Abstract
Microscale thermophoresis is a relatively new technique used by an increasing number of academic laboratories to estimate relative binding affinities between ligand (analyte) that is titrated and a target (generally protein) that is either fluorescently labeled exogenously in the red or blue channel (labeled thermophoresis) or endogenously labeled via the presence of sufficient aromatic amino acid residues such as tryptophan (label-free thermophoresis). There are advantages and disadvantages to each technique; however, one major disadvantage of label-free thermophoresis is that protein-protein interactions cannot be measured, as generally most proteins have enough aromatic residues to generate an interfering signal. Thermophoresis can be used to determine steady-state binding affinities as between SNAREs and relevant binding partners of SNAREs and labeled thermophoresis is increasingly becoming a reliable technique to screen binding partners of fusion machinery to determine relevance in terms of direct biochemical interactions.
Keywords: Phosphatidic acid, Sec18, NSF, Nanodisc, SNARE
1. Introduction
Commercially available microscale thermophoresis (MST) is a recent advance in determining disassociation constants (KD) between biochemically relevant ligands, and the leading commercial retailer is NanoTemper [1]. NanoTemper retails two popular MST technologies, one requiring a fluorescent tag and another based on using inherent fluorescence of proteins due to presence of aromatic residues [2]. One major disadvantage of label-free thermophoresis is that it does not allow for the measurement of protein-protein interactions unless one of the proteins to be measured does not contain enough aromatic residues for measurable signal. Generally, thermophoresis does not have as strict of a buffer matching restriction as surface plasmon resonance; however, exact buffer matching is generally preferred [3].
MST generally measures the movement of particles in response to microscopic temperature fields and can be used to measure a wide variety of biochemicals [2]. MST allows measurement of proteins binding to small molecules and even small ions such as calcium. MST can be used to measure protein-protein interactions, membrane-protein to membrane-protein interactions including when such proteins are SNAREs, competitive inhibition, protein-lipid interactions where the lipid is in micelle, short-chain or relatively flat bilayer (e.g., nanodisc), and even kinetic (NanoTemper Application Note NT-MO-014) or thermodynamic parameters (NanoTemper Application Note NT-MO-004) in some instances.
Thermophoresis, however, is most often used for obtaining binding affinities or KDs in lieu of isothermal calorimetry (ITC) and surface plasmon resonance (SPR) because KDs can be procured using MST at a lower raw material cost and in less time generally than SPR or ITC. Thermophoresis is generally not used for obtaining thermodynamic parameters nor kinetic parameters, with ITC and SPR being the methods of choice for these measurements, respectively [1].
Scientific Background adapted from User Manual for the Monolith NT.115: Thermophoresis measurements depend on local temperature (T) difference ΔT of a solution generated from an infrared laser (IR). The solution for MST experiments is placed in a uniformly homogeneous capillary such that a fluorescence detector coupled to the IR beam reflected against a hot mirror can pick up changes in fluorescence due to changes in local concentration of the fluorescent molecule at the microscopic point where both the IR present head and the detector detect fluorescence. Changes in temperature ΔT correspond to depletion of molecules at the point of heat cHot and accumulation of molecules surrounding the beam of the IR laser cCOLD according to the formula ST = cHOT/cCOLD = e−St ΔT. Therefore, measured fluorescence generally decreases closer to the IR laser as T increases, and this effect is based on differences in charge, size, or solvation entropy due to a difference in chemical composition of the capillary as buffer is kept constant. The microscopic T gradient results in two measurable changes in fluorescence: (1) an intrinsic fluorescence difference because of temperature dependence of a fluorescent molecule, and (2) movement of molecules from hot to colder parts of the capillary.
An MST experiment therefore takes advantage of these properties to allow the user to titrate in two potential binding partners in a uniform buffer, one kept constant (the fluorescent molecule) and another titrated in to measure potential chemical differences between the two samples. If the two molecules are independent at a given concentration, one would expect similar thermophoresis patterns to ensue at those concentrations; however, if those molecules interact, they may move differently together and this can be measured by measuring the movement of the fluorescent molecule away from or toward the fluorescent detector, which is aimed at the spot where a microscopic temperature gradient exists due to the presence of IR laser in that area.
Analysis of MST can be challenging depending upon the nature of the chemical interaction you are measuring. This stems from the fact that though a given thermophoretic measurement may be robust, a researcher may not be able to exactly pinpoint the exact reason for the production of said signal, as it may stem from size, charge, solvation entropy, or conformational change of the ligand being measured. This is further exacerbated when using label-free thermophoresis due to the additional noise present in measuring fluorescence in ranges where inherent fluorescence of the protein itself is measured.
2. Materials
Phosphatidylcholine (POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine); phosphatidylethanolamine (POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine); phosphatidic acid (PA, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate).
Phosphate-buffered saline (PBS): 137 mM NaCl, 2.5 mM KCl, 10 mM NaH2PO4, and 2 mM KH2PO4, pH 7.4 (see Note 1).
Atto 647 dye: Stock Atto 647 Ni-NTA dye is diluted to 100 nM from a 100% DMSO solution into PBS without Tween (see Note 1).
PA nanodiscs: 5% PA nanodiscs are generated using MSP1D1 membrane scaffold protein with 5% POPA, 20% POPC, and 75% POPC as described [4, 5].
Sec18 Monomer: Sec18-GST can be expressed as a fusion protein in E. coli and purified using glutathione resin. The GST tag is cleaved using TEV protease and monomeric Sec18 was purified using size-exclusion chromatography.
His6-Sec18: His6-Sec18 can be expressed as a fusion protein in E. coli and purified using Ni-NTA and size-exclusion chromatography [6]. Hexamer and monomer pools are separated by size-exclusion chromatography.
3. Methods
3.1. Atto 647-Labeled Sec18-Binding PA Liposomes
Dilute His6-Sec18 to 200 nM in PBS.
Dilute Atto 647 dye to 100 nM in PBS.
Mix His6-Sec18 and Atto 647 dye in a 1:1 volumetric ratio (e.g., 100 μL 200 nM protein and 100 μL 100 nM dye) and incubate for 30 min at room temperature (see Note 2).
Centrifuge mixture of Atto 647 and protein for 10 min in a darkroom using a tabletop centrifuge at 11,000 × g.
Wrap tube in tin foil to prevent photobleaching and store on ice or at 4 °C.
Titrate 1:1 PA liposomes starting at 1 mM down to 30.5 nM.
Mix 5 μL of PA liposome at the highest concentration with 5 μL Atto 647-labeled His6-Sec18 and let stand at room temperature for 10 min.
Turn on NanoTemper Monolith NT.115 machine, press upward arrow to open machine, and slide out capillary rack.
Take solution containing the highest concentration PA liposome and load capillary tubes by letting solution flow up the capillary tube avoiding hand contact in the middle of the tube, taking note not to allow liquid at either end of the tube to be flush with capillary opening.
Place capillary in slot 1 on capillary rack.
Repeat steps 9 and 10 moving from high to low concentration PA liposome until capillary rack is filled.
Run NT.control software on computer attached to NanoTemper Monolith device.
Enter Fluo settings for this experiment. Before set to 5 s, MST on 30 s, and Fluo after 5 s (see Note 3).
Enter concentration, position, and name information in Table of Capillaries for each capillary tube. For example, enter the highest concentration of 500 for concentration for capillary tube 1 and PA liposome for name and drag down to auto-fill slots 2–16.
Run a capillary scan by hitting Start Cap Scan at 20% LED (preset) (see Note 4).
Select a range of MST power (see Note 5).
Start CapScan + MST Measurement (see Note 6).
Analyze using M.O. Affinity Analysis software provided by NanoTemper (see Note 7).
3.2. Atto 647-Labeled PA Nanodiscs Binding Sec18
Perform labeling procedure as in Subheading 3.1, steps 1–5 above, on 5% PA nanodiscs (see Note 8).
Titrate Sec18 monomer (cleaved GST) from 200 μM to 6 nM in PBS.
Mix 5 μL of Sec18 monomer at the highest concentration of 200 nM with 5 μL Atto 647-labeled PA nanodiscs and let stand at room temperature for 10 min (see Note 5).
Perform Monolith.115 startup procedure as in Subheading 3.1, steps 8–13.
Enter 100 for concentration of capillary 1 and Sec18 monomer for name and drag down concentration ensuring that capillary 16 reads 3 nM.
Perform CapScan and run CapScan + MST Measurement at a range of MST as in Subheading 3.1, steps 15–17.
Analyze using M.O. Affinity Analysis software provided by NanoTemper and analyze for changes in initial fluorescence with thermophoresis and T-Jump (see Notes 8 and 9).
3.3. Analysis
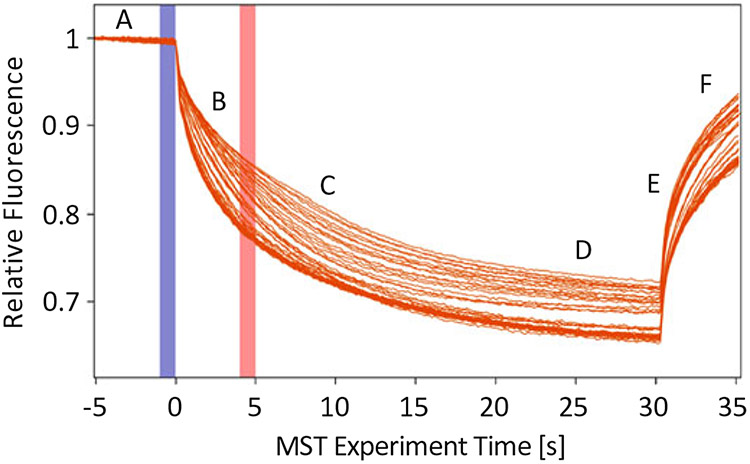
Use M.O. Affinity Analysis Software to determine binding constants. Figure 1 presents the thermophoretic output from three trials of Sec18 monomer titrated against PA nanodiscs as described in Subheading 3.2 using thermophoresis + T-Jump setting from M.O. Affinity Analysis V.2.1.5 software preset. The blue vertical rectangle represents the starting point of the relative fluorescence calculation (see Note 8) and the orange vertical rectangle represents the time point for the thermophoresis portion of the analysis. T-jump is calculated immediately after the IR laser is turned on. The output from this calculation can then be plotted using Fnorm or be set in the Compare Results screen to Percent Bound. Figure 2 shows the exported data for the three trials analyzed using thermophoresis + T-Jump for the titration of Sec18 monomer against PA nanodiscs, indicating a robust binding affinity curve.
Fig. 1.
Thermophoretic movement of Sec18 monomer bound to PA nanodiscs labeled with Atto 647 dye. This is a sample output using M.O. Affinity Analysis software V2.1.5 of Sec18 Monomer titrated against PA nanodiscs as described in Subheading 3.2 exporting chart from the Dose Response Fit Screen and defined according to [1]. (A) Initial fluorescence: Measurement taken at room temperature prior to turning on IR laser. (B) T-Jump: Fluorescence measurement differences present when IR laser is first turned on prior to thermophoretic movement of particles being measured. (C) Thermophoresis: Fluorescence change due to thermophoretic motion differences due to factors such as size, charge, solvation entropy, or conformational change of particle being measured with respect to titrated analyte. (D) Steady state: Equilibrium achieved between all capillaries containing different concentrations of analyte, generally occurring between 10 and 30 s. (E) Inverse T-Jump: Fluorescence measurement differences present when IR laser is first turned off after thermophoretic equilibrium or near equilibrium. (F) Back diffusion: Returning of relative fluorescence back to initial, however, may not reach the same value due to photobleaching (n = 3)
Fig. 2.
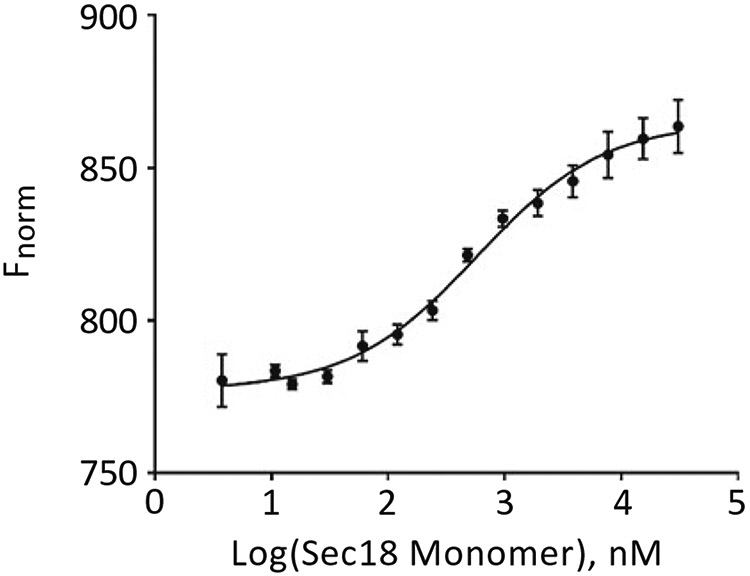
Normalized fluorescence of Sec18 monomer bound to PA nanodiscs evaluated using M.O. Affinity Analysis thermophoresis with T-Jump. Analysis of exported data relating to Fig. 1 from Compare Results screen of M.O. Affinity Analysis v2.1.5 using normalization F norm [%]. Analysis using Graphpad Prism v.7 using Sigmoidal, 4PL, X is log(concentration) with titrated Sec18 monomer concentration on the X-axis and normalized fluorescence on the Y-axis (n = 3)
Acknowledgments
This work was supported in part by NIH grant GM101132 to RAF.
Footnotes
Oftentimes PBS-T is used for labeling with the addition of 0.05% Tween-20; however, Tween-20 is generally avoided for lipid micelle or nanodisc protocols.
Generally, a label-free capillary tube can hold up to 10 μL of solution; however one can reasonably achieve accurate measurements down to 5 μL solution if solution centered in capillary with no bubbles. For 16 points in triplicate at 10 μL it would require 480 μL so one could at minimum mix 250 μL Atto 647 dye and 250 μL protein of interest for publishable results.
The detection settings for MST correspond to the initial fluorescence (before), the fluorescence detection once infrared laser is turned on (MST), and the fluorescence detection after infrared laser is turned off (MST off). Depending on the interaction you are measuring you may require longer or shorter periods of time to get accurate readings for these values.
For labeled thermophoresis it is optimal to have a fluorescence signal above 200 and below 2000 fluorescence units. It may be necessary to either increase or decrease LED to an optimal range; however, generally the Atto 647 dye yields robust signal at 20% LED, and if possible it is often better to use a lower LED for better results. It may be possible to use a lower volume of labeled ligand depending on the measured initial fluorescence of the ligand at a given LED. Generally, for labeled NanoTemper experiments, fluorescence should be above 200 for all capillary scans. A fluorescence above 200 can be achieved using lower labeled ligand concentrations by raising the LED or by using ligands that have multiple attachment sites for fluorophores. For instance, PA nanodiscs labeled with Atto 647 Ni-NTA generate a higher signal due to the presence of two 6X-His tags.
This can be done in the table of runs such that you can set up three runs at different MSTs, i.e., MST 30, 60, and 90 with the appropriate conditions from step 13. Depending on your given interaction, it may be that data is more or less robust at a given MST, so oftentimes it is necessary to run titrations at different MSTs at the onset of an experiment to determine which MST power will yield the best results.
It is important to have the CapScan with the MST measurement for reference when the final results are outputted. The run should take approximately 30 min. Depending upon the software package you may be able to view the points as they come in and begin to analyze data either through NT.Control or M.O. Affinity Analysis. If a point looks like an outlier, the researcher can check the CapScan to determine if it has a large initial fluorescence difference from the rest of the capillaries or an irregular shape, in order to decide whether there is a reason to throw out that point.
Analysis software provided by NanoTemper is proprietary and is generally performed using M.O. Affinity Analysis. Generally, newer versions of this software are preset to automatically evaluate data using thermophoresis with T-Jump using an algorithm to take advantage of the two measurement strategies. The Evaluation Strategy in M.O. Affinity Analysis can be set to manual to take advantage of the additional three settings of thermophoresis, T-Jump, and manual. Depending on a given chemical interaction the measurement may better fit traditional binding curves using any of the three additional settings; however, it is challenging to decipher when to do so. Additionally, switching between thermophoresis and T-Jump or measuring different reactions can lead to inverse curves, such that the maximum rel. fluorescence may be either at the lowest or at the highest concentration. When analyzing the data, it is possible in the Compare Results screen to switch the Y-axis to Percent Bound, allowing all interactions to have the same standard curve. Importantly, directionality of a curve is not requisite for robust data [1]; however, it can be difficult for a researcher to feel comfortable when experimental data differs depending on the nature of the chemical reaction to be measured.
Because PA nanodiscs have two 6X-His per particle, it is theoretically possible to use up to 200 nM Atto 647 dye to maximize fluorescent signal as needed.
Certain chemical reactions result in changes in initial fluorescence prior to the IR laser being turned on. In these cases robust fits to traditional binding curves can be made in M.O. Affinity Analysis software by creating a new analysis in the Data Selection screen and selecting Initial Fluorescence Analysis. This option presets analysis to be determined based on initial fluorescence; however, setting Evaluation Strategy to manual allows for additional options of measuring bleaching rate, slope of linear regression, and average of region.
References
- 1.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9:342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jerabek-Willemsen M, André T, Randy Wanner R, Roth HM, Duhr S, Baaske P et al. (2014) MicroScale thermophoresis: interaction analysis and beyond. J Mol Struct 1077:101–113 [Google Scholar]
- 3.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:100. [DOI] [PubMed] [Google Scholar]
- 4.Bayburt TH, Sligar SG (2003) Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci 12:2476–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc 126:3477–3487 [DOI] [PubMed] [Google Scholar]
- 6.Starr ML, Hurst LR, Fratti RA (2016) Phosphatidic acid sequesters Sec18p from cis-SNARE complexes to inhibit priming. Traffic 17:1091–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]