Abstract
Acanthopanax sessiliflorus (Araliaceae) have been reported to exhibit many pharmacological activities. Our preliminary study suggested that A. sessiliflorus fruits include many bioactive 3,4-seco-triterpenoids. A. sessiliflorus fruits were extracted in aqueous EtOH and fractionated into EtOAc, n-BuOH, and H2O fractions. Repeated column chromatographies for the organic fractions led to the isolation of 3,4-seco-triterpenoid glycosides, including new compounds. Ultra-high-performance liquid chromatography (UPLC) mass spectrometry (MS) systems were used for quantitation and quantification. BV2 and RAW264.7 cells were induced by LPS, and the levels of pro-inflammatory cytokines and mediators and their underlying mechanisms were measured by ELISA and Western blotting. NMR, IR, and HR-MS analyses revealed the chemical structures of the nine noble 3,4-seco-triterpenoid glycosides, acanthosessilioside G–O, and two known ones. The amounts of the compounds were 0.01–2.806 mg/g, respectively. Acanthosessilioside K, L, and M were the most effective in inhibiting NO, PGE2, TNF-α, IL-1β, and IL-6 production and reducing iNOS and COX-2 expression. In addition, it had inhibitory effects on the LPS-induced p38 and ERK MAPK phosphorylation in both BV2 and RAW264.7 cells. Nine noble 3,4-seco-triterpenoid glycosides were isolated from A. sessiliflorus fruits, and acanthosessilioside K, L, and M showed high anti-inflammatory and anti-neuroinflammatory effects.
Keywords: Acanthopanax sessiliflorus fruit, acanthosessiliosides, anti-inflammation, anti-neuroinflammation, NMR, quantification
1. Introduction
Oxidative stress in the human body is caused by excess reactive oxygen or nitrogen species (ROS or RNS), including superoxide (O2−), hydrogen peroxide (H2O2), and nitric oxide (NO), and it triggers certain types of apoptotic cell death, such as neuronal cell death and macrophage immune injury. If ROS or RNS are not appropriately removed, oxidative stress can cause the lipid peroxidation of the cellular membrane, leading to inordinate cell death [1].
Recently, the number of patients with degenerative brain disease has increased due to the aging society [2]. Therefore, the investigation of the cause and drug development for degenerative brain disease is rapidly progressing. The microglia in the central nerve system (CNS) plays an important role in the immunity, degeneration, and inflammation of the CNS, so it can prevent degenerative brain diseases by regulating the inflammatory response that occurs in microglials [3]. If the microglia overwhelmed by stimulation such as lipopolysaccharide (LPS), interferon-gamma (IFN-α), or tumor necrosis factor-alpha (TNF-α), NO, excessive cytokines, and ROS are secreted, causing neurotoxicity and destroying brain tissue, causing degenerative brain diseases such as Alzheimer’s, Parkinson’s disease, and Croyfeldt–Jakob disease [4,5,6]. Therefore, to prevent brain damage caused by ROS, RNS (NO), and excessive cytokines, it is useful to take anti-inflammatory and anti-neuroinflammatory supplements. However, synthetic anti-inflammatory agents can be toxic and carcinogenic and can interfere with the metabolic and respiratory activities of cells. Therefore, there is a need to develop alternative natural anti-inflammatory drugs without side effects.
The Acanthopanax genus is the deciduous broadleaf shrub in the family Araliaceae. Acanthopanax species are native to eastern Asia, from southeast Siberia and Japan to the Philippines and Vietnam [7]. Acanthopanax species are used commonly in traditional oriental medicine to treat cerebrovascular diseases, tumors, rheumatoid arthritis, diabetes, and hypertension [8]. A. sessiliflorus (Rupr. Et Maxim) is a deciduous broadleaf shrub that grows from 3 to 4 m in length, with sharp thorns on its branches. The barks are gray and the leaves are palmate leaves with five small leaves and have long petioles. Flowers are yellow-green and, in spring, hang in an umbel at the end of a long peduncle, and each flower has five petals. Fruits are ball-shaped, 6–7 mm in diameter, and ripen black around September [8,9]. Previous research has reported that triterpenoids and lignans are thought to be the active constituents in this plant [10,11,12]. Our previous study reported the isolation and identification of 3,4-seco-lupane triterpenes and its glycosides from an EtOAc fraction of A. sessiliflorus fruits, and their cytotoxicity against six human cancer cell lines and their ability to inhibit LPS-induced NO production in RAW 264.7 macrophages were shown [8]. Therefore, it is predictable that A. sessiliflorus fruits contain more various bioactive triterpene glycosides. Furthermore, recent reports have stated that an ethanol extract of A. sessiliflorus has antioxidant, antibacterial [13], antihypertensive [14], and anti-inflammatory [15] effects by these components.
In this study, we isolated and identified the 3,4-seco triterpenoid glycosides in A. sessiliflorus fruits and the isolated compounds were evaluated for their anti-inflammatory and anti-neuroinflammatory activities in RAW 264.7 macrophage and BV2 microglia.
2. Materials and Methods
2.1. Plant Materials
A. sessiliflorus fruits were harvested from the Pyeongchang county, Gangwon province, Korea (latitude 37°6′ N, longitude 128°3′ E). A voucher specimen (MPS006299) was deposited at the Herbarium of the Department of Herbal Crop Research, National Institute of Horticultural and Herbal Science, Rural Development Administration, Eumseong, Korea.
2.2. General Experimental Procedures
The materials and equipment we used for the isolation and structure determination of constituents are described in a previous study [16]. Tissue culture reagents, such as Roswell Park Memorial Institute 1640 (RPMI1640) and fetal bovine serum, were purchased from Gibco BRL Co. (Grand Island, NY, USA). All chemicals were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Primary antibodies, including anti-iNOS, anti-COX-2, and anti-β-actin, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-p-JNK, anti-JNK, anti-p-ERK, anti-ERK, anti-p-p38, and anti-p38 were purchased from Cell Signaling Technology (Danvers, MA, USA); anti-rabbit and anti-mouse secondary antibodies were purchased from Millipore (Billerica, MA, USA). Enzyme-linked immunosorbent assay (ELISA) kits for PGE2, IL-6, and TNF-α assessments were purchased from R&D Systems, Inc. (Minneapolis, MN, USA). The compound structure was determined by analyzing various spectroscopic data, including mass spectroscopy and nuclear magnetic resonance, which were consistent with those previously reported [17].
2.3. Extraction and Isolation
The dried and powdered fruits of A. sessiliflorus (5 kg) were extracted with 70% aqueous ethanol (MeOH, 10 L × 2) for 24 h, which yielded a concentrated extract (ASFE, 1211 g). The concentrated extract was suspended in water (2 L) and successively extracted with EtOAc (2 L × 2) and n-BuOH (1.6 L × 2) to yield concentrated extracts of the EtOAc (AFE), n-BuOH (AFB, 88 g), and H2O (AFH) fractions. The concentrated n-BuOH fraction (AFB, 80 g) was subjected to silica gel CC (15 × 25 cm) using a gradient of CHCl3-MeOH-H2O (15:3:1→12:3:1→7:3:1→65:35:10) to yield ten fractions (B1 to 10). Fraction B1 (5 g) was subjected to silica gel CC (4 × 18 cm, CHCl3-MeOH-H2O (12:3:1→9:3:1)) to yield twenty-two subfractions (FAB1-1 to 22). Subfractions (FAB1-17, 50 mg) were separated by CC (RP-10 (2 × 8 cm), MeOH-H2O (2:1)) to yield Compound 1 (21 mg). Subfractions (FAB1-22, 80 mg) were separated by CC (RP-10 (2 × 10 cm), MeOH-H2O (2:1)) to yield Compounds 2 (8 mg) and 4 (17 mg). Fraction B4 (11 g) was subjected to silica gel CC (5 × 15 cm, CHCl3-MeOH-H2O (9:3:1→7:3:1)) to yield twenty subfractions (FAB4-1 to 20). Subfractions (FAB4-1, 180 mg) were fractionated using silica gel CC (3.5 × 20 cm, CHCl3-MeOH-H2O (7:3:1)) and yielded seven subfractions (FAB4-1-1 to 7). Subfractions (FAB4-1-7, 40 mg) were purified using CC (RP-18 (2 × 6 cm), MeOH-H2O (3:1)) and yielded Compound 10 (14 mg). Subfractions (FAB4-12-24, 21 mg) were separated by CC (RP-10 (1 × 10 cm), MeOH-H2O (1:1)) to yield Compound 5 (12 mg). Subfractions (FAB4-12-26, 105 mg) were separated by CC [RP-10 (3 × 10 cm), MeOH-H2O (2:1)] to yield Compounds 8 (25 mg) and 9 (13 mg). Subfractions (FAB4-12-27, 33 mg) were separated by CC (RP-10 (1.5 × 8 cm), MeOH-H2O (1:1)) to yield Compound 6 (8.5 mg). Subfractions (FAB4-12-29, 25 mg) were separated by CC (RP-10 (1.5 × 8 cm), MeOH-H2O (1.5:1)) to yield Compound 3 (12.3 mg). Subfractions (FAB4-12-29, 70 mg) were chromatographed (RP-18 (3.5 × 6.5 cm), MeOH-H2O (2:1)) to yield Compound 7 (28 mg). Fraction B7 (3.3 g) was fractionated using silica gel CC (4 × 12 cm, CHCl3-MeOH-H2O (7:3:1)) and yielded nine subfractions (FAB7-1 to 9). Subfractions (FAB7-5, 330 mg) were purified using CC (RP-18 (3.5 × 10 cm), MeOH-H2O (3:1)) and yielded ten subfractions (FAB7-5-1 to 10). Purification of Subfractions FAB7-5-6 (80 mg) using CC (RP-18 (3 × 6 cm), MeOH-H2O (2:1)) yielded Compound 11 (25 mg).
Acanthosessilioside G [22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 28-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranose] (1): White amorphous powder; IR (CaF2 window) 3360, 1735, 1650 cm−1; negative ESI-QTOF/MS m/z 809.4448 [M−H]− (calculated for C42H65O15, 809.4323); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Table 1.
1H-NMR data of acanthosessilioside G–O.
| No. | δH, Coupling Pattern, J in Hza) | ||||
|---|---|---|---|---|---|
| G (1) | H (2) | I (3) | J (5) | K (7) | |
| 1 | 1.82, overlap | 1.91, overlap | 1.91, overlap | 2.17, overlap 1.54, overlap |
3.70, overlap |
| 2 | 1.39, overlap 1.36, overlap |
1.37, overlap 1.33, overlap |
1.37, overlap 1.33, overlap |
1.80, overlap 1.24, overlap |
2.86, overlap 2.58, overlap |
| 9 | 1.73, overlap | 1.77, overlap | 1.77, overlap | 1.75, overlap | 2.63, d, 9.2 |
| 11 | 1.47, overlap | 1.51, overlap | 1.51, overlap | 1.25, overlap | 4.55, overlap |
| 18 | 2.69, overlap | 2.69, overlap | 2.69, overlap | 1.84, overlap | 2.51, overlap |
| 19 | 3.70, m | 3.57, m | 3.60, m | 3.48, m | 3.52, m |
| 22 | 4.83, overlap | 4.83, overlap | 4.84, overlap | 1.73, overlap 1.36, overlap |
4.80, overlap |
| 23 | 5.12, d, 2.0 4.90, d, 2.0 |
5.04, d, 2.0 4.81, d, 2.0 |
5.00, d, 2.0 4.81, d, 2.0 |
4.94, br. s 4.78, br. s |
5.00, d, 2.0 4.80, d, 2.0 |
| 24 | 1.71, s | 1.81, s | 1.71, s | 1.71, s | 1.74, s |
| 25 | 1.14, s | 1.22, s | 1.15, s | 1.03, s | 1.17, s |
| 26 | 1.32, s | 1.24, s | 1.17, s | 1.13, s | 1.19, s |
| 27 | 0.81, s | 0.88, s | 0.79, s | 0.77, s | 0.82, s |
| 29 | 2.13, s | 1.99, s | 1.95, s | 1.72, s | 1.95, s |
| 30a | 4.90, overlap 4.79, overlap |
4.95, overlap 4.89, overlap |
4.90, overlap 4.81, overlap |
4.79, overlap 4.75, overlap |
4.96, overlap 4.89, overlap |
| OCH3 | - | - | 3.63, s | 3.64, s | - |
| 1′ | 6.37, d, 8.0 | 6.38, d, 8.0 | 6.32, d, 8.0 | 6.31, d, 8.0 | 6.33, d, 8.0 |
| 2′ | 4.30, overlap | 4.09, overlap | 4.10, overlap | 4.09, overlap | 4.10, overlap |
| 3′ | 4.37, overlap | 4.38, overlap | 4.32, overlap | 4.38, overlap | 4.35, overlap |
| 4′ | 4.30, overlap | 4.30, overlap | 4.28, overlap | 4.30, overlap | 4.29, overlap |
| 5′ | 4.00, overlap | 4.13, overlap | 4.14, overlap | 4.13, overlap | 4.15, overlap |
| 6′ | 4.58, dd, 12.0, 2.4 4.46, dd, 12.0, 4.8 |
4.69, overlap 4.30, overlap |
4.64, overlap 4.30, overlap |
4.69, overlap 4.30, overlap |
4.66, overlap 4.29, overlap |
| 1′′ | 5.35, d, 8.0 | 4.94, d, 8.0 | 4.83, d, 8.0 | 4.96, d, 8.0 | 4.94, d, 8.0 |
| 2′′ | 4.07, dd, 8.0, 8.0 | 4.10, overlap | 4.10, overlap | 4.10, overlap | 4.10, overlap |
| 3′′ | 4.27, overlap | 4.25, overlap | 4.25, overlap | 4.25, overlap | 4.25, overlap |
| 4′′ | 4.30, overlap | 4.38, overlap | 4.31, overlap | 4.38, overlap | 4.35, overlap |
| 5′′ | 4.00, overlap | 4.12, overlap | 4.15, overlap | 4.12, overlap | 4.15, overlap |
| 6′′ | 4.42, dd, 12.0, 2.0 4.34, dd, 12.0, 5.2 |
4.19, overlap 4.09, overlap |
4.21, overlap 4.08, overlap |
4.19, overlap 4.09, overlap |
4.18, overlap 4.06, overlap |
| 1‴ | - | 5.83, br. s | 5.75, br. s | 5.79, br. s | 5.79, br. s |
| 2‴ | - | 4.66, overlap | 4.61, overlap | 4.66, overlap | 4.66, overlap |
| 3‴ | - | 4.53, dd, 8.4, 3.2 | 4.47, dd, 8.4, 3.2 | 4.53, dd, 8.4, 3.2 | 4.55, dd, 8.4, 3.2 |
| 4‴ | - | 3.93, dd, 8.4, 8.4 | 3.88, dd, 8.4, 8.4 | 3.93, dd, 8.4, 8.4 | 3.89, dd, 8.4, 8.4 |
| 5‴ | - | 4.93, overlap | 4.92, overlap | 4.93, overlap | 4.93, overlap |
| 6‴ | - | 1.71, d, 6.0 | 1.65, d, 6.0 | 1.67, d, 6.4 | 1.67, d, 6.0 |
| 1⁗ | 4.42, overlap | ||||
| 2⁗ | 2.05, overlap | ||||
| 3⁗ | 0.79, overlap | ||||
| No. | δH, Coupling Pattern, J in Hza) | ||||
| L (8) | M (9) | N (10) | O (11) | ||
| 1 | 4.86, dd, 12.0, 2.4 | 4.86, dd, 12.0, 2.8 | 4.87, overlap | 4.86, overlap | |
| 2 | 3.65, overlap 2.66, dd, 12.0, 8.0 |
3.65, dd, 12.0, 2.8 2.66, dd, 12.0, 8.0 |
3.65, overlap 2.66, overlap |
3.65, overlap 2.64, overlap |
|
| 9 | 1.88, d, 11.2 | 1.90, d, 10.8 | 1.95, overlap | 1.95, overlap | |
| 11 | 1.25, overlap | 4.07, overlap | 4.07, overlap | 4.07, overlap | |
| 18 | 1.72, overlap | 1.80, dd, 12.0, 11.2 | 1.72, overlap | 1.72, overlap | |
| 19 | 3.50, m | 3.34, m | 3.58, m | 3.49, m | |
| 22 | 2.45, overlap 1.42, overlap |
2.35, overlap 1.44, overlap |
4.80, overlap | 4.79, overlap | |
| 23 | 1.24, s | 1.16, s | 1.24, s | 4.93, d, 2.0 4.80, d, 2.0 |
|
| 24 | 1.37, s | 1.39, s | 1.37, s | 1.37, s | |
| 25 | 1.32, s | 1.31, s | 1.33, s | 1.32, s | |
| 26 | 1.21, s | 1.15, s | 1.21, s | 1.24, s | |
| 27 | 1.10, s | 1.13, s | 1.15, s | 1.15, s | |
| 29 | 1.93, s | 1.68, s | 1.96, s | 1.93, s | |
| 30 | 4.91, br. s 4.66, br. s |
4.80, br. s 4.64, overlap |
4.83, overlap 4.63, overlap |
4.87, overlap 4.66, overlap |
|
| OCH3 | 3.60, s | 3.58, s | 3.60, s | 3.60, s | |
| 1′ | 6.37, d, 8.4 | 6.29, d, 8.4 | 6.31, d, 8.4 | 6.31, d, 8.0 | |
| 2′ | 4.15, overlap | 4.16, overlap | 4.16, overlap | 4.17, overlap | |
| 3′ | 4.27, overlap | 4.27, overlap | 4.27, overlap | 4.28, overlap | |
| 4′ | 4.32, overlap | 4.34, overlap | 4.33, overlap | 4.32, overlap | |
| 5′ | 4.10, overlap | 4.09, overlap | 4.08, overlap | 4.09, overlap | |
| 6′ | 4.41, br. d, 12.0 4.38, dd, 12.0, 6.0 |
4.66, overlap 4.29, overlap |
4.67, overlap 4.29, overlap |
4.66, overlap 4.30, overlap |
|
| 1′′ | 4.92, d, 8.0 | 4.92, d, 7.6 | 4.90, d, 8.0 | 4.90, d, 8.0 | |
| 2′′ | 4.12, overlap | 4.10, overlap | 4.10, overlap | 4.10, overlap | |
| 3′′ | 4.25, overlap | 4.25, overlap | 4.25, overlap | 4.25, overlap | |
| 4′′ | 4.35, overlap | 4.34, overlap | 4.33, overlap | 4.34, overlap | |
| 5′′ | 4.15, overlap | 4.16, overlap | 4.16, overlap | 4.17, overlap | |
| 6′′ | 4.19, overlap 4.07, overlap |
4.19, overlap 4.05, overlap |
4.18, overlap 4.06, overlap |
4.18, overlap 4.03, overlap |
|
| 1‴ | 5.85, br. s | 5.79, d, 1.2 | 5.78, br. s | 5.77, br. s | |
| 2‴ | 4.68, overlap | 4.64, overlap | 4.63, overlap | 4.62, overlap | |
| 3‴ | 4.54, dd, 8.4, 3.2 | 4.49, dd, 8.4, 3.2 | 4.48, dd, 8.4, 3.2 | 4.49, dd, 8.4, 3.2 | |
| 4‴ | 3.91, dd, 8.4, 8.4 | 3.89, dd, 8.4, 8.4 | 3.88, dd, 8.4, 8.4 | 3.88, dd, 8.4, 8.4 | |
| 5‴ | 4.93, overlap | 4.88, overlap | 4.92, overlap | 4.92, overlap | |
| 6‴ | 1.69, d, 6.0 | 1.67, d, 6.0 | 1.66, d, 6.0 | 1.64, d, 6.0 | |
| 1⁗ | - | 4.37, overlap | - | - | |
| 2⁗ | - | 2.01, overlap | - | - | |
| 3⁗ | - | 0.76, overlap | - | - | |
1H-NMR spectrum was measured in pyridine-d5 at 400 MHza).
Table 2.
13C-NMR data of acanthosessilioside G–O.
| No. | δCa) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| G (1) | H (2) | I (3) | J (5) | K (7) | L (8) | M (9) | N (10) | O (11) | |
| 1 | 35.1 | 35.8 | 34.7 | 30.9 | 34.8 | 87.1 | 87.3 | 87.2 | 69.7 |
| 2 | 29.7 | 29.9 | 28.6 | 30.1 | 29.9 | 41.8 | 38.7 | 41.9 | 37.6 |
| 3 | 173.6 | 175.2 | 174.3 | 178.5 | 174.8 | 173.4 | 173.0 | 173.4 | 175.2 |
| 4 | 148.1 | 148.5 | 147.9 | 148.6 | 148.0 | 79.3 | 79.2 | 79.2 | 148.4 |
| 5 | 50.6 | 50.8 | 50.3 | 50.3 | 50.4 | 55.9 | 56.1 | 56.1 | 51.4 |
| 6 | 27.4 | 27.1 | 26.6 | 25.9 | 28.9 | 17.9 | 19.2 | 18.8 | 31.0 |
| 7 | 33.5 | 33.4 | 32.9 | 34.7 | 33.0 | 35.3 | 35.4 | 35.4 | 33.8 |
| 8 | 40.1 | 40.0 | 39.5 | 39.5 | 39.6 | 42.8 | 42.9 | 42.9 | 42.4 |
| 9 | 41.6 | 41.6 | 41.1 | 41.0 | 41.2 | 48.8 | 48.9 | 49.0 | 49.5 |
| 10 | 43.7 | 43.7 | 43.2 | 43.2 | 43.2 | 43.9 | 47.0 | 47.0 | 45.9 |
| 11 | 22.3 | 22.3 | 21.7 | 21.6 | 64.2 | 22.8 | 67.6 | 67.7 | 64.2 |
| 12 | 25.4 | 25.5 | 24.9 | 24.9 | 25.9 | 37.2 | 37.0 | 37.3 | 36.7 |
| 13 | 38.9 | 38.7 | 38.2 | 38.4 | 38.3 | 38.5 | 37.5 | 38.5 | 37.4 |
| 14 | 41.2 | 41.3 | 40.8 | 40.8 | 40.8 | 42.7 | 42.8 | 42.8 | 43.0 |
| 15 | 30.2 | 30.1 | 29.6 | 28.6 | 31.0 | 29.8 | 30.4 | 29.8 | 32.6 |
| 16 | 26.3 | 26.4 | 25.9 | 32.2 | 26.6 | 32.5 | 31.1 | 32.6 | 32.2 |
| 17 | 63.3 | 63.6 | 63.1 | 57.0 | 63.1 | 49.6 | 57.0 | 58.1 | 57.1 |
| 18 | 44.6 | 44.7 | 44.2 | 49.8 | 44.3 | 47.4 | 49.5 | 49.6 | 49.7 |
| 19 | 48.3 | 48.0 | 47.5 | 47.3 | 47.6 | 46.9 | 47.3 | 47.4 | 47.8 |
| 20 | 152.1 | 151.7 | 151.2 | 150.8 | 151.2 | 150.8 | 150.5 | 150.9 | 150.4 |
| 21 | 42.6 | 42.4 | 41.9 | 32.9 | 41.9 | 30.6 | 30.9 | 30.2 | 41.7 |
| 22 | 76.0 | 75.7 | 76.4 | 36.8 | 76.5 | 36.8 | 36.7 | 76.4 | 76.5 |
| 23 | 114.4 | 114.0 | 113.6 | 113.7 | 113.7 | 24.8 | 24.9 | 24.9 | 113.9 |
| 24 | 23.9 | 24.1 | 23.4 | 23.4 | 23.4 | 32.4 | 32.6 | 32.5 | 23.7 |
| 25 | 20.7 | 21.0 | 20.3 | 20.3 | 20.4 | 18.5 | 18.8 | 19.2 | 20.9 |
| 26 | 16.8 | 16.8 | 16.3 | 16.3 | 16.3 | 17.9 | 17.9 | 17.9 | 17.4 |
| 27 | 15.3 | 15.3 | 14.8 | 14.7 | 14.9 | 15.1 | 15.2 | 15.2 | 14.8 |
| 28 | 179.0 | 177.0 | 174.8 | 174.9 | 174.0 | 174.7 | 174.9 | 174.7 | 175.0 |
| 29 | 19.7 | 19.7 | 19.2 | 19.5 | 19.3 | 19.2 | 19.6 | 19.3 | 19.6 |
| 30 | 110.9 | 111.0 | 110.5 | 110.0 | 110.6 | 110.9 | 110.2 | 110.7 | 110.0 |
| OCH3 | - | - | 51.3 | 51.3 | - | 51.1 | 49.6 | 51.0 | 51.1 |
| 1′ | 94.3 | 95.8 | 95.3 | 95.3 | 95.4 | 95.4 | 95.3 | 95.4 | 95.4 |
| 2′ | 83.6 | 74.5 | 73.9 | 74.0 | 74.0 | 73.9 | 74.0 | 73.9 | 74.0 |
| 3′ | 78.6 | 77.5 | 77.9 | 78.0 | 78.0 | 77.9 | 78.1 | 78.2 | 78.1 |
| 4′ | 71.1 | 71.4 | 70.3 | 70.3 | 71.0 | 70.8 | 71.0 | 71.0 | 71.1 |
| 5′ | 79.2 | 78.8 | 78.4 | 78.4 | 78.4 | 78.0 | 78.5 | 78.4 | 78.5 |
| 6′ | 62.5 | 70.0 | 69.5 | 69.5 | 69.6 | 69.5 | 69.5 | 69.6 | 69.6 |
| 1′′ | 107.1 | 105.6 | 105.0 | 105.1 | 105.1 | 105.1 | 105.1 | 105.1 | 105.1 |
| 2′′ | 76.8 | 75.4 | 74.0 | 74.1 | 74.1 | 74.0 | 74.1 | 74.1 | 74.1 |
| 3′′ | 78.5 | 78.4 | 77.0 | 77.0 | 77.1 | 77.1 | 77.1 | 77.1 | 77.2 |
| 4′′ | 71.8 | 76.9 | 76.4 | 76.5 | 76.5 | 76.3 | 76.5 | 76.4 | 76.5 |
| 5′′ | 79.7 | 79.1 | 78.6 | 78.7 | 78.7 | 78.6 | 78.7 | 78.7 | 78.7 |
| 6′′ | 62.4 | 61.8 | 61.3 | 61.4 | 61.4 | 61.1 | 61.4 | 61.4 | 61.4 |
| 1‴ | - | 103.2 | 102.6 | 102.7 | 102.7 | 102.6 | 102.7 | 102.7 | 102.8 |
| 2‴ | - | 72.9 | 72.5 | 72.5 | 72.5 | 72.5 | 72.5 | 72.5 | 72.5 |
| 3‴ | - | 73.1 | 72.7 | 72.7 | 72.7 | 72.7 | 72.7 | 72.7 | 72.8 |
| 4‴ | - | 74.4 | 75.2 | 75.3 | 75.2 | 75.2 | 75.3 | 75.2 | 75.3 |
| 5‴ | - | 70.7 | 70.9 | 71.0 | 70.3 | 70.2 | 70.3 | 70.3 | 70.3 |
| 6‴ | - | 18.9 | 18.4 | 18.5 | 18.4 | 18.5 | 18.4 | 18.4 | 18.5 |
| 1⁗ | - | - | - | - | 69.5 | - | 69.4 | - | - |
| 2⁗ | - | - | - | - | 19.4 | - | 19.3 | - | - |
| 3⁗ | - | - | - | - | 13.7 | - | 13.7 | - | - |
13C-NMR spectrum was measured in pyridine-d5 at 100 MHza).
Acanthosessilioside H [22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (2): White amorphous powder; IR (CaF2 window) 3356, 1715, 1609 cm−1; negative ESI-QTOF/MS m/z 955.5059 [M−H]− (calculated for C48H75O19, 955.4902); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Acanthosessilioside I [22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (3): White amorphous powder; IR (CaF2 window) 3354, 1751, 1639 cm−1; negative ESI-QTOF/MS m/z 969.5034 [M−H]− (calculated for C49H77O19, 969.5059); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Sessiloside (4): White amorphous powder; IR (CaF2 window) 3355, 1715, 1640 cm−1; negative ESI-QTOF/MS m/z 939.5123 [M−H]−; 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) data were consistent with those in [18].
Acanthosessilioside J [3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (5): White amorphous powder; IR (CaF2 window) 3349, 1710, 1648 cm−1; negative ESI-QTOF/MS m/z 953.5128 [M−H]− (calculated for C49H77O18, 953.5109); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Inermoside (6): White amorphous powder; IR (CaF2 window) 3351, 1725, 1677 cm−1; negative FAB-MS m/z 969 [M−H]−; 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) data were consistent with those in [19].
Acanthosessilioside K [(1R)-22α-propoxy-1α, 11α-dihydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3, 11-lactone 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (7): White amorphous powder; IR (CaF2 window) 3360, 1714, 1627 cm−1; negative ESI-QTOF/MS m/z 1011.5530 [M−H]− (calculated for C51H79O20, 1011.5165); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Acanthosessilioside L [(1R)-1,4-epoxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (8): White amorphous powder; IR (CaF2 window) 3360, 1720, 1615 cm−1; negative ESI-QTOF/MS m/z 969.5034 [M−H]− (calculated for C49H77O19, 969.5059); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Acanthosessilioside M [(1R)-1,4-epoxy-11α-propoxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (9): White amorphous powder; IR (CaF2 window) 3348, 1736, 1648 cm−1; negative ESI-QTOF/MS m/z 1027.5483 [M−H]− (calculated for C52H83O20, 1027.5478); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Acanthosessilioside N [(1R)-1,4-epoxy-11α, 22α-dihydroxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (10): White amorphous powder; IR (CaF2 window) 3351, 1715, 1640 cm−1; negative ESI-QTOF/MS m/z 1001.4948 [M−H]− (calculated for C49H77O21, 1001.4957); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
Acanthosessilioside O [1, 11α, 22α-trihydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose]] (11): White amorphous powder; IR (CaF2 window) 3356, 1750, 1615 cm−1; negative ESI-QTOF/MS m/z 1001.4948 [M−H]− (calculated for C49H77O21, 1001.1957); 1H-NMR (400 MHz, pyridine-d5, δH) and 13C-NMR (100 MHz, pyridine-d5, δC) refer to Table 1 and Table 2.
2.4. UPLC-QTOF/MS and UPLC-MS/MS Analyses
UPLC was performed using a Waters ACUITY I-CLASS UPLC (Waters Corp., Milford, MA, USA) with a Thermo Hypersil GOLD column (2.1 mm × 100 mm; 1.9 μm). The mobile phases were composed with Solvent A (water and 0.1% formic acid (v/v)) and Solvent B (acetonitrile and 0.1% formic acid (v/v)). The flow rate was 450 μL/min and the injection volume was 2 μL. The elution conditions were as follows: 0–4 min, B 10–30%; 4–15 min, B 30–60%; 15–16 min, B 60–100%; 16–18 min, B 100–10. The column oven and sample tray were maintained at 40 ℃ and 4 ℃, respectively. MS analysis was performed using a Waters Xevo G2-S QTOF MS (Waters Corp., Milford, MA, USA) operating in negative ion mode. Accurate mass measurements were obtained by an automated calibration delivery system that contained an internal reference (Leucine-enkephalin, m/z 554.262 (ESI-)). The operating parameters were set by modifying those from the preceding research [20]. UPLC with tandem mass spectrometry (MS/MS) was carried out for the quantitative analysis of compounds in the A. sessiliflorus fruits. Mass spectrometric detection was carried out using 3200 QTRAP mass spectrometer (AB SCIEX, Framingham, MA, USA) in multiple reaction monitoring (MRM) mode. Precursor ions of the compounds were selected. They and product ions were generated by applying collision energies to selected precursor ions, and product ions were used for quantification. The optimal operating parameters were set as shown in Table 3.
Table 3.
LC–MS/MS MRM conditions for the simultaneous quantification of acanthosessilioside G–O.
| Acanthosessiliosides (No.) | MRM a | Time b | DP c | EP d | CEP e | CE f | CXP g |
|---|---|---|---|---|---|---|---|
| G (1) | 809.364/467.4 | 150 | –55 | –10.5 | –36 | –46 | –6 |
| H (2) | 955.436/485.4 | 150 | –60 | –11 | –42 | –50 | –6 |
| I (3) | 969.464/499.4 | 150 | –70 | –10 | –26 | –48 | –6 |
| J (5) | 953.473/483.3 | 150 | –75 | –10 | –42 | –56 | –8 |
| K (7) | 1011.509/541.5 | 150 | –110 | –10.5 | –28 | –44 | –6 |
| L (8) | 969.431/499.4 | 150 | –75 | –9.5 | –38 | –52 | –6 |
| M (9) | 1027.45/557.4 | 150 | –75 | –10.5 | –42 | –54 | –8 |
| N (10) | 1001.44/531.4 | 150 | –70 | –8.5 | –26 | –46 | –6 |
| O (11) | 1001.45/531.5 | 150 | –70 | –12 | –46 | –50 | –6 |
a MRM: multiple reaction monitoring. b Mass scan time. c DP: declustering potential. d EP: entrance potential. e CEP: collision cell entrance potential. f CE: collision energy. g CXP: collision cell exit potential.
2.5. Cell Culture and Viability Assays
BV2 and RAW264.7 cells were incubated at 5 × 105 cells/mL in RPMI1640 containing 1% antibiotic (Penicillin-Streptomycin) and 10% heat-inactivated FBS at 37 °C in a humidified 5% CO2 and a 95% air atmosphere. These cells were incubated according to a previously described method [21,22].
2.6. Determination of Nitrite
As an indicator of nitrite oxide (NO) production in cells, the production of nitrite, a stable end-product of NO oxidation, was measured. Briefly, the concentration of nitrite in the conditioned media was determined by a method based on the Griess reaction [23]. The details of the assay have been described previously [21].
2.7. PGE2 Assay
The concentration of PGE2 in each sample was measured with the use of a commercially available kit from R&D Systems, Inc (Minneapolis, MN, USA), according to a method described previously [21]. Briefly, BV2 and RAW264.7 cells treated with compounds were cultured in 48-well plates, followed by a pre-incubation with different concentrations of compounds for 3 h. Subsequently, stimulation was performed for 24 h with LPS stimulation (1 μg/mL). The resulting cell culture supernatants were collected and centrifuged at 13,000× g for 2 min to remove particulate matter. At the end of the procedure, the samples were added to a 96-well plate pre-coated with polyclonal antibodies specific to PGE2. Enzyme-linked polyclonal antibodies were added to the wells and allowed to react for 20 h, followed by a final washing step to remove any unbound antibody-enzyme reagent. A substrate solution was added, after which the intensity of the color produced was measured at 450 nm (a correction wavelength set at 540 or 570 nm), which was proportional to the amount of PGE2 present.
2.8. Assays for IL-1β, IL-6, and TNF-α
The culture medium was collected to determine the levels of IL-β, IL-6, and TNF-α present in each sample using ELISA kits tailored to each collection process (R&D Systems, Inc.), as per the manufacturer’s instructions. Briefly, BV2 and RAW264.7 cells were seeded in 48-well culture plates at a density of 5 × 105 cells/well. After incubation, the supernatant was collected and used in the cytokine ELISA kits for measuring the concentrations of IL-1β, IL-6, and TNF-α.
2.9. Western Blotting Analysis
The pelleted BV2 and RAW264.7 cells were washed with PBS and then lysed in RIPA buffer. Equal amounts of proteins quantified by Protein Assay Dye Reagent Concentrate obtained from Bio-Rad Laboratories (#5000006; Hercules, CA, USA), mixed in the sample loading buffer and separated by SDS-PAGE. The separated proteins were transferred to a nitrocellulose membrane. Non-specific binding to the membrane was blocked by incubation in a solution of skimmed milk. The membrane was incubated with primary antibodies at 4 °C overnight and then reacted with a horseradish peroxidase-conjugated secondary antibody from Millipore.
2.10. Statistical Analysis
Data are presented as the mean ± standard deviation of three independent experiments. A one-way analysis of variance, followed by Dunnett’s comparison tests, was used to compare the two groups. Statistical analyses were performed using GraphPad Prism software, Version 5.01 (GraphPad Software Inc., San Diego, CA, USA).
3. Results and Discussion
3.1. Chemical Structure Elucidation of Compounds 1–11
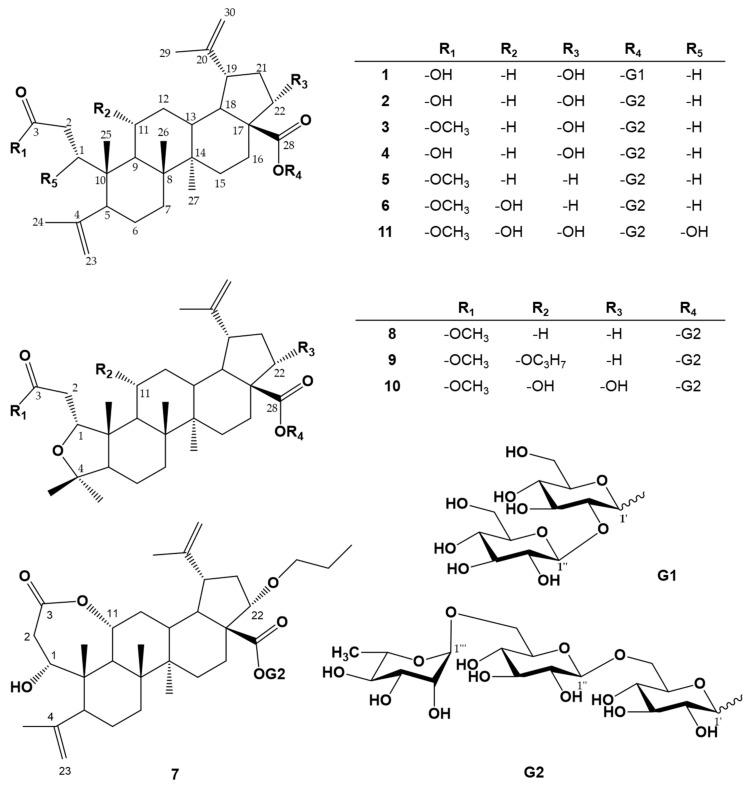
A 70% ethanolic extract of dried A. sessiliflorus fruits was suspended in H2O and extracted successively with EtOAc and n-BuOH. The EtOAc- and n-BuOH-soluble fractions were concentrated under reduced pressure to produce a residue that was subjected to multiple chromatographic steps, using Diaion HP-20, Sephadex LH-20, silica gel, and reversed-phase C18 silica gel, yielding compounds 1–11. Comparing the 1D- and 2D-NMR and QTOF/MS data with reported values allowed us to identify known compounds sessiloside (4) [18] and inermoside (6) [19]. The other nine noble compounds are newly reported here (Figure 1).
Figure 1.
Chemical structures of compounds 1–11 from the fruits of Acanthopanax sessiliflorus.
The infrared spectrum (cm−1) of compounds 1–3, 5, and 7–11 suggested the presence of a double bond (1665, 1640), a carbonyl (1746, 1710), and a hydroxyl (3462, 3342). Compound 1 was obtained as a white, amorphous powder. The molecular formula was determined to be C42H66O15 from the pseudomolecular ion peak [M−H]− at m/z 809.4448 (calculated for C42H65O15, 809.4323) in the negative HR-QTOF/MS. In the 1H NMR spectrum (Table 1), the observation of signals for two allyl methyl protons (chemical shift, coupling pattern, J in Hz, proton number; δH 1.71, H-24; δH 2.13, H-29) located at sp2 carbons and four olefinic methine protons (δH 5.12, d, 2.0, H-23a, 4.83, 2.0, H-23b; δH 4.90, overlap, H-30a, 4.79, overlap, H-30b), of which the chemical shifts and small coupling constants (or broad singlet) were typical of two exomethylene units, confirmed the presence of two isopropenyl moieties. In addition, signals for three tertiary methyl protons (δH 1.21, s, H-26; δH 1.12, s, H-25, δH 0.80, s, H-27) and an oxygenated methine proton (δH 4.80, overlap, H-22) were observed. The 1H NMR signals of the two sugars included two hemiacetals (δH 6.37, d, 8.0, H-1′; δH 5.35, d, 8.0, H-1′′), eight oxygenated methines (δH 4.00, overlap, H-5″; δH 4.00, overlap, H-5′; δH 4.07, overlap, H-2″; δH 4.27, overlap, H-3″; δH 4.30, overlap, H-4″; δH 4.30, overlap, H-4′; δH 4.30, overlap, H-2′; δH 4.37, overlap, H-3′), and two oxygenated methylenes (δH 4.34, overlap, H-6″b, 4.42, 1H, br. d, 12.0, H-6″a; δH 4.46, overlap, H-6′b, 4.58, 1H, br. d, 12.0, H-6′a). The 13C NMR spectrum of 1 (Table 2) supported by DEPT experiments indicated the presence of 30 carbons including two carbonyl carbons (δC 173.6, C-3; δC 179.0, C-28), two olefinic quaternary carbons (δC 152.1, C-20; δC 148.1, C-4), two exomethylene carbons (δC 114.4, C-23; δC 110.9, C-30), an oxygenated methine carbon (δC 76.0, C-22), five methyl carbons (δC 23.6, C-24; δC 20.5, C-25; δC 19.5, C-29; δC 16.5, C-26; δC 15.0, C-27), and 18 other carbon signals. The chemical shifts in the 13C NMR signals due to two hemiacetals (δC 94.3, C-1″; δC 107.1, C-1′), eight oxygenated methines (δC 71.1, C-4′; δC 71.8, C-4′′; δC 76.8, C-2″; δC 78.5, C-3″; δC 78.6, C-3′; δC 79.2, C-5′; δC 79.7, C-5″; δC 83.6, C-2′), and two oxygenated methylenes (δC 62.5, C-6′; δC 62.4, C-6″) and the coupling constant of the anomer proton signal (J = 8.0 Hz) revealed that the two sugars were β-glucopyranosyl-(1→2)-β-glucopyranose. The oxygenated methine carbon signal (C-2′) was down-shifted at δC 83.6 owing to the glycosidation effect from their usual detection at δC 74.5 in β-d-glucopyranose [16]. In addition, the anomer carbon signal (C-1′) was up-shifted at δC 94.3 owing to the esterification effect from their usual detection at δC 105.0 in β-d-glucopyranose [8]. Several cross-peaks in the 1H−1H COSY spectrum confirmed a key connection among the proton signals. The HMBC spectrum showed crucial long-range correlations between H-23a, H-23b/C-5; H-2a, H-2b/C-3; and H-1/C-3. Therefore, 1 was assigned as a 3,4-secolupane-type triterpenoid. In addition, two anomer proton signals (δH 6.33, H-1′; δH 4.94, H-1″) showed cross peaks with the oxygenated olefin quaternary carbon signal (δC 179.0, C-28) and the one oxygenated methine (δC 83.6, C-2′). On further analysis of the HSQC and DEPT 135 spectra of 1, the assignments of the proton and carbon NMR signals (Table 1 and Table 2) were confirmed unambiguously. The coupling constant between H-22 and H-21 was 4.4 Hz, indicating that OH-22 is α-oriented. Therefore, the structure of compound 1 was determined as 22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 28-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranose, which has not been previously reported. This compound was named acanthosessilioside G (Figure S1).
Compound 2 was obtained as a white, amorphous powder. The molecular formula was determined to be C48H76O19 from the pseudomolecular ion peak [M−H]− at m/z 955.5059 (calculated for C48H75O19, 955.4902) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of compound 2 were similar to those of compound 1, except for signals indicating an additional monosaccharide moiety. The 1H NMR signals of the monosaccharide included a hemiacetal (δH 5.83, br. s, H-1‴), four oxygenated methines (δH 3.93, dd, 8.4, 8.4, H-4‴; δH 4.53, dd, 8.4, 3.2, H-3‴; δH 4.66, overlap, H-2‴; δH 4.93, overlap, H-5‴), and one methyl (δH 1.71, d, 6.0, H-6‴). The chemical shifts in the 13C NMR signals due to a hemiacetal (δC 103.2, C-1‴), four oxygenated methines (δC 70.7, C-5″; δC 72.9, C-2‴; δC 73.1, C-3‴; δC 74.4, C-4‴), and one methyl (δC 18.9, C-6‴) and the coupling constant of the anomer proton signal (br. s) revealed that the monosaccharide was a α-rhamnopyranose. In the gHMBC spectrum, three anomer proton signals (δH 6.38, H-1′; δH 5.70, H-1‴; δH 4.94, H-1″) showed cross peaks with the ester carbon signal (δC 177.0, C-28), the oxygenated methylene carbon signal (δC 70.0, C-6′), and the oxygenated methine carbon signal (δC 76.9, C-4′′), respectively, which revealed that the three sugars were α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose. Consequently, the structure of compound 2 was determined as 22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside H (Figure S2).
Compound 3 was obtained as a white, amorphous powder. The molecular formula was determined to be C49H78O19 from the pseudomolecular ion peak [M−H]− at m/z 969.5034 (calculated for C49H77O19, 969.5059) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of compound 3 were similar to those of compound 2, except for signals indicating an additional one methoxy group signal (δH 3.63, s; δC 51.3, 3-OCH3). In the gHMBC spectrum, the methoxy proton signal (δH 3.63) displayed cross peaks with the ester carbon signal (δC 174.3, C-3), suggesting that the methoxy groups were at the C-3 positions. When compared with a previously reported compound, acanthosessiligenins I [5], the structure of compound 3 was determined as 22α-hydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside I (Figure S3).
Compound 5 was obtained as a white, amorphous powder. The molecular formula was determined to be C49H78O18 from the pseudomolecular ion peak [M−H]− at m/z 953.5128 (calculated for C49H77O18, 953.5109) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of Compound 5 were similar to those of Compound 3, except for signals indicating an exceptional one hydroxy group signal (δH 1.73, overlap, H-22a, 1.36, overlap, H-22b; δC 36.8, C-22). When compared with a previously reported compound, acanthosessiliosides A [8], the structure of compound 5 was determined as 3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside J (Figure S4).
Compound 7 was obtained as a white, amorphous powder. The molecular formula was determined to be C51H80O20 from the pseudomolecular ion peak [M−H]− at m/z 1011.5530 (calculated for C51H79O20, 1011.5165) in the negative HR-QTOF/MS. Its 1H NMR and 13C NMR spectra (Table 1 and Table 2) were similar to those of 22α-hydroxychiisanoside [12], except for signals indicating an additional propoxy group signal (δH 4.80, overlap, H-22; δC 67.7, C-11; δH 4.42, overlap, H-1⁗; δC 69.5, C-1⁗; δH 2.05, overlap, H-2⁗; δC 19.4, C-2⁗; δH 0.79, overlap, H-3⁗; δC 13.7, C-3⁗). The sugar moieties were similar to those of the sugars in compound 5. In the gHMBC spectrum, the oxygenated methine proton signal (δH 4.80, H-22) displayed a cross peak with one ester carbon signals (δC 174.0, C-28) and one oxygenated methylene carbon signal (δC 69.5, C-1⁗), suggesting that the oxygenated methine group was at the C-22 positions and attached with a propoxy group. Consequently, the structure of compound 7 was determined as (1R)-22α-propoxy-1α, 11α-dihydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3,11-lactone 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside K (Figure S5).
Compound 8 was obtained as a white, amorphous powder. The molecular formula was determined to be C49H78O19 from the pseudomolecular ion peak [M−H]− at m/z 969.5034 (calculated for C49H77O19, 969.5059) in the negative HR-QTOF/MS. The 1H and 13C NMR spectra (Table 1 and Table 2) exhibited signals for five tertiary methyl groups (δH 1.10, 1.21, 1.24, 1.32, 1.37), an allyl methyl group (δH 1.93), an exomethlyene group (δH 4.91/4.66) due to an isopropenyl moiety, one oxygenated methine group (δH 4.86, dd, 12.0, 2.4, H-1; δC 86.6, C-1), a methoxy group (δH 3.60, s; δC 51.1, 3-OCH3), and two carbonyl groups (δC 173.4, C-3; δC 174.7, C-28). When compared with a previously reported compound, acanthosessiligenin II [8], 8 was found to lack a hydroxyl group at the C-22 position but to possess an additional α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose group. In the gHMBC spectrum, three anomer proton signals (δH 6.37, H-1′; δH 5.85, H-1‴; δH 4.92, H-1″) showed cross peaks with the ester carbon signal (δC 174.7, C-28), the oxygenated methylene carbon signal (δC 69.5, C-6′), and the oxygenated methine carbon signal (δC 76.3, C-4′′), respectively. In addition, the long-range correlations between the methoxy protons signal (δH 3.60) and the carbonyl carbon signal at C-3 (δC 173.4) revealed that the methoxy group is affixed to C-3. Consequently, the structure of compound 8 was determined as (1R)-1,4-epoxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside L (Figure S6).
Compound 9 was obtained as a white, amorphous powder. The molecular formula was determined to be C52H84O20 from the pseudomolecular ion peak [M−H]− at m/z 1027.5483 (calculated for C52H83O20, 1027.5478) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of compound 9 were similar to those of compound 8, except for signals indicating an additional one propoxy group signals (δH 4.07, overlap, H-11; δC 67.7, C-11; δH 4.37, overlap, H-1⁗; δC 69.4, C-1⁗; δH 2.01, overlap, H-2⁗; δC 19.3, C-2⁗; δH 0.76, overlap, H-3⁗; δC 13.7, C-3⁗). In the gHMBC spectrum, one oxygenated methine proton signals (δH 4.07, H-11) displayed cross peaks with two methyl carbon signals (δC 18.8, C-25; δC 17.9, C-26) and one oxygenated methylene carbon signal (δC 69.4, C-1⁗), suggesting that one oxygenated methine groups were at the C-11 position and attached with the propoxy group. Consequently, the structure of compound 9 was determined as (1R)-1,4-epoxy-11α-propoxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside M (Figure S7).
Compound 10 was obtained as a white, amorphous powder. The molecular formula was determined to be C49H78O21 from the pseudomolecular ion peak [M−H]− at m/z 1001.4948 (calculated for C49H77O21, 1001.4957) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of compound 10 were similar to those of compound 8, except for signals indicating two additional hydroxy group signals (δH 4.07, overlap, H-11; δC 67.7, C-11; δH 4.80, overlap, H-22; δC 76.4, C-22). In the gHMBC spectrum, two oxygenated methine proton signals (δH 4.07, H-11; δH 4.80, H-22) displayed cross peaks with two methyl carbon signals (δC 20.4, C-25; δC 16.3, C-26) and one ester carbon signal (δC 174.7, C-28), suggesting that two oxygenated methine groups were at the C-11 and C-22 positions. Consequently, the structure of compound 10 was determined as (1R)-1,4-epoxy-11α, 22α-dihydroxy-3,4-seco-lup-20(30)-ene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside N (Figure S8).
Compound 11 was obtained as a white, amorphous powder. The molecular formula was determined to be C49H78O21 from the pseudomolecular ion peak [M−H]− at m/z 1001.4948 (calculated for C49H77O21, 1001.4957) in the negative HR-QTOF/MS. The 1H NMR and 13C NMR of compound 11 were similar to those of compound 7, except for signals indicating an additional one hydroxy group signal (δH 4.86, overlap, H-1; δC 69.7, C-1) and one methoxy group signal (δH 3.60, s; δC 51.1, 3-OCH3). In the gHMBC spectrum, the oxygenated methine proton signal (δH 4.86, H-1) displayed cross peaks with one methyl carbon signal (δC 20.9, C-25) and one quaternary carbon signal (δC 45.9, C-10), suggesting that one oxygenated methine group was at the C-1 position. In addition, the long-range correlations between the methoxy protons signal (δH 3.60) and the carbonyl carbon signal at C-3 (δC 175.2) revealed that the methoxy group is affixed to C-3. Consequently, the structure of compound 11 was determined as 1,11α,22α-trihydroxy-3,4-seco-lupa-4(23),20(30)-diene-3,28-dioic acid 3-methyl ester 28-[α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-glucopyranose], which has not been previously reported. This compound was named acanthosessilioside O (Figure S9).
3.2. Analyses of Compounds 1–11 in ASFE by UPLC-QTOF/MS and Tandem Mass Spectrometry (MS/MS)
Compounds 1–11 were analyzed using ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-QTOF/MS). The extract of A. sessiliflorus fruits (ASFEx) was also subjected to UPLC-QTOF/MS. Four compounds, namely compound 1–3 and 5, were determined in the extract but the intensities in the extract were too low. As a result of this, fractionation was carried out. A typical base peak intensity (BPI) chromatogram of the extract and the n-butanol fraction (AFB) are shown in Figure 2. After fractionation, eight compounds, 1, 3–7, 9, and 10, were detected in the UPLC chromatogram of the AFB. However, the intensities of compounds were low in the n-butanol fraction for quantitative analysis using UPLC-QTOF/MS. Hence, QTRAP® tandem mass spectrometry (MS/MS) was used for quantifying compounds 1–11. The precursor ions of noble compounds, acanthosessilioside G–O were selected under optimized multiple reaction monitoring (MRM) conditions, and collision energy was applied to the selected precursor ions. After that, the product ions with the highest intensities were selected for quantitative analysis (Table 4). The linear calibration curves were based on regression analysis of the values measured with various concentration of compounds 1–11. The values of the calibration plots are shown in Table 5. The result of quantitative analysis was measured in the range of 0.01–2.806 mg/g.
Figure 2.
Representative negative ion mode mass chromatogram of (A) ASFEx, (B) AFB, and (C) isolated compounds (1–11) based on a UPLC(ESI)-QTOF-MS.
Table 4.
UPLC–MS/MS data-negative ion mode for acanthosessilioside G–O identified in ASFEx.
| Acanthosessilioside | Molecular Formula | MW | Measured Value a [M–H] |
MS/MS Fragmentation a |
|---|---|---|---|---|
| G | C42H66O15 | 810.44 | 809.2 | 647.2 [M–H–Glc]–; 587.2 [M–Glu–C2H3O2]–; 485.2 [M–H–Glc–Glc]–; 467.4 [M–H–Glc–Glc–H2O]–; 423.0 [M–H–Glc–Glc–HCOO–H2O]–; 405.4 [M–H–Glc–Glc–HCOO– 2H2O]–; 179.2 [M–H–Glu–3,4-seco-Triterpenoid–C2H3O2–2C3H5–H2O–HCOO]–; 161.2[M–H–3,4-seco-Triterpenoid–Glu–C2H3O2–2C3H5–2H2O-HCOO]– |
| H | C48H76O19 | 956.50 | 955.6 | 485.4 [M–H–Rha–Glc–Glc]–; 469.4 [M–H–Rha–Glc–Glc–H2O]–; 439.4 [M–H–Rha–Glc–Glc–HCOO]–; 423.4 [M–H–Rha–Glc–Glc–HCOO–H2O]–; 405.2 [M–H–Rha–Glc–Glc–HCOO–2H2O]–; 367.0 [M–H–Rha–Glc–Glc–HCOO–H2O–2C3H5]–; 325.0 [M–H–Rha-3,4-seco-Triterpenoid–C2H3O2–H2O–2C3H5–HCOO]–; 161.2 [M–H–Rha–Glu–3,4-seco-Triterpenoid – C2H3O2–H2O–2C3H5–HCOO]– |
| I | C49H78O19 | 970.51 | 969.6 | 808.0 [M–H–Rha]–; 499.4 [M–H–Rha–Glc–Glc]–; 469.2 [M–H–3,4-seco-Triterpenoid –C3H5O2–2C3H5–H2O–HCOO]–; 455.0 [M–H–Rha–Glc–Glc–HCOO]–; 365.4 [M–H–Rha–Glc–Glc–HCOO–C3H5O2–H2O]–; 325.0 [M–H–Rha–3,4-seco-Triterpenoid–C3H5O2–2C3H5–H2O–HCOO]–; 160.6 [M–H–Rha–Glu–3,4-seco-Triterpenoid–C3H5O2–2C3H5–2H2O–HCOO]– |
| J | C49H78O18 | 954.52 | 953.6 | 483.2 [M–H–Rha–Glc–Glc]–; 469.2 [M–H–Rha–Glc–Glc–H2O]–; 409.2 [M–H–Rha–Glc–Glc–HCOO–C3H6O2]–; 367.2 [M–H–Rha–Glc–Glc–HCOO–C3H6O2–C3H5]–; 323.4 [M–H–Rha–Glc–Glc–HCOO–C3H6O2–C3H5–HCOO]-; 160.8 [M–H–Rha–Glu–3,4-seco-Triterpenoid–C3H5O2–2C3H5–HCOO–H2O]– |
| K | C51H80O20 | 1012.52 | 1011.5 | 849.6 [M–H–Rha–H2O]–; 541.4 [M–H–Rha–Glc–Glc]–; 479.6 [M–H–Rha–Glu–Glu–HCOO–H2O]–; 469.2 [M–H–3,4-seco-Triterpenoid–HCOO]–; 454.8 [M–H–Rha–Glu–Glu–HCOO–C3H5]–; 405.4 [M–H–Rha–Glu–Glu–C2H2O2–C3H7O–H2O]–; 364.8 [M–H–Rha–Glu–Glu–C2H2O2–C3H7O–H2O–C3H5]–; 325.0 [M–H–Rha–Glu–Glu–C2H2O2–C3H7O–H2O–2C3H5]–; 160.8 [M–H–Rha–Glu–3,4-seco-Triterpenoid–C2H2O2–2H2O–2C3H5–C3H7O–HCOO]– |
| L | C49H78O19 | 970.51 | 969.6 | 499.4 [M–H–Rha–Glc–Glc]–; 469.2 [M–H–3,4-seco-Triterpenoid–HCOO]–; 323.0[M–H–Rha–3,4-seco-Triterpenoid–C3H5O2–C3H5–HCOO]–;160.8[M–H–Rha–Glu–3,4-seco-Triterpenoid–C3H5O2–C3H5–HCOO]– |
| M | C52H84O20 | 1028.56 | 1027.6 | 557.4 [M–H–Rha–Glc–Glc]–; 468.8 [M–H–3,4-seco-Triterpenoid]–; 367.2 [M–H–Rha–Glc–Glc–C3H6O2–C3H8O–C3H6]–; 323.2 [M–H–Rha–3,4-seco-Triterpenoid–C3H5O2–C3H7O–C3H5–H2O–HCOO]–; 161.2 [M–H–Rha–Glu–3,4-seco-Triterpenoid–C3H5O2–C3H7O–C3H5–H2O–HCOO]- |
| N | C49H78O21 | 1002.50 | 1001.6 | 531.2 [M–H–Rha–Glc–Glc]–; 486.8 [M–H–Rha–Glc–Glc–HCOO]–; 469.2[M–H–Rha–Glu–Glu–HCOO–2H2O]–; 437.2 [M–H–Rha–Glu–Glu–2H2O–C2H3O2]–; 393.4 [M–H–Rha–Glu–Glu–HCOO–H2O–C2H3O2]–; 379.4 [M–H–Rha–Glu–Glu–HCOO–2H2O–C3H5O2]–; 367.4 [M–H–Rha–3,4-seco-Triterpenoid–2H2O–C3H5O2]–; 325.2 [M–H–3,4-seco-Triterpenoid–2H2O–C3H5O2–HCOO–Rha]–; 161.0[M–H–Rha–Glu–3,4-seco-Triterpenoid–2H2O–C3H5O2–HCOO]– |
| O | C49H78O21 | 1002.50 | 1001.6 | 531.4 [M–H–Rha–Glc–Glc]–; 469.0 [M–H–3,4-seco-Triterpenoid–C3H5O2–2H2O–2C3H5]–; 437.4 [M–H–Rha–Glc–Glc–C2H3O2–2H2O]–; 379.2 [M–H–Rha–Glu–Glu–C3H5O2–2H2O–HCOO]–; 367.2[M–H–Rha–3,4-seco-Triterpenoid–C3H5O2–3H2O–2C3H5]–; 325.2[M–H–Rha–3,4-seco-Triterpenoid–C3H5O2–3H2O–2C3H5–HCOO]−; 161.2[M–H–Rha–Glu–3,4-seco-Triterpenoid–C3H5O2–4H2O–2C3H5–HCOO]– |
a Mass accuracy < 5 ppm.
Table 5.
Quantification, linear range, regression equation, coefficient of determination LOD and LOQ for LC–MS/MS MRM analysis of compounds 1–11 a.
| Compounds | Rt b (min) |
Calibration Curve c | R 2 d | Linear Range (μg/mL) | LOD e (ppm) |
LOQ f (ppm) |
Amount (mg/g) |
|---|---|---|---|---|---|---|---|
| 1 | 6.20 | y = 27.8x − 952 | 0.9988 | 0.1–2.5 | 0.0117 | 0.0357 | 0.010 |
| 2 | 6.20 | y = 161x − 2.31 × 104 | 0.9974 | 0.1–2.5 | 0.0028 | 0.0086 | 2.806 |
| 3 | 9.68 | y = 146x − 5.45 × 103 | 0.9982 | 0.1–2.5 | 0.0025 | 0.0077 | 0.059 |
| 4 | 7.77 | y = 198x − 1.46 × 104 | 0.9989 | 0.1–2.5 | 0.0014 | 0.0044 | 0.43 |
| 5 | 11.49 | y = 203x − 1.99 × 104 | 0.9987 | 0.1–2.5 | 0.0016 | 0.0049 | 0.026 |
| 6 | 10.12 | y = 231x − 1.63e × 104 | 0.9954 | 0.1–2.5 | 0.0026 | 0.008 | 0.009 |
| 7 | 12.75 | y = 3.21x − 16.2 | 0.9998 | 0.05–2.5 | 0.0263 | 0.0799 | 0.023 |
| 8 | 9.19 | y = 41.7x − 1.64 × 103 | 0.9888 | 0.1–2.5 | 0.0237 | 0.0718 | 0.102 |
| 9 | 10.15 | y = 97.5x − 1.02 × 104 | 0.9965 | 0.1–2.5 | 0.0054 | 0.0165 | 0.017 |
| 10 | 4.67 | y = 65.1x − 6.4 × 103 | 0.9988 | 0.1–2.5 | 0.0048 | 0.0146 | 0.031 |
| 11 | 4.67 | y = 91.9x − 1.38 × 104 | 0.9958 | 0.1–2.5 | 0.0063 | 0.0193 | 0.016 |
a Mean values of samples (n = 3). b Rt: retention time. c y: logarithmic value of peak area; x: logarithmic value of amount injected. d R2: linearity. e LOD: limit of detection. f LOQ: limit of quantitation.
3.3. Effects of Compounds 1–11 on Cell Viability and Nitrite Contents in BV2 and RAW264.7 Cells
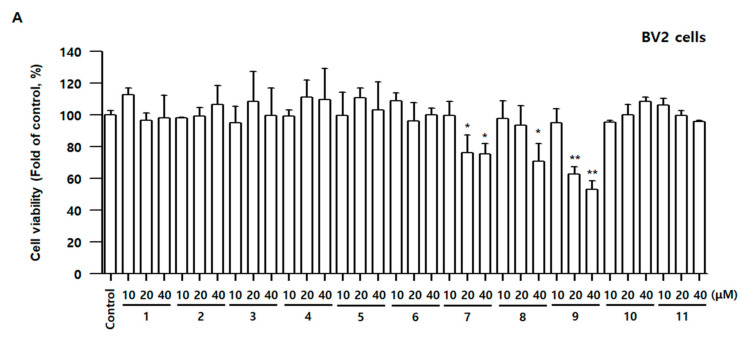
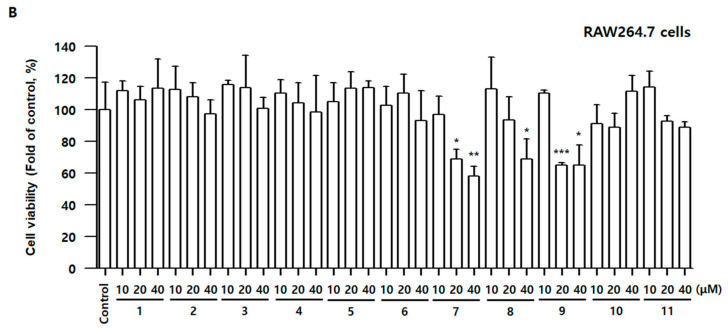
To examine whether compounds 1–11 have cell toxicity, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was conducted in BV2 and RAW264.7 cells. The cells were incubated for 48 h with various concentrations of compounds 1–11. The cell viability did not alter following the addition of 40 μM concentrations of compounds 1–6, 10, and 11 for 48 h; however, compound 7 at 20 μM, Compound 8 at 40 μM, and compound 9 at 20 μM demonstrated cell toxicity in BV2 and RAW264.7 cells. Therefore, adequate concentrations of compounds 1–11 in subsequent experiments were determined based on the results of Figure 3.
Figure 3.
Effects of compounds 1–11 on cell viability in BV2 (A) and RAW264.7 (B) cells. The cells were incubated for 48 h with various concentrations of compounds 1–11. Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Bars represent means ± standard deviation of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 compared with control group.
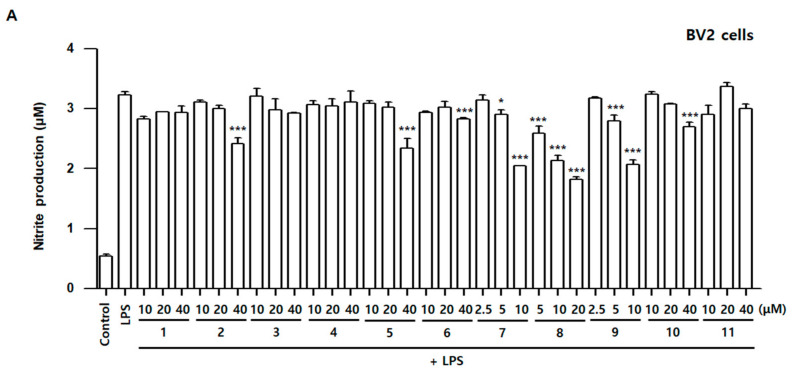
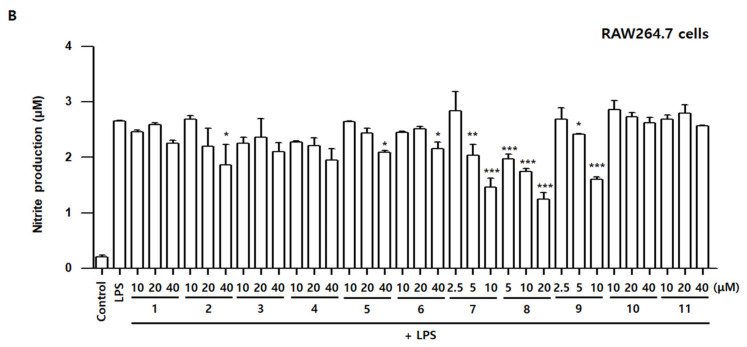
To assess the anti-inflammatory and neuroinflammatory effects of compounds 1–11 in LPS-stimulated BV2 and RAW264.7 cells, nitrite concentration was measured using the Griess reagents. Over-production of iNOS-derived NO may promote the formation of RNS, aggravate the inflammatory response, and can even lead to neuronal cell death. LPS stimulates macrophage or/and microglia and advances the production of ROS [24]. Therefore, reducing ROS production in microglia and macrophage cells might be an effective strategy to protect against inflammatory damage. In other previous studies, LPS treatment significantly increased ROS production [25]. LPS-treated groups significantly increased nitrite concentration compared with the control group in both BV2 and RAW264.7 cells. In BV2 cells, the increased nitrite concentration was significantly inhibited by compounds 2, 5, 6, 7, 8, 9, and 10. In RAW264.7 cells, the increased nitrite concentration was significantly inhibited by compounds 2, 5, 6, 7, 8, and 9 (Figure 4). Among them, compounds 7, 8, and 9 were the most effective in inhibiting nitrite production in both BV2 and RAW264.7 cells. These results showed that, among the eleven compounds, compounds 7, 8, and 9 are suitable for further experiments aimed at determining the biological mechanism.
Figure 4.
Effects of Compounds 1–11 on nitrite contents in LPS-stimulated BV2 (A) and RAW264.7 (B) cells. Cells were pretreated for 3 h with indicated concentrations of compounds 1–11 and stimulated for 24 h with LPS (1 μg/mL). Nitrite concentration was performed as described in Materials and Methods. Bars represent means ± standard deviation of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 compared with the LPS-treated group.
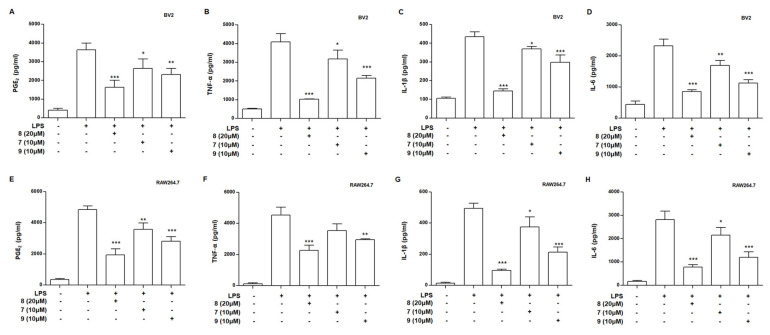
3.4. Effects of Compounds 7–9 on Levels of Prostaglandin E2 (PGE2), Tumor Necrosis Factor (TNF)-α, Interleukin (IL)-1β, and Interleukin (IL)-6 in LPS-Stimulated BV2 and RAW264.7 Cells
Macrophages and microglia play an important role in maintaining homeostasis of our body, they can be activated by many stimulates, such as ROS, external stimuli, and pro-inflammatory mediators. Cytokines have a complex regulatory action on inflammatory and immune responses. Pro-inflammatory cytokines, which contain both IL-6 and TNF-α, are mainly produced by various types of activated immune cells and are associated with all inflammatory properties of inflammatory diseases [26]. Therefore, we assessed the inhibitory effects of compounds 7–9 on the LPS-induced production of NO, PGE2, TNF-α, IL-1β, and IL-6. BV2 and RAW264.7 cells were incubated with compounds 7–9 for 3 h and then stimulated with LPS for 24 h. The production of PGE2, TNF-α, IL-1β, and IL-6 was significantly inhibited by compounds 7–9 in BV2 cells (Figure 5A–D). Figure 5E–H show that compounds 7–9 also significantly repressed PGE2, TNF-α, IL-1β, and IL-6 in BV2 cells. These results suggest that compounds 7–9 exert anti-neuroinflammatory and anti-inflammatory effects by modulating inflammatory mediators and cytokines.
Figure 5.
Effects of compounds 7–9 on levels of PGE2 (A,E), tumor necrosis TNF-α (B,F), IL-1β (C,G), and IL-6 (D,H) in lipopolysaccharide (LPS)-stimulated BV2 and RAW264.7 cells. Cells were pretreated for 3 h with indicated concentrations of compounds 7–9 and stimulated for 24 h with LPS (1 μg/mL). PGE2 assay and ELISA analysis were performed as described in Materials and Methods. Bars represent means ± standard deviation of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 compared with LPS-treated group.
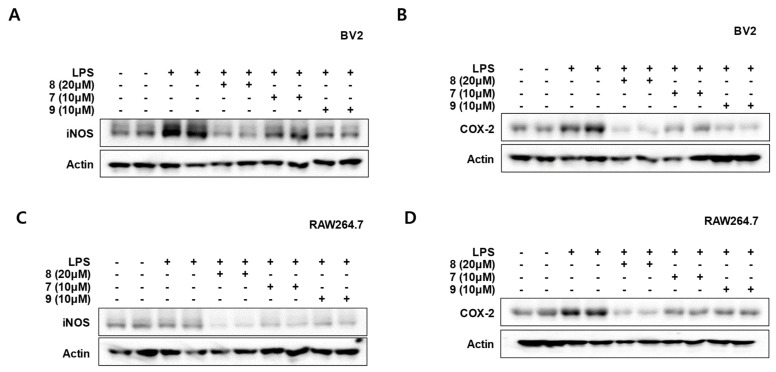
3.5. Effects of Compounds 7–9 on the Protein Expression Levels of Inducible Nitric Oxide Synthase (iNOS) and Cyclooxygenase-2 (COX-2) in LPS-Stimulated BV2 and RAW264.7 Cells
Inflammatory cytokines and mediators are rapidly increased after macrophages and microglia are activated. The expression of pro-inflammatory proteins can also be increased. In BV2 and RAW264.7 cells, LPS induces NO production through activating iNOS expression. LPS also increases COX-2 expression, which mediates the synthesis of prostaglandins and cytokines. NO or prostaglandins regulate various biological functions of immune responses [27]. Therefore, we assessed iNOS and COX-2 expression in BV2 and RAW264.7 cells that affect the production of inflammatory mediators and cytokines, respectively. Cells were pretreated for 3 h with indicated concentrations of compounds 7–9 and stimulated for 24 h with LPS. The LPS-induced expression of iNOS and COX-2 in both types of cells was significantly inhibited by compounds 7–9 (Figure 6). In this result, compounds 7–9 significantly regulated iNOS and COX-2 protein expression, suggesting that it exhibits inhibitory effects on inflammatory mediators and cytokines.
Figure 6.
Protein expression levels of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) by compounds 7–9 in LPS-stimulated BV2 (A,B) and RAW264.7 (C,D) cells. Cells were pretreated for 3 h with indicated concentrations of compounds 7–9 and stimulated for 24 h with LPS (1 μg/mL). Western blot analysis was performed as described in Materials and Methods. Representative blots from three independent experiments are shown.
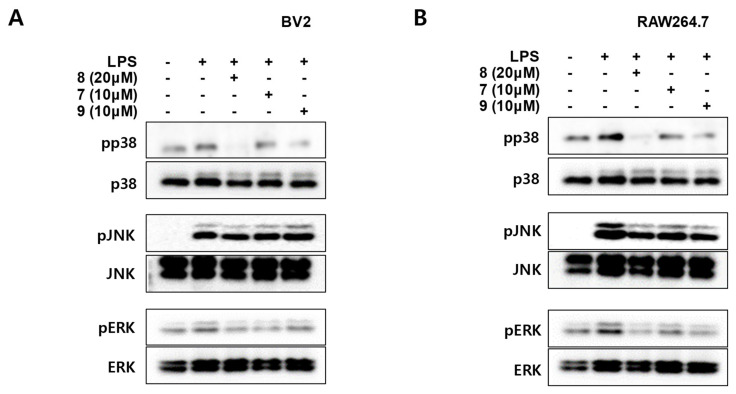
3.6. Effects of Compounds 7–9 on p38, c-Jun N-Terminal Kinase (JNK)-1/2, and Extracellular Signal-Regulated Kinase (ERK)-1/2 Phosphorylation in BV2 and RAW264.7 Cells
In mammalian cells and tissues, many physiological and pathological responses are mediated by the mitogen-activated protein kinase (MAPK) signaling pathway, including stress responses, inflammation, and apoptosis. ROS modify the gene expression of pro-inflammatory mediators by altering MAPK cascades [28]. MAPKs are activated during the process, releasing ILs, TNFs, and various inflammatory mediators such as NO, PGE2, histamine, and lysosome granules [29]. Therefore, we explored the effects of compounds 7–9 on the MAPK pathway activation in BV2 and RAW264.7 cells. Cells were incubated with compounds 7–9 for 3 h and stimulated with LPS for 30 min. First, compound 7 inhibited the LPS-induced phosphorylation of p38 and ERK MAPKs, but did not reduce the phosphorylation of JNK in both BV2 and RAW264.7 cells (Figure 7). Compound 8 showed an excellent inhibitory effect on the LPS-induced phosphorylation of p38 and ERK MAPK in both BV2 and RAW264.7 cells, but not the phosphorylation of JNK (Figure 7). Compound 9 showed a significant inhibitory effect on LPS-induced phosphorylation of p38 MAPK and showed a tendency to inhibit ERK MAPK phosphorylation in BV2 cells. In addition, Compound 9 significantly inhibited the phosphorylation of p38 and ERK MAPK in RAW264.7 cells. However, it did not decrease the phosphorylation of JNK in both BV2 and RAW264.7 cells (Figure 7). Taken together, in both BV2 and RAW264.7 cells, compounds 7–9 (acanthosessilioside K, L, and M) had inhibitory effects on the phosphorylation of p38 and ERK MAPKs, but not the phosphorylation of JNK. In addition, compound 8 (acanthosessilioside L) had the most effects on the inhibition of MAPK phosphorylation. These results indicate that acanthosessilioside L significantly regulated the inflammatory mediators and cytokines through the inhibiting p38 and ERK MAPK.
Figure 7.
Effects of compounds 7–9 on p38, JNK-1/2, and ERK-1/2 phosphorylation in BV2 (A) and RAW264.7 (B) cells. Cells were pretreated with indicated concentrations of compounds 7–9 for 3 h and stimulated for 30 min with LPS (1 μg/mL). Cell extracts were analyzed using Western blotting with antibodies specific for phosphorylated p-p38, p-JNK1/2, and p-ERK1/2. Membranes were stripped and re-probed to measure total abundance of each mitogen-activated protein kinase (MAPK) as a control measurement. Representative blots from three independent experiments are shown.
4. Conclusions
Nine noble 3,4-seco-triterpenoid glycosides, acanthosessilioside G–O, along with two previously known triterpenoid glycosides were isolated from A. sessiliflorus fruits, and the chemical structures were determined without ambiguity based on the intensive analysis of 1D-NMR, 2D-NMR, UV, IR, and MS data. In this study, the LC–MS/MS MRM analysis method for quality control of the A. sessiliflorus fruit was first developed using the isolated compounds 1–11. The advantages of hybrid LC–QTOF mass spectrometry include not only accurate and sensitivity, but also fast LC–MS/MS MRM analysis, making structural elucidations easier. It can be used for the qualitative and quantitative determination of minor or novel compounds, which is helpful in improving the quality control of A. sessiliflorus fruits. Acanthosessilioside K, L, and M (7–9) were the most effective in inhibiting NO, PGE2, TNF-α, IL-1β, and IL-6 production and reducing iNOS and COX-2 expression. In addition, these compounds had inhibitory effects on p38 and ERK MAPK phosphorylation in both BV2 and RAW264.7 cells. Our results suggest that acanthosessilioside K, L, and M could be good candidates for the development of therapeutic agents for inflammatory and neuroinflammatory diseases.
Abbreviations
| AFB | Acanthopanax sessiliflorus fruits n-butanol fraction |
| AFE | Acanthopanax sessiliflorus fruits ethyl acetate fraction |
| AFH | Acanthopanax sessiliflorus fruits aqueous fraction |
| ASFEx | Acanthopanax sessiliflorus fruits extraction |
| CC | column chromatography |
| DW | dry weight |
| EtOAc | ethyl acetate |
| FAB | subfractions from Acanthopanax sessiliflorus fruits n-butanol fraction |
| gHMBC | gradient heteronuclear multiple bond coherence |
| IFN- γ | interferon-gamma |
| LPS | lipopolysaccharide |
| MeOH | methanol |
| n-BuOH | n-butanol |
| NMR | nuclear magnetic resonance |
| NO | nitric oxide |
| ODS | octadecyl silica gel |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SiO2 | silica gel |
| TLC | thin layer chromatography |
| TNF-α | tumor necrosis factor-alpha |
| UV | ultraviolet |
| 13C-NMR | carbon-13 nuclear magnetic resonance |
| 1D-NMR | one-dimensional (1D) NMR |
| 2D-NMR | two-dimensional (2D) NMR |
| 1H-NMR | proton-1 nuclear magnetic resonance |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antiox10091334/s1, Figure S1: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside G. Figure S2: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside H. Figure S3: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside I. Figure S4: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside J. Figure S5: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside K. Figure S6: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside L. Figure S7: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside M. Figure S8: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside N. Figure S9: 1H, 13C NMR, Qtrap/MSMS, and HRESIMS of Acanthosessilioside O.
Author Contributions
Conceptualization and visualization: D.Y.L.; material preparation: Y.-S.L.; compound isolation: H.-G.K., N.-I.B., and D.Y.L.; compound identification (UV, IR, NMR, and MS): H.-G.K., D.Y.L., and N.-I.B.; UPLC-QTOF/MS analysis: B.-R.C. and S.M.O.; Q-trap/MS analysis: B.-R.C. and D.Y.; anti-inflammatory assays: Y.-S.L., W.K., L.D., and D.-S.L.; writing—original draft preparation: B.-R.C., H.-G.K., D.-S.L., and D.Y.L.; writing—review and editing: D.Y.L.; funding acquisition: D.Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the “Cooperative Research Program for Agriculture Science and Technology Development” (Project No. PJ01420402), Rural Development Administration, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rha C.S., Jeong H.W., Park S., Lee S., Jung Y.S., Kim D.O. Antioxidative, anti-Inflammatory, and anticancer effects of purified flavonol glycosides and aglycones in green tea. Antioxidants. 2019;8:278. doi: 10.3390/antiox8080278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohn E.S., Sohn E.H. A study on the R&D trend and patent analysis of treatments for degenerative brain diseases. J. Korea Acad. Industr. Coop. Soc. 2011;12:4411–4417. [Google Scholar]
- 3.Nakamura Y., Si Q.S., Kataoka K. Lipopolysaccharide-induced microglial activation in culture: Temporal profiles of morphological change and release of cytokines and nitric oxide. Neurosci. Res. 1999;35:95–100. doi: 10.1016/S0168-0102(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 4.Yan Q., Zhang J., Liu H., Babu-Khan S., Vassar R., Biere A.L., Citron M., Landreth G. Anti-inflammatory drug therapy alters â-amyloid processing and deposition in an animal model of Alzheimer’s disease. J. Neurosci. 2003;23:7504–7509. doi: 10.1523/JNEUROSCI.23-20-07504.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tremblay M.È., Stevens B., Sierra A., Wake H., Bessis A., Nimmerjahn A. The role of microglia in the healthy brain. J. Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreutzberg G.W. Microglia: A sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 7.Tumilowics J., Banaszczak P. Woody species of Araliaceae at the Rogow Arboretum. Rocz. Dendrologiczny. 2006;54:35–50. [Google Scholar]
- 8.Lee D.Y., Seo K.H., Lee D.S., Kim Y.C., Chung I.S., Kim G.W., Cheoi D.S., Baek N.I. Bioactive 3,4-seco-Triterpenoids from the Fruits of Acanthopanax sessiliflorus. J. Nat. Prod. 2012;75:1138–1144. doi: 10.1021/np3002173. [DOI] [PubMed] [Google Scholar]
- 9.Park J.K., Kim C.K., Gong S.K., Yu A.R., Lee M.Y., Park S.K. Acanthopanax sessiliflorus stem confers increased resistance to environmental stresses and lifespan extension in Caenorhabditis elegans. Nutr. Res. Pract. 2014;8:526–532. doi: 10.4162/nrp.2014.8.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F., Li W., Fu H., Zhang Q., Koike K. Pancreatic Lipase-Inhibiting Triterpenoid Saponins from Fruits of Acanthopanax senticosus. Chem. Pharm. Bull. 2007;55:1087–1089. doi: 10.1248/cpb.55.1087. [DOI] [PubMed] [Google Scholar]
- 11.Lee S.H., Son D.W., Ryu J.Y., Lee Y.S., Jung S.H., Kang J.G., Lee S.Y., Kim H.S., Shin K.H. Anti-oxidant activities of Acanthopanax senticosus stems and their lignan components. Arch. Pharm. Res. 2004;27:106–110. doi: 10.1007/BF02980055. [DOI] [PubMed] [Google Scholar]
- 12.Yoshizumi K., Hirano K., Ando H., Hirai Y., Ida Y., Tsuji T., Tanaka T., Satouchi K., Terao J. Lupane-Type Saponins from Leaves of Acanthopanax sessiliflorus and Their Inhibitory Activity on Pancreatic Lipase. J. Agric. Food Chem. 2006;54:335–341. doi: 10.1021/jf052047f. [DOI] [PubMed] [Google Scholar]
- 13.Hwang E., Kim G.W., Song K.D., Lee H.K., Kim S.J. The enhancing effect of Acanthopanax sessiliflorus fruit extract on the antibacterial activity of porcine alveolar 3D4/31 macrophages via nuclear factor kappa Bl and lipid metabolism regulation. Asian-Australas. J. Anim. Sci. 2019;32:1776–1788. doi: 10.5713/ajas.18.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung I.H., Kim S.E., Lee Y.G., Kim D.H., Kim H., Kim G.S., Baek N.I., Lee D.Y. Antihypertensive Effect of Ethanolic Extract from Acanthopanax sessiliflorus Fruits and Quality Control of Active Compounds. Oxid. Med. Cell Longev. 2018;2018:5158243. doi: 10.1155/2018/5158243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung H.J., Nam J.H., Choi J.W., Lee K.T., Park H.J. Anti-inflammatory effects of chiisanoside and chiisanogenin obtained from the leaves of Acanthopanax chiisanensis in the carrageenan- and Freund’s complete adjuvant-induced rats. J. Ethnopharmacol. 2005;97:359–367. doi: 10.1016/j.jep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Kim H.G., Oh H.J., Ko J.H., Song H.S., Lee Y.G., Kang S.C., Lee D.Y., Baek N.I. Lanceoleins A–G, hydroxychalcones, from the flowers of Coreopsis lanceolata and their chemopreventive effects against human colon cancer cells. Bioorg. Chem. 2019;85:274–281. doi: 10.1016/j.bioorg.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Quang T.H., Ngan N.T., Yoon C.S., Cho K.H., Kang D.G., Lee H.S., Kim Y.C., Oh H. Protein tyrosine phosphatase 1B inhibitors from the Roots of Cudrania tricuspidata. Molecules. 2015;20:11173–11183. doi: 10.3390/molecules200611173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S.Y., Chang S.Y., Yook C.S., Nohara T. New 3,4-seco-lupane-type triterpene glycosides from Acanthopanax senticosus forma inermis. J. Nat. Prod. 2000;63:1630–1633. doi: 10.1021/np000277c. [DOI] [PubMed] [Google Scholar]
- 19.Shirasuna K., Miyakoshi M., Mimoto S., Isoda S., Satoh Y., Hirai Y., Ida Y., Shoji J. Lupane triterpenoid glycosyl esters from leaves of Acanthopanax divaricatus. Phytochemistry. 1997;45:579–584. doi: 10.1016/S0031-9422(97)00017-4. [DOI] [Google Scholar]
- 20.Lee D.Y., Kim M.-J., Yoon D., Lee Y.-S., Kim G.-S., Yoo Y.C. Ginseng Berry Prevents Alcohol-Induced Liver Damage by Improving the Anti-Inflammatory System Damage in Mice and Quality Control of Active Compounds. Int. J. Mol. Sci. 2019;20:3522. doi: 10.3390/ijms20143522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko W., Sohn J.H., Jang J.H., Ahn J.S., Kang D.G., Lee H.S., Kim J.S., Kim Y.C., Oh H. Inhibitory effects of alternaramide on inflammatory mediator expression through TLR4-MyD88-mediated inhibition of NF-κB and MAPK pathway signaling in lipopolysaccharide-stimulated RAW264.7 and BV2 cells. Chem. Biol. Interact. 2016;244:16–26. doi: 10.1016/j.cbi.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Lee H., Ko W., Chowdhury A., Li B., Kim S.C., Oh H., Kim Y.C., Woo E.R., Baek N.I., Lee D.S. Brassicaphenanthrene A from Brassica rapa protects HT22 neuronal cells through the regulation of Nrf2-mediated heme oxygenase-1 expression. Mol. Med. Rep. 2020;21:493–500. doi: 10.3892/mmr.2019.10824. [DOI] [PubMed] [Google Scholar]
- 23.Titheradge M.A. The enzymatic measurement of nitrate and nitrite. Methods Mol. Biol. 1998;100:83–91. doi: 10.1385/1-59259-749-1:83. [DOI] [PubMed] [Google Scholar]
- 24.Pandur E., Varga E., Tamasi K., Pap R., Nagy J., Sipos K. Effect of inflammatory mediators lipopolysaccharide and lipoteichoic acid on iron metabolism of differentiated SH-SY5Y cells alters in the presence of BV-2 microglia. Int. J. Mol. Sci. 2018;20:17. doi: 10.3390/ijms20010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S.K., Ko Y.H., Lee Y., Lee S.Y., Jang C.G. Antineuroinflammatory Effects of 7,3′,4′-Trihydroxyisoflavone in Lipopolysaccharide-Stimulated BV2 Microglial Cells through MAPK and NF-κB Signaling Suppression. Biomol. Ther. 2021;29:127–134. doi: 10.4062/biomolther.2020.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boka G., Anglade P., Wallach D., Javoy-Agid F., Agid Y., Hirsch E.C. Immunocytochemical analysis of tumor necrosis factor and its receptors in Parkinson’s disease. Neurosci. Lett. 1994;172:151–154. doi: 10.1016/0304-3940(94)90684-X. [DOI] [PubMed] [Google Scholar]
- 27.Ricciotti E., FitzGerald G.A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Min J.S., Kim B., Chae U.B., Yun J.W., Choi M.S., Kong I.K., Chang K.T., Lee D.S. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-kappaB pathways. Neurosci. Lett. 2015;584:191–196. doi: 10.1016/j.neulet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Ripmeester E.G.J., Timur U.T., Caron M.M.J., Welting T.J.M. Recent insights into the contribution of the changing hypertrophic chondrocyte phenotype in the development and progression of osteoarthritis. Front. Bioeng. Biotechnol. 2018;6:18. doi: 10.3389/fbioe.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article.