analysis were similar to those measured by Northern blotting. In general, when discrepancies were observed, larger fold changes were detected by Northern blotting. These results suggest that HDOA analysis is a reasonable but not a perfect predictor of the magnitudes of change.
We know of at least two UPF-dependent mRNAs identified in previous studies, CTF13 (9) and PPR1 (15, 24), that had unexpectedly low CKI scores that would generally not be indicative of a dependence on the UPF genes. For this reason, these two mRNAs were not included in the list of UPF-dependent mRNAs predicted by HDOA analysis despite three- to fourfold increases in abundance demonstrated by Northern blotting. These mRNAs may have escaped detection by HDOA analysis because their levels of abundance in wild-type strains are near the threshold of detection. When mRNAs are present at threshold levels, errors in predicting relative abundance are more likely to occur. Consequently, greater inconsistencies between trials can lower the index score and cause an erroneous call. In addition to these false-negative calls, false-positive calls are possible at some frequency, especially for mRNAs with CKI scores that reflect borderline consistency across trials (scores near +0.44). Assuming that false-negative and false-positive calls occur with similar frequencies, on balance our results indicate that well over 500 mRNAs change in abundance when the UPF genes are inactivated; 63% of the mRNAs exhibited greater than twofold increases in abundance. The largest change in abundance was 11-fold.
The best-described function for the Upf proteins is in their role in promoting the accelerated decay of nonsense mRNAs. These mRNAs are targeted for rapid decay by the presence of a premature stop codon caused either by a mutation or by an error in gene expression. However, the Upf proteins could also cause a reduction in the overall decay rate of any mRNA as part of the normal repertoire of gene expression for that mRNA. Although naturally occurring mRNAs do not typically contain a premature stop codon, they could be targeted for rapid decay by an alternate mechanism. For example, they might contain a stop codon at the end of a translatable upstream ORF or some other sequence element that serves a targeting function, or the normal stop codon at the end of the ORF might have the atypical property of triggering rapid decay. In any case, it seems likely that the Upf proteins cause changes in the abundance of naturally occurring mRNAs through a mechanism involving RNA decay.
If this is so, then the inactivation of a UPF gene should cause increased mRNA abundance of a selective group of targeted mRNAs. While most of the UPF-dependent mRNAs exhibited increased abundance, we observed some declines in abundance and confirmed one of these (for PHO84 mRNA) by Northern blotting. Changes in abundance in both directions could be explained if mRNAs coding for either positive or negative regulatory proteins served as direct targets for accelerated decay.
In support of this view, we found that the mRNA coding for the transcriptional activator Ppr1p was increased in upf− strains as were two mRNAs (URA1 and URA10) coding for enzymes in uracil biosynthesis that are transcriptionally activated by Ppr1p. A third mRNA, URA4, which has been reported to be regulated by PPR1, did not respond to loss of UPF gene function for unknown reasons. It was reported previously that the half-life of PPR1 mRNA increases threefold, commensurate with a threefold increase in PPR1 mRNA abundance (24). One mRNA (URA3) that is regulated by PPR1 was shown to increase in abundance due to an increased rate of transcription (15, 16). This example illustrates one way that the altered half-life of a single mRNA coding for a regulatory protein could indirectly influence the abundance of additional mRNAs.
Using similar logic, we reason that increased accumulation of a transcriptional repressor should cause a decrease in the accumulation of mRNAs regulated by a repressor. The five UPF-dependent mRNAs involved in phosphate utilization could involve negative regulation by one or more repressors given that one of the mRNAs (PHO80) was increased whereas four others (PHO84, PHO5, PHO86, and PHO8) all declined in abundance. PHO80 codes for a cyclin-dependent protein kinase that represses transcription of the PHO5 gene coding for secreted acid phosphatase by phosphorylating transcription factors encoded by PHO2 and PHO4 (2, 5, 6). Thus, the observed decline in PHO5 mRNA accumulation could be due to the increased accumulation of PHO80 mRNA. Further studies will be required to establish whether the PHO mRNAs change in abundance as a group through independent direct targeting of multiple mRNAs or indirect targeting of regulators that influence the abundance of the other mRNAs. To further support of the idea that mRNAs coding for regulatory proteins may serve as targets of UPF-mediated decay, we identified a host of additional UPF-dependent mRNAs coding for positively and negatively acting factors that influence transcription, most notably PDR3 (CKI = +0.47), RMS1 (CKI = +0.78), FZF1 (CKI = +0.63), KSS1 (CKI = +0.94), and HST1 (CKI = +0.41), and HST2 (CKI = +0.53).
We measured the half-lives of nine mRNAs selected among those that had a CKI score of ≥+0.44 and where the increased abundance was confirmed by Northern blotting. None of these mRNAs appeared to have an altered half-life (data not shown), which suggests that indirect targets may predominant over direct targets and that the Upf proteins may cause a change in the mRNA half-life of a small subset of the UPF-dependent mRNAs. Further studies are in progress to identify the direct targets of accelerated decay among naturally occurring mRNAs and to establish the mechanism for recruiting these mRNAs into the UPF-mediated pathway for rapid decay.
ACKNOWLEDGMENTS
We are indebted to members of the Affymetrix Academic User’s Center, notably Chris Harrington and Sumathi Venkatapathy, for valuable technical expertise. We thank Renee Shirley, Amanda Ford, and Judith Berman for critical reading of the manuscript and Jeff Dahlsied and Erin O’Shea for helpful discussions.
Microarray analysis was performed by M.J.L. at the Affymetrix Academic User’s Center, which is funded by NIH grant PO1 HG01323. The research was supported by the College of Agricultural and Life Sciences, University of Wisconsin, Madison, under NSF grant MCB-9870313 (M.R.C.). M.J.L. was supported by NRSA postdoctoral fellowship NIH GM19070. Additional funding was provided by the Research Committee of the University of Wisconsin Medical School.
Footnotes
Laboratory of Genetics paper 3529.
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jack H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima K, Oshima T, Kubota I, Nakamura N, Mizunaga T, Toh-e A. The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 1983;11:1657–1672. doi: 10.1093/nar/11.6.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 4.Badcock, K., C. M. Churcher, B. G. Barrell, M. A. Rajandream, and S. V. Walsh. 1997. Swiss-Prot accession no. Q09820, gene name SPAC16C9.06C.
- 5.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 7.Culbertson M R. RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 8.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlseid J N, J. P, Shirley R L, Atkin A L, Hieter P, Culbertson M R. Accumulation of mRNA coding for the Ctf13p kinetochore subunit of Saccharomyces cerevisiae depends on the same factors that promote rapid decay of nonsense mRNAs. Genetics. 1998;150:1019–1035. doi: 10.1093/genetics/150.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [PubMed] [Google Scholar]
- 11.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 12.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko Y, Toh-e A, Oshima Y. Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:127–137. doi: 10.1128/mcb.2.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 16.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–77. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lew J, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1998;18:6121–6130. doi: 10.1128/mcb.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 19.Long R M, Elliott D J, Stutz F, Rosbash M, Singer R H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 20.Losson R, Fuchs R P, Lacroute F. Yeast promoters URA1 and URA3. Examples of positive control. J Mol Biol. 1985;185:65–81. doi: 10.1016/0022-2836(85)90183-4. [DOI] [PubMed] [Google Scholar]
- 21.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 23a.Munich Information Centre for Protein Sequences. February 1999, revision date. [Online.] http://mips.biochem.mpg.de/. [July 1999. last date accessed.]
- 23b.NMD Database. October 1999, posting date. [Online.] http://144.92.19.47/default.htm. University of Wisconsin, Madison.
- 24.Peltz S, Jacobson A. mRNA turnover in Saccharomyces cerevisiae. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. pp. 291–328. [Google Scholar]
- 25.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Proteome, Inc. May 1999, revision date. [Online.] http://www.proteome.com/. [July 1999, last date accessed.]
- 26.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 27.Roy A, Exinger F, Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990;10:5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hoof A, Green P J. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- 30.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 31.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]