Abstract
Simple Summary
Liquid biopsy provides real-time monitoring of tumor evolution and response to therapy through analysis of circulating tumor cells (CTCs) and plasma-circulating tumor DNA (ctDNA). USP44 is a member of family proteins deubiquitinases, and plays an important role in cell growth; however, its accurate role in other cellular networks is under research. In this study, we examined for the first time USP44 promoter methylation in plasma cell-free DNA (cfDNA) of patients with prostate cancer (early stage n = 32, metastatic n = 39) and 10 healthy donors (HD). USP44 promoter methylation was detected in plasma cell-free DNA by a newly developed highly specific and sensitive real-time MSP method. We report for the first time that detection of USP44 promoter methylation in plasma cell free DNA provides significant prognostic information in metastatic prostate cancer.
Abstract
Liquid biopsy provides real-time monitoring of tumor evolution and response to therapy through analysis of circulating tumor cells (CTCs) and plasma-circulating tumor DNA (ctDNA). USP44 is a critical gene which plays an important role in cell proliferation; however, its accurate role in other cellular networks is under research. USP44 promoter methylation has been so far reported in colorectal neoplasia and metastatic breast cancer. In this study, we examined for the first time USP44 promoter methylation in plasma cell-free DNA (cfDNA) of patients with prostate cancer (early stage n = 32, metastatic n = 39) and 10 healthy donors (HD). USP44 promoter methylation was detected in plasma cell-free DNA by a newly developed highly specific and sensitive real-time MSP method. Our findings indicate that USP44 promoter is methylated in plasma cell-free DNA of metastatic prostate cancer patients and that detection of USP44 promoter methylation is significantly associated with overall survival (OS) (p = 0.008). We report for the first time that detection of USP44 promoter methylation in plasma cell free DNA provides significant prognostic information in metastatic prostate cancer.
Keywords: cfDNA, liquid biopsy, real-time methylation specific PCR (MSP), USP44
1. Introduction
Ubiquitination comprises a cellular post-translation modification, in which proteins can be modified through Lys-linked isopeptide bonds [1]. This signal leads proteins to their degradation, ubiquitin is attached to substrates by a sophisticated three-step enzymatic cascade utilizing E1 ubiquitin activating, E2 ubiquitin conjugating, and a variety of E3 ubiquitin ligating enzymes [2,3]. On the other hand, deubiquitinases can remove ubiquitin modification through the antagonism of E3 ligase and induce protein degradation [4]. USP44 is a member of family proteins deubiquitinases, and plays an important role in cell growth. USP44 regulates the separation of chromosomes in anaphase through deubiquitinasion of cdc20, a co-factor of APC gene [5,6]. Experiments in mouse models revealed that USP44 modulated the mitotic checkpoint by regulating centrosome positioning and spindle geometry through a direct interaction with the centriole protein centrin. The function of USP44 at the centrosome at least in part involves regulating the events surrounding centrosome separation, as its loss leads to an increase in cells with incomplete centrosome separation [7]. A second distinct tumor suppressive function of USP44 is that it may play an important role in nucleotide excision repair (NER) by stabilizing DDB2; experiments in cell lines which were exposed to UVC radiation have shown that DDB2, a gene that plays role in NER mechanism, remains ubiquitin-free due to USP44 function until XPC (Xeroderma pigmentosum) arrives and its activity further exists for subsequent rounds of damage recognition [8].
Epigenetic modifications consist of a genomic mechanism that influences gene expression without changes in the genome sequence [9]. In cancer, methylation plays an important role in many cellular pathways like apoptosis, differentiation, and cell growth [10]. DNA methylation is the first interaction between the genome and environmental factors [11]. USP44 methylation is reported so far in a small number of studies in breast cancer, colorectal neoplasia, and non-small cell lung cancer [12,13,14]. RNA-sequencing (RNA-Seq) transcriptome data and DNA methylation data of the TCGA-BRCA revealed that USP44 methylation could be potential biomarker of breast cancer [15]. Experiments in breast cancer cell lines revealed that USP44 hypermethylation promotes cell proliferation and metastasis [12].
Analysis of TCGA data in breast cancer has shown that USP44 expression was significantly decreased when compared to normal [12]. In xenograft models and cells lines, USP44 expression suppresses multipolarity and forms a bipolar spindle during mitosis. In KD USP44 cells the multipolarity spindle pattern was induced, and the vasculogenic mimicry (VM) formation was inhibited, thereby suggesting that the planar-like and apico-basal–like spindle patterns promoted by USP44 expression played a key role in VM, plasticity of aggressive cancer cells, and cancer metastasis [8]. USP44 methylation is correlated with its low expression in tissue samples of colorectal neoplasia. The same results were observed in colorectal adenocarcinoma cell lines but the methylation of USP44 did not correlate with aneuploidy [13]. Moreover, recent experiments in colorectal cancer using decitabine (a demethylating agent) has shown re-expression of USP44 [13]. In contrast, according to the only study so far that reported on the USP44 activity in prostate cancer, the expression of USP44 appeared to promote tumor growth through the expression of the EZH2 in prostate cancer cell lines, a histone-modifying enzyme which modulates the expression of genes involved in cell cycle and DNA repair. Immunoprecipitation assay in cell lines has shown an interaction between USP44 and EZH2 in catalytic activity. USP44 KD cells have shown decreased levels of EZH2 protein [16].
Liquid biopsy, a minimally invasive blood-based testing, offers the possibility to follow-up tumor evolution in real time, and presents a significant approach that may soon change the management of cancer patients in diagnosis, prognosis, and therapeutic interventions. Liquid biopsy is based on the analysis of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating miRNAs, and tumor-derived extracellular vesicles (EVs) that are shed from primary tumors and their metastatic sites into peripheral blood [17]. In clinical routine, multiple histological biopsies are required to monitor the response of cancer patients to treatment. However, tissue biopsies are invasive procedures and in some cases cannot be performed; for this reason, the scientific community has high expectations that liquid biopsy will surrogate tissue biopsies for the detection, prognosis, and therapeutic management of cancer patients [18,19,20].
In recent years, DNA methylation analysis in CTCs and ctDNA has shown strong potential, since it provides a valuable source of novel circulating epigenetic biomarkers for diagnosis, prognosis, risk assessment, and disease monitoring in many types of cancer [20]. Our group was the first to demonstrate epigenetic alterations in CTCs and corresponding ctDNA [21,22]. In breast cancer we reported that ESR1 methylation in CTCs and ctDNA was correlated with lack of response to treatment [23,24]. Moreover, we reported that in breast cancer, CST6 and BRMS1 methylation was detected in ctDNA and CTCs, in ovarian cancer RASSFIA and ESR1 methylation was detected in ctDNA, while in NSCLC BRMS1 and SOX17 methylation was detected in ctDNA [20].
In the present study we first investigated the molecular profile of prostate adenocarcinomas that are characterized by USP44 promoter methylation through meta-analysis of TCGA data. We further evaluated for the first time the clinical significance of the detection of USP44 methylation in plasma cell-free DNA of prostate cancer patients and we report a statistically significant association between USP44 promoter methylation and overall survival.
2. Results
2.1. USP44 Promoter Methylation in Prostate Cancer
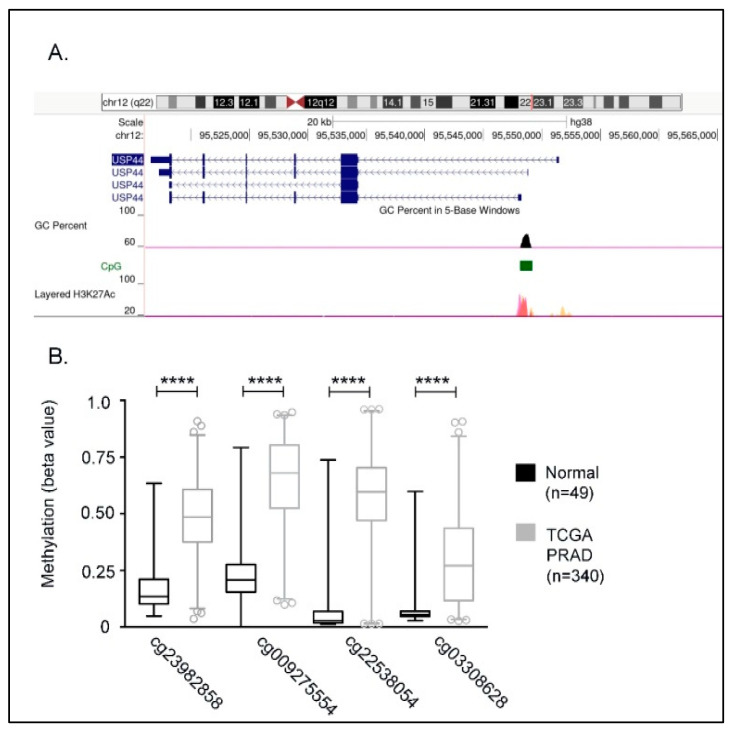
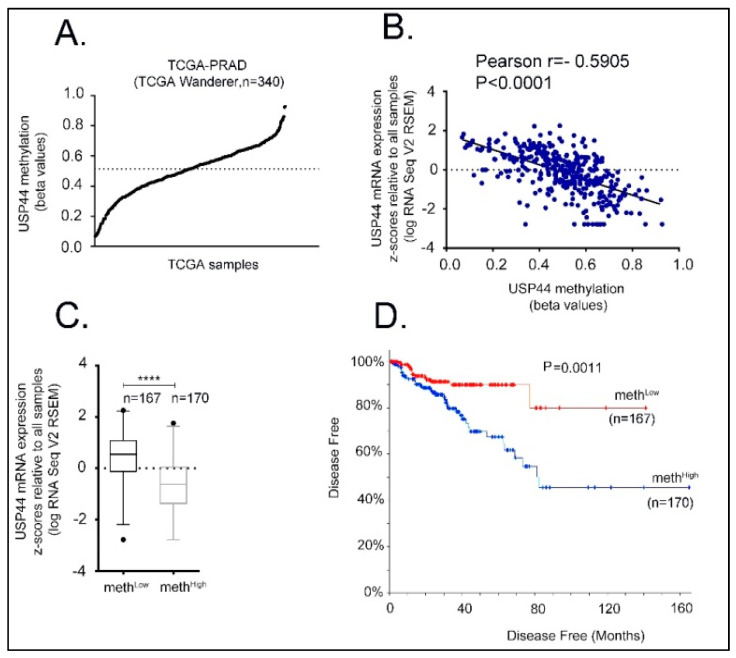
According to ENCODE data, the proximal USP44 promoter (hg19, chr1: 28735-29810) is characterized by the presence of a 1074 bp CG rich region (CpG 116, 73.2% GC content). This CpG island is enriched in H3K27Ac histone marks, indicating a critical role of this region in the transcriptional regulation of USP44 locus (Figure 1A). To investigate whether prostate carcinogenesis is associated with DNA methylation in this CpG island, we first compared its methylation profile between prostate adenocarcinoma samples and normal prostate tissues using publicly available methylation data (TCGA-PRAD) obtained by the TCGA Wanderer database. As a result, we observed a higher level of methylation in prostate cancer tissues compared to that in normal prostate tissues (Figure 1B). More specifically, our analysis showed that among the Illumina methylation detection probes that are spanning the CpG island, the probes cg23982858, cg009275554, cg22538054, and cg03308628 display the higher difference on methylation (expressed as β values) compared to normal tissues. In a next step, the average methylation value of these probes was used to stratify the cohort of TCGA prostate adenocarcinoma samples with available methylation data (Figure 2A, TCGA-PRAD, TCGA Wanderer, n = 340). Based on the median value (β = 0.5144), the cohort samples were grouped either as methHigh (n = 170, β > 0.5144) or methLow (n = 167, β < 0.5144). Furthermore, we observed a negative association between USP44 mRNA expression and the average methylation value of cg23982858, cg009275554, cg22538054, and cg03308628 probes in TCGA-PRAD tumors (Firehose Legacy)(Pearson r = −0.5905, p < 0.0001; Figure 2B). As expected, USP44 mRNA levels were significantly lower in methHigh group of TCGA-PRAD tumors, compared to methLow group of tumors (p < 0.0001; Figure 2C). Notably, disease-free analysis of TCGA-PRAD (Firehose Legacy) patient data showed that disease free survival (DFS) was significantly shorter in methHigh compared to methLow group of patients (log-rank p = 0.0011; Figure 2D), indicating an association of USP44 loss by methylation with aggressive disease.
Figure 1.
USP44 promoter methylation. (A) Structure of USP44 gene isoforms by ENCODE genome viewer. Promoter regions with high CpG context (black peaks) is indicated with green color. Transcriptional start site is indicated by peaks of H3K27Ac histone marks according to ENCODE data. (B) Cytosine sites within the indicated CpG island that display significant methylation in prostate adenocarcinoma samples as compared to normal samples. Methylation data were obtained from TCGA Wanderer database and Mann-Whitney test was used to analyze methylation levels between cancerous and normal samples. p values < 0.0001 are indicated with four asterisks (**** indicates p < 0.0001).
Figure 2.
USP44 promoter methylation and its correlation to prostate adenocarcinoma patient data. (A) USP44 promoter methylation calculated by the average methylation values of cg23982858, cg009275554, cg22538054, and cg03308628 sites in TCGA prostate adenocarcinoma samples (TCGA-PRAD). Data were obtained from TCGA Wanderer database. Median USP44 promoter methylation among TCGA samples is indicated by the horizontal line. (B) Correlation between USP44 promoter methylation and mRNA expression. Expression data were obtained by TCGA-PRAD Firehose Legacy. (C) Boxplot of USP44 mRNA levels in methHigh and methLow groups. p < 0.0001. (D) Disease-free curve comparing methHigh and methLow groups. Patient data were obtained by TCGA-PRAD Firehose Legacy through cbioportal. Log-rank test p = 0.0011 (**** indicates p < 0.0001).
2.2. USP44 Methylation and Its Correlation with Genomic Deletions That Affect the Molecular Properties of Prostate Adenocarcinomas
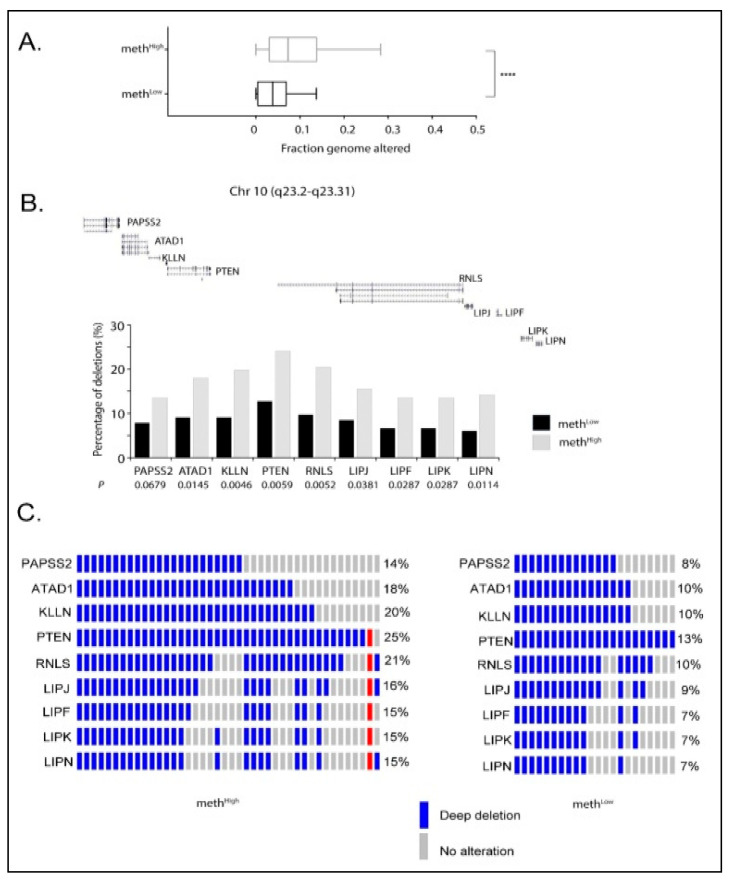
The comparative analysis of copy number variation data for methHigh and methLow groups (TCGA-PRAD; Firehose Legacy) showed that prostate tumors with higher levels of USP44 promoter methylation are characterized by a higher fraction of genome altered (p < 0.0001; Figure 3A). Moreover, this analysis revealed that loss of USP44 expression by promoter methylation is highly correlated with deep deletions in the genomic region chr10q23.2-q23.3. As a result, the frequency of deletions in PAPSS2, ATAD1, KLLN, PTEN, RNLS, LIPJ, LIPF, LIPK, LIPN genes, which are located within this genomic region, is significantly higher in methHigh group compared to methLow group (Figure 3B). Notably, the tumor suppressor PTEN is deleted in 25% of methHigh and in 13% methLow group of tumors. As shown in Figure 3C, deletions of the tumor suppressor PTEN in methHigh tumors is associated with co-deletions of the other genes in close proximity within the chr 10q23.2-q23.3 region (PAPSS2, ATAD1, KLLN, RNLS, LIPJ, LIPF, LIPK, LIPN), indicating that this chromosomic region is sensitive to USP44 loss associated genomic instability.
Figure 3.
USP44 methylation and its correlation to PTEN deletions in prostate adenocarcinoma patients. (A) Fraction of genome altered between methHigh and methLow groups. Data were obtained by TCGA-PRAD (Firehose Legacy). p > 0.0001. (B) USP44 promoter methylation and correlation to gene deletions in chromosomal region Chr 10 (q23.2-q23.31). Copy number variation data (CNVs) were obtained by TCGA-PRAD (Firehose Legacy). p values underneath the gene names were calculated with Mann-Whitney test. (C) Oncoprints indicating patients with gene deletion events within the chromosomal region Chr 10 (q23.2-q23.31) in methHigh (left) and methLow (right) group. Gene deletion frequencies (%) within the group are indicated on the right side of the oncoprint. Oncoprints were generated through cbioportal using CNV data from TCGA-PRAD (Firehose Legacy).
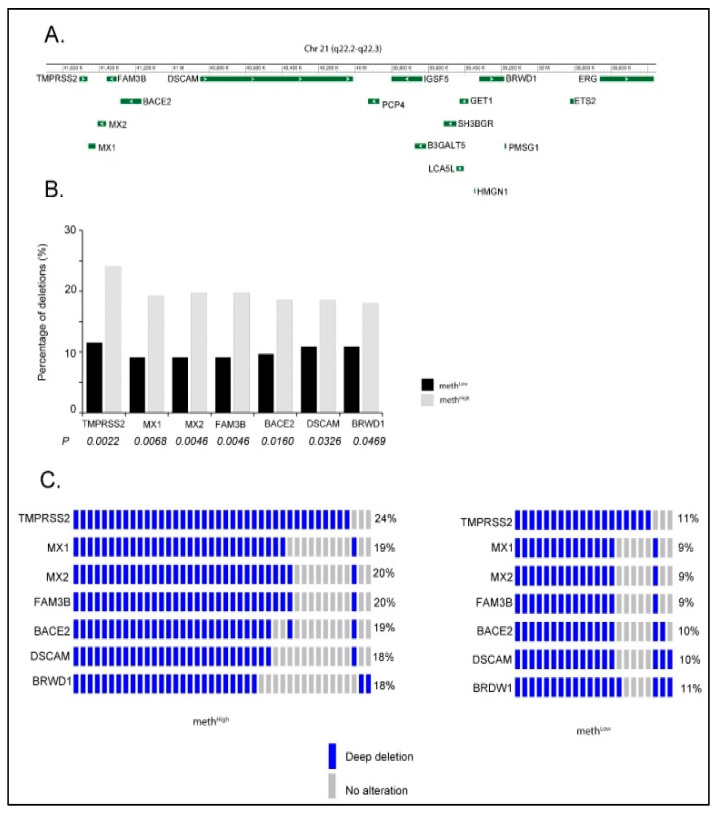
Interestingly, the chr 21q22.2-22.3 genomic region (Figure 4A) was also identified to be associated with deep deletion events in prostate tumors with high levels of USP44 promoter methylation. As a result, the frequency of deletions in TMPRSS2, MX1, MX2, FAM3B, BACE2, DSCAM, BRWD1, and ERG genes that are located within this region is significantly increased in methHigh compared to methLow group of tumors (Figure 4B). Moreover, as shown in Figure 4C, the copy number data indicate a profile of co-deletions for these genes in methHigh tumors, which is probably induced by the loss of the main body of the chr 21q22.2-22.3 genomic region in these samples. Notably, the deletion of the interstitial region between the TMPRSS2 and ERG genes has been described as a mechanism for generation of TMPRSS2-ERG fusions (30).
Figure 4.
USP44 promoter methylation and its correlation to deletions in the genomic region between TMPRSS2 and ERG genes in prostate adenocarcinoma patients. (A) Genome view of the chromosomal region Chr 21 (q22.2-q22.3). (B) Copy number variation data for Chr 21 (q22.2-q22.3) for methHigh and methLow groups. CNV data were obtained by TCGA-PRAD (Firehose Legacy). p values underneath the gene names were calculated with Mann-Whitney test. (C) Oncoprints indicating patients with gene deletion events within the chromosomal region Chr 21 (q22.2-q22.3) in methHigh (left) and methLow (right) group. Gene deletion frequencies (%) within each group are indicated on the right side of the oncoprint. Oncoprints were generated through cbioportal using CNV data from TCGA-PRAD (Firehose Legacy).
2.3. Analytical Validation of the Developed MSP Assay for USP44
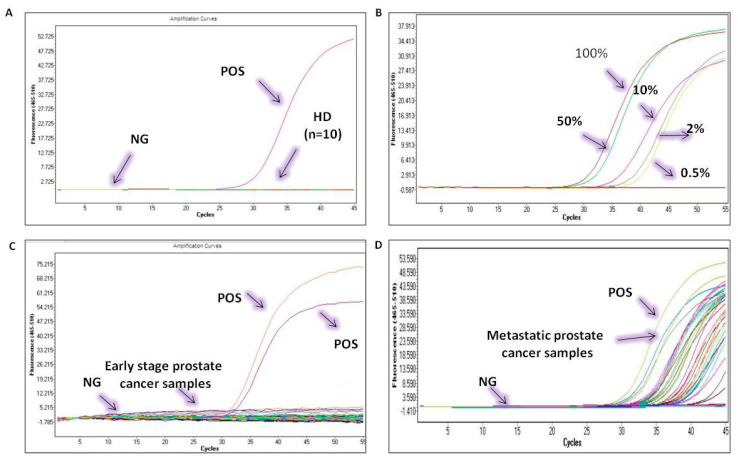
Analytical specificity: We initially evaluated the analytical specificity of the USP44 real-time MSP, by testing the primers in silico and then in PCR, using SB-modified human placental gDNA samples that were not methylated; no amplification of the USP44 was observed. In contrast, amplification was observed only when SB-treated DNA from the 100% methylated standard was used. The developed assay is highly specific since it detects only SB-treated USP44 methylated sequences (Figure 5A).
Figure 5.
Analytical validation of theUSP44 promoter methylation real time MSP assay. (A) analytical specificity, (B) analytical sensitivity, (C) USP44 RT-MSP in early-stage prostate cancer samples, (D) USP44 RT-MSP in metastatic prostate cancer samples.
Analytical sensitivity: The analytical sensitivity of the developed real-time MSP was evaluated by using synthetic mixtures based on serial dilutions of SB-converted DNA control samples (0% and 100% methylated) at various percentages of methylation (0.5%, 1%, 10%, and 50%). The developed real-time MSP assay for USP44 methylation could specifically and reliably detect the presence of 1% methylated USP44 sequences in the presence of 99% non-methylated USP44 sequences (Figure 5B).
2.4. USP44 Methylation in Plasma Cell-Free DNA of Prostate Cancer Patients
The developed assay was first applied in 10 HD plasma samples to evaluate specificity; none of the HD plasma samples was found methylated at the USP44 promoter (0/10, 0%) (Figure 5A).
We then examined the methylation status of USP44 in plasma samples from 32 early prostate cancer patients; we did not observe methylation of USP44 in this group (0%) (Figure 5C).
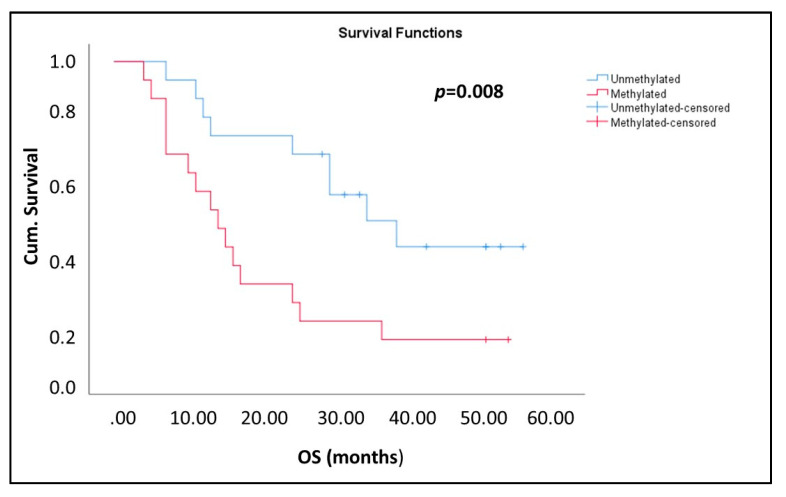
We then examined the methylation status of USP44 in plasma cfDNA samples from 39 prostate cancer patients with verified metastasis. In this group, 20/39 samples tested positive for USP44 methylation (51.3%) (Figure 5D). Kaplan-Meier analysis has shown that in the group of metastatic prostate cancer patients, the detection of USP44 methylation in plasma cfDNA was significantly correlated with worse OS (p = 0.008; Figure 6).
Figure 6.
Kaplan–Meier estimates of patients with metastatic prostate cancer in relation to USP44 methylation in ctDNA (n = 39).
3. Discussion
In this study we report for the first time that detection of USP44 promoter methylation in plasma cell free DNA provides significant prognostic information in metastatic prostate cancer, since it was significantly correlated with a worse overall survival. USP44 is a member of family proteins deubiquitinases and plays an important role in cell growth. Targeted ubiquitination plays an essential role in the orderly progression of cells through mitosis. Our meta-analysis of TCGA prostate methylation and patient data indicate that high levels of USP44 promoter methylation are associated with a more aggressive disease. USP44 was recently found to play an important role in genome integrity maintenance, as it is an essential component of DNA damage response and mitotic checkpoint machinery [7,24,25,26]. Therefore, loss of USP44 expression by methylation is expected to increase genomic instability in cancer. Towards this direction, our meta-analysis of TCGA copy variation data demonstrated that USP44 promoter methylation is associated with deletions in genes that are located in chr10q23.2-q23.3 and chr 21q22.2-22.3 genomic regions including the PTEN locus and the interstitial region between TMPRSS2 and ERG genes.
Numerous studies have shown that genomic deletion of PTEN is linked to prostate cancer recurrence and castration resistance [27,28,29,30]. On the other hand, several studies have demonstrated that deletion of the chr 21q22.2-22.3 genomic region and more specifically deletions of the interstitial genes between the androgen-regulated gene TMPRSS2 and the ETS transcription factor family member ERG leads to the generation of a TMPRSS2-ERG fusion gene that promotes prostate cancer progression [31,32,33]. Therefore, the association of USP44 promoter methylation with genomic alterations related to PTEN loss and TMPRSS2-ERG fusion formation provides further evidence for the prognostic value of this epigenetic alteration.
Few studies have attempted so far to elucidate the clinical significance of USP44 promoter methylation in cancer patients. The first studies on USP44 methylation were performed in mouse models and revealed that USP44 loss caused spontaneous tumors (31). USP44 expression analysis in human T-cell acute lymphoblastic leukemia (ALL) has shown overexpression of USP44 compared to healthy donors (32). In vitro experiments in breast cancer and TCGA analysis suggested that USP44 low expression was caused by promoter hypermethylation (12). The same group reported that USP44 overexpression significantly suppressed cancer cell proliferation, migration, and invasion and induced apoptosis (12). In colorectal neoplasia an interaction between USP44 promoter methylation and cancer was shown (13). Experiments in cell lines have shown that when USP44 promoter was methylated, USP44 mRNA transcripts were undetectable. It was also reported that USP44 promoter was methylated in 89% of colorectal adenomas studied (n = 89), while in the matched normal colon mucosa samples only 3% (n = 51) were methylated. However, loss of USP44 did not correlate with aneuploidy in colorectal adenomas (13). In NSCLC, there is only one study so far on USP44 (14). In this study, USP44 downregulation in 16 NSCLC tissues and in 4 cell lines was reported (14).
We evaluated for the first time the prognostic significance of USP44 promoter methylation in plasma cfDNA from patients with prostate cancer. Towards this we firstly developed and analytically validated a novel highly specific and sensitive real-time MSP assay. Our findings indicate that in the early stage of the disease USP44 promoter methylation was not detected in plasma cfDNA. On the contrary, in the metastatic setting, a high percentage (51.4%) of plasma cfDNA samples was positive for USP44 promoter methylation, indicating that USP44 promoter methylation may play an important role in this stage of prostate cancer. Moreover, according to our findings the detection of USP44 promoter methylation was significantly associated with OS in these patients. So far, there is only one study on USP44 in prostate cancer, based on the protein expression of this gene by immunofluorescence (33). According to the results presented in this study, USP44 regulates enhancer of zeste homolog 2 (EZH2), a histone H3 lysine 27 methyltransferase, which is involved in the development and progression of various cancers (33). In vitro experiments performed in this study suggested that the loss of USP44 reduced both the EZH2 protein levels and oncogenic activity of prostate cancer cells. Our results indicate on the contrary that loss of USP44 through promoter methylation results in a worse OS in patients with metastatic disease. However, this could be explained by the fact that this study was based on prostate cancer cell lines and no clinical samples were used.
In conclusion, we report for the first time that USP44 promoter is methylated at a high percentage in plasma cfDNA of metastatic prostate cancer patients but not in healthy donors and that detection of USP44 promoter methylation in plasma cell free DNA provides significant prognostic information in metastatic prostate cancer.
4. Materials and Methods
4.1. Prostate Adenocarcinoma TCGA Data and Statistical Analysis
All meta-analyses performed in this manuscript used data generated by The Cancer Genome Atlas Research (TCGA) Network and retrieved from cBioPortal (http://cbioportal.org (accessed on 20 April 2021) and TCGA Wanderer (http://maplab.imppc.org/wanderer/ (accessed on 25 April 2021) databases. Our analyses relied exclusively upon patient data which are publicly available. More specifically, methylation values (Illumina 450k Infinium chip data) from prostate tumor (n = 340) and normal samples (n = 49) were obtained from TCGA Wanderer. RNAseq, copy number variation, and clinical data for the prostate tumor samples with available methylation data were retrieved from the TCGA-PRAD Firehose legacy cohort.
Statistical differences in methylation (expressed as β values) of CG probes between normal and prostate tumors were determined by the non-parametric Mann-Whitney test. Pearson correlation coefficient was calculated to measure the linear relationship between USP44 mRNA expression and the mean methylation value of cg23982858, cg009275554, cg22538054, and cg03308628 sites among the TCGA samples. Based on the median value of methylation in these sites (β = 0.5144), the cohort samples were grouped either as methHigh (n = 170, β > 0.5144) or methLow (n = 167, β < 0.5144). The long-rank test was used to compare the disease-free survival between methHigh and methLow groups. Statistical significance of group enrichments in copy number alterations was accompanied by p values derived from chi-square test (X2). Statistical analyses were performed using GraphPadPrism ver.7.0. A p value of less than 0.05 was considered statistically significant.
4.2. Clinical Samples
Our study material consisted of a total of 71 clinical samples: (a) 32 plasma samples of patients with early stage prostate cancer; (b) 39 plasma samples of patients with metastatic prostate cancer from the second Department of Surgery at the Aretaieio University Hospital of Athens—Medical School, the National Kapodistrian University of Athens, and the Mutual Health Fund of National Bank of Greece Personnel; and (c) 10 plasma samples from healthy donors (HD). The study was conducted in accordance with the 1964 Declaration of Helsinki and was approved by the ethics and scientific committees of the participating institutions. All participating patients gave their signed informed consent in order to participate in the study.
4.3. Sample Preparation
To avoid contamination, different rooms, dedicated labware, and dedicated areas were used for all procedures. All DNA preparation and handling steps took place in specific laminar flow hoods under DNase-free conditions. DNA concentration in all cases was measured with a NanoDrop-1000 spectrophotometer (Thermo Scientific Waltham, Massachusetts, USA); isolated gDNA.
4.4. cfDNA Isolation from Plasma
Peripheral blood samples were collected into venous blood collection tubes using EDTA as a coagulant. Samples were mixed thoroughly and plasma was isolated within 2 to 4 h from sample collection by centrifugation at 530× g for 10 min at room temperature. Once isolated, plasma samples were centrifuged again at 2000× g for 10 min, before transferring into clean 2-mL tubes and freezing at −70 °C until time of processing. cfDNA was extracted from 2,00 mL plasma using the QIAamp® Circulating Nucleic Acid kit 50 (Qiagen®, Hilden, Germany), according to the manufacturer’s instructions. Quality control checks were performed in every step before every experimental procedure. Cell free DNA integrity in all plasma samples was checked by amplifying a region in exon 20 of the PIK3CA gene as previously described [34].
4.5. Sodium Bisulfite Conversion
All samples with good quality of cell-free DNA (verified by positive amplification for PIK3CA) were further processed to Sodium Bisulfite (SB) treatment using the EZ DNA Methylation Gold Kit (ZYMO Research). Up to 500 ng cfDNA were used in each SB-reaction. The Universal Methylated Human DNA Standard (ZYMO Research) was used as fully methylated (100%) positive control. The quality of SB-treated DNA was checked by a real-time PCR assay for β-actin (ACTB). SB-converted DNA samples were stored at −70 °C until further use.
4.6. In Silico Design of MSP Primers
We first designed MSP primers for USP44 in silico using the Primer Premier 5 software (Premier Biosoft International, San Francisco, CA, USA) avoiding the formation of stable hairpin structures, primer dimers, cross dimers, and false priming sites. The in silico validation was carried out using BLAST tool, in order to check their specificity and eliminate the risk of amplifying undesired sequences. The gene region of interest was selected by using the database TCGA Wanderer which provides gene expression and methylation data derived from TCGA database [35] (http://maplab.cat/wanderer (accessed on 20 April 2021)).
4.7. Optimization of Experimental Conditions
The experimental conditions of real-time MSP for USP44 promoter methylation were first optimized in detail for the annealing temperature and time, then for the optimum concentrations of the primer pair, and finally for buffer, MgCl2, dNTPs, and BSA concentrations (data not shown). Each MSP reaction was performed in the 96-well plate LightCycler® 480 System (IVD; Roche Molecular Diagnostics) (Mannheim, Germany) in a total volume of 10 μL. One microliter of SB-converted DNA was added to 9 μL reaction mixture containing 0.05 U·μL−1 GoTaq® Hot Start Polymerase (Promega, Maddison, WI, USA), 0.2× of the supplied PCR buffer, 2 mM of MgCl2, 0.15 mM of each dNTP (Thermo Fisher Scientific, Waltham, Massachusetts, USA), 0.15 μg·μL−1 BSA, 0.2 mM of the forward and reverse primers, and 1× LC Green Plus Dye (Idaho Technology, Salt Lake City, UT, USA). Finally, deionized water was added to a final volume of 10 μL. Real-Time MSP protocol began with one cycle at 95 °C for 2 min followed by 45 cycles of 95 °C for 10 s, 65 °C for 20 s, and 72 °C for 20 s. Immediately after amplification, a rapid cooling cycle to 40 °C for 30 s was introduced in order to prepare the melting curve acquisition step. Real-time fluorescence acquisition was set at the elongation step (72 °C). The following melting curve analysis included the steps of 55 °C for 10 s, 92 °C for 0 s with a ramp rate 0.11 °C·s−1 (acquisition mode: continuous), 92 °C for 1 min, and 40 °C for 1 min.
5. Conclusions
In conclusion, we report for the first time that USP44 promoter is methylated at a high percentage in plasma cfDNA of metastatic prostate cancer patients but not in healthy donors and that detection of USP44 promoter methylation in plasma cell free DNA provides significant prognostic information in metastatic prostate cancer.
Author Contributions
Conceptualization, E.S.L. and T.R.; methodology, D.L.; S.M.; validation, D.L., E.S.L.; formal analysis, D.L., M.Z., T.R. and E.S.L.; investigation, D.L., T.R.; resources, E.B., A.T. and E.S.L.; writing—original draft preparation, D.L., T.R. and E.S.L.; writing—review and editing D.L., T.R. and E.S.L.; visualization, D.L. and E.S.L.; supervision, E.S.L.; project administration, E.S.L.; funding acquisition, E.S.L. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was co-funded by the European Union and Greek national funds through the Operational Program “Competitiveness, Entrepreneurship and Innovation”, under the TRANSCAN-2 3rd JOINT TRANSNATIONAL call (project code: Τ8ΕΡA2-00036).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics and Scientific Committee of Aretaieio University Hospital and Mutual Health Fund of National Bank of Greece Personnel. (Ethical code: 31/18.12.2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Morreale F.E., Walden H. Types of Ubiquitin Ligases. Cell. 2016;165:248. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Argiles-Castillo D., Kane E.I., Zhou A., Spratt D.E. Erratum: HECT E3 ubiquitin ligases—Emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020;133:jcs228072. doi: 10.1242/jcs.228072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clague M.J., Urbé S., Komander D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019;20:338–352. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 5.Simonetti G., Bruno S., Padella A., Tenti E., Martinelli G. Aneuploidy: Cancer strength or vulnerability? Int. J. Cancer. 2019;144:8–25. doi: 10.1002/ijc.31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland A.J., Cleveland D.W. The deubiquitinase USP44 is a tumor suppressor that protects against chromosome missegregation. J. Clin. Investig. 2012;122:4325–4328. doi: 10.1172/JCI66420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Mandemaker I.K., Matsumoto S., Foreman O., Holland C.P., Lloyd W.R., Sugasawa K., Vermeulen W., Marteijn J.A., Galardy P.J. USP44 Stabilizes DDB2 to Facilitate Nucleotide Excision Repair and Prevent Tumors. Front. Cell Dev. Biol. 2021;9:959. doi: 10.3389/fcell.2021.663411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T., Sun B., Zhao X., Li Y., Zhao X., Liu Y., Yao Z., Gu Q., Dong X., Shao B., et al. USP44+ cancer stem cell subclones contribute to breast cancer aggressiveness by promoting vasculogenic mimicry. Mol. Cancer Ther. 2015;14:2121–2131. doi: 10.1158/1535-7163.MCT-15-0114-T. [DOI] [PubMed] [Google Scholar]
- 9.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berdasco M., Esteller M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Wu X., Lei W. USP44 hypermethylation promotes cell proliferation and metastasis in breast cancer. Future Oncol. 2021;17:279–289. doi: 10.2217/fon-2020-0415. [DOI] [PubMed] [Google Scholar]
- 13.Sloane M.A., Wong J.W.H., Perera D., Nunez A.C., Pimanda J.E., Hawkins N.J., Sieber O.M., Bourke M.J., Hesson L.B., Ward R.L. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics. 2014;9:1092–1100. doi: 10.4161/epi.29222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y.K., Tian W.Z., Zhang R.S., Zhang Y.J., Ma H.T. Ubiquitin-specific protease 44 inhibits cell growth by suppressing AKT signaling in non-small cell lung cancer. Kaohsiung J. Med. Sci. 2019;35:535–541. doi: 10.1002/kjm2.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong X., Zhong G. Prognostic biomarker identification and tumor classification in breast cancer patients by methylation and transcriptome analysis. FEBS Open Bio. 2021;11:2139–2151. doi: 10.1002/2211-5463.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J.M., Lee J.E., Park C.M., Kim J.H. Usp44 promotes the tumorigenesis of prostate cancer cells through ezh2 protein stabilization. Mol. Cells. 2019;42:17–27. doi: 10.14348/molcells.2018.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lianidou E., Pantel K. Liquid Biopsies. Genes Chromosomes Cancer. 2018;58:219–232. doi: 10.1002/gcc.22695. [DOI] [PubMed] [Google Scholar]
- 18.Lianidou E., Hoon D. Circulating tumor cells and circulating tumor DNA. In: Nader R., Horvath A.R., Wittwer C.T., Park J., editors. Principles and Applications of Molecular Diagnostics. 1st ed. Elsevier; Amsterdam, The Netherlands: 2018. pp. 235–281. [Google Scholar]
- 19.Kilgour E., Rothwell D.G., Brady G., Dive C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell. 2020;37:485–495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Lianidou E. Detection and relevance of epigenetic markers on ctDNA: Recent advances and future outlook. Mol. Oncol. 2021;15:1683–1700. doi: 10.1002/1878-0261.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chimonidou M., Strati A., Malamos N., Georgoulias V., Lianidou E.S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin. Chem. 2013;59:270–279. doi: 10.1373/clinchem.2012.191551. [DOI] [PubMed] [Google Scholar]
- 22.Chimonidou M., Strati A., Tzitzira A., Sotiropoulou G., Malamos N., Georgoulias V., Lianidou E.S. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin. Chem. 2011;57:1169–1177. doi: 10.1373/clinchem.2011.165902. [DOI] [PubMed] [Google Scholar]
- 23.Mastoraki S., Strati A., Tzanikou E., Chimonidou M., Politaki E., Voutsina A., Psyrri A., Georgoulias V., Lianidou E. ESR1 methylation: A Liquid biopsy-based epigenetic assay for the follow up of patients with metastatic breast cancer receiving endocrine treatment: Short running title: Liquid biopsy: ESR1 methylation in CTCs and paired ctDNA. Clin. Cancer Res. 2018;24:1500–1510. doi: 10.1158/1078-0432.CCR-17-1181. [DOI] [PubMed] [Google Scholar]
- 24.Mosbech A., Lukas C., Bekker-Jensen S., Mailand N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J. Biol. Chem. 2013;288:16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Foreman O., Wigle D.A., Kosari F., Vasmatzis G., Salisbury J.L., van Deursen J., Galardy P.J. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Investig. 2012;122:4362–4373. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stegmeier F., Rape M., Draviam V.M., Nalepa G., Sowa M.E., Ang X.L., McDonald III E.R., Li M.Z., Hannon G.J., Sorger P.K., et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 27.Kroh A., Diedler T., Burkhardt L., Mayer P.S., De Silva C., Meyer-Kornblum M., Kötschau D., Tennstedt P., Huang J., Gerhäuser C., et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol. 2012;181:401–412. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Sircar K., Yoshimoto M., Monzon F.A., Koumakpayi I.H., Katz R.L., Khanna A., Alvarez K., Chen G., Darnel A.D., Aprikian A.G., et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 2009;218:505–513. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 29.Choucair K., Ejdelman J., Brimo F., Aprikian A., Chevalier S., Lapointe J. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC Cancer. 2012;12:543. doi: 10.1186/1471-2407-12-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carver B.S., Chapinski C., Wongvipat J., Hieronymus H., Chen Y., Chandarlaparty S., Arora V.K., Le C., Koutcher J., Scher H., et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermans K.G., van Marion R., van Dekken H., Jenster G., van Weerden W.M., Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 32.Mehra R., Tomlins S.A., Yu J., Cao X., Wang L., Menon A., Rubin M.A., Pienta K.J., Shah R.B., Chinnaiyan A.M. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linn D.E., Penney K.L., Bronson R.T., Mucci L.A., Li Z. Deletion of Interstitial Genes between TMPRSS2 and ERG Promotes Prostate Cancer Progression. Cancer Res. 2016;76:1869–1881. doi: 10.1158/0008-5472.CAN-15-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vorkas P.A., Poumpouridou N., Agelaki S., Kroupis C., Georgoulias V., Lianidou E.S. PIK3CA hotspot mutation scanning by a novel and highly sensitive high-resolution small amplicon melting analysis method. J. Mol. Diagn. 2010;12:697–704. doi: 10.2353/jmoldx.2010.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Díez-Villanueva A., Mallona I., Peinado M.A. Wanderer, An interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin. 2015;8:1–8. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical restrictions.