Abstract
In Saccharomyces cerevisiae, two transcription factors, SBF (SCB binding factor) and MBF (MCB binding factor), promote the induction of gene expression at the G1/S-phase transition of the mitotic cell cycle. Swi4 and Mbp1 are the DNA binding components of SBF and MBF, respectively. The Swi6 protein is a common subunit of both transcription factors and is presumed to play a regulatory role. SBF binding to its target sequences, the SCBs, is a highly regulated event and requires the association of Swi4 with Swi6 through their C-terminal domains. Swi4 binding to SCBs is restricted to the late M and G1 phases, when Swi6 is localized to the nucleus. We show that in contrast to Swi6, Swi4 remains nuclear throughout the cell cycle. This finding suggests that the DNA binding domain of Swi4 is inaccessible in the full-length protein when not complexed with Swi6. To explore this hypothesis, we expressed Swi4 and Swi6 in insect cells by using the baculovirus system. We determined that partially purified Swi4 cannot bind SCBs in the absence of Swi6. However, Swi4 derivatives carrying point mutations or alterations in the extreme C terminus were able to bind DNA or activate transcription in the absence of Swi6, and the C terminus of Swi4 inhibited Swi4 derivatives from binding DNA in trans. Full-length Swi4 was determined to be monomeric in solution, suggesting an intramolecular mechanism for auto-inhibition of binding to DNA by Swi4. We detected a direct in vitro interaction between a C-terminal fragment of Swi4 and the N-terminal 197 amino acids of Swi4, which contain the DNA binding domain. Together, our data suggest that intramolecular interactions involving the C-terminal region of Swi4 physically prevent the DNA binding domain from binding SCBs. The interaction of the carboxy-terminal region of Swi4 with Swi6 alleviates this inhibition, allowing Swi4 to bind DNA.
In budding yeast, commitment to enter the mitotic cell cycle occurs in the late G1 phase at a point called Start, which is analogous to the restriction point in mammalian cells (32, 35). In either case, cell cycle commitment is controlled by cyclin-dependent kinases (Cdks), whose activation requires association with positive regulatory subunits called cyclins. In Saccharomyces cerevisiae, passage through Start requires activation of the Cdk Cdc28 by association with the G1 cyclins Cln1, Cln2, and Cln3 (reviewed in reference 32). The Cdk Pho85, in association with the G1 cyclins Pcl1 and Pcl2, has also been implicated in regulating events at Start (15, 30). It is presumed that the association of Cdks with different cyclins allows the phosphorylation of substrates that are crucial for cell cycle entry. Due to the pivotal role of cyclins and Cdks in coordinating the cell cycle, cyclin-Cdk activation is highly regulated. One important mechanism of controlling Cdk activation at Start is the G1-periodic transcription of G1 cyclin genes.
Maximal expression of the G1 cyclin genes CLN1, CLN2, PCL1, and PCL2 at Start requires the activity of a transcription factor, SBF (SCB binding factor) (15, 30, 33, 34). SBF is a complex composed of at least two proteins, Swi4 and Swi6, which bind the repeated upstream regulatory sequence CACGAAA (SCB [Swi4/Swi6-dependent cell cycle box]) (2, 3, 34, 47). SBF is also required for the G1-specific expression of the HO gene and various cell wall biosynthetic genes (2, 9, 24). Biochemical studies have revealed that Swi4 is the component of SBF that specifically binds SCB sequences (4, 37). Swi4 contains an N-terminal DNA binding domain that is sufficient for the specific recognition of SCB sequences in vitro (37). The DNA binding region is highly homologous to that of Mbp1, and crystallographic studies of the Mbp1 DNA binding domain have revealed a helix-turn-helix structure (8, 48, 50). In contrast, Swi6 has no DNA binding activity but is present in the SBF complex because of its interaction with Swi4 via the carboxy-terminal regions (CTRs) of the two proteins (4, 27, 37, 42).
The timing of SBF-mediated gene expression is tightly controlled and requires multiple levels of regulation of SBF activity: the binding of SBF to SCBs in early G1, the activation of SBF at Start, and the dissociation of SBF from SCBs after the S phase. In vivo footprinting studies with both an SCB reporter plasmid and the CLN2 promoter as well as chromatin immunoprecipitation experiments show that SBF is bound to SCBs in the late M and G1 phases (11, 23, 28). Interestingly, the binding of SCBs by SBF is not coincident with SBF-mediated transcription; a secondary event must occur in order to activate SBF-dependent transcription. The activation of SBF is dependent on the activity of Cln3/Cdc28 kinase at Start (14, 46). However, the mechanism of Cln3-dependent activation of SBF remains a mystery, and a direct interaction of Cln3 with SBF has not been reported. In the G2 phase, the Clb/Cdc28 kinases become active and are required for the repression of SBF-dependent transcription (1, 28). The repression of SBF by the Clb kinases may involve the interaction of Clb2 with Swi4 and/or the phosphorylation of Swi4 (1). Clb2 interacts with the central ankyrin domain of Swi4 in vitro (44). It has been postulated that upon exit from mitosis, the rapid proteolysis of the B-type cyclins allows SBF to once again bind SCBs (28).
SBF activity is also regulated by changes in the subcellular localization of Swi6. Swi6 is largely cytoplasmic during the S, G2, and early M phases and is predominantly nuclear during the late M and G1 phases (43). The localization of Swi6 is dependent on the phosphorylation of serine-160, which is located next to a nuclear localization signal. Serine-160 is phosphorylated during the late G1, S, and M phases and may “hide” the nuclear localization signal, preventing the nuclear localization of Swi6. The relocalization of Swi6 to the nucleus is coincident with the in vivo footprinting of SCBs in the late M phase (23, 28).
Although, until this study, the subcellular localization of Swi4 was unknown, several lines of evidence suggested that additional control over SBF activity occurred through the regulation of Swi4 DNA binding. There is no evidence that full-length Swi4 can bind SCBs independently of Swi6. In swi6Δ strains, SCB-driven expression of CLN1 and CLN2 is severely reduced and the expression of HO is eliminated despite the fact that Swi4 protein is present and stable in swi6Δ mutants (2, 9, 33, 34). Further, in vivo footprinting studies have shown that in the absence of Swi6, protection of SCBs cannot be detected (23, 28). While endogenous levels of Swi4 in the absence of Swi6 cannot active SCB reporter genes, overexpression of C-terminal truncations of Swi4 in vivo can promote Swi6-independent transcription from SCB elements (4, 41). Ectopic expression of wild-type Swi4 also allows some activation of SBF-dependent gene expression, but this activation has been attributed to C-terminal degradation of Swi4 due to overexpression (42). Together, these observations suggest a model in which the DNA binding domain of Swi4 is inaccessible in the full-length protein when not complexed with Swi6. In this paper, we explore this model through a series of in vivo and in vitro experiments.
MATERIALS AND METHODS
Strains and plasmids.
Standard methods for yeast culturing and transformation were followed (21). Standard rich medium and supplemented minimal medium were used (26). Yeast strains are shown in Table 1.
TABLE 1.
Yeast strains used in the study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY263 | MATa trp1Δ63 GAL2+ ura3-52 lys2-801 ade2-107 his3Δ200 leu2-Δ1 | 30 |
| BY107a | MATα swi6Δ HIS3 | 34 |
| BY108a | MATα swi4Δ HIS3 | 34 |
| BY184a | MATα swi4Δ HIS3 SCB::lacZ | J. Ogas |
| BY185a | MATα swi6Δ HIS3 SCB::lacZ | J. Ogas |
| BY289a | MATa TRP+ | This study |
Isogenic relative to BY263 except as noted.
To construct a full-length clone of Swi6 with convenient restriction enzyme sites at the 5′ end, an NcoI site was introduced at the ATG of the SWI6 open reading frame by PCR amplification from a SWI6 template with the following primers: 5′CCGGCCATGGCGTTGGAAGAAGTGG3′ and 5′CCGTCTCATTGTCATCAGTGCC3′. The 630-bp PCR product was digested with NcoI and ApaI and cloned into NcoI-ApaI-digested pSL1180 (Pharmacia) to create plasmid pBA786. An ApaI-BglII fragment carrying the remainder of the SWI6 gene was then cloned into ApaI-BglII-digested pBA786 to reconstitute the entire gene (pBA788). The BglII-SalI genomic fragment containing SWI4 was cloned into the BamHI-SalI site of modified pUC18 in which the BglII polylinker had been incorporated at the EcoRI-HindIII site. The vector expressing a fusion of glutathione S-transferase (GST) to the C-terminal 144 amino acids of Swi4 was generated by use of PCR to amplify the 3′ end of Swi4. The primers used were 5′EcoRISwi4 (5′GTGCAGATCTTCGATATCAGAT′3) and 3′EndSwi4 (5′GACTGTCGACCATGGTTATGCGTTTGCCCTC′3). The PCR product was digested with EcoRI and SalI and cloned into the EcoRI-SalI sites of vector pGEX-4T-2 (Pharmacia) to create pBA1248. The integrity of all PCR products was confirmed by sequence analysis. To construct a vector for expression of Swi4 from the constitutive glycerol-3-phosphate dehydrogenase promoter, a BglII-SalI fragment containing the SWI4 gene was cloned from vector pBA476 into the BamHI-SalI sites of vector p424 GPD (ATCC 87357) to create pBA1262. Plasmids used for in vitro transcription-translation of Swi4 and Swi6 have been previously described (pBA462 for full-length Swi4, pBA513 for Swi6, and pBA586, a derivative of Swi4 which has an internal deletion of amino acids 198 to 745) (4).
Immunofluorescence.
For indirect immunofluorescence with Swi4 antiserum, a Swi4 polyclonal antibody was affinity purified and preadsorbed to a 1:1 mixture of fixed yeast cells and spheroplasts of the yeast strain BY184 essentially as described elsewhere (17, 34, 38, 51). Strain BY184 (200 ml) was grown to the log phase in standard rich medium at 30°C, harvested, and fixed by the addition of formaldehyde to 3.7% for 1 h at 30°C with shaking. The fixed cells were washed twice with 1.2 M sorbitol–50 mM K2POH4 (pH 7.5) and resuspended in 4 ml of 1.2 M sorbitol–50 mM K2POH4 (pH 7.5). Spheroplasts were made from 2 ml of fixed cells by the addition of β-mercaptoethanol to 0.1% and Zymolyase 20000T to 0.25 mg/ml, followed by incubation for 1 h at 30°C. The spheroplasts were washed twice with 1.2 M sorbitol–50 mM K2POH4 (pH 7.5) and added to the remaining 2 ml of whole fixed yeast cells. The cell mixture was washed and resuspended in 4 ml of phosphate-buffered saline (PBS). The cell mixture (200 μl) was incubated with 200 μl of affinity-purified Swi4 antibody at 4°C for 1 h with shaking. Cells were pelleted, and the antibody supernatant was transferred to another tube containing 200 μl of fresh cell mixture. Antibody-cell incubation was repeated seven times, including an overnight incubation, resulting in a fivefold dilution of the pretreated affinity-purified Swi4 antibody.
Wild-type cells (BY263) and swi4Δ cells (BY184) were grown to the early log phase and fixed in 3.6% formaldehyde at 30°C for 2 h. Cells were washed twice with 100 mM KH2PO4 (pH 7.4), resuspended in 100 mM KH2PO4 (pH 7.4)–0.1% β-mercaptoethanol–0.25 mg of Zymolyase 20000T per ml, and incubated at 30°C for 30 min to digest the cell wall. The cells were washed twice in PBS containing 1.2 M sorbitol. The cells were incubated with PBS plus 2% bovine serum albumin (BSA) (Sigma) for 30 min at room temperature. The cells were pelleted, washed once in PBS–0.2% BSA, and incubated for 2 days at 4°C in a 1/20 dilution of affinity-purified and preadsorbed anti-Swi4 antibody in PBS–0.2% BSA. The cells were washed three times for 10 min each time with PBS–0.2% BSA. A 1/20 dilution of preadsorbed sheep anti-rabbit CY3-conjugated secondary antibody (a gift from M. Synder) in PBS–0.2% BSA was added and incubated for 2 h at room temperature. The cells were washed again as outlined above, followed by a final wash with PBS containing 0.05 μg of diamidophenylindole (DAPI) per ml. One drop of the cell suspension was deposited on a polylysine-coated slide (Flow Laboratories). Once the cells had settled, the adherent cells were rinsed with PBS–0.1% BSA and mounted in 90% glycerol in PBS containing 0.1% p-phenylamine diamine. Cells were observed at a magnification of ×630 by use of a Leica DM-LB microscope with Nomarski optics and a Princeton charge-coupled device camera. Swi4 staining was visualized with rhodamine fluorescence optics. A total of 800 wild-type cells were scored for nuclear staining and position in the cell cycle as judged by bud size.
Construction of baculovirus vectors.
We used the Bac-N-Blue baculovirus system (Invitrogen) to express both Swi6 and Swi4 derivatives in insect cells. To construct a baculovirus expressing Swi6, the NcoI-HindIII fragment from pBA788 containing full-length SWI6 was cloned into the NcoI-HindIII sites of the baculovirus transfer vector pBlueBacIII (Invitrogen). To construct a Swi4 expression vector, a BglII fragment from pBA476 containing full-length SWI4 was cloned into the BamHI site of pBlueBacIII. The SWI4ΔAnkyrin motif (Swi4ΔAA) transfer vector was constructed by digestion of pBA476 with NsiI followed by religation to create a Swi4 derivative with a 1,047-bp internal deletion. The deleted SWI4 fragment was then cloned into the BamHI site of the baculovirus transfer vector pBACHISA (Invitrogen). The Swi4Δ144 transfer vector was constructed by cloning the BglII-EcoRI fragment from pBA476 into the BamHI-EcoRI site of baculovirus transfer vector pBlueBac4.5 (Invitrogen). The transfer vectors were cotransfected with baculovirus genomic DNA into Sf9 insect cells, and recombinant baculoviruses were isolated and plaque purified as outlined in the Invitrogen Bac-N-Blue transfection kit manual.
Expression and purification of Swi6 from insect cells.
Monolayers of High Five cells (Invitrogen) (107 cells in 150-cm2 flasks) were infected at a multiplicity of infection (MOI) of 5 PFU per cell. At 45 h after infection, the cells were harvested by centrifugation at 1,000 rpm for 5 min in a Sorvall GLC-1 centrifuge, washed with cold water, and resuspended in cold lysis buffer 1 (50 mM Tris-HCl [pH 7.4], 0.5 mM EDTA, 50 mM NaCl, 1 mM phenylmethyl sulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml) at 1 ml/flask. Lysis was done by drawing the resuspended cells eight times through a 27.5-gauge needle, and the lysates were clarified by centrifugation at 13,000 × g for 20 min. The lysates typically contained 10 mg of total protein/ml. Swi6 was purified from the insect lysates as described for Swi6 expressed in bacteria (42). Proteins were precipitated with 20 to 35% ammonium sulfate, and the precipitate was resuspended and dialyzed overnight in lysis buffer 1, further purified over a 1-ml DEAE-Sepharose column (Pharmacia), and eluted with a salt gradient. Peak fractions were dialyzed overnight in lysis buffer 1 with the addition of 20% glycerol and stored at −80°C. Swi6 protein was estimated to be at least 80% pure, as judged by Coomassie blue staining of fractions resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Expression and partial purification of SBF and Swi4 derivatives from insect cells.
For the production of SBF (Swi6 and Swi4) in insect cells, monolayers of High Five cells (five 150-cm2 flasks of 107 cells each) were infected with both full-length Swi4- and Swi6-containing baculoviruses at an MOI of 5 PFU of each virus per cell. At 45 h after infection, the cells were harvested and lysed as outlined above for Swi6-infected cells. SBF lysates typically contained 10 mg of total protein/ml. The lysates were loaded on a 5-ml HiTrap heparin column (Pharmacia) preequilibrated with buffer A (50 mM Tris-HCl [pH 7.4], 0.5 mM EDTA, 100 mM NaCl). The column was washed with 15 ml of buffer A, and the bound proteins were eluted with a 40-ml linear salt gradient (0.1 to 1.0 M NaCl) and collected in 2-ml fractions. SBF peak elution occurred in fractions 12 and 13, which contained 450 to 550 mM salt, as determined by conductivity. The peak fractions were pooled, glycerol was added to 20%, and the fractions were stored at −80°C. Peak fractions typically contained 0.5 mg of protein/ml.
For the production of full-length Swi4, Swi4Δ144, or Swi4ΔAA in insect cells, monolayers of High Five cells (five 150-cm2 flasks of 107 cells each) were infected with the baculovirus at an MOI of 5 PFU per cell. The purification procedure for Swi4, Swi4ΔAA, or Swi4Δ144 was identical to that described above for SBF, with peak elution in fraction 12. However, the peak for Swi4ΔAA was broader. The production of Swi4 and the Swi4 derivatives in insect cells was significantly reduced in the absence of coexpression with Swi6. Using Western blot and gel shift assays, we estimated that this procedure caused an eightfold enrichment of Swi4 and significantly decreased the levels of insect cell proteins that bound nonspecifically to SCB-containing probes (see Results).
Gel retardation assay.
Yeast extracts were prepared and gel shift assays were performed with an SCB-containing probe as previously described (2). Binding reactions were performed with 20 μl of assay buffer (25 mM Tris-HCl [pH 7.4], 10% glycerol, 3 mM MgCl2, 0.2 mM EDTA). Poly(dI · dC)-poly(dI · dC) (Pharmacia) was added at 5 μg/reaction mixture for crude yeast and crude insect cell extracts and at 1 μg/reaction mixture for partially purified protein samples. Unlabeled wild-type and mutant SCB competitors were prepared from annealed oligonucleotides as described previously and added to the binding reaction mixtures as indicated elsewhere (4). For competition experiments with the Swi4 C terminus, a recombinant protein composed of the last 144 amino acids of Swi4 fused to GST (GST-4CTR) was purified from Escherichia coli harboring the appropriate expression plasmid as previously described (31). The GST-4CTR protein was incubated with thrombin while still bound to glutathione–S-Sepharose 4B beads (Pharmacia). The C-terminal fragment of Swi4 was eluted from bound GST and dialyzed in buffer A. The final concentration of the C-terminal fragment of Swi4 (the CTR) was 0.2 μg/μl. Gel retardation assays with this fragment were performed as described above, except that the reaction mixtures were incubated at 4°C for 20 min prior to the addition of the probe and for 10 minutes at room temperature after the addition of the probe.
Screen for Swi4 CTR mutants.
To generate random mutations in the Swi4 CTR, the last 432 nucleotides of SWI4 were amplified by PCR with Taq DNA polymerase, the primers 5′EcoRI (5′GTGCAGATCTTCGATATCAGAT3′) and 3′MutSalI (5′CCTAGACTTCAGGTTGTCTT3′), and the SWI4 gene as a template. The PCR product was digested with EcoRI and SalI and cloned into vector pBA1262 that had been digested with EcoRI and SalI. The resulting pool of mutagenized SWI4 plasmids was used to transform BY185 (swi6Δ SCB::lacZ) (20). The colonies were transferred to nitrocellulose filters and assayed for β-galactosidase activity as described previously (7). Transformants that turned blue before the BY185 transformant containing the vector alone were selected. Mutant plasmids were isolated from the yeast strains, passaged through E. coli, and used to retransform BY185 to confirm that the increase in β-galactosidase activity was due to the plasmid-borne SWI4 gene (40). Once increased β-galactosidase activity was confirmed, the mutated SWI4 genes were sequenced. To determine whether the Swi4 proteins encoded by the mutated SWI4 genes could still interact with Swi6 in vivo, the mutated SWI4 genes were used to transform BY184 (swi4Δ SCB::lacZ), yeast extracts were prepared, and gel shift assays were performed with an SCB-containing probe as described above.
Glycerol gradients.
Sedimentation in glycerol gradients was performed essentially as described previously (12). Glycerol gradients (4 ml of 40 to 10% [vol/vol] glycerol in buffer A) were poured in seven steps of 500 μl and allowed to equilibrate for 1 h at room temperature, followed by 1 h at 4°C. Partially purified Swi4, Swi4Δ144, or SBF preparations (50 μg) were layered on top of the gradient along with 100 μg of an internal control protein (catalase, from the Pharmacia gel filtration high-molecular-weight calibration kit). A control gradient with 100 μl of protein markers containing 100 μg each of catalase, aldolase, and albumin (Pharmacia) was run in parallel. The gradients were centrifuged in a SW60.1 rotor at 55,000 rpm for 13 h. Two-drop fractions (90 to 100 μl) were collected from the bottom of the tube with a syringe needle. The Bio-Rad protein assay was used to detect molecular weight standards and the internal control protein catalase. To assay fractions containing Swi4 or Swi4 derivatives, 50 μl of each fraction was analyzed by immunoblotting with anti-Swi4 antibodies and visualized by chemiluminescence.
In vitro transcription and translation of Swi4 and Swi6.
To produce full-length Swi4, full-length Swi6, and Swi4 with an internal deletion, the plasmid templates pBA462, pBA513, and pBA548, respectively, were used as recommended in the T7 “TnT” coupled reticulocyte lysate system (Promega). To produce Swi4Δ421 and Swi4Δ896, pBA462 was linearized with XbaI and NsiI, respectively, used as a template in the TnT system.
Batch affinity chromatography.
GST and GST-4CTR were purified from E. coli harboring the appropriate expression plasmids as previously described (31). For affinity chromatography with insect cell-derived Swi4 and Swi6, GST and GST-4CTR were bound to glutathione S-Sepharose 4B beads at a concentration of 1 μg/μl of beads. Either GST or GST-4CTR beads (10 μl) were incubated with 10 μg of partially purified Swi4, Swi4Δ144, or Swi6 for 45 min at 4°C. The beads were harvested, and the unbound supernatant was collected. The beads were washed in 1 ml of lysis buffer 2 (50 mM Tris-HCl [pH 7.4], 0.5 mM EDTA, 50 mM NaCl) four times for 5 min each time followed by four washes for 5 min each time in RIPA-500 buffer (50 mM Tris-HCl [pH 7.5], 0.1% SDS, 0.5% deoxycholate, 500 mM NaCl, 1% Triton X-100). After the final wash, the beads were resuspended in 25 μl of 1× SDS-polyacrylamide gel loading dye and boiled. The bound and one-half of the unbound supernatant fractions were separated by SDS–6% PAGE. The proteins were transferred to nitrocellulose, and the Swi4 derivatives were detected by Western blotting as outlined above. For affinity chromatography with in vitro-translated and -transcribed Swi4 and Swi6, 15 μl of either GST or GST-4CTR beads was incubated with 30 μl of PBS and 7 μl of either in vitro-translated Swi4, Swi6, Swi4Δ421, Swi4ΔAnks, or Swi4Δ896 for 2 h at 4°C. The beads were harvested, and the unbound supernatant was collected. The beads incubated with Swi4, Swi6, Swi4Δ421, and Swi4ΔAnks were washed three times for 2 min each time in 100 μl of RIPA-500 buffer followed by a wash in PBS. The beads incubated with Swi4Δ896 were washed three times for 2 min each time in 100 μl of buffer A followed by a wash in PBS. After the final wash, the beads were resuspended in 30 μl of 1× SDS-polyacrylamide gel loading dye and boiled. The bound and one-half of the unbound supernatant fractions were separated by SDS–10% PAGE. The gels were fixed, treated with Amplify (Amersham), dried, and exposed to X-ray film.
RESULTS
Nuclear localization of Swi4 throughout the cell cycle.
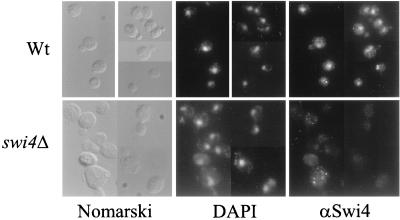
In vivo footprinting analysis and chromatin immunoprecipitation assays showed that SCBs are bound by SBF throughout the G1 phase, indicating that Swi4 and Swi6 must be nuclear at least in G1 (11, 23, 28). Indeed, Swi6 localization studies have confirmed that the majority of Swi6 is nuclear throughout the late M and G1 phases but is largely cytoplasmic during the rest of the cell cycle (43). The nuclear localization of Swi6 is coincident with the binding of SBF to SCBs, implying that Swi4 must be present in the nucleus at the time of Swi6 localization. To investigate the subcellular localization of Swi4 throughout the cell cycle, we developed an indirect immunofluorescence assay with Swi4 antibodies on wild-type and swi4Δ log-phase cells. No distinct staining was seen when swi4Δ cells were stained with Swi4 antiserum (Fig. 1). To assay wild-type cells for Swi4 staining, 800 wild-type cells were scored for nuclear staining (with DAPI) and position in the cell cycle by assessing bud morphology. We found that 60% of all cells scored had a distinct Swi4 staining signal, suggesting that our protocol or antibodies were not optimized to achieve 100% staining. However, for the 60% of cells that were stained, nuclear staining was seen in cells at all stages of the cell cycle, in both unbudded and budded forms. We conclude that, unlike Swi6, whose localization changes throughout the cell cycle, Swi4 remains nuclear throughout the cell cycle.
FIG. 1.
Subcellular localization of Swi4. Swi4 localization was assayed by indirect immunofluorescence with Swi4 antiserum and a fluorescein isothiocyanate-conjugated secondary antibody. Wild-type (Wt) and swi4Δ cells were photographed at a magnification of ×630 with an imaging system (see Materials and Methods). Photographs of the same fields of cells viewed with Nomarski optics and stained with DAPI to visualize cell nuclei are also shown.
The Swi4-Swi6 complex from insect cells can bind SCBs in vitro.
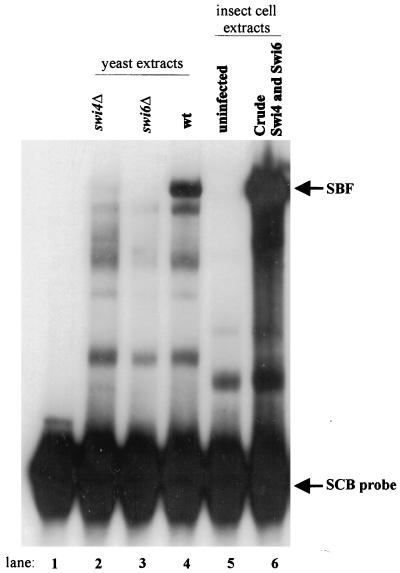
Since Swi4 is nuclear in the S, G2, and M phases but fails to bind SCBs, we next sought to investigate the mechanism regulating Swi4 binding to DNA. To generate reagents useful for our studies, we constructed vectors for expressing Swi4 and Swi6 in insect cells. While Swi6 can be purified from E. coli, attempts at expressing Swi4 in bacteria have met with limited success (42, 47). To determine whether Swi4 and Swi6 produced in insect cells formed a functional SBF complex, we performed gel retardation analysis by using an SCB-containing probe. Incubation of the probe with crude yeast extracts from wild-type, swi4Δ, and swi6Δ cells showed the formation of SBF in the wild-type extracts (Fig. 2, lanes 2 to 4), as previously described (2, 4, 33, 34, 42, 47). Incubation of the probe with crude insect cell lysates from cells coinfected with baculovirus vectors expressing Swi4 and Swi6 led to the formation of a major complex that comigrated with SBF from yeast extracts (Fig. 2, lane 6). This complex was not seen when extracts from uninfected insect cells were used in the assay (Fig. 2, lane 5). Since the SBF complex formed from insect cells and yeast extracts migrated at the same position, SBF is likely composed of only Swi4 and Swi6 proteins. We cannot exclude the possibility that other proteins are present in this complex; however, the proteins would need to be present in both yeast and insect cells. We conclude that SBF can be functionally reconstituted by the expression of Swi4 and Swi6 in insect cells.
FIG. 2.
Reconstitution of SBF in insect cells. A gel retardation assay with an SCB-containing probe and either crude yeast or insect cell lysates is shown. The labeled probe contained three SCB sequences from the upstream region of the HO gene (see reference 2). The following extracts were used in the binding assays: lane 1, probe alone; lanes 2 to 4, 10 μg of crude yeast extract from swi4Δ, swi6Δ, and wild-type strains, respectively; lane 5, 10 μg of crude cell lysate from uninfected insect cells; and lane 6, 10 μg of crude cell lysate from insect cells coinfected with Swi4- and Swi6-expressing baculovirus vectors. The migration positions of the SBF complex and the unbound probe are indicated to the right.
Inhibition of Swi4 binding to DNA in the absence of Swi6.
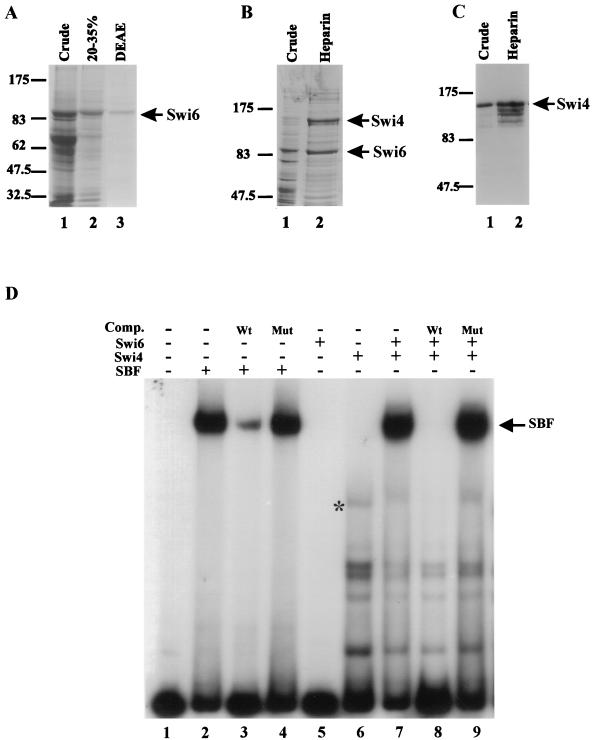
Although it is clear that Swi4 binds DNA in the context of SBF, there is no evidence that Swi4 can bind DNA on its own. Two observations led us to hypothesize that the DNA binding domain of Swi4 is inaccessible in the full-length protein when not complexed with Swi6. First, SCB-driven gene expression is reduced or eliminated in a swi6Δ strain even though Swi4 is present and nuclear throughout the cell cycle (this study, 2, 9, 33, 34). Second, in vivo footprinting studies have shown that in the absence of Swi6, the binding of Swi4 to SCB sequences cannot be detected (23, 28). In order to rigorously test the hypothesis that the DNA binding domain of Swi4 is inaccessible in the full-length protein, we partially purified both Swi4 and Swi6 from insect cells. We adapted previously established procedures to purify Swi6 produced in insect cells (Fig. 3A, lane 3) (42, 47). We used a heparin column to greatly enrich both Swi4 and SBF expressed in insect cells (Fig. 3B and C). Coexpression of Swi4 with Swi6-containing baculoviruses significantly increased the production of Swi4, suggesting that a Swi4-Swi6 interaction may stabilize Swi4 in insect cells. In contrast, Swi4 was poorly produced when expressed in the absence of Swi6 in insect cells. However, by using heparin-agarose chromatography, we were able to obtain an eightfold purification of Swi4, as assessed by densitometry of Swi4 Western blots and by measurement of the specific activity of Swi4 in a gel shift assay (Fig. 3C and data not shown). Further, the purification procedure reduced the levels of nonspecific DNA binding proteins in the insect cell extracts. Problems with insolubility were encountered when the Swi4 heparin fractions were subjected to additional purification steps.
FIG. 3.
SCB binding activity of partially purified Swi4, Swi6, and SBF. (A) Purification of Swi6 protein expressed in insect cells. Swi6-containing fractions obtained during purification were analyzed by SDS–6% PAGE followed by Coomassie blue staining. Lane 1, crude lysate from insect cells expressing Swi6 protein; lane 2, 20 to 35% ammonium sulfate precipitation; lane 3, DEAE-Sepharose fraction. A 10-μl aliquot of each fraction was loaded per lane. (B) Purification of SBF expressed in insect cells. SBF-containing fractions obtained during purification were analyzed as described in panel A for Swi6. Lane 1, crude lysate from insect cells infected with both SWI4- and SWI6-expressing baculoviruses; lane 2, heparin-agarose fraction. A 10-μl aliquot each of the crude and partially purified fractions was loaded. (C) Enrichment of Swi4 expressed in insect cells. Swi4-containing fractions obtained during purification were separated by SDS–6% PAGE and analyzed by Western blotting with affinity-purified Swi4 antiserum. Lane 1, crude lysate from insect cells infected with a SWI4-expressing baculovirus vector (10 μg); lane 2, heparin-agarose fraction (10 μg). For panels A through C, the migration positions of molecular weight markers are indicated to the left (in thousands). (D) Gel retardation assay with partially purified SBF, Swi4, and Swi6. A labeled SCB-containing probe (see the legend to Fig. 2) was incubated with the following protein preparations: lane 1, no extract; lanes 2 to 4, SBF heparin-agarose fraction (1 μg); lane 5, Swi6 DEAE-Sepharose fraction (3 μg); lane 6, Swi4 heparin-agarose fraction (5 μg); and lanes 7 to 9, both partially purified Swi4 and Swi6 fractions. Where indicated above the lanes, a 100-fold molar excess of either wild-type SCB competitor (Comp.) DNA (Wt) or mutated SCB competitor DNA (Mut) was added. The migration position of SBF is shown to the right. The asterisk in lane 6 marks the migration position of a complex composed of the SCB-containing probe and either full-length Swi4 or a small C-terminal truncation of Swi4.
We used our purified Swi6 and Swi4 preparations in gel retardation assays to assess the binding of full-length Swi4 to SCBs in the absence of Swi6. First, we confirmed that our partially purified SBF fractions supported SBF complex formation in vitro (Fig. 3D, lane 2). Binding was specific for SCB DNA, since the SBF complex was inhibited by a wild-type SCB oligonucleotide competitor and not by a mutant SCB oligonucleotide competitor, confirming that the purification procedure did not alter the SCB binding ability of SBF produced in insect cells (Fig. 3D, lanes 3 and 4). Incubation of partially purified Swi4 with purified Swi6 allowed for efficient reconstitution of the SBF complex in vitro (Fig. 3D, lane 7). Incubation of the C-terminal half of Swi6 fused to GST with partially purified Swi4 also produced an SBF complex (data not shown). The reconstituted SBF complex was inhibited by wild-type SCB competitor and not by a mutant SCB competitor (Fig. 3D, lanes 8 and 9). In contrast, incubation of full-length Swi4 with the SCB-containing probe led to the formation of a series of faster-migrating complexes (Fig. 3D). These complexes likely represent SCB binding by C-terminal truncations of Swi4, since they were inhibited by wild-type SCB DNA and were unaffected by the addition of Swi6. One minor complex, indicated by an asterisk in Fig. 3D, may reflect binding of the SCB-containing probe by full-length Swi4. We used phosphorimager analysis to compare the amount of this minor complex to the amount of the SBF complex formed upon the addition of excess Swi6. Our analysis suggested that less than 5% of the Swi4 protein in the assay participated in the minor complex (data not shown). Therefore, if the minor product indeed contains full-length Swi4, the ability of intact Swi4 to bind SCBs in the absence of Swi6 must be severely compromised. Since we were using partially purified components derived from a heterologous system, our results suggest that full-length Swi4 cannot bind DNA efficiently in the absence of Swi6. No experiments with recombinant full-length Swi4 have been previously reported. The mechanism of inhibition appears intrinsic to Swi4 and is alleviated upon the interaction of Swi6 with Swi4 through the Swi4 CTR.
Domains required for auto-inhibition of Swi4 binding to DNA.
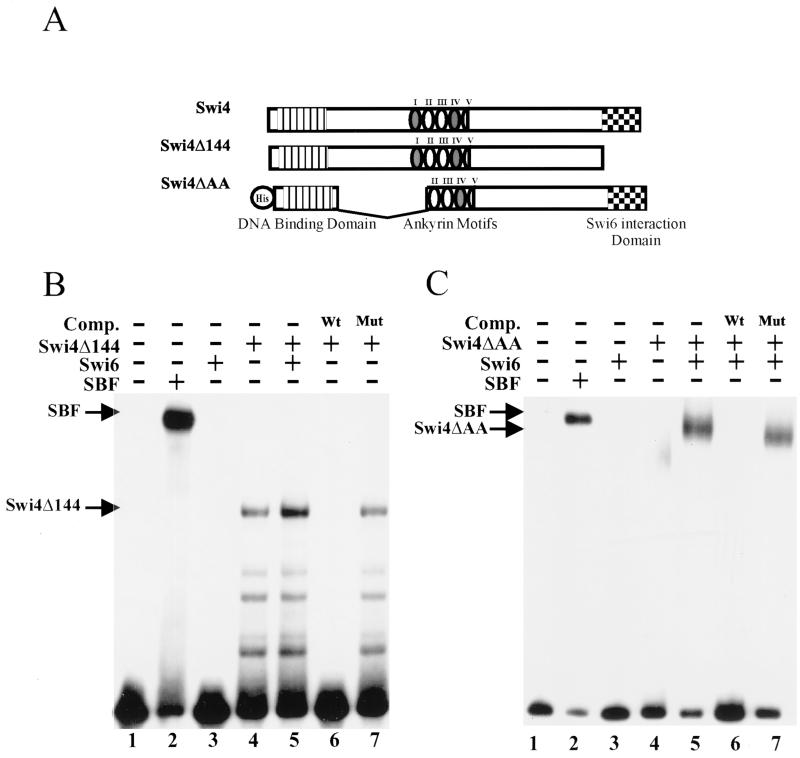
As outlined earlier, several lines of evidence suggested that the C terminus of Swi4 is necessary for the regulation of Swi4 binding to DNA. To confirm this hypothesis, we expressed a truncated version of Swi4, Swi4Δ144, lacking the C-terminal 144 amino acids, in insect cells. We had previously used gel retardation assays to show that Swi4Δ144 in crude yeast extracts formed a Swi6-independent complex with SCB DNA (4). A similar result was obtained with an in vitro-translated truncation of Swi4 (37). We used Swi4Δ144 that had been partially purified from insect cells in a gel shift assay with an SCB-containing probe. Swi4Δ144 formed a specific complex with DNA that was efficiently competed by wild-type but not by mutated SCB-containing DNA (Fig. 4B, lanes 4, 6, and 7). Since Swi6 interacts with Swi4 through the last 78 amino acids of Swi4, the addition of Swi6 did not affect Swi4Δ144 DNA binding (Fig. 4B, lane 6) (44). A larger Swi4 truncation, Swi4Δ421, behaved in a similar manner (data not shown). Our results confirm the hypothesis that the C-terminal 144 amino acids of Swi4 are involved in the inhibition of Swi4 binding to DNA.
FIG. 4.
Analysis of SCB binding by deletion derivatives of Swi4. (A) Schematic of the Swi4 derivatives expressed in insect cells. The relative positions of the N-terminal DNA binding domain, the multiple ankyrin repeats, and the C-terminal Swi6 interaction domain are indicated. His indicates the presence of an N-terminal histidine tag. (B) Gel retardation assay with partially purified SBF or C-terminally truncated Swi4 (Swi4Δ144). A labeled SCB-containing probe (see the legend to Fig. 2) was incubated with the following protein preparations: lane 1, no extract; lane 2, SBF heparin-agarose fraction (1 μg); lane 3, 3 μg of purified Swi6; and lanes 4 to 7, 5 μg of partially purified Swi4Δ144. Lane 5 also contains 3 μg of a Swi6 DEAE-Sepharose fraction. In lanes 6 and 7, a 100-fold molar excess of either wild-type SCB competitor (Comp.) DNA (Wt) or mutated SCB competitor DNA (Mut) was added. (C) Gel retardation assay with partially purified SBF or a Swi4 internal deletion derivative (Swi4ΔAA). The labeled SCB-containing probe was incubated with the following protein preparations: lane 1, no extract; lane 2, SBF heparin-agarose fraction (1 μg); lane 3, 3 μg of purified Swi6; lane 4, 5 μg of partially purified Swi4ΔAA; and lanes 5 to 7, 5 μg of partially purified Swi4ΔAA and 3 μg of purified Swi6. In lanes 6 and 7, a 100-fold molar excess of either wild-type SCB competitor DNA (Wt) or mutated SCB competitor DNA (Mut) was added.
Ankyrin motifs have been implicated in the auto-inhibition of numerous transcription factors, including NF-κB (reviewed in reference 18). We next used convenient restriction sites in the Swi4 gene to construct a baculoviral Swi4 derivative, Swi4ΔAA, with an internal deletion of 349 amino acids. This deletion disrupts the first ankyrin domain of Swi4 along with a significant region between the DNA binding domain and the ankyrin domain. The first and fourth ankyrin domains of Swi4 were first identified due to their similarity to other ankyrin domains. Upon closer inspection, it became apparent that Swi4 has three other degenerate ankyrin repeats (6). Single amino acid changes in the ankyrin repeats of both Swi6 and Swi4 result in proteins that are temperature sensitive for function (14a, 16, 42). This finding suggests that deletion of the first ankyrin repeat in Swi4ΔAA should greatly reduce, if not abolish, the function of the ankyrin domains. Unlike Swi4Δ144, Swi4ΔAA had only a limited ability to bind DNA in the absence of Swi6 (Fig. 4C, lane 4). Full binding was restored upon the addition of Swi6 (Fig. 4C, lanes 5 to 7). We conclude that the inhibition of Swi4 binding to DNA does not involve amino acids 199 to 547 of Swi4.
Point mutations in the Swi4 CTR allow Swi6-independent activation of SBF-dependent transcription.
Our in vitro experiments showed that the inhibition of Swi4 binding to DNA was relieved by deletion of the C-terminal 144 amino acids of Swi4. The C-terminal 78 amino acids of Swi4 are required for interaction with Swi6 (44). This region of Swi4 is conserved in other members of the Swi4 family of transcription factors and is predicted to be highly α-helical in structure (Fig. 5A). To determine whether the region of the Swi4 C terminus involved in the inhibition of Swi4 DNA binding was separable from the Swi6 interaction domain, we undertook a screen for point mutations in the CTR-encoding portion of SWI4 that alleviate the DNA binding inhibition of Swi4. We used PCR-mediated mutagenesis to introduce random mutations into the region encoding the C-terminal 144 amino acids of Swi4. The mutagenized SWI4 fragments were cloned into a SWI4 gene on a 2μm plasmid to allow expression from the constitutive GPD promoter. We transformed the pool of CTR mutants into a swi6Δ strain carrying an integrated SCB::lacZ reporter gene. Using a β-galactosidase filter test, we identified Swi4 mutants which, in the absence of Swi6, allowed a higher level of SCB::lacZ expression than that seen with the wild-type SWI4 gene expressed from the same vector.
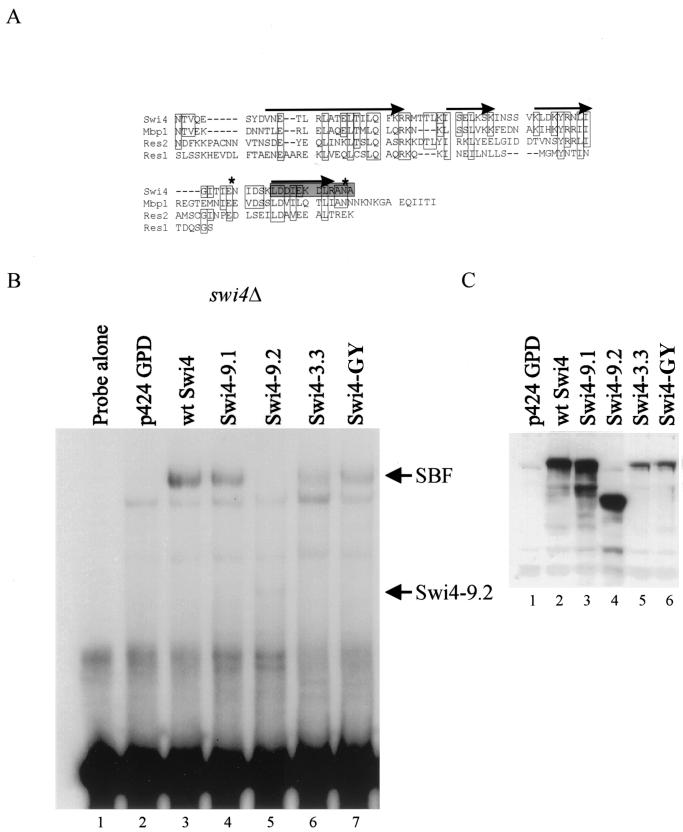
FIG. 5.
Gel retardation analysis of wild-type SWI4 and mutant swi4 alleles with yeast cell extracts. (A) Alignment of the extreme CTRs of Swi4 family members. Residues identical to those in Swi4 or conservative substitutions are boxed. Putative alpha helices are indicated by arrows, as predicted by the PHD protein structure algorithm. The shaded box shows the amino acids that were deleted in Swi4-3.3. The asterisks indicate the positions of the point mutations E1076G and N1092Y in mutant Swi4-GY. (B) A labeled SCB-containing probe was incubated with 10 μg of crude extract from a swi4Δ yeast strain (BY184) transformed with the following plasmids: lane 1, no extract; lane 2, empty vector, p424 GPD; lane 3, p424 GPD-Swi4 (wild-type [wt] SWI4); lane 4, p424 GPD-Swi4-9.1; lane 5, p424 GPD-Swi4-9.2; lane 6, p424 GPD-Swi4-3.3; and lane 7, p424 GPD-Swi4-GYr. The SWI4 mutations in the various plasmids are described in Table 2 and in the text. The migration positions of SBF and a complex of Swi4-9.2 and the SCB-containing probe are indicated to the right. (C) Western blot analysis of extracts used in the binding assay shown in panel B. Fifty micrograms of the crude yeast lysates used in the gel retardation analysis were separated by SDS–6% PAGE, and the Swi4 protein in the extracts was visualized with Swi4 antiserum. The Swi4 protein present in each extract is indicated above the lanes (see panel B).
Using this screen, we identified four new SWI4 mutants (Table 2). Two mutants, Swi4-9.1 and Swi4-9.2, were the result of improper ligation of the PCR product into the SWI4-containing vector. Mutant Swi4-9.1 had an addition of 12 amino acids to the C terminus of Swi4 (A-N-F-N-K-I-L-T-L-T-I-S). This addition resulted in a fourfold increase in SCB-dependent expression in the absence of Swi6. Mutant Swi4-9.2 had a large C-terminal truncation at amino acid 803 that allowed for a threefold activation of SCB::lacZ expression in the absence of Swi6. These results support our in vitro data demonstrating that the C terminus of Swi4 inhibits Swi4 DNA binding. We also identified a smaller C-terminal truncation in our screen. In Swi4-3.3, a 1-bp deletion in the codon for L1081 resulted in the truncation of 12 amino acids from the C terminus of Swi4 and the addition of 4 amino acids (N-W-T-I). This smaller alteration resulted in a modest but reproducible induction of SCB::lacZ expression (Table 2). Finally, we isolated a fourth mutant, Swi4-C26, which had three point mutations: N995H, E1076G, and N1092Y. By separating the N995H mutation from the E1076G and N1092Y mutations, we were able to determine that the twofold induction in SCB::lacZ activity was due to the two most C-terminal mutations. The mutant carrying these two mutations was named Swi4-GY.
TABLE 2.
β-Galactosidase activities of Swi4 mutants
| Plasmid | β-Galactosidase activity (Miller units)a | Description |
|---|---|---|
| p424 GPD | 0.1 | Vector control |
| p424 GPD-Swi4 | 11.4 | Wild-type Swi4 |
| p424 GPD-Swi4-9.1 | 41.3 | Addition of 12 amino acids to the C terminus of Swi4 |
| p424 GPD-Swi4-9.2 | 33.8 | C-terminal truncation at amino acid 803 |
| p424 GPD-Swi4-3.3 | 15.6 | 1-bp deletion at amino acid 1081 yielding a 12-amino-acid truncation + 4 amino acids |
| p424 GPD-Swi4-GY | 22.4 | E1076G + N1092Y |
See Materials Methods and reference 40; average of three independent transformations.
Since the CTR of Swi4 is also required for the interaction with Swi6, we next examined whether our new Swi4 mutants could still interact with Swi6. To answer this question, we performed DNA binding assays with an SCB-containing probe and crude yeast extracts from a swi4Δ strain transformed with plasmids encoding the Swi4 mutants. Western blot analysis with an anti-Swi4 antibody showed that all the Swi4 mutant proteins were expressed (Fig. 5C). Crude lysates from cells expressing mutant Swi4-9.2, which lacks the Swi6 interaction domain, did not support SBF complex formation; however, a distinct, faster-migrating species was formed (Fig. 5B, lane 5). In contrast, crude lysates from cells expressing wild-type Swi4, or the Swi4 mutants Swi4-9.1, Swi4-3.3, and Swi4-GY, supported SBF complex formation (Fig. 5B, lanes 4, 6, and 7). The mutants Swi4-3.3 and Swi4-GY appeared to form the SBF-DNA complex less efficiently, a result which correlated with decreased expression levels, as determined by Western blot analysis (Fig. 5C, lanes 5 and 6). Although we saw SBF complex formation by the Swi4 CTR mutants in the presence of Swi6, we did not see a lower-molecular-weight complex that might correspond to binding of the Swi4 CTR mutant proteins to SCBs in the absence of Swi6 (data not shown). We presume that the ability of the Swi4 CTR mutants to bind SCBs in the absence of Swi6 is sufficient to yield increased SCB-dependent transcription but may be undetectable by our biochemical assay. Our results suggest that small alterations in the extreme C terminus of Swi4 may alleviate DNA binding inhibition but do not affect the interaction with Swi6.
A C-terminal fragment of Swi4 can inhibit Swi4 DNA binding in trans.
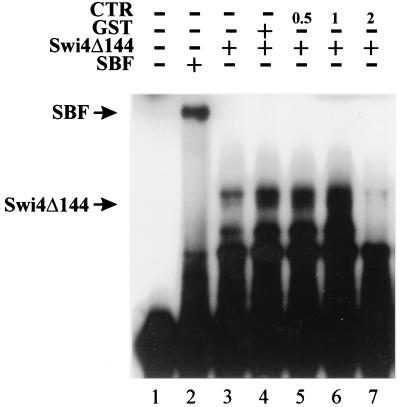
Both truncations and point mutations in the C terminus of Swi4 appear to alleviate the inhibition of Swi4 DNA binding. Our results suggest that the C terminus of Swi4 may inhibit the DNA binding of a C-terminal truncation of Swi4. To test this model, we performed gel retardation assays with an SCB-containing probe, partially purified Swi4Δ144, and the C-terminal 144 amino acids of Swi4 (the CTR) (Fig. 6). The CTR fragment was purified by thrombin cleavage of a GST-CTR fusion protein (see Materials and Methods). Incubation of Swi4Δ144 with increasing amounts of the CTR prior to the addition of the SCB-containing probe resulted in an inhibition of Swi4Δ144 binding to the SCB-containing probe (Fig. 6, lane 7). At low concentrations of the CTR, the amount of the Swi4Δ144-SCB complex appeared to increase slightly. This increase may have been due to the small amounts of GST which were present in the CTR preparation. We found that the use of GST alone also increased the amount of the Swi4Δ144-SCB complex (Fig. 6, lane 4). Presumably, GST acts to nonspecifically stabilize complex formation in our assay. Nonetheless, our gel shift assays show that the CTR of Swi4 can functionally interact with an N-terminal region of Swi4 to inhibit Swi4 binding to DNA.
FIG. 6.
Inhibition of Swi4Δ144-SCB complex formation by the CTR of Swi4. A gel retardation assay with an SCB-containing probe is shown (see the legend to Fig. 2). The probe was incubated with the following protein preparations: lane 1, no extract; lane 2, SBF heparin-agarose fraction (1 μg); and lanes 3 to 7, 2 μg of partially purified Swi4Δ144. Lane 4 also contained 1 μg of GST, and lanes 5 to 7 also contained increasing amounts (in micrograms) of purified Swi4 CTR (C-terminal 144 amino acids of Swi4). The migration positions of the SBF-SCB and Swi4Δ144-SCB complexes are indicated on the left.
Analysis of Swi4 complexes on glycerol gradients.
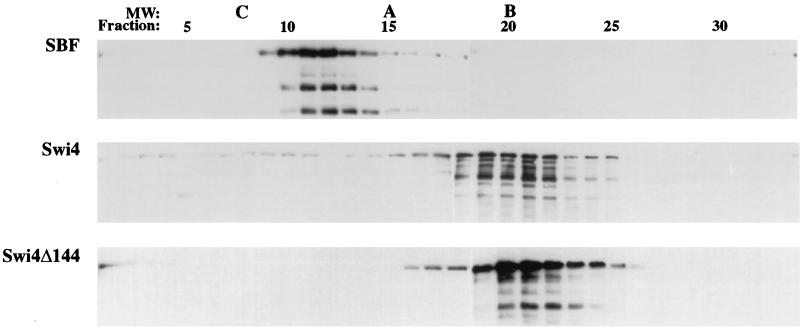
Since our in vitro studies were performed with partially purified reagents expressed in insect cells, it is unlikely that Swi4 autoinhibition depends on another protein binding the CTR of Swi4. Rather, our results suggest that the CTR of Swi4 may be involved in an inter- or intramolecular interaction with another region or molecule of Swi4 to inhibit DNA binding. To determine whether Swi4 forms dimers or multimers in solution, we analyzed protein size by glycerol gradient sedimentation. Swi4 ran at approximately 67 kDa, a mass which is drastically smaller than its predicted mass of 123 kDa (Fig. 7). This result suggests that, in solution, Swi4 must be highly asymmetric in shape and monomeric in nature. Truncating the C-terminal 144 amino acids of Swi4 did not significantly change the sedimentation of Swi4. In contrast, the addition of Swi6 to Swi4 greatly increased the sedimentation of Swi4 (Fig. 7, top panel). SBF ran at 180 kDa, a size which is close to the predicted size of a heterodimer of Swi4 (123 kDa) and Swi6 (91 kDa).
FIG. 7.
Glycerol gradient sedimentation of Swi4, Swi4Δ144, and SBF. Fifty micrograms of partially purified Swi4, Swi4Δ144, and SBF was analyzed by glycerol gradient sedimentation. Glycerol gradients (4 ml of 10 to 40% [vol/vol] glycerol) were centrifuged in an SW60.1 rotor for 13 h at 55,000 rpm. Fractions from the gradients were analyzed on Western blots with anti-Swi4 antibody. The peak fractions of molecular weight (MW) standards run in parallel gradients are indicated by C (catalase, 232,000), A (aldolase, 158,000), and B (bovine albumin, 67,000). Fraction numbers are shown at the top of the figure, with the bottom of the gradients on the left.
Interaction of the CTR of Swi4 with N-terminal domains.
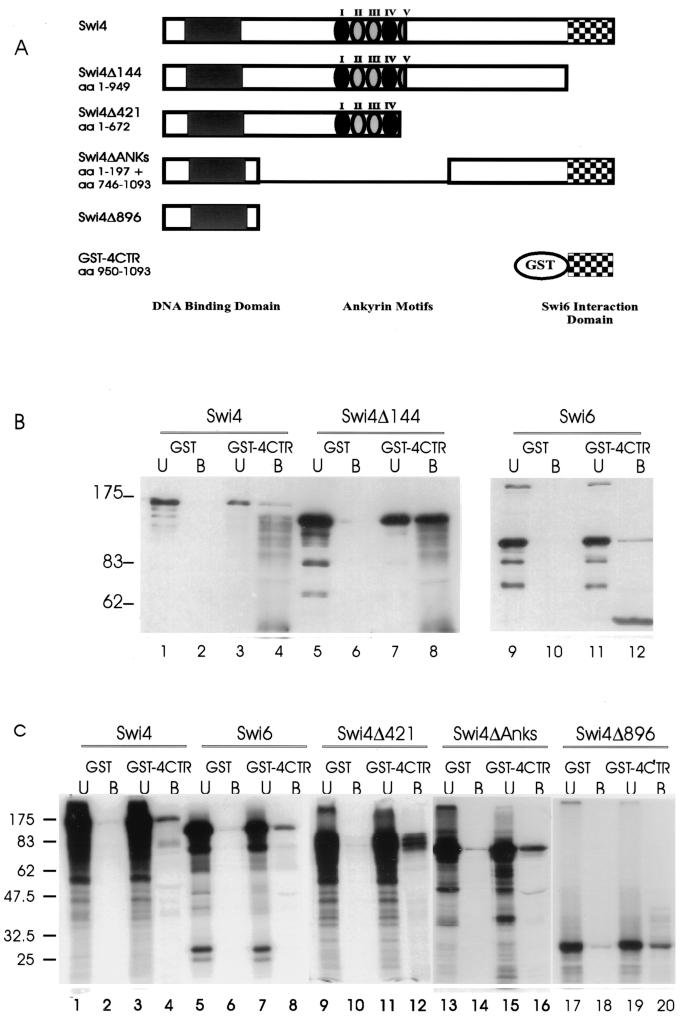
Our glycerol gradient assays revealed that Swi4 was monomeric in solution, suggesting that the inhibition of Swi4 binding to DNA involves an intramolecular interaction. In order to determine whether the C-terminal 144 amino acids of Swi4 were capable of protein-protein interactions within Swi4, we used GST-4CTR (Fig. 8A) and performed batch affinity chromatography assays with either insect cell-produced Swi4 derivatives or in vitro-translated Swi4 derivatives. GST or GST-4CTR was incubated with full-length Swi4, Swi4Δ144, or Swi6 produced from insect cells (Fig. 8B). GST-4CTR efficiently bound Swi6 (Fig. 8B, lane 12), as previously demonstrated (42). GST-4CTR also bound full-length Swi4 and Swi4Δ144 (Fig. 8B, lanes 4 and 8), showing that the C-terminal 144 amino acids of Swi4 can interact in vitro with the first 949 amino acids of Swi4. These results are consistent with our gel shift analysis results showing a functional inhibition of Swi4Δ144 binding to SCBs by the CTR. To further define the N-terminal interaction domain of Swi4, affinity chromatography experiments were conducted with a series of in vitro-transcribed and -translated Swi4 derivatives (Swi4, Swi4Δ421, Swi4ΔAnks, and Swi4Δ896) (Fig. 8A). Like the insect cell-produced proteins, the C-terminal 144 amino acids of Swi4 interacted with in vitro-translated full-length Swi4 (Fig. 8C, lane 4). GST-4CTR also bound both Swi4Δ421 and Swi4ΔAnks, indicating that the Swi4-Swi4 interaction does not require the ankyrin motifs or the C terminus of Swi4 (Fig. 8C, lanes 12 and 16). Further, GST-4CTR interacted directly with the N-terminal 197 amino acids of Swi4, which contain the DNA binding domain of Swi4 (Fig. 8C, lane 20). This interaction appeared weaker than interactions between GST-4CTR and larger N-terminal fragments of Swi4, suggesting that other parts of Swi4 may provide structural support for this interaction. We have previously found that Swi4 fails to interact in vitro with C-terminally truncated Swi6 (4), showing that protein derivatives containing the Swi4 CTR are not indiscriminate ligands. Together with our evidence that Swi4 is monomeric in solution, our data suggest that the C terminus of Swi4 is involved in an intramolecular interaction with an N-terminal DNA binding region of Swi4 and that, in the absence of Swi6, this interaction causes an inhibition of Swi4 DNA binding.
FIG. 8.
Binding of the C-terminal 144 amino acids of Swi4 to Swi6 and N-terminal regions of Swi4 in vitro. (A) Schematic of the Swi4 proteins used in the assay. The relative positions of the DNA binding domain, ankyrin motifs, and C-terminal domain (Swi6 interaction domain) are depicted. aa, amino acids. (B) Ten micrograms of partially purified Swi4, Swi4Δ144, or Swi6 derived from insect cell extracts (see Fig. 2) was incubated with either GST or GST-4CTR immobilized on glutathione beads. The unbound (U) and bound (B) fractions were separated by SDS–6% PAGE. The gels were then blotted and incubated with Swi4 antiserum (lanes 1 to 8) or Swi6 antiserum (lanes 9 to 12) to identify the Swi4 or Swi6 proteins. The migration positions of molecular weight markers are indicated to the left (in thousands). (C) Seven microliters of in vitro-translated Swi4, Swi6, Swi4Δ421, Swi4ΔAnks, and Swi4Δ896 was incubated with either GST or GST-4CTR immobilized on glutathione beads. The unbound (U) and bound (B) fractions were separated by SDS–10% PAGE. The migration positions of molecular weight markers are indicated to the left (in thousands).
DISCUSSION
We found that Swi4 is nuclear throughout the cell cycle and yet is incapable of promoting transcription in the absence of Swi6. We reconstituted active SBF in vitro from Swi4 and Swi6 expressed in a baculovirus system in insect cells. Partially purified full-length Swi4 could not bind SCBs in the absence of Swi6; however, Swi4 derivatives truncated at the C terminus or carrying point mutations in the extreme C terminus were able to bind DNA or activate transcription in the absence of Swi6. Further, the binding of a C-terminally truncated Swi4 protein to SCBs was inhibited by the addition of a C-terminal fragment of Swi4 in trans. Full-length Swi4 was monomeric in solution, suggesting an intramolecular mechanism for auto-inhibition of binding to DNA by Swi4. We detected a direct interaction between a C-terminal fragment of Swi4 and the N-terminal 197 amino acids of Swi4, which contain the DNA binding domain of Swi4. Our data suggest that the interaction of the CTR of Swi4 with the N-terminal DNA binding domain of Swi4 physically inhibits the DNA binding domain from binding SCBs. Interaction of the CTR of Swi4 with Swi6 alleviates this inhibition, allowing Swi4 to bind DNA.
The C terminus of Swi4 inhibits the binding of Swi4 to SCBs.
Our experiments implicate the extreme C terminus of Swi4 in the auto-inhibition of Swi4 binding to DNA. The C terminus of Swi4 also contains the Swi6 interaction domain, which has been localized to the last 78 amino acids of Swi4 (44). As depicted in Fig. 5A, this region of Swi4 is very similar to comparable regions in other members of the Swi4 family and has been predicted to contain alpha helices with an amphipathic character (8, 42, 44). While our screen for mutations in the CTR of Swi4 was not saturating, we isolated a large truncation of Swi4 and three isolates with different mutations that affect the extreme C terminus of Swi4. One mutant, Swi4-GY, carried two mutations in the extreme C terminus of Swi4, E1076G and N1092Y. While we have yet to separate the mutations, it is interesting to note that both map to conserved residues (Fig. 5A). In another mutant, Swi4-3.3, a predicted alpha helix is deleted from the Swi4 C terminus. Notably, both of these latter mutants could still bind Swi6 to form SBF in vitro. This result suggests that the domain or residues responsible for the Swi4-Swi6 interaction may be distinct from those necessary for Swi4 auto-inhibition. Alternatively, the domains or residues required for the Swi4 auto-inhibitory function and the Swi4-Swi6 interaction function may be shared and our SBF assay was not sufficiently sensitive to detect a decrease in the Swi4-Swi6 interaction. It will be interesting to determine whether the Swi4-Swi6 and Swi4-Swi4 interaction domains are fully separable. We suspect that the Swi6 interaction domain may localize to the conserved residues or putative alpha helices N terminal to the more C-terminal region that our data implicate in Swi4 auto-inhibition.
Model for Swi4 DNA binding inhibition.
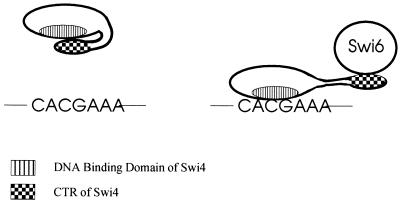
We found that Swi4 was largely monomeric in solution and that the CTR of Swi4 could interact with the N-terminal 197 amino acids containing the DNA binding domain of Swi4 in vitro. Further, the addition of the CTR of Swi4 to a binding reaction mixture containing a C-terminally truncated Swi4 protein (Swi4Δ144) inhibited DNA binding by Swi4Δ144. Together, our data suggest an intramolecular model for the inhibition of Swi4 binding to DNA. Interestingly, intramolecular interactions within the Swi6 protein have recently been reported (41). The internal ankyrin domains of Swi6 form a stabilized central structure with which adjacent transcriptional activation domains interact. We propose that the intramolecular interaction of the CTR of Swi4 with the N-terminal region of Swi4 results in the DNA binding domain of Swi4 becoming inaccessible or incapable of binding DNA in the absence of Swi6 (Fig. 9). Upon the addition of Swi6, the Swi4-Swi4 interaction is disrupted, alleviating the inhibition.
FIG. 9.
Model of the auto-inhibition of Swi4 binding to DNA. (Left) An intramolecular interaction involving the extreme C terminus of Swi4 (CTR) and a more N-terminal region of the protein is depicted. Our data suggest that when Swi4 is not bound to Swi6, the CTR of Swi4 is free to form an intramolecular interaction with the DNA binding domain of Swi4, preventing Swi4 binding to SCBs (CACGAAA). (Right) The binding of Swi6 to the CTR of Swi4 disrupts the intramolecular interaction of Swi4, allowing Swi4 to bind to SCBs.
Our batch affinity chromatography analysis showed that the CTR of Swi4 can interact directly with the N-terminal 197 amino acids of Swi4. This result suggests that the inhibition of Swi4 DNA binding by the CTR may be due to a direct masking of the DNA binding domain. Alternatively, the CTR of Swi4 may interact with amino acids outside the core DNA binding domain, causing a conformational change in the DNA binding domain. Swi4 proteins containing a fusion of the minimal DNA binding domain (amino acids 36 to 170) to the C-terminal 146 amino acids or 65 amino acids of Swi4 were reported to form a complex with DNA in the absence of Swi6 (37). If an intramolecular interaction is the mechanism for Swi4 auto-inhibition, then the CTR of Swi4 may interact either with the first 36 amino acids of Swi4, which are N-terminal to the DNA binding domain, or with amino acids 170 to 197, which lie C-terminal to the DNA binding domain. Alternatively, direct fusion of the CTR with the DNA binding domain may cause a conformational rigidity that prevents the intramolecular interaction. Indeed, it has been suggested that both Swi4 and Swi6 have inherent modularity and flexibility that are crucial for their in vivo function (41). Hydrodynamic analysis and proteolytic cleavage studies of Swi6 have determined that the N-terminal 15-kDa domain of Swi6 is connected to the central ankyrin region of Swi6 by a long and potentially flexible linker region. This 15-kDa region appears to have no function, and it is thought to represent a nonfunctional remnant of a DNA binding domain from a common ancestor which has remained active in some family members (reviewed in reference 8). Members of the Swi4-Swi6 family of transcription factors have a similar domain structure, and the flexible arm adjoining the N-terminal domain of Swi6 with the central core ankyrin domain may be a conserved feature of the entire family. This arm may provide the flexibility needed for the N-terminal DNA binding domain of Swi4 to interact with the C-terminal domain.
Intramolecular interactions have been implicated in the regulation of DNA binding for many transcription factors, including numerous Ets family members, such as Ets-1 and GABPα (reviewed in reference 5). The ability of Ets-1 to bind DNA is negatively regulated by at least two domains: an N-terminal region and a C-terminal region (22, 29). Recent studies revealed that direct interactions between the Ets-1 inhibitory N-terminal region and both the Ets domain and the Ets-1 inhibitory C-terminal region are responsible for the intramolecular inhibition of Ets-1 DNA binding activity (25, 36, 45). In the full-length protein, the two inhibitory domains interact allosterically, placing stress on the Ets domain and destabilizing DNA contacts. Loss of coupling between the two domains leads to an altered conformation in the N-terminal inhibitory region, allowing the Ets domain to make stable contacts with DNA. However, this relaxed conformation is transient, and reestablishment of the interaction between the inhibitory regions causes the repression of DNA binding. Stable DNA–Ets-1 interactions are established through several mechanisms which disrupt the Ets-1 intramolecular interaction, including phosphorylation of the N-terminal inhibitory region and a direct protein-protein interaction with the N-terminal inhibitory region (19, 39). A similar method of DNA binding inhibition has also been established for GABPα, whose interaction with the ankyrin-containing protein GABPβ allows GABPα to bind DNA (10, 13, 49). Although there is little sequence homology between Swi4 and the Ets family of proteins, the recent determination of the crystal structure of the DNA binding domain of the Swi4 family member Mbp1 revealed that the Swi4 family and the Ets family proteins do share a common fold in their core DNA binding domains. The core consists of a short strand N terminal to the helix-turn-helix and a β-hairpin C terminal to the helix-turn-helix (48, 50). Although the structure of Mbp1 outside the core diverged from that of the Ets proteins, similar allosteric forces may contribute to the DNA binding inhibition of the Swi4 family of transcription factors.
Role of Swi4 DNA binding inhibition in the regulation of SBF.
Clb/Cdc28 activity is necessary for the dissociation of SBF from the CLN2 promoter in the G2 phase and mitosis (23, 28). Clb2 immunoprecipitation experiments have shown that Swi4 can interact with Clb2 during the M phase and that Swi4 is phosphorylated in vivo (1, 44). Interestingly, the interaction of Clb2 with Swi4 appears to be independent of Swi6. These observations have led to the suggestion that a Clb interaction with Swi4 is necessary for preventing Swi4 from binding DNA. Our data suggest that the inhibition of Swi4 DNA binding is intrinsic to Swi4 and does not require any other proteins. The role of Clb may not be to inhibit Swi4 DNA binding but rather to promote the dissociation of SBF from SCBs. Clb-dependent regulation of SBF may occur through a disruption of the Swi4-Swi6 interaction. Once Swi4 and Swi6 are dissociated from each other, Swi6 is transported from the nucleus, allowing the CTR of Swi4 to form an intramolecular interaction with the DNA binding domain of Swi4, thereby inhibiting Swi4 DNA binding. Interestingly, gel retardation assays with whole-cell extracts of synchronized cells arrested at different stages of the cell cycle show that the SBF-DNA complex can form at all stages of the cell cycle (47). This result suggests that the inhibition of Swi4 DNA binding is immediately relieved upon the addition of Swi6. To test this model, it will be important to determine the relative affinities of Swi4-Swi4 and Swi4-Swi6 interactions.
It is unlikely that Swi6 is fully excluded from the nucleus throughout the M and G2 phases, although SBF footprinting is not detected. It is possible that the formation of the SBF-DNA complex is undetectable in the M and G2 phases because Clb/Cdc28 is continually disrupting the SBF complex and Swi6 is actively transported from the nucleus. Alternatively, in addition to the auto-inhibition of Swi4 DNA binding, there may be another mechanism regulating SBF-DNA complex formation during the G2 and M phases. Both the role and sites of Swi4 phosphorylation by Clb2/Cdc28 have yet to be established. One possibility is that the phosphorylation of Swi4 may alter the affinity of the Swi4-Swi6 interaction or the stability of Swi4 auto-inhibition. There is one consensus Cdc28 phosphorylation site in the CTR of Swi4 (S1007) and numerous other potential sites in the CTR and the N-terminal DNA binding domain. It will be interesting to determine whether Clb/Cdc28 phosphorylates these sites and whether phosphorylation contributes to the regulation of Swi4 DNA binding.
ACKNOWLEDGMENTS
We thank B. Funnell, H. Friesen, J. Moffatt, and D. McCallum for critical comments on the manuscript.
K.B. is a research student of the National Cancer Institute of Canada and is supported with funds provided by the Terry Fox Run. This work was supported by a grant from the Medical Research Council of Canada to B.A., who is a scientist of the Medical Research Council of Canada.
REFERENCES
- 1.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 2.Andrews B J, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 3.Andrews B J, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 4.Andrews B J, Moore L A. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc Natl Acad Sci USA. 1992;89:11852–11856. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassuk A G, Leiden J M. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64:65–104. doi: 10.1016/s0065-2776(08)60887-1. [DOI] [PubMed] [Google Scholar]
- 6.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins; mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 7.Breeden L, Nasmyth K. Regulation of HO. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 8.Breeden L L, editor. Start-specific transcription in yeast. Curr Top Microbiol Immunol. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- 9.Breeden L L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 10.Brown T, McKnight S. Specificities of protein-protein and protein-DNA interaction of GABPα and two newly defined Ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 11.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 12.Davey M J, Funnell B E. The P1 plasmid partition protein ParA: a role for ATP in site specific DNA binding. J Biol Chem. 1994;269:29908–29913. [PubMed] [Google Scholar]
- 13.de la Brousse F, Birkenmeier E, King D, Rowe B, McKnight S. Molecular and genetic characterization of GABPβ. Genes Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- 14.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln/Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Donoviel, M. Personal communication.
- 15.Espinoza F H, Ogas J, Herskowitz I, Morgan D O. Cell cycle control by a complex of the cyclin HCS26(PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 16.Ewaskow S P, Sidorova J M, Hendle J, Emery J C, Lycan D E, Zhang K Y J, Breeden L L. Mutation and modeling analysis of the Saccharomyces cerevisiae Swi6 ankyrin repeats. Biochemistry. 1998;37:4437–4450. doi: 10.1021/bi972652e. [DOI] [PubMed] [Google Scholar]
- 17.Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 19.Giese K, Kingsley C, Kirshner J, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 20.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 22.Hagman J, Grosschedl R. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci USA. 1992;89:8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington L A, Andrews B J. Binding to the yeast Swi4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igual J, Johnson A, Johnston L. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsen M D, Peterson J M, Xu Q P, Graves B J. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 27.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 28.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 29.Lim F, Kraut N, Frampton J, Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992;11:643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2(ORF2)-PHO85 cyclin dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 31.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. A family of cyclin-like proteins that interact with Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 33.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 34.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2 and a putative new G1 cyclin (HCS26) by Swi4, a positive regulator of G1 specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 35.Pardee A B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 36.Petersen J, Skalicky J, Donaldson L, McIntosh L, Alber T, Graves B. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of α helix. Science. 1995;269:1866–1868. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 37.Primig M, Sockanathan S, Auer H, Nasmyth K. Anatomy of a transcription factor important for the Start of the cell cycle in Saccharomyces cerevisiae. Nature. 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- 38.Pringle J R, Adams A E M, Drubins D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–601. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 39.Rabault B, Ghysdael J. Calcium-induced phosphorylation of ETS1 inhibits its specific DNA binding activity. J Biol Chem. 1994;269:28143–28151. [PubMed] [Google Scholar]
- 40.Rose M, Botstein D. Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 41.Sedgwick S G, Taylor I A, Adam A C, Spanos A, Howell S, Morgan B A, Treiber M K, Kanuga N, Banks G R, Foord R, Smerdon S J. Structural and functional architecture of the yeast cell-cycle transcription factor Swi6. J Mol Biol. 1998;281:763–775. doi: 10.1006/jmbi.1998.1996. [DOI] [PubMed] [Google Scholar]
- 42.Sidorova J M, Breeden L L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidorova J M, Mikesell G E, Breeden L L. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Cell Biol. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegmund R F, Nasmyth K. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol Cell Biol. 1996;16:2647–2655. doi: 10.1128/mcb.16.6.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skalicky J, Donaldson L, Petersen J, Graves B, McIntosh L. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart D, Wittenberg C. CLN3, not positive feedback, determines CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 47.Taba M R M, Muroff I, Lydall D, Tebb G, Nasmyth K. Changes in a Swi4,6-DNA binding complex occur at the time of HO activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- 48.Taylor I A, Treiber M K, Olivi L, Smerdon S J. The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1A resolution. J Mol Biol. 1997;272:1–8. doi: 10.1006/jmbi.1997.1229. [DOI] [PubMed] [Google Scholar]
- 49.Thompson C, Brown A, McKnight S. Convergence of Ets- and Notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- 50.Xu R M, Koch C, Liu Y, Horton J R, Knapp D, Nasmyth K, Cheng X. Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis. Structure. 1997;5:349–358. doi: 10.1016/s0969-2126(97)00192-5. [DOI] [PubMed] [Google Scholar]
- 51.YGAC Website. [Online.] http://ycmi.med.yale.edu/YGAC/antibodycleanup.html. [3 August 1999, last date accessed.]