Abstract
A hallmark of inflammation is the burst-like formation of certain proteins, initiated by cellular stress and proinflammatory cytokines like interleukin 1 (IL-1) and tumor necrosis factor, stimuli which simultaneously activate different mitogen-activated protein (MAP) kinases and NF-κB. Cooperation of these signaling pathways to induce formation of IL-8, a prototype chemokine which causes leukocyte migration and activation, was investigated by expressing active and inactive forms of protein kinases. Constitutively active MAP kinase kinase 7 (MKK7), an activator of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) pathway, induced IL-8 synthesis and transcription from a minimal IL-8 promoter. Furthermore, MKK7 synergized in both effects with NF-κB-inducing kinase (NIK). Activation of the IL-8 promoter by either of the kinases required functional NF-κB and AP-1 sites. While NIK and MKK7 did not affect degradation of IL-8 mRNA, an active form of MKK6, which selectively activates p38 MAP kinase, induced marked stabilization of the transcript and further increased IL-8 protein formation induced by NIK plus MKK7. Consistently, the MAP kinase kinase kinase MEKK1, which can activate NF-κB, SAPK/JNK, and p38 MAP kinases, most potently induced IL-8 formation. These results provide evidence that maximal IL-8 gene expression requires the coordinate action of at least three different signal transduction pathways which cooperate to induce mRNA synthesis and suppress mRNA degradation.
Interleukin-8 (IL-8) is a member of the still-growing family of chemokines, cytokines whose main function is to attract and activate leukocytes (2). It plays a significant role in recruiting leukocytes at sites of acute inflammation. On the other hand, excessive amounts of locally produced IL-8 can have deleterious effects (2, 45). Expectedly, therefore, IL-8 gene expression is tightly controlled at several levels (45). IL-8 synthesis, low or undetectable in normal noninflamed tissue, can be induced in vivo as well as in a wide variety of cells in vitro by proinflammatory cytokines such as IL-1 or tumor necrosis factor (TNF) (21, 5) or as a direct consequence of contact with pathogens like bacteria (1, 18), viruses (35, 46), and cell-stressing agents (10, 30, 54, 57). Stimulus-dependent activation of IL-8 gene transcription has been demonstrated in nuclear run-on experiments (5, 21). In a number of studies, it was found that a sequence spanning nucleotides −1 to −133 within the 5′ flanking region of the IL-8 gene is essential and sufficient for transcriptional regulation of the gene (16, 43; reviewed in reference 45). As demonstrated by mutational and deletional analysis, this promoter element is regulated in a highly cell-type-specific fashion. The promoter contains an NF-κB element that is required for activation in all cell types studied, as well as AP-1 and C/EBP binding sites. The latter two sites are dispensable for transcriptional activation in some cells but contribute to activation in others. Thus, unlike the NF-κB site, the AP-1 and C/EBP sites are not essential for induction (1, 5, 16, 18, 21, 25, 30, 35, 36, 44–46, 64).
Formation of cytokines may also be restricted by mechanisms regulating mRNA half-life. Rapid degradation of cytokine transcripts has been ascribed to AU-rich sequences in their 3′ untranslated regions (UTRs) and distinct proteins interacting with them (7, 55). AU-rich sequences are also present in the 3′ UTR of IL-8 mRNA. Several reports indicate that IL-8 mRNA degradation is subject to modulation (6, 23, 58, 59, 61), and it has been suggested that the proinflammatory cytokines IL-1 and TNF also control IL-8 formation on this level (6, 21, 58, 59, 61).
Stimuli that induce IL-8 production, like IL-1 and TNF, simultaneously activate stress protein kinase cascades that regulate the activity of transcription factors which can bind to NF-κB, AP-1, and C/EBP binding sites. NF-κB is a dimeric transcription factor retained in the cytoplasm by its binding to IκB proteins. Recently two IκB kinases (IκBKα and -β) which specifically phosphorylate two adjacent serines in IκB proteins have been identified (12, 37, 51, 53, 63, 67). This phosphorylation results in ubiquitination and rapid degradation of IκBs by the proteasome, allowing NF-κB to translocate to the nucleus and bind to DNA. This process is critical for NF-κB activation, but enhanced NF-κB-induced transcriptional activity might additionally require phosphorylation of the subunits as well as binding of coactivators (3, 4, 60). IκB kinases α and β are phosphorylated by NF-κB-inducing kinase (NIK), a recently identified protein activated by IL-1, TNF, and Fas (33). IκB kinases can also be directly activated by MEKK1 (28, 29, 47), a mitogen-activated protein (MAP) kinase kinase kinase which activates the three best-characterized MAP kinases, namely, extracellular regulated kinase (ERK) (26), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) (31, 66), and p38 MAP kinase (15, 22, 31). The transcriptional regulator AP-1 is a dimer composed of Fos, Jun, ATF-2, and other family members (reviewed in references 20 and 62). In contrast to NF-κB, AP-1 proteins are usually constitutively bound to their cognate DNA elements. Transcriptional activity of AP-1 proteins is regulated by their abundance, by phosphorylation of transactivation domains, and by their binding to protein kinases (20, 62). Protein kinases activating AP-1 include the ERKs, SAPK/JNK, p38 MAP kinases (20, 62), and a partially characterized Fos kinase (11).
Despite the rapid progress in identifying stress-induced signaling pathways and, on the other hand, structural elements important in transcriptional activation, there is little information on how different signaling pathways interact with each other in order to mediate a particular biological response, such as expression of a gene like that encoding IL-8. In that context, it is of importance to determine not only how stress kinase pathways cooperate to regulate promoter activity but also how they affect steps other than transcription in the overall process of gene expression.
We have recently identified the IL-6 and IL-8 genes as new target genes regulated by SAPK/JNK (24). This result raised the question by which molecular mechanism this pathway contributes to IL-8 gene expression and how this compares to the activation of NF-κB, which plays a major role in IL-8 transcription.
In this study, we investigated the contribution of NF-κB and stress-activated protein kinase cascades to IL-8 transcription, mRNA stability, and protein formation by overexpressing selective upstream activators for each pathway. We provide evidence for coordinated but distinct function of each of the pathways in IL-8 gene expression.
MATERIALS AND METHODS
Cells and Materials.
KB and HEK-293 cells were obtained from the American Type Culture Collection. HeLa cells stably transfected with plasmid pUHD15-1 expressing the tet transactivator protein (14) were obtained from Hermann Bujard, University of Heidelberg. Cell lines were cultured in Dulbecco’s modified Eagle medium complemented with 10% fetal calf serum. E64 [trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane], pepstatin, leupeptin, PMSF (phenylmethanesulfonyl fluoride), and all other chemicals were from Sigma; [γ-32P]ATP was purchased from Hartmann Analytics. Antiserum SAK14 to the N terminus of NIK, raised in rabbits immunized with the peptide VMEMAYPGAPGSAVGQQKELC, was a kind gift of Jeremy Saklatvala, Kennedy Institute of Rheumatology, London, England. M2 antibodies against the Flag epitope (M2 agarose beads and Bio-M2) were from Kodak; antibodies 12CA5 against the hemagglutinin (HA) epitope and 9E10 against the c-Myc epitope were from Boehringer Mannheim. Epidermal growth factor (EGF) and horseradish peroxidase-coupled secondary antibodies against mouse, rabbit, and rat immunoglobulin G (IgG) were from Sigma. Protein A-, protein G-, and glutathione (GSH)-Sepharose were from Pharmacia. Human recombinant IL-1α was produced as described previously (24).
Plasmids.
The expression plasmid for glutathione S-transferase (GST)–Jun (amino acids 1 to 135) was a kind gift of J. R. Woodgett, The Ontario Cancer Research Institute. GST fusion proteins were expressed and purified from Escherichia coli by standard methods. PCS3MT-MKK7 encodes Myc-tagged MKK7 (19). Mutations were introduced to replace amino acids serine 271, threonine 275, and serine 277 with glutamic acid in pCS3MT-MKK73E and with alanine in pCS3MT-MKK73A. pCS3MT-MKK7K149M was mutated to replace the ATP-binding lysine at position 149 in kinase domain II with methionine. pCDNA3flagNIK and pCDNA3flagNIK(KK429-430AA) encode N-terminally Flag-tagged wild-type and dominant negative NIK, respectively (33). BamHI/XhoI fragments of both plasmids were subcloned into the BglII/XhoI sites of pCS3MT to generate pCS3MT-NIK and pCS3MT-NIK(KK429-430AA) encoding the N-terminally Myc-tagged proteins. The cDNAs of human MKK6 (GenBank accession no. U39656) and JNK2 (GenBank accession no. L31951) were amplified from KB cell RNA by reverse transcription (RT)-PCR and cloned into the KpnI site of plasmid peVHA (24), which adds an N-terminal HA epitope tag. Serine 207 and threonine 211 (according to reference 50) in MKK6 were mutated to glutamic acid to generate peVHA-MKK62E. In MKK6K82A, the ATP-binding lysine at position 82 was mutated to alanine. Plasmid pFC-MEKK1 encoding amino acids 360 to 672 of MEKK1 was obtained from Stratagene. A 180-nucleotide fragment of the human IL-8 promoter (nucleotides 1348 to 1527 in GenBank accession no. M28130) was amplified from genomic DNA by PCR. To generate the IL-8 promoter-driven luciferase reporter plasmid pUHC13-3-IL-8pr, the fragment was cloned into the XhoI/SalI sites of plasmid pUHC13-3 (14), replacing the tet transactivator-controlled and cytomegalovirus promoter sequences. Site-directed mutagenesis of AP-1 and NF-κB sites was performed as described by others (64), using the following oligonucleotides (binding sites in capital letters; point mutations underlined): AP-1, 5′gaagtgtgaTATCTCAggtttgccc3′; and NF-κB, 5′gggccatcagttgcaaatcgTTAACTTTCCtctgacataatg3′.
The human IL-8 cDNA (nucleotides 20 to 1554; GenBank accession no. M28130) was amplified by RT-PCR and cloned into the BamHI site of pUHD10-3 downstream of the tet transactivator-controlled promoter (14). A fragment (nucleotides 13 to 206) of chloramphenicol acetyltransferase (CAT) cDNA, in which the start codon was mutated to ATC, was inserted 5′ of the IL-8 cDNA into the EcoRI site of the plasmid to generate pUHD10-3-CATIL-8. All mutations described above were introduced by using a Quick Change site-directed mutagenesis kit (Stratagene). Primer sequences used for PCR are available upon request. Sequences were confirmed by automated DNA sequencing on an ABI 310 sequencer (Applied Biosystems).
Transfections and preparation of cell extracts.
Cells (1 × 105 to 2.5 × 105/well) were seeded into six-well plates. The next day, transfections were performed in triplicate by the calcium phosphate method. KB cells were transfected by using Dosper (Boehringer Mannheim) according to the manufacturer’s instructions. In all transfections, DNA amounts were kept constant by adding empty expression plasmids. After 24 h, the medium was changed and cells were incubated further for 24 h. Cells from one triplicate transfection were placed on ice; the medium was removed, and cells were washed once in phosphate-buffered saline and scraped in phosphate-buffered saline. For determination of reporter gene activity, cells were lysed in ice-cold potassium phosphate buffer (100 mM, pH 7.4), containing 0.2% Triton X-100, 1 μg of pepstatin per ml, 10 μg of leupeptin per ml, and 1 mM PMSF. Luciferase activity was determined by using reagents from Promega. For preparation of whole-cell extracts, cells were lysed in 10 mM Tris (pH 7.05)–30 mM NaPPi–50 mM NaCl–1% Triton X-100–2 mM Na3VO4–50 mM NaF–20 mM β-glycerophosphate with freshly added 0.5 mM PMSF–0.5 μg leupeptin per ml–0.5 μg of pepstatin per ml–10 mM p-nitrophenyl phosphate–400 nM okadaic acid (whole-cell lysis buffer). After 10 min on ice, lysates were cleared by centrifugation at 10,000 × g for 15 min at 4°C. Nuclear and cytosolic extracts were prepared as described previously (24). Briefly, cells were suspended and pelleted in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.3 mM Na3VO4, freshly added 200 μM leupeptin, 10 μM E64, 300 μM PMSF, 0.5 μg of pepstatin per ml, 5 mM dithiothreitol [DTT], 400 nM okadaic acid, 20 mM β-glycerophosphate). The pellet was resuspended in buffer A containing 0.1% Nonidet P-40. After centrifugation at 10,000, × g for 5 min at 4°C, supernatants were taken as cytosolic extracts. Pellets were resuspended in buffer B (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.3 mM Na3VO4, 20 mM β-glycerophosphate, 200 μM leupeptin, 10 μM E64, 300 μM PMSF, 0.5 μg of pepstatin per ml, 5 mM DTT, 400 nM okadaic acid). After 1 h on ice, nuclear extracts were cleared at 10,000 × g for 5 min at 4°C and supernatants were collected. Protein concentration of cell extracts was determined by the Bradford method, and samples were stored at −80°C.
Immunoprecipitation and Western blotting.
One milligram of whole-cell extract protein from cells transfected with plasmids encoding Flag-tagged or Myc-tagged proteins was diluted in 500 μl of immunoprecipitation buffer (20 mM Tris [pH 7.3], 154 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100). Samples were incubated for 4 h with 20 μl of M2-agarose beads or with 2 μg of anti-Myc antibody 9E10 to which 20 μl of protein G-Sepharose was then added. Beads were spun down, washed three times in 500 μl of immunoprecipitation buffer, and resuspended in 40 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (2% SDS, 25 mM Tris [pH 6.8], 1% β-mercaptoethanol, 6% glycerol, 0.02% bromophenol blue). Proteins were eluted from the beads by boiling for 5 min, separated by SDS-PAGE on a 7.5 or 10% gel and electrophoretically transferred to polyvinylidene difluoride membranes (Immobilon; Millipore). After blocking with 5% dried milk in Tris-buffered saline overnight, membranes were incubated for 4 to 24 h with primary antibodies, washed in Tris-buffered saline, and incubated for 2 to 4 h with the peroxidase-coupled secondary antibody. Proteins were detected by using the Amersham enhanced chemiluminescence system.
SAPK/JNK assay.
The assay was performed as previously described (24, 49). Briefly, 10 μl of whole-cell extract containing 30 μg of protein was incubated with 10 μl of GST-Jun (1 μg) and 10 μl of kinase buffer (150 mM Tris, [pH 7.4], 30 mM MgCl2, 60 μM ATP, 4 μCi of [γ-32P]ATP). After 15 min at room temperature, 10 μl of GSH-beads, equilibrated in whole-cell lysis buffer containing 1 mM DTT, was added. Samples were agitated for 30 min at room temperature. Beads were recovered by centrifugation at 10,000 × g for 5 min and washed twice in 200 μl of whole-cell lysis buffer. Bound GST-Jun was eluted from the beads by boiling for 5 min in SDS-PAGE sample buffer. After centrifugation at 10,000 × g for 5 min, supernatants were separated by SDS-PAGE on a 10% gel. Equal recovery of GST-Jun was confirmed by Coomassie staining.
Electrophoretic mobility shift assay (EMSA).
A double-stranded oligonucleotide containing (in capitals) the NF-κB consensus sequence (5′tgacagagGGGACTTTCCagaga3′) was end labeled by using [γ-32P]ATP and T4 polynucleotide kinase and purified by gel filtration on S-200 spin columns (Pharmacia). Protein-DNA binding reactions were performed with 5 to 20 μg of whole-cell or nuclear extract protein, labeled oligonucleotide, and 1 μg of poly(dI-dC) in 10 mM Tris (pH 7.4)–10 mM EDTA–0.5% (wt/vol) dried nonfat milk–0.5 M NaCl–10 mM DTT–50% glycerol in a total volume of 10 μl. After incubation at room temperature for 30 min, protein-DNA complexes were resolved by PAGE on a 4% gel and visualized by autoradiography.
RNA stability measurements.
HeLa cells constitutively expressing the tet transactivator protein (14) were seeded into 9-cm-diameter petri dishes (5 × 106 cells per dish). The next day, cells were transfected by the calcium phosphate method as described above. After 8 h, cells from each dish were divided into five 25-cm2 flasks for assaying the time course of RNA decay. The next day, transcription from the tet transactivator-controlled promoter was stopped by adding the tetracycline analog doxycycline (3 μg/ml) to the culture medium. At indicated times thereafter, total RNA was isolated by using a Qiagen RNA extraction kit according to the manufacturer’s instructions. Then 10 μg of RNA of each sample was separated by denaturing 1% agarose gel electrophoresis in 20 mM morpholine propanesulfonic acid (pH 7.0)–1 mM EDTA–5 mM sodium acetate–6.8% formaldehyde. RNA was blotted onto nitrocellulose Hybond-N membranes (Amersham) by capillary transfer. The membranes were incubated in prehybridization buffer (50% formamide, 20% blocking reagent [Boehringer Mannheim], 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.02% SDS, 0.1% N-lauryl sarcosine) for 2 h at 68°C, followed by overnight hybridization in the same buffer containing an IL-8 antisense RNA probe, which was transcribed from the IL-8 cDNA inserted in Bluescript vector and labeled with digoxigenin by using commercial kits (Boehringer Mannheim). Thereafter the membranes were washed twice in 2× SSC–0.1% SDS at room temperature and twice in 0.1× SSC–0.1% SDS at 68°C. Blots were then incubated with an anti-digoxigenin-alkaline phosphatase-coupled antibody and developed by using CSPD {disodium3-[4-methoxyspiro (1,2-dioxetane-3,2′-(5′chloro)tricyclo(3.3.1.1.3,7)decan)-4-yl]phenyl phosphate} as the substrate, and chemiluminescence was visualized on X-ray films (X-Omat; Kodak). Films were scanned with the GelDoc100system and quantified with the Molecular Analyst program (Bio-Rad).
Enzyme-linked immunosorbent assay (ELISA).
IL-8 protein concentrations in the cell culture medium, collected between 24 and 48 h after transfection unless stated otherwise, were determined by using the human IL-8 duo set kit (Genzyme) exactly as instructed by the manufacturer.
Statistics.
Samples from ELISA and luciferase reporter determinations were analyzed by paired Student t-test. Results are presented as means ± standard errors of the means (SEM).
RESULTS
The SAPK/JNK-activating kinase MKK7 induces IL-8 alone and in synergy with NIK.
We have previously demonstrated that inhibition of SAPK/JNK results in impaired formation of IL-8 in response to IL-1, indicating an essential role of this signaling pathway (24). In the present study, we have further analyzed which pathways activated by IL-1 contribute to expression of IL-8. SAPK/JNK require phosphorylation of tyrosine and threonine within the conserved motif TGY by dual-specificity MAP kinase kinases (39). Recently, a novel MAP kinase kinase, MKK7, also called JNKK2 or SKK4, which specifically activates SAPK/JNK was identified (13, 19, 27, 41, 65). IL-1 activates MKK7 (27, 65) and we have shown that this enzyme is a physiologically relevant activator of SAPK/JNK utilized by IL-1 in vivo (13).
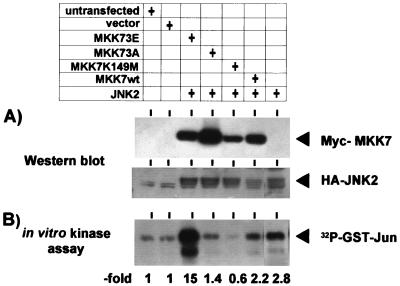
All MAP kinase kinases require phosphorylation at conserved Ser/Thr residues in subdomain XIII of the protein for activation. Substitution of these Ser/Thr residues with charged or uncharged amino acids generates constitutively active or inactive forms of MAP kinase kinases, respectively, as shown for MKK1, MKK3, and MKK6 (34, 50). An active MKK7 mutant was constructed by replacing S271, T275, and S277 with glutamic acid (MKK73E). To obtain an inactive mutant, these amino acids were replaced by alanine (MKK73A). A second inactive mutant was generated by mutating the ATP binding site (MKK7K149M). The activity of these mutants and wild-type MKK7 toward JNK2 was analyzed in cotransfection experiments. As shown in Fig. 1, only the MKK73E mutant showed significant activation of coexpressed HA-JNK2 in intact cells (about fivefold increase in GST-Jun phosphorylation compared to cells transfected with HA-JNK2 only), whereas MKK73A and MKK7K149M were inactive.
FIG. 1.
Activation of JNK2 by MKK7 mutants. HEK-293 cells were transiently transfected with 2.5 μg of plasmids encoding Myc-tagged forms of the indicated kinases (pCS3MT-MKK7, pCS3MT-MKK73E, pCS3MT-MKK73A, and pCS3MT-MKK7K149M) and with peVHA-JNK2. Empty pCS3MT vector was added to a total of 7.5 μg of DNA per transfection; 48 h later, cells were lysed in whole-cell lysis buffer. (A) A 100-μg aliquot of protein from each cell extract was separated by SDS-PAGE on a 10% gel, and expression of Myc epitope-tagged MKK7 proteins and HA-JNK2 was analyzed by Western blotting using anti-Myc and anti-HA antibodies, respectively. (B) GST-Jun kinase activity of cell extracts was assayed in vitro, using GST-Jun (amino acids 1 to 135) and [γ-32P]-ATP as the substrate. GST-Jun was then purified from the reaction mixture and resolved by SDS-PAGE on a 10% gel, and phosphorylation was visualized by autoradiography. See Materials and Methods for details. MKK7wt, wild-type MKK7.
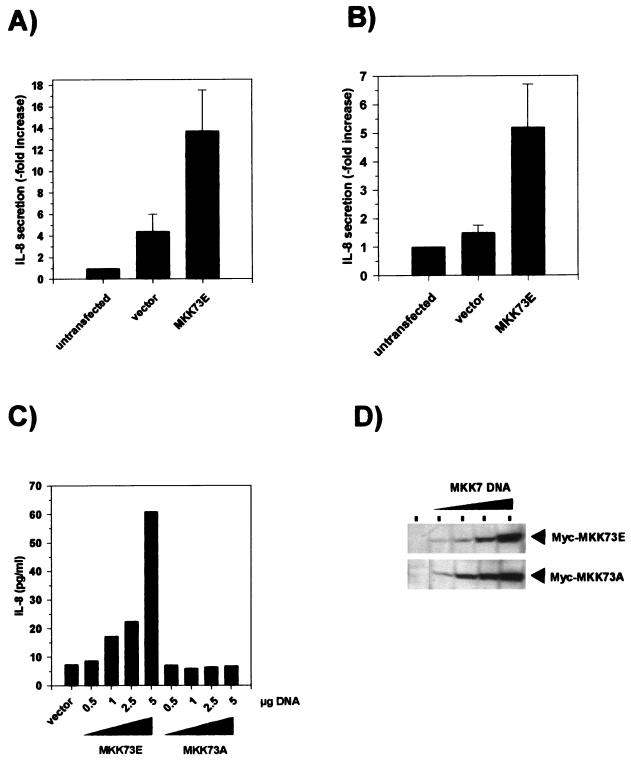
The effect of MKK7 mutants on IL-8 formation, assayed by specific ELISA of cell culture supernatants, was tested by transient transfection of KB and HEK-293 cells. In most experiments, transfection with empty vector alone induced some increase in IL-8 levels, in particular in the KB cells. This is apparently due to the transfection procedure itself. Expression of MKK73E in KB (Fig. 2A) and HEK-293 (Fig. 2B) cells resulted in significantly increased release of IL-8 into the culture medium compared to vector alone. In HEK-293 cells, the amount of IL-8 increased dose dependently with increasing concentrations of transfected plasmid and correlated closely with the amount of kinase expressed, as detected by Western blotting (Fig. 2C and D). No increase of IL-8 formation above the level of vector-transfected cells was observed in cells expressing an inactive form of the enzyme, MKK73A. This result suggests that activation of the SAPK/JNK pathway is sufficient to induce the endogenous IL-8 gene in these cells.
FIG. 2.
Transient expression of active MKK7 is sufficient to induce IL-8 secretion. KB (A) or HEK-293 (B) cells were transfected with 5 μg of empty vector pCS3MT-MKK73E or left untransfected; 24 h after transfection, the medium was changed. Cells were incubated for a further 24 h, and the amount of IL-8 in the medium was determined by specific ELISA as described in Materials and Methods. Shown is the fold increase (mean ± SEM) of IL-8 secretion compared to untransfected cells from three (A) or eight (B) independent experiments, each performed in triplicate (P < 0.01 for comparison of MKK73E versus vector). (C) HEK-293 cells were transfected with increasing amounts of the expression plasmid pCS3MT-MKK73E or pCS3MT-MKK73A or empty vector. The total DNA amount in each transfection (5 μg) was kept constant by adding empty pCS3MT. Supernatants were collected, and IL-8 protein concentrations were determined as for panel A. (C) The cells were lysed in whole-cell lysis buffer as described in Materials and Methods; 100-μg aliquots of proteins from lysates were separated by SDS-PAGE on 10% gels, and expression of Myc-tagged MKK7 protein kinases was analyzed by Western blotting using anti-Myc antibodies.
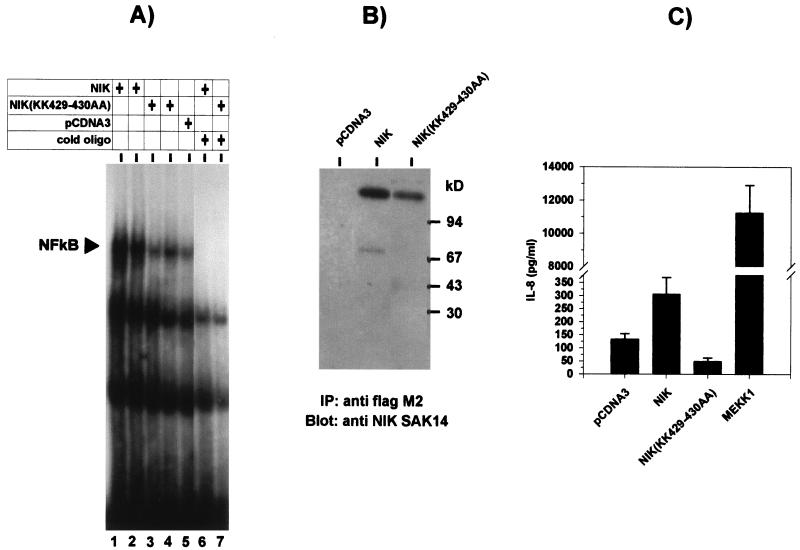
Several studies have demonstrated the involvement of NF-κB in the activation of the IL-8 promoter in response to extracellular stimuli (1, 5, 10, 16, 18, 21, 30, 35, 43, 45, 46, 57, 64). Expression of NIK in HEK-293 cells strongly activated NF-κB (Fig. 3A and B). IL-8 secretion in those cells was increased about threefold compared to vector-transfected cells (Fig. 3C). A kinase-inactive form of NIK, NIK(KK429-430AA), which did not affect NF-κB activity (Fig. 3A) did not induce but rather suppressed IL-8 synthesis compared with the vector control (Fig. 3C). This is not reflected in decreased NF-κB activity, possibly because total NF-κB activity in the vector-transfected cells may be contributed only to a small part by endogenous NIK. The extent of IL-8 induction and NF-κB activation by empty pCDNA3 vector reproducibly surpassed that of empty pCS3MT. Therefore, for subsequent experiments NIK and NIK(KK429-430AA) were recloned into pCS3MT.
FIG. 3.
Transient expression of NIK activates NF-κB and induces IL-8 secretion. (A) HEK-293 cells were transfected with 2.5 (lanes 1 and 3) or 5 (lanes 2 and 4) μg of expression plasmid pCDNA3flagNIK or pCDNA3flagNIK(KK429-430AA) or 5 μg of empty pCDNA3 vector (lane 5). In lanes 1 and 3, 2.5 μg of pCDNA3 was added to keep the DNA amount constant. At 48 h after transfection, cells were lysed in whole-cell lysis buffer. Activation of NF-κB in whole-cell extracts was analyzed by binding to a radiolabeled NF-κB consensus oligonucleotide as described in Materials and Methods. Protein-DNA complexes were resolved by nondenaturing PAGE on a 5% gel and visualized by autoradiography. The migration position of NF-κB complexed with labeled DNA, which was competed by excess unlabeled oligonucleotide (cold oligo; lanes 6 and 7), is indicated. (B) Expression of NIK was analyzed in extracts from transfected cells by immunoprecipitation (IP) with anti-Flag antibodies followed by Western blotting with polyclonal anti-NIK antiserum (SAK14). (C) IL-8 concentrations were determined by ELISA in supernatants of cells 48 h after transfection with pCDNA3flagNIK, pCDNA3flagNIK(KK429-430AA), or pCDNA3 (5 μg of each) or with 50 ng pFcMEKK1 plus 4.95 μg of pCDNA3. Shown are the means values ± SEM from eight transfection experiments performed in triplicate (P < 0.01 for comparison of all kinases to vector).
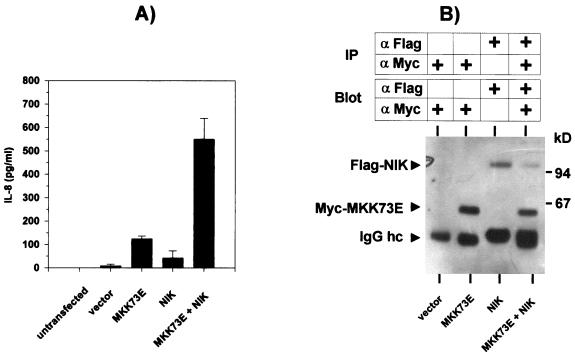
Levels of IL-8 formed in response to activation of the SAPK/JNK and NF-κB pathways by MKK73E (Fig. 2) and NIK (Fig. 3), respectively, were lower than those in cultures of cells transfected with an expression vector for a constitutively active form of MEKK1 (Fig. 3C). The low induction of IL-8 by NIK could not be ascribed to insufficient activation of NF-κB, since its extent was similar to that induced by MEKK1 (see Fig. 5C). Considering that MEKK1 can activate SAPK/JNK as well as NF-κB pathways, we asked whether both pathways might synergize to induce IL-8. As shown in Fig. 4A, coexpressing NIK and MKK73E induced supra-additive formation of IL-8 protein. This could not be ascribed to increased expression levels of NIK and MKK7 (Fig. 4B), since amounts of both kinases were similar in single and combined transfections (weaker intensity of the NIK band in the cotransfection in the particular experiment shown in Fig. 4B was not reproduced in other experiments [see also Fig. 5 and 7).
FIG. 5.
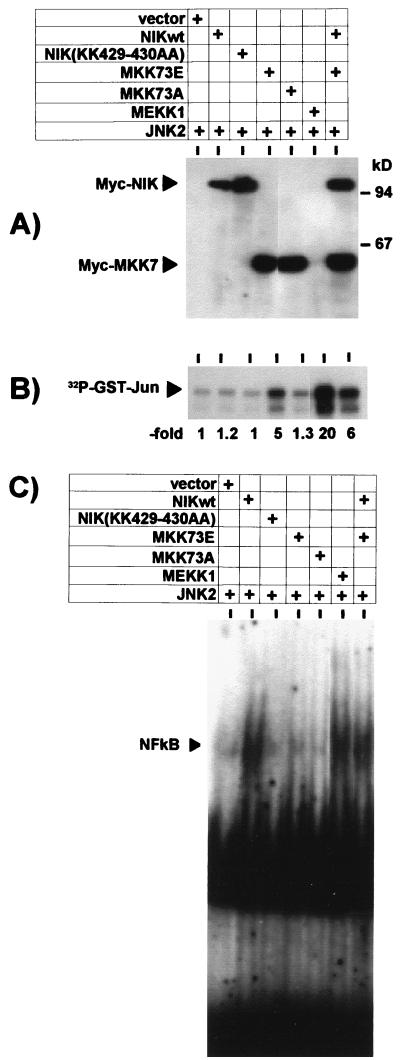
Activation of SAPK/JNK or NF-κB by overexpression of MKK73E or NIK. HEK-293 cells were cotransfected with 2.5 μg of expression plasmids for the indicated kinases and empty vector (7.5 μg of total DNA in each sample); 48 h later, cells were lysed, and cytosolic and nuclear extracts were prepared. (A) Ectopically expressed proteins were detected in the lysates (100 μg of protein) by Western blotting using antibodies against the epitope tags. (B) Activation of SAPK/JNK was assayed in cytosolic extracts as described for Fig. 1. (C) Activation of NF-κB was tested by EMSA in nuclear extracts as described for Fig. 3. Shown are the data for one of four independent experiments with essentially similar results. NIKwt, wild-type NIK.
FIG. 4.
Synergistic activation of IL-8 secretion by coexpression of NIK and MKK73E. (A) HEK-293 cells were transfected with 5 μg of empty vector, pCS3MT-MKK73E, pCDNAflag3NIK, or both; 48 h later, IL-8 secretion into the cell culture supernatant was determined by ELISA. Shown are means ± SEM from three independent experiments performed in triplicate. (B) Expression levels of MKK7 and NIK from one experiment were analyzed by immunoprecipitation (IP) from 1 mg of cell extract protein followed by Western blotting using antibodies against the Myc and Flag epitope tags, respectively (IgG hc, IgG heavy chain).
FIG. 7.
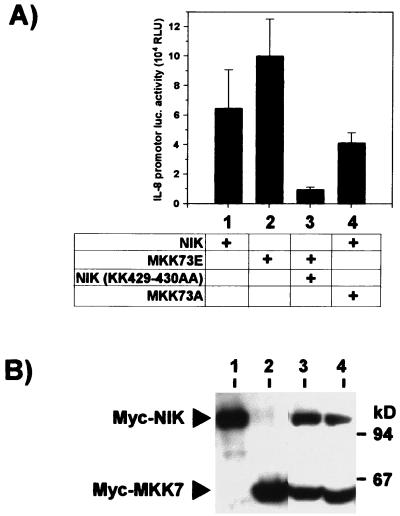
MKK73E-induced activation of the IL-8 promoter is suppressed by coexpression of dominant negative NIK. HEK-293 cells were transfected with 5 μg of pCS3MT-MKK73E, pCS3MT-MKK73A, pCS3MT-NIK, or pCS3MT-NIK(KK429-430AA) in the indicated combinations together with pUHC13-3-IL-8pr. Where required, empty pCS3MT was added to keep total DNA amounts constant. (A) At 48 h after transfection, cells were lysed and luciferase reporter gene activity was determined as for Fig. 6 (mean ± SEM from three independent experiments); (B) 100 μg of lysate proteins from one experiment was separated by SDS-PAGE and Western blotted to confirm equal expression of Myc-epitope tagged protein kinases MKK7 and NIK. RLU, relative light units.
MKK7 and NIK selectively activate SAPK/JNK and NF-κB, respectively.
Since both NIK and MKK73E triggered IL-8 formation, it was important to determine whether they acted via the same or different downstream effector molecules. Furthermore, since the combined effect of NIK and MKK73E on IL-8 formation was still far below that of MEKK1 (compare IL-8 concentrations in Fig. 3C and 4A), it was of interest to determine whether this was based on different intensities of signals induced. Therefore, activation of signaling mechanisms by MKK7 and NIK alone and in combination, as well as by MEKK1, were assayed. Compared to cells transfected with vector alone, expression of MKK73E resulted in marked activation of SAPK/JNK2 (Fig. 5A and B). No significant influence on SAPK/JNK activity was observed by expressing inactive MKK73A or active or inactive forms of NIK. Cotransfection of NIK did not significantly influence MKK73E-induced SAPK/JNK activation. Of note, MEKK1 clearly is more active than MKK73E in activating SAPK/JNK, suggesting that a more efficient trigger of that pathway, in combination with NIK, would give rise to stronger formation of IL-8. As no other selective activator for SAPK/JNK is available at present, this cannot be tested directly. Determination of NF-κB activity in EMSA, performed in parallel for the same cultures (Fig. 5C), showed that the active form of NIK strongly induced complex formation with the labeled oligonucleotide, while NIK(K429-430AA) as well as both forms of MKK7 were inactive in that respect. Furthermore, the active MKK73E did not affect the extent of NF-κB activation by NIK. Note that MEKK1-induced NF-κB activation is not stronger but comparable to NIK-induced activation. This argues against insufficient NF-κB activation by NIK as an explanation for its low IL-8 induction. Taken together, these results confirm selective activation of the SAPK/JNK pathway by MKK7 and of the NF-κB pathway by NIK, thus arguing against induction of IL-8 by MKK7 through cross-activation of NF-κB.
MKK7 and NIK each require NF-κB and AP-1 cis elements and synergize to activate a minimal IL-8 promoter.
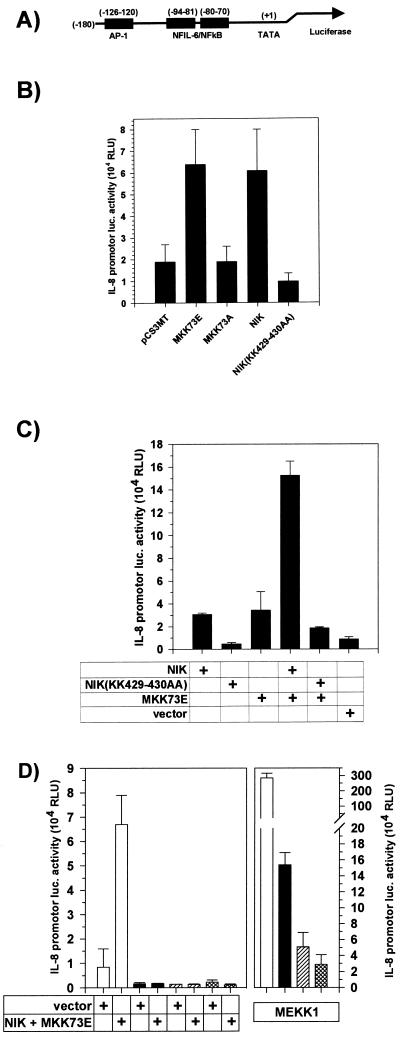
To further delineate the mechanisms involved in MKK7-induced IL-8 formation, we studied its effect on the transcriptional activity of a minimal IL-8 promoter, containing the AP-1 and NF-κB binding sequences (43, 64), placed upstream of a luciferase cDNA (Fig. 6A). MKK73E induced a threefold increase in luciferase activity, comparable to that induced by active NIK (Fig. 6B). In agreement with other studies, the induction of a synthetic promoter consisting of a 5-fold repeat of a consensus NF-κB site by NIK was much more pronounced (about 10-fold [data not shown]), arguing for distinct requirements for induction of the minimal IL-8 promoter. The dominant negative mutant NIK(KK429-430AA) slightly but reproducibly suppressed activity compared to vector-transfected cells (Fig. 6B and C), consistent with its suppression of IL-8 formation (Fig. 3C). The inactive MKK73A did not have a significant effect (Fig. 6B). Coexpression of both NIK and MKK73E had a synergistic effect (Fig. 6C). Consistent with its induction of high levels of IL-8 (Fig. 3C), MEKK1 induced much higher levels of luciferase activity than the active form of MKK7 or NIK (Fig. 6D). Mutation of the AP-1 site or the NF-κB site or both resulted in a strong decrease in basal activity and in a loss of inducibility by a combination of NIK and MKK73E (Fig. 6D), as well as by each of them alone (not shown). Thus, unexpectedly, each of the kinases assayed requires the presence of both sites for efficient stimulation of transcription. Furthermore, basal activity in this system appears to involve NF-κB activity and both sites as well. Activation of the mutated promoters in MEKK1-transfected cells was also strongly reduced but still clearly discernible. It is not clear at present whether this is due to quantitative differences in SAPK/JNK activation or triggering of additional signaling mechanisms by MEKK1.
FIG. 6.
MKK73E and NIK each require NF-κB and AP-1 cis elements and synergize to activate a minimal IL-8 promoter. (A) Schematic representation of the minimal IL-8 promoter cloned 5′ of the luciferase cDNA in pUHC13-3-IL- 8pr. (B) HEK-293 cells were cotransfected with 0.25 μg of pUHC13-3-IL-8pr and with 5 μg of pCS3MT-MKK73E, pCS3MT-MKK73A, pCS3MT-NIK, or pCS3MT-NIK(KK429-430AA), 0.1 μg of pFcMEKK1 plus 4.9 μg of pCS3MT, or pCS3MT alone; 48 h after transfection, cells were lysed and luciferase (luc.) reporter gene activity was determined as described in Materials and Methods. Results are expressed as relative light units [RLU; mean ± SEM from four independent experiments performed in triplicate; P < 0.01 for comparing vector versus MKK73E and NIK). (C) HEK-293 cells were cotransfected with 0.25 μg of pUHC13-3-IL-8pr and either pCS3MT-NIK or pCS3MT-NIK(KK429-430AA) (0.5 μg of each), 5 μg pCS3MT-MKK73E alone, or a combination thereof as indicated. Amounts of DNA were kept equal by adding empty pCS3MT. Reporter gene activity (mean ± SEM from three independent experiments) was determined as for panel B. (D) HEK-293 cells were cotransfected with 0.25 μg of pUHC13-3-IL-8pr (open bars) or of mutants thereof in which the AP-1 (black bars) or NF-κB (hatched bars) site or both sites (cross-hatched bars) were mutated and a combination of pCS3MT-MKK73E (5 μg) plus pCS3MT-NIK (0.5 μg) or pFcMEKK1 (0.1 μg) plus pCS3MT (4.9 μg). Reporter gene activity (mean ± SEM from four independent experiments) was determined as for panel B. For each experiment, equal expression of protein kinases was confirmed by Western blotting (not shown).
Considering the evidence for some basal NF-κB-dependent IL-8 transcription (Fig. 6B and C) and protein formation (Fig. 3C), its role in MKK7-induced promoter activation was tested in a more direct way. As shown in Fig. 7, cotransfection of the dominant negative form of NIK(KK429-430AA) resulted in marked inhibition of the MKK73E-induced luciferase activity. On the other hand, dominant negative MKK73A only marginally interfered with active NIK-induced transcription. These data support a model in which NF-κB-induced activation of IL-8 transcription is enhanced by SAPK/JNK-induced signaling.
Low induction of IL-8 by EGF correlates with insufficient SAPK/JNK activation.
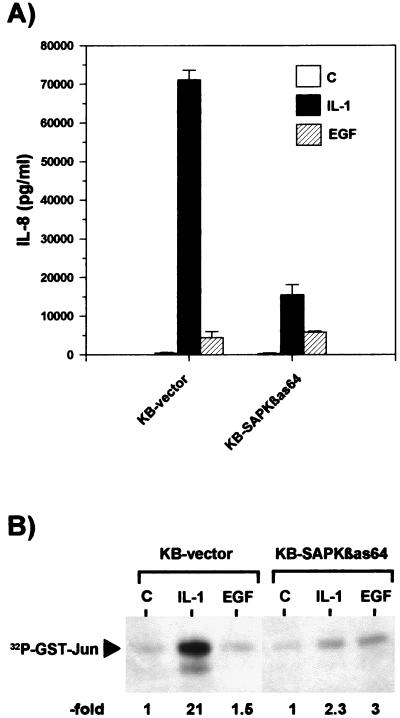
The hypothesis that cooperation of the SAPK/JNK and NF-κB pathways is required for maximal IL-8 gene expression is further supported by observations in human KB cells. In these cells, IL-1 induced a more than 100-fold increase in IL-8 secretion (Fig. 8A), as well as strong activation of SAPK/JNK (Fig. 8B) and NF-κB (24). Overexpression of SAPKβ antisense RNA resulted in a strong suppression of IL-1-induced IL-8 secretion (Fig. 8A and reference 24) without affecting activation of NF-κB (24). In the same cells, EGF induced only a 10-fold increase in IL-8 secretion (Fig. 8A). EGF did not activate SAPK/JNK (Fig. 8B). Accordingly, the EGF-induced IL-8 secretion was not decreased in the cell line overexpressing SAPKβ antisense RNA (Fig. 8B). This finding suggests that the extent of IL-8 induction by EGF is limited due to its inability to sufficiently activate SAPK/JNK.
FIG. 8.
Weak induction of IL-8 secretion by EGF correlates with its lack of SAPK/JNK activation. Human KB epithelial cells stably transfected with vector (KB-vector) or with antisense RNA to SAPKβ (KB-SAPKβas64, clone 64) as described in detail in reference 24 were left untreated (control [C]) or stimulated for 24 h with IL-1α (IL-1; 10 ng/ml) or with EGF (50 ng/ml). (A) IL-8 concentrations in the cell culture supernatant were determined by ELISA (means ± SEM from two independent experiments performed in duplicate). (B) The cells were stimulated with IL-1α (10 ng/ml) or EGF (50 ng/ml) or left untreated for 15 min and lysed, and SAPK/JNK activity was determined as described for Fig. 1. Shown is a result representative for three independent experiments.
Taken together, the data obtained so far suggest that signals generated by NIK and MKK73E cooperate on the transcriptional level to generate IL-8 formation.
MKK6 contributes to IL-8 induction by stabilizing its mRNA.
In addition to activation of transcription, posttranscriptional mechanisms contribute to the induction of IL-8 gene expression (6, 21, 23, 58, 59, 61) and may be regulated by these protein kinase pathways. We therefore investigated the role of NIK and MKK7 in IL-8 mRNA degradation. To compare the half-life of IL-8 mRNA in kinase-activated cells to that in control cells (which express only spurious amounts of the mRNA), it was necessary to transfect cells with a plasmid expressing the IL-8 mRNA. Fusion of a 194-nucleotide fragment of the CAT gene to its 5′ UTR allowed us to distinguish the ectopically expressed mRNA by size from the endogenous IL-8 mRNA induced by active kinases. To avoid the use of a general transcriptional inhibitor like actinomycin D, the cDNA was placed under the control of a tetracycline-regulated promoter, which allows rapid and selective inhibition of transcription (14).
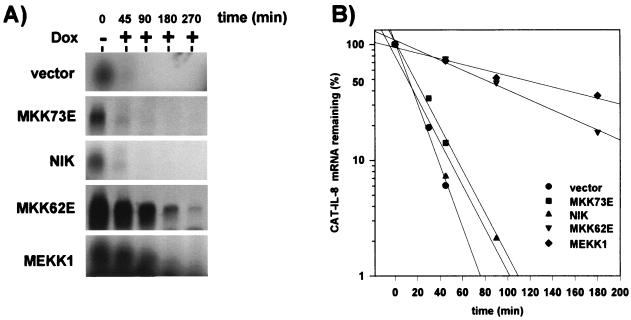
HeLa cells constitutively expressing the tet transactivator (14) were transiently transfected with the CAT–IL-8 plasmid together with empty vector or with expression vectors for the different active forms of kinases. Following inhibition of transcription by adding the tetracycline analogue doxycycline, the CAT–IL-8 mRNA rapidly decayed in cells cotransfected with empty vector (half-life of <20 min [Fig. 9]). Cotransfection of plasmids encoding active forms of MKK7 or NIK did not affect RNA degradation. In sharp contrast, cotransfection with MEKK1 resulted in pronounced stabilization of the RNA (half-life of >80 min [Fig. 9]). In addition to activation of NF-κB and SAPK/JNK pathways, MEKK1 has been shown to activate p38 MAP kinase through the MAP kinase kinase MKK4/SEK1 (15, 22). MKK4/SEK1 also activates SAPK/JNK (66). For that reason, we tested the involvement of p38 MAP kinase in IL-8 mRNA degradation by using an active form of the p38-activating kinase MKK6, MKK62E. MKK6 specifically activates p38 MAP kinase but not ERK or SAPK/JNK MAP kinases (50). Expression of MKK62E increased the stability of the CAT–IL-8 mRNA comparable to that induced by MEKK1 (Fig. 9). Similar results were obtained with authentic IL-8 mRNA lacking the CAT cDNA insertion, when the amount of endogenous IL-8 mRNA (which comigrates with it in Northern blots) was subtracted (data not shown). Thus, the p38 MAP kinase pathway contributes to induction of IL-8 synthesis by stabilizing its mRNA.
FIG. 9.
MEKK1 and MKK6, but not NIK or MKK7, induce IL-8 mRNA stabilization. HeLa cells stably expressing the tet transactivator protein were cotransfected with 4 μg of the IL-8 mRNA reporter plasmid pUHD10-3-CATIL-8 and 12 μg of expression plasmids for the indicated kinases or empty vector. After 24 h, doxycyline (Dox; 3 μg/ml) was added to stop tet transactivator-dependent transcription. (A) Total RNA was prepared at the indicated times thereafter, and CAT-tagged IL-8 mRNA was detected by Northern blotting as described in Materials and Methods. (B) Intensity of CAT–IL-8 mRNA bands (as shown in panel A and from additional experiments) was quantified, and values are expressed relative to CAT–IL-8 mRNA amount measured at the time of doxycycline addition (=100%).
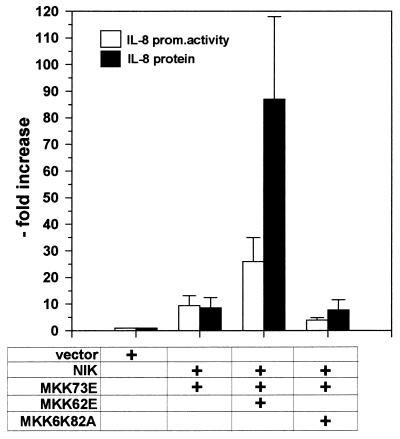
In agreement with this finding, coexpression of the active MKK62E strongly enhanced NIK- and MKK7-induced IL-8 protein secretion while only moderately enhancing transcription. The kinase-inactive mutant MKK6K82A had no effect (Fig. 10).
FIG. 10.
MKK62E potentiates NIK and MKK73E-induced IL-8 protein secretion. HEK-293 cells were transfected with 0.5 μg of pCS3MT-NIK, 2.5 μg of pCS3MT-MKK73E, 2.5 μg of peVHA-MKK62E, and 2.5 μg of peVHA-MKK6K82A in the combinations indicated; 0.25 μg of pUHC13-3-IL-8pr was co-transfected. Total amount of DNA was kept constant by adding empty pCS3MT. The cell culture medium was changed 24 h after transfection; 24 h later, cell culture supernatants were analyzed for IL-8 protein content by ELISA. Cells were lysed, and IL-8 promoter (prom.) activity was determined as described in Materials and Methods. Results are expressed as fold increase (mean ± SEM from two independent experiments).
Our data suggest that rapid accumulation of high levels of IL-8 transcript, a prerequisite for massive production of the protein, involves the combined effects of the SAPK/JNK and NF-κB pathways on IL-8 promoter activity and the mRNA-stabilizing effect of the p38 MAP kinase pathway.
DISCUSSION
Leukocyte recruitment and migration toward sites of trauma or infection is essential for innate and adaptive immune reactions. It is initiated by a family of extracellular signaling molecules, termed chemokines (2), of which IL-8 was among the first to be cloned. Control of chemokine production is a crucial step in regulating leukocyte infiltration and hence the intensity of an inflammatory process. This is reflected in the fact that IL-8 is low or absent under normal conditions but highly inducible by a wide range of extracellular stimuli, such as the proinflammatory cytokines IL-1 and TNF (5, 21, 45).
While the IL-8 gene contains a well-characterized promoter region, information on postreceptor events triggered by inflammatory cytokines to activate transcription of IL-8 is lacking. Furthermore, only limited information is available on the contribution of posttranscriptional mechanisms to IL-8 formation. In this report, we show that three distinct protein kinase cascades cooperate on different mechanistic levels to induce IL-8 expression. Appropriate forms of the upstream activators NIK, MKK7, and MKK6 were used to selectively activate the NF-κB, SAPK/JNK, and p38 MAP kinase pathways, respectively.
Transient ectopic expression of the NF-κB inducing kinase NIK was sufficient to induce secretion of IL-8 (Fig. 3 and 4) and transcription from a minimal IL-8 promoter (Fig. 6). These results complement previous data in which deletion or mutation of binding sites for NF-κB abolished responsiveness of an IL-8 promoter to IL-1, TNF, or other stimuli. However, the extent to which transfected NIK induces IL-8 expression is low compared to its strong activation of NF-κB (Fig. 3). NIK activates NF-κB as strongly as MEKK1 (Fig. 5C), by activating IKKs to comparable extents (28, 29, 47, 56). Yet MEKK1 induces a much stronger expression of IL-8 (Fig. 3 and 6). This observation suggests that additional MEKK1-activated pathways contribute to IL-8 induction.
SAPK/JNK are part of another MEKK1-activated pathway. NIK did not activate JNK2 in our experiments (Fig. 5), which is in agreement with two recent reports showing that NIK failed to activate coexpressed JNK1 (48, 56). Importantly, a gain-of-function mutant of the upstream activator of the SAPK/JNK pathway, MKK73E, was as effective as NIK in inducing IL-8 secretion (Fig. 4) and transcription (Fig. 6 and 7). This observation is not totally unexpected, since we previously reported that the SAPK/JNK pathway provides an essential signal for IL-1-induced IL-8 formation in KB cells (24).
While AP-1 represents a major nuclear target for SAPK/JNK in general, previous studies have disagreed as to the importance of the AP-1 site in the IL-8 promoter. In contrast to the NF-κB site, which is essential, the AP-1 site was dispensable in some studies (45, 64), contributed only partially to IL-8 transcription (1, 16, 46, 57), or was equally important (18, 30, 35). From these observations a model has emerged where the AP-1 site is required in addition to the NF-κB site for maximal transcription from the IL-8 promoter (1, 16, 18, 25, 30, 35, 36, 43, 44, 46, 57, 64). In support of this model, simultaneous triggering of NF-κB and SAPK/JNK by NIK and MKK7 resulted in synergistic activation of IL-8 transcription and secretion (Fig. 4 and 6). MEKK1 was still far more effective in inducing IL-8 transcription and secretion than the combined NIK and MKK73E. This correlates with a much stronger activation of SAPK/JNK by MEKK1 than by MKK73E (Fig. 5B). MEKK1 activates SAPK/JNK through stimulation of both MKK7 and MKK4 (17, 27, 32, 56, 65). The combination of MKK4 and MKK7 might result in stronger activation of the SAPK/JNK pathway and consequently IL-8 gene expression than was achievable with the active MKK7 mutant alone. However, it is also possible that MEKK1 activates a third pathway enhancing IL-8 expression.
The IL-8 promoter provided a model with which to study the relative contribution of NIK- and MKK7-induced pathways to activation of a natural promoter containing a single NF-κB site and a single AP-1 site. We found that NIK and MKK7 acted through separate immediate downstream events, since NIK did not activate SAPK/JNK and MKK7 did not activate NF-κB (Fig. 5). Mutational analysis of the IL-8 promoter showed that NIK and MKK73E each required functional AP-1 and NF-κB sites for IL-8 transcriptional activation (Fig. 6). These data suggest that signals from the NF-κB and the SAPK/JNK pathways converge at the same sites on the IL-8 promoter. Two observations indicate that the MKK7 signal may serve to further enhance transcription which is activated by NF-κB, rather than inducing transcription independently: First, MKK7-induced transcription is inhibited by coexpression of dominant negative NIK (Fig. 7). Second, basal IL-8 transcription is also inhibited by dominant negative NIK, indicating that some NF-κB activity is involved. Basal NF-κB activity may be necessary to observe transcriptional activation by MKK7. On the other hand, neither basal nor NF-κB-activated transcription was sensitive to coexpression of dominant negative MKK7, arguing against basal activity of that pathway.
These data can be explained by a model in which NIK induces translocation of the NF-κB dimer to the IL-8 promoter, where it binds in close proximity to AP-1 proteins. Activated SAPK/JNK molecules bound to AP-1 may phosphorylate NF-κB subunits or other regulatory components in addition to phosphorylating AP-1. This could lead to enhanced IL-8 promoter activity.
In that model, the SAPK/JNK pathway is used by the cell to boost IL-8 transcription initiated by NF-κB. Support for a crucial role of the SAPK/JNKs in IL-8 (and IL-6) formation comes from experiments in which IL-1-induced cytokine secretion was strongly reduced by inhibiting SAPK/JNK, but activation of NF-κB was unimpaired (24). The fact that EGF, a poor activator of SAPK/JNK, induced only low levels of IL-8 secretion also supports the notion that SAPK/JNK is required for the formation of this cytokine. EGF is also a weak activator of NF-κB (23a).
Several studies have suggested that IL-8 mRNA stabilization may be induced by IL-1 or TNF, but the signaling pathways involved have not been identified (6, 21, 58, 59, 61). By using an inducible expression system which allows rapid transcriptional shutoff, we found no effect of NIK or MKK7 on IL-8 mRNA degradation. However, an active mutant of MKK6, a specific activator of p38 but not SAPK/JNK or ERK MAP kinases (50), stabilized the IL-8 mRNA (Fig. 9). These data define a novel function for MKK6, namely, regulating IL-8 mRNA stability. During further analysis of this effect (62a), we also observed MKK6-induced stabilization of IL-6 mRNA and of β-globin-reporter mRNAs carrying AU-rich sequences of different cytokine mRNAs in their 3′ UTRs. An unrelated transcript (of the CAT gene) was not affected. This finding suggests that AU-rich elements are involved in the observed regulation. Recently it was shown that the SAPK/JNK pathway regulates the stability of the IL-2 (8) and IL-3 (42) mRNAs. JNKK2 (MKK7) and MEKK1, but not MKK6, enhanced IL-2 mRNA stability, suggesting that the SAPK/JNK pathway was involved (8). In discordance with the latter results, MKK7 did not affect IL-8 mRNA decay in our study. We do not know whether this discrepancy is related to the difference in the transcripts and/or the cell types studied (T cells versus epithelial cells). In support of our results, it was recently shown that the p38 MAP kinase inhibitor SB203580 suppressed IL-1- or lipopolysaccharide-induced stabilization of cyclo-oxygenase II (9, 52) and IL-6 (40) mRNAs. In summary, our data suggest that NIK- and MKK7-dependent pathways cooperatively regulate IL-8 transcription, whereas a third protein kinase cascade involving p38 MAP kinase regulates IL-8 mRNA stability. Thus, high expression of IL-8 requires at least three distinct protein kinase cascades (Fig. 10). Stimuli which are capable of activating NF-κB, SAPK/JNK, and p38 MAP kinase cascades, such as TNF, IL-1, or the upstream kinase MEKK1, consequently result in maximal IL-8 production and secretion.
In conclusion, our results provide a striking example of usage of three signal transduction pathways for regulating the expression level of an endogenous gene.
ACKNOWLEDGMENTS
This work was supported by grants Kr1143/2-1, SFB 244/B15, and SFB 244/B18 from the Deutsche Forschungsgemeinschaft to H.H. and M.K.
We thank Hermann Bujard for providing plasmids pUHD10-3 and pUHC13-3 and HeLa cells expressing the tet transactivator protein, J. R. Woodgett for his gift of expression plasmid for GST-Jun, and Jeremy Saklatvala for providing antiserum SAK14 against NIK and for helpful discussions. We gratefully acknowledge the skillful technical assistance of Birgit Ritter.
REFERENCES
- 1.Aihara M, Tsuchimoto D, Takizawa H, Azuma A, Wakebe H, Ohmoto Y, Imagawa K, Kikuchi M, Mukaida N, Matsushima K. Mechanisms involved in Helicobacter pylori-induced interleukin-8 production by a gastric cancer cell line, MKN45. Infect Immun. 1997;65:3218–3224. doi: 10.1128/iai.65.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann M, Hart L, Lindsay M, Barnes P J, Newton R. IκB degradation and nuclear factor-κB DNA binding are insufficient for interleukin-1β and tumor necrosis factor-α-induced κB-dependent transcription. J Biol Chem. 1998;273:6607–6610. doi: 10.1074/jbc.273.12.6607. [DOI] [PubMed] [Google Scholar]
- 4.Bird T A, Schooley K, Dower S K, Hagen H, Virca G D. Activation of nuclear transcription factor NFκB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J Biol Chem. 1997;272:32606–32612. doi: 10.1074/jbc.272.51.32606. [DOI] [PubMed] [Google Scholar]
- 5.Brasier A R, Jamaluddin M, Casola A, Duan W, Shen Q, Garofalo R P. A promoter recruitment mechanism for tumor necrosis factor-α-induced interleukin-8 transcription in type II pulmonary epithelial cells. J Biol Chem. 1998;273:3551–3561. doi: 10.1074/jbc.273.6.3551. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary L R, Avioli L V. Regulation of interleukin-8 gene expression by interleukin-1β, osteotropic hormones, and protein kinase inhibitors in normal human bone marrow stromal cells. J Biol Chem. 1996;271:16591–16596. doi: 10.1074/jbc.271.28.16591. [DOI] [PubMed] [Google Scholar]
- 7.Chen C-Y A, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 9.Dean J L E, Brook M, Clark A R, Saklatvala J. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J Biol Chem. 1999;274:264–269. doi: 10.1074/jbc.274.1.264. [DOI] [PubMed] [Google Scholar]
- 10.DeForge L E, Preston A M, Takeuchi E, Kenney J, Boxer L A, Remick D G. Regulation of the interleukin-8 gene expression by oxidant stress. J Biol Chem. 1993;268:25568–25576. [PubMed] [Google Scholar]
- 11.Deng T, Karin M. c-Fos transcriptional activity stimulated by H-ras-activated protein kinase distinct from JNK and ERK. Nature. 1994;371:171–175. doi: 10.1038/371171a0. [DOI] [PubMed] [Google Scholar]
- 12.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NFκB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 13.Finch A, Holland P, Cooper J, Saklatvala J, Kracht M. Selective activation of JNK/SAPK by interleukin-1 in rabbit liver is mediated by MKK7. FEBS Lett. 1997;418:144–148. doi: 10.1016/s0014-5793(97)01364-1. [DOI] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Z, Buckman S Y, Pentland A P, Templeton D J, Morrison A R. Induction of cyclooxygenase-2 by the activated MEKK1α→SEK1/MKK4α→p38 mitogen-activated protein kinase pathway. J Biol Chem. 1998;273:12901–12908. doi: 10.1074/jbc.273.21.12901. [DOI] [PubMed] [Google Scholar]
- 16.Harant H, deMartin R, Andrew P J, Foglar E, Dittrich C, Lindley I J D. Synergistic activation of interleukin-8 gene transcription by all-trans-retinoic acid and tumor necrosis factor-α involves the transcription factor NFκB. J Biol Chem. 1996;271:26954–26961. doi: 10.1074/jbc.271.43.26954. [DOI] [PubMed] [Google Scholar]
- 17.Hirai S, Noda K, Moriguchi T, Nishida E, Yamashita A, Deyama T, Fukuyama K, Ohno S. Differential activation of two JNK activators, MKK7 and SEK1, by MKN28-derived nonreceptor serine/threonine kinase/mixed lineage kinase 2. J Biol Chem. 1998;273:7406–7412. doi: 10.1074/jbc.273.13.7406. [DOI] [PubMed] [Google Scholar]
- 18.Hobbie S, Chen L M, Davis R J, Gala J E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 19.Holland P, Suzanne M, Campbell J S, Noselli S, Cooper J A. MKK7 is a stress-activated mitogen-activated protein kinase functionally related to hemipterous. J Biol Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 20.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 21.Kasahara T, Mukaida N, Yamashita K, Yagisawa H, Akahoshi T, Matsushima K. IL-1 and TNF-α induction of IL-8 and monocyte chemotactic and activating factor (MCAF) mRNA expression in a human astrocytoma cell line. Immunology. 1991;74:60–67. [PMC free article] [PubMed] [Google Scholar]
- 22.Khokhatchev A, Xu S, English J, Wu P, Schaefer E, Cobb M. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. J Biol Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 23.Kowalski J, Denhardt D T. Regulation of the mRNA for monocyte-derived neutrophil-activating peptide in differentiating HL60 promyelocytes. Mol Cell Biol. 1989;9:1946–1957. doi: 10.1128/mcb.9.5.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Kracht, M. Unpublished data.
- 24.Krause A, Holtmann H, Eickemeier S, Winzen R, Szamel M, Resch K, Saklatvala J, Kracht M. Stress-activated protein kinase/Jun-N-terminal kinase is required for interleukin-1-induced IL-6 and IL-8 gene expression in the human epidermal carcinoma cell line KB. J Biol Chem. 1998;273:23681–23689. doi: 10.1074/jbc.273.37.23681. [DOI] [PubMed] [Google Scholar]
- 25.Kunsch C, Lang R K, Rosen C A, Shannon M F. Synergistic transcriptional activation of the IL-8 gene by NFκB p65 (RelA) and NF-IL6. J Immunol. 1994;153:153–163. [PubMed] [Google Scholar]
- 26.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 27.Lawler S, Cuenda A, Goedert M, Cohen P. SKK4, a novel activator of stress-activated protein kinase-1 (SAPK/JNK) FEBS Lett. 1997;414:153–158. doi: 10.1016/s0014-5793(97)00990-3. [DOI] [PubMed] [Google Scholar]
- 28.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 29.Lee F S, Peters R T, Dang L C, Maniatis T. MEKK1 activates both IκB kinase α and IκB kinase β. Proc Natl Acad Sci USA. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee L-F, Haskill S J, Mukaida N, Matsushima K, Ting J P-Y. Identification of tumor-specific paclitaxel (Taxol)-responsive regulatory elements in the interleukin-8 promoter. Mol Cell Biol. 1997;17:5097–5105. doi: 10.1128/mcb.17.9.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin A, Minden A, Martinetto H, Claret F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the jun kinases and p38-mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 32.Lu X, Nemoto S, Lin A. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J Biol Chem. 1997;272:24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 33.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. MAP3K-related kinase involved in NFκB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 34.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 35.Mastronarde J G, Monick M M, Mukaida N, Matsushima K, Hunninghake G W. Activator protein-1 is the preferred transcription factor for cooperative interaction with nuclear factor-κB in respiratory syncytial virus-induced interleukin-8 gene expression in airway epithelium. J Infect Dis. 1998;177:1275–1281. doi: 10.1086/515279. [DOI] [PubMed] [Google Scholar]
- 36.Matsukaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NFκB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J W, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NFκB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 38.Meyer C F, Wang X, Chang C, Templeton D, Tan T-H. Interaction between c-rel and the mitogen-activated protein kinase kinase kinase 1 signaling cascade in mediating κB enhancer activation. J Biol Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 39.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 40.Miyazawa K, Mori A, Miyata H, Akahane M, Ajisawa Y, Okudaira H. Regulation of interleukin-1β-induced interleukin-6 gene expression in human fibroblast-like synoviocytes by p38 mitogen-activated protein kinase. J Biol Chem. 1998;273:24832–24838. doi: 10.1074/jbc.273.38.24832. [DOI] [PubMed] [Google Scholar]
- 41.Moriguchi T, Toyoshima F, Masuyama N, Hanafuda H, Gotoh Y, Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFα and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ming X-F, Kaiser M, Moroni C. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 1998;17:6039–6048. doi: 10.1093/emboj/17.20.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 44.Mukaida N, Morita M, Ishikawa Y, Rice N, Okamoto S, Kasahara T, Matsushima K. Novel mechanism of glucocorticoid-mediated gene repression. J Biol Chem. 1994;269:13289–13295. [PubMed] [Google Scholar]
- 45.Mukaida N, Okamoto S, Ishikawa Y, Matsushima K. Molecular mechanism of interleukin-8 gene expression. J Leukoc Biol. 1994;56:554–558. [PubMed] [Google Scholar]
- 46.Murayama T, Ohara Y, Obuchi M, Khabar K S A, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1 and NFκB-binding sites of the interleukin-8 gene. J Virol. 1997;71:5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Differential regulation of IκB kinase α and β by two upstream kinases, NFκB-inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natoli G, Costanzo A, Moretti F, Fulco M, Balsano C, Levrero M. Tumor necrosis factor (TNF) receptor 1 signaling downstream of TNF receptor-associated factor 2. J Biol Chem. 1997;272:26079–26082. doi: 10.1074/jbc.272.42.26079. [DOI] [PubMed] [Google Scholar]
- 49.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 50.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 52.Ridley S H, Dean J L E, Sarsfield S J, Brook M, Clark A R, Saklatvala J. A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett. 1998;439:75–80. doi: 10.1016/s0014-5793(98)01342-8. [DOI] [PubMed] [Google Scholar]
- 53.Rothwarf D M, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro L, Dinarello C A. Osmotic regulation of cytokine synthesis in vitro. Proc Natl Acad Sci USA. 1995;92:12230–12234. doi: 10.1073/pnas.92.26.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 56.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-κB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonoda Y, Kasahara T, Yamaguchi Y, Kuno K, Matsushima K, Mukaida N. Stimulation of interleukin-8 production by okadaic acid and vanadate in a human promyelocyte cell line, and HL-60 subline. J Biol Chem. 1997;272:15366–15372. doi: 10.1074/jbc.272.24.15366. [DOI] [PubMed] [Google Scholar]
- 58.Stoeckle M Y. Post-transcriptional regulation of gro α, β, γ, and IL-8 mRNAs by IL-1β. Nucleic Acids Res. 1991;19:917–920. doi: 10.1093/nar/19.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tobler A, Meier R, Seitz M, Dewald B, Baggiolini M, Frey M F. Glucocorticoids downregulate gene expression of GM-CSF, NAP-1/IL-8, and IL-6, but not of M-CSF in human fibroblasts. Blood. 1992;79:45–51. [PubMed] [Google Scholar]
- 60.Van den Berghe W, Plaisance S, Boone E, De Bosscher K, Schmitz M L, Fiers W, Haegeman G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-κB p65 transactivation mediated by tumor necrosis factor. J Biol Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 61.Villarete L H, Remick D G. Transcriptional and post-transcriptional regulation of interleukin-8. Am J Pathol. 1996;149:1685–1693. [PMC free article] [PubMed] [Google Scholar]
- 62.Whitmarsh A J, Davis R J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 62a.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C.-Y. A. Chen, A.-B. Shyu, M. Müller, M. Gaestel, K. Resch, and H. Holtmann. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 63.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D. IκB kinase-β: NFκB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 64.Wu G D, Lai E J, Huang N, Wen X. Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. J Biol Chem. 1997;272:2396–2403. [PubMed] [Google Scholar]
- 65.Wu Z, Wu J, Jacinto E, Karin M. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan M, Dal T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 67.Zandi E, Rothwarf D M, Delhase M, Hyakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NFκB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]