Abstract
The Arabidopsis WD40 repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) regulates cell fate determination, including trichome initiation and root hair formation, as well as secondary metabolism such as flavonoid biosynthesis and seed coat mucilage production. TTG1 regulates different processes via regulating the expression of its downstream target genes by forming MYB-bHLH-WD40 (MBW) activator complexes with different R2R3 MYB and bHLH transcription factors. Here, we report the identification of the carboxyl (C)-terminus as a critical domain for TTG1′s functions in Arabidopsis. We found that the ttg1Δ15aa mutant shows pleiotropic phenotypes identical to a TTG1 loss-of-function mutant. Gene sequencing indicates that a single nucleotide substitution in TTG1 led to a premature stop at the W327 residue, leading to the production of a truncated TTG1 protein with a deletion of the last 15 C-terminal amino acids. The expression of TTG1 under the control of its native promoter fully restored the ttg1Δ15aa mutant phenotypes. Consistent with these observations, the expression levels of TTG1 downstream genes such as GLABRA2 (GL2) and CAPRICE (CPC) were reduced in the ttg1Δ15aa mutant. Assays in Arabidopsis protoplast show that TTG1Δ15aa failed to interact with the bHLH transcription factor GL3, and the deletion of the last 3 C-terminal amino acids or the 339L amino acid alone fully abolished the interaction of TTG1 with GL3. Furthermore, the expression of TTG1Δ3aa under the control of TTG1 native promoter failed to restore the ttg1Δ15aa mutant phenotypes. Taken together, our results suggest that the C-terminal domain of TTG1 is required for its proper function in Arabidopsis.
Keywords: TTG1, trichome formation, root hair formation, secondary metabolism, transcription factor, CRISPR/Cas9 gene editing, Arabidopsis
1. Introduction
TRANSPARENT TESTA GLABRA1 (TTG1) is a WD40 repeat protein with four WD40 motifs [1]. It is well known that TTG1 regulates cell fate determination including trichome and root hair formation and secondary metabolism such as flavonoid biosynthesis and seed coat mucilage production [2]. Arabidopsis loss-of-function mutants of TTG1 show a pleiotropic phenotype including glabrous leaves, increased root hairs, transparent testa seed coat, reduced anthocyanin accumulation, and seed coat mucilage production [1,3,4,5,6]. In addition, studies in recent years have also shown that TTG1 is involved in the regulation of other biological processes, such as the accumulation of fatty acids and proteins during the seed maturation [7,8], flowering time [9], as well as plant responses to biotic and abiotic stresses [10,11,12].
The presence of WD40 motifs is the only common feature of the WD40 proteins, and the WD40 motifs are able to provide a platform for the interactions of WD40 proteins and other proteins [13,14,15]. Indeed, the available evidence suggests that, at least in the terms of regulating cell fate determination and secondary metabolism, TTG1 is able to interact with different basic helix-loop-helix (bHLH) transcription factors and with different R2R3 MYB transcription factors to form different MYB-bHLH-WD40 (MBW) activator complexes, thereby regulating the expression of its downstream target genes involved in cell fate determination or secondary metabolism [16,17,18,19,20].
In regulating trichome formation, MBW activator complexes formed by TTG1, the bHLH transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3), and the R2R3 MYB transcription factor GLABRA1 (GL1) activate the expression of trichome regulator genes, including the homeodomain gene GLABRA2 (GL2,) and some of the R3 MYB genes, including TRYPTICHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), and ETC3 [17,18,19,21,22]. In regulating root hair formation, MBW activator complexes formed by TTG1, the bHLH transcription factor GL3 or EGL3, and the R2R3 MYB transcription factor WERWOLF (WER) activate the expression of root hair regulator gene GL2 [19,23,24]. In regulating anthocyanin and proanthocyanidin biosynthesis, MBW activator complexes formed by TTG1, the bHLH transcription factors TRANSPARENT TESTA 8 (TT8), GL3 or EGL3, and the R2R3 MYB transcription factors PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), PAP2, MYB113, MYB114, or TT2 activate the expression of the late biosynthesis genes in the flavonoid biosynthesis pathway, including DIHYDROFLAVONOL 4-REDUCTASE (DFR), ANTHOCYANIDIN SYNTHASE (ANS) and UDP-GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UF3GT), BANYULS (BAN), TT19, and TT12 [16,18,25,26,27,28,29,30,31]. However, in regulating seed coat mucilage production, the MBW activator complexes formed by TTG1, the bHLH transcription factor TT8 or EGL3, and the R2R3 MYB transcription factor MYB5 or TT2 activate the expression of mucilage biosynthesis genes TTG2 and GL2 [16,32,33,34,35,36].
Some experiments suggest that the formation of the MBW activator complexes may not be required for the activation of TTG1 downstream genes. For example, it has also been shown that the interaction of a R2R3 MYB and a bHLH transcription factor are required and sufficient to activate the expression of GL2 and some R3 MYB genes [37], and instead to form MBW activator complexes, TTG1 may compete with GL1 for binding GL3, therefore to form GL1-GL3 or GL3-TTG1 dimers [38]. Yet experiments in both yeast and plant cells show that TTG1 is able to interact with bHLH transcription factors [39,40], and the presence of TTG1 enhanced the interactions between R2R3 MYB and bHLH transcription factors [25,39].
The domains required for the interaction of R2R3 MYB and bHLH transcription factors have been well studied. It has been shown that R2R3 MYB proteins interact with bHLH transcription factors via their R3 domains, and the [D/E]L × 2[R/K] × 3L × 6L × 3R in R3 domain is the amino acid signature required for these interactions [39,40,41]. On the other hands, the N-terminal domain of bHLH transcription factors is required for the interaction of bHLH transcription factors with R2R3 MYB proteins [40]. However, the domain required for TTG1 to interact with bHLH transcription factor remains largely unstudied. So far, more than 20 mutants of TTG1 gene have been identified. Some loss-of-function mutants are caused by amino acid substitution, and the premature stop of TTG1 [2] suggests that the interaction of TTG1 and bHLH proteins may be affected in those mutants. However, it has only been shown that TTG1 with a deletion of the 25 carboxyl (C)-terminal amino acids, a truncated TTG1 protein corresponding to that in the ttg1-1 mutant [1], failed to interact with GL3 [39], indicating that the C-terminal domain is required for the interaction of TTG1 with bHLH transcription factors.
We report here the identification of ttg1Δ15aa, a new mutant of TTG1, and the characterization of C-terminus in the interaction of TTG1 with the bHLH transcription GL3. We found that ttg1Δ15aa is a TTG1 loss-of-function mutant, and assays in transfected protoplasts show that TTG1Δ15aa failed to interact with GL3. Further assays show that the deletion of the last three C-terminal amino acids or the 339L amino acid alone fully abolished the interaction of TTG1 with GL3, and the expression of TTG1Δ3aa under the control of TTG1 native promoter failed to restore the ttg1Δ15aa mutant phenotypes. There results suggest that the last few C-terminal amino acids are critical for TTG1′s functions in Arabidopsis.
2. Results
2.1. Phenotypes in the ttg1δ15aa Mutant Are Identical to That in A TTG1 Loss-of-Function Mutant
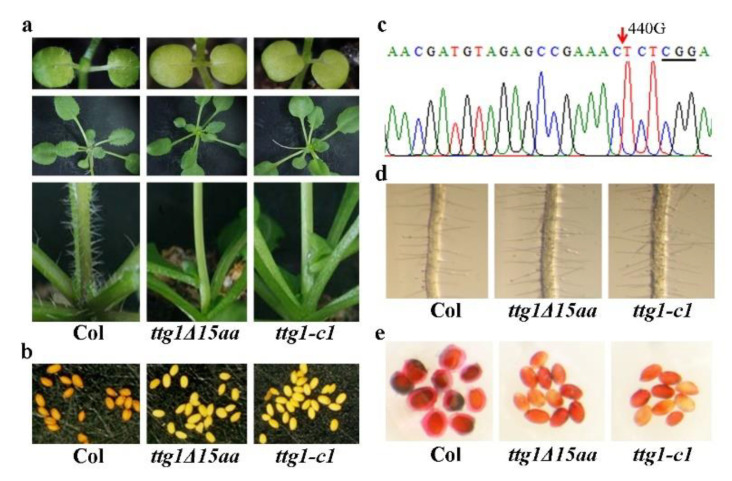
From an ethyl methanesulfonate (EMS) mutant pool in the Col ecotype background, we identified a mutant with glabrous phenotype (Figure 1a). The mutant was named ttg1Δ15aa, as a single nucleotide substation in TTG1 led to production of a truncated TTG1 protein with a deletion of the last 15 C-terminal amino acids (see Section 2.2 for details). In addition to glabrous leaves and stems, the ttg1Δ15aa mutant also has defects in anthocyanin accumulation (Figure 1a), as well as seed proanthocyanidin production (Figure 1b) and typical phenotypes observed in the TTG1 loss-of-function mutants [1,3,4,5,16].
Figure 1.
Phenotypes of the ttg1Δ15aa mutant and a TTG1 gene-edited mutant. (a) Trichome formation on leaves of 10-day-old seedlings (up panel), 4-week-old plants (middle panel), and stems of 5-week-old plants (lower panel) of the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. Seeds of the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants were germinated and grown in soil pots. Pictures were taken directly or under a Motic K microscope by using an EOS 1100D digital camera at indicated stages. (b) Seed color of the Col wild type, the ttg1Δ15aa and the ttg1-c1 mutants. Pictures were taken under a Motic K microscope by using an EOS 1100D digital camera. (c) Editing status of TTG1 in the ttg1-c1 mutant. The arrow indicates the 440 G deletion in the ttg1-c1 mutant. Underline indicates the PAM site. The mutant was obtained by transforming the Col wild type plants with the pHDE-TTG1 CRISPR/Cas9 construct. Editing status of TTG1 was examined in T1 plants, and Cas9-free homozygous mutant plants were obtained in T2 generation. (d) Root hair formation in the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. Seeds of Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants were germinated and grown on half MS plates vertically. Pictures were taken from 7-day-old seedlings under a Motic K microscope by using an EOS 1100D digital camera. (e) Mucilage production in the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. Seeds were stained with 0.01% (w/v) Ruthenium red for 2 h, and pictures were taken under a Motic K microscope by using an EOS 1100D digital camera.
To examine if ttg1Δ15aa mutant is a TTG1 loss-of-function mutant, we generated CRISPR/Cas9 gene edited mutant in the Col wild type background and compare their phenotypes side by side. In the ttg1-c1 mutant, a single G nucleotide deletion occurred at the position of 440 relative to the start codon of TTG1 (Figure 1c). As shown in Figure 1a,b, the ttg1Δ15aa mutant is identical to the ttg1-c1 mutant in terms of trichome formation, anthocyanin accumulation, and seed proanthocyanidin production. Both of the ttg1Δ15aa and the ttg1-c1 mutants also produced more root hairs (Figure 1d) and have defects on seed coat mucilage biosynthesis (Figure 1e).
2.2. A Single Nucleotide Substitution Led to the Production of A Truncated TTG1 Protein in the ttg1δ15aa Mutant
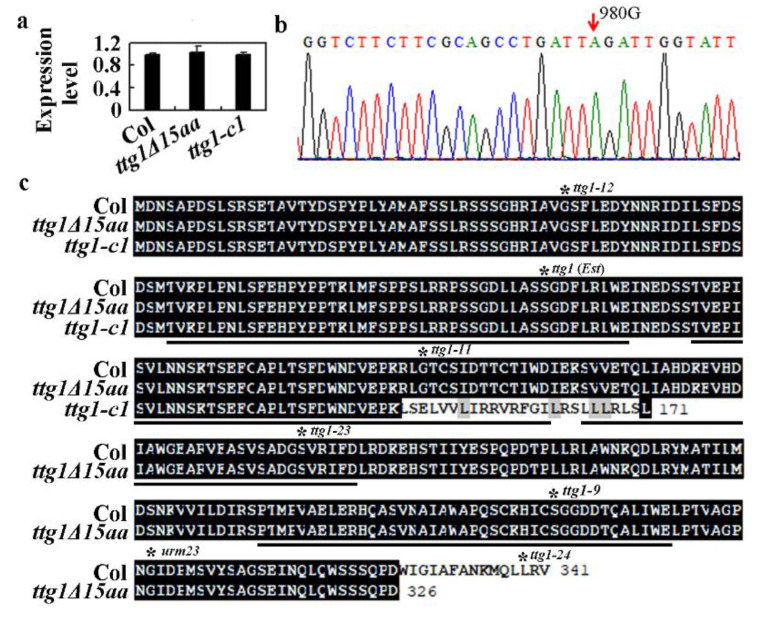
The above results suggest that ttg1Δ15aa is likely a loss-of-function mutant of TTG1. To examine if altered expression level of TTG1 may be responsible for the phenotype’s observation in the ttg1Δ15aa mutant, we examined the expression of TTG1 by using qRT-PCR. As show in Figure 2a, the expression level of TTG1 in seedlings of the ttg1Δ15aa mutant is largely indistinguishable from that in the Col wild type seedlings. Consistent with the fact of ttg1-c1 as a CRISPR/Cas9-edited mutant with only a single nucleotide deletion in TTG1, the expression level of TTG1 in the ttg1-c1 mutant is also similar to that in the Col wild type seedlings (Figure 2a).
Figure 2.
A single nucleotide substation in TTG1 led to premature stop in the ttg1Δ15aa mutant. (a) Expression of TTG1 in the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. RNA was isolated from 10-day-old seedlings, and qRT-PCR was used to examine the expression of TTG1. The expression of ACT2 was used as an inner control, and the expression level of TTG1 in the Col wild type was set as 1. Data represent the mean ± SD of three replicates. (b) Sequence of TTG1 in the ttg1Δ15aa mutant. The arrow indicates the 980G-A single-base substitution. (c) Amino acid alignment of TTG1 in the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. ORFs of TTG1 in the ttg1Δ15aa and the ttg1-c1 mutants were identified by using DNAMAN, and the predicted full-length amino acid sequences were used for alignment with the full-length amino acid sequence of TTG1 in the Col wild type. The numbers at the C-terminal indicate the numbers of total amino acids of TTG1 in the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants. Solid underlines indicate the WD40 domains. Stars indicate the amino acids that were substituted in the corresponding mutants as summarized previously [2].
Having shown that the phenotypes in the ttg1Δ15aa mutant are not caused by a reduced expression of TTG1 (Figure 2a), we then amplified and sequenced the genome sequence of TTG1 in the ttg1Δ15aa mutant. We found that the 980 G nucleotide was substituted by a nucleotide A (Figure 2b), therefore creating a TAG stop codon at the W327 residue, resulting in the production of truncated TTG1 protein with a deletion of the 15 C-terminal amino acids (Figure 2c). We thus named the mutant ttg1Δ15aa. As shown in Figure 2c, the deletion of the 15 amino acids did not affect any of the 4 WD40 repeats in TTG1 protein, whereas the nucleotide deletion in the ttg1-c1 mutant led to a few amino acid substitutions in the second WD40 repeat, and a premature stop occurred after the 171 amino acid residue. For comparison, mutations in all the known mutants with an amino acid substitution in TTG1, as summarized recently [2], were indicated in Figure 2c.
2.3. Expression of TTG1 under the Control of its Native Promoter Restored the Ttg1δ15aa Mutant Phenotypes
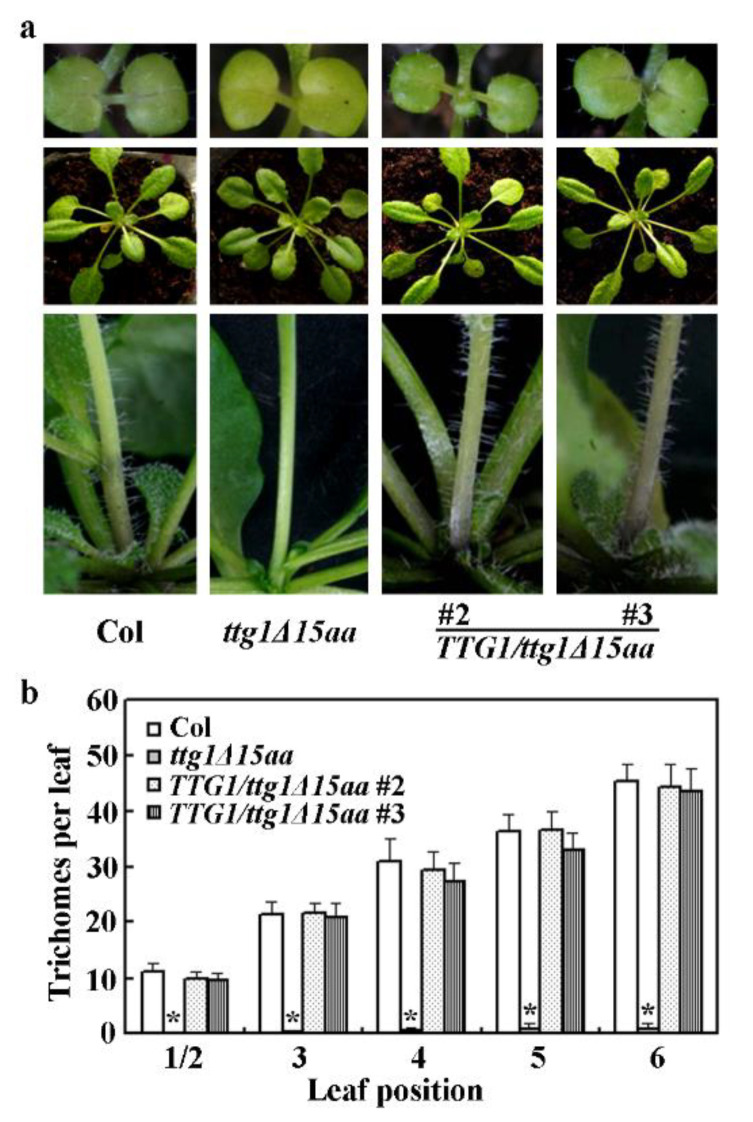
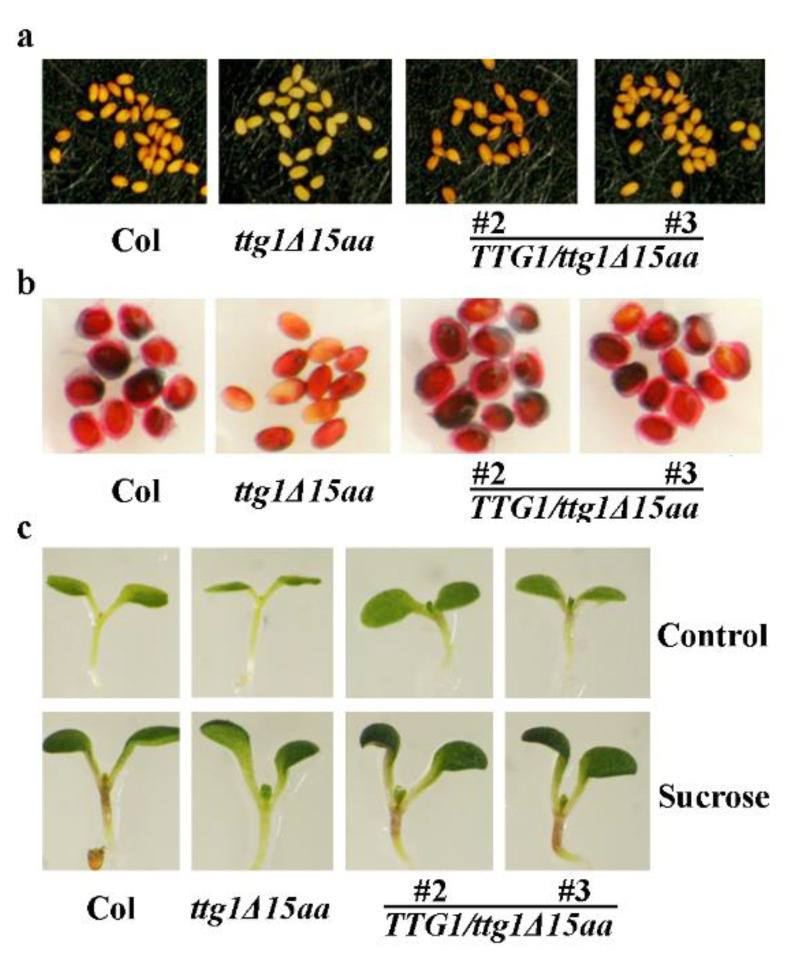
To further examine if ttg1Δ15aa is a TTG1 lost-of-function mutant, we performed phenotypic complementation experiment. Transgenic plants expressing TTG1 under the control of its native promoter were generated in the ttg1Δ15aa mutant. We found that trichome formation in the transgenic plants was restored (Figure 3a), and quantitative analysis show that trichome numbers in the transgenic plants examined were restored to a Col wild type level (Figure 3b). In addition, anthocyanin biosynthesis phenotype observed in the ttg1Δ15aa mutant was also restored in the transgenic plants (Figure 3a).
Figure 3.
Expression of TTG1 under the control of its native promoter restores trichome formation in the ttg1Δ15aa mutant. (a) Trichome formation on leaves of 10-day-old seedlings (up panel), 4-week-old plants (middle panel), and stems of 5-week-old plants (lower panel) of the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants were germinated and grown in soil pots. Pictures were taken directly or under a Motic K microscope by using an EOS 1100D digital camera at indicated stages. (b) Trichome numbers on the first two, third, fourth, fifth and sixth rosette leaves of the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. Trichome numbers on the indicated leaves were count under a Motic K microscope. Date represence mean ± SD of 10–14 leaves. *, Significantly different from that in the Col wild type (student’s t test, p < 0.0001).
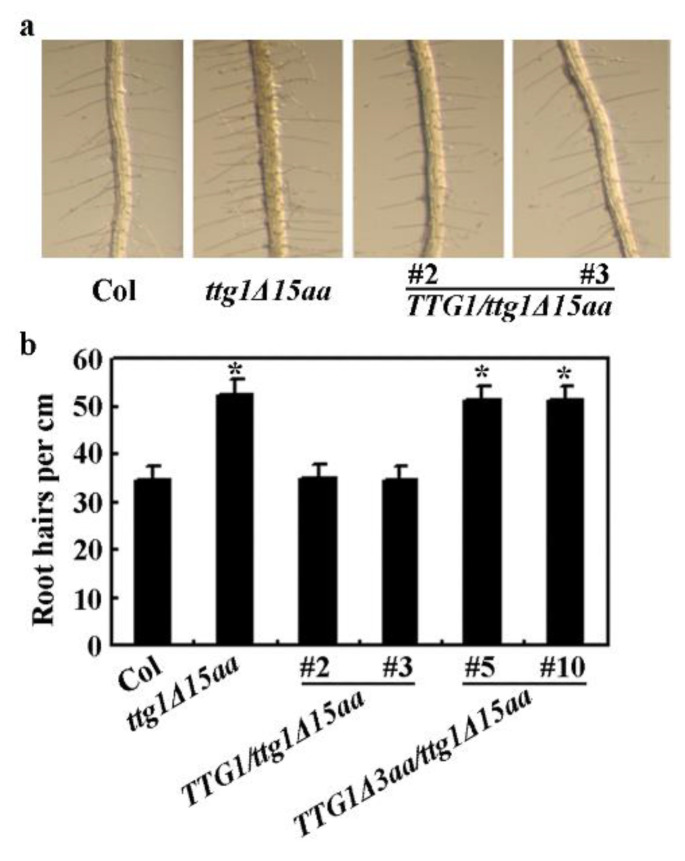
We also examined root hair formation in the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. We found that root hair formation is restored in the TTG1/ttg1Δ15aa transgenic plant seedlings (Figure 4a). Quantitative analysis showed that root hair density in the seedlings of the TTG1/ttg1Δ15aa, but not the TTG1Δ3aa/ttg1Δ15aa transgenic plants, is similar to that of the Col wild type seedlings (Figure 4b). TTG1Δ3aa is a truncated TTG1 protein with a deletion of the last three c-terminal amino acids (see Section 2.5 for details).
Figure 4.
Expression of TTG1 under the control of its native promoter restores root hair formation in the ttg1Δ15aa mutant. (a) Root hair formation in the Col wild type, the ttg1Δ15aa mutant and the TTG1/ttg1Δ15aa transgenic plants. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants were germinated and grown on ½ MS plates vertically. Pictures were taken from 7-day-old seedlings under a Motic K microscope by using an EOS 1100D digital camera. (b) Root hair density in seedlings of the Col wild type, the ttg1Δ15aa mutant, the TTG1/ttg1Δ15aa, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. Root numbers were count under a Motic K microscope, and root hair density was calculated. Date represence mean ± SD of 17–18 seedlings. *, Significantly different from that in the Col wuld type (student’s t test, p < 0.0001).
We further examined if the metabolism phenotypes in the TTG1/ttg1Δ15aa transgenic plants including seed proanthocyanidin and mucilage production were recovered in the TTG1/ttg1Δ15aa transgenic plants. As shown in Figure 5a, the seed color in the transgenic plants is similar to the Col wild type plants, and the seed coat mucilage biosynthesis was also restored in the TTG1/ttg1Δ15aa transgenic plants (Figure 5b). We also examined anthocyanin biosynthesis in seedlings in response to sucrose. We found that the ttg1Δ15aa mutant seedlings failed to produce anthocyanin, whereas the TTG1/ttg1Δ15aa transgenic plant seedlings accumulated more anthocyanin, similar to that of the Col wild type seedlings (Figure 5c).
Figure 5.
Expression of TTG1 under the control of its native promoter restores secondary metabolism in the ttg1Δ15aa mutant. (a) Seed coat color in the Col wild type, the ttg1Δ15aa mutant and the TTG1/ttg1Δ15aa transgenic plants. Pictures were taken under a Motic K microscope by using an EOS 1100D digital camera. (b) Mucilage production in the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. Seeds were stained with 0.01% (w/v) Ruthenium red for 2 h, and pictures were taken under a Motic K microscope by using an EOS 1100D digital camera. (c) Anthocyanin accumulation in seedlings of the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants in response to sucrose. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants were germinated and grown on half MS plates with or without 3% sucrose. Pictures were taken from 6-day-old seedlings.
2.4. TTG1Δ15aa Is Unable to Interact with GL3 and with Regulated Downstream Target Genes
It is well known that TTG1 directly interacts with bHLH transcription factors and together with different R2R3 MYB transcription factors to form MBW activator complexes to regulate downstream gene expression, thereby to regulate different processes including trichome formation, root hair formation, anthocyanin and proanthocyanidin biosynthesis, and seed coat mucilage production [2]. Since the above results suggest that ttg1Δ15aa is a TTG1 loss-of-function mutant, we examined if TTG1Δ15aa may be still able to interact with bHLH transcription factors.
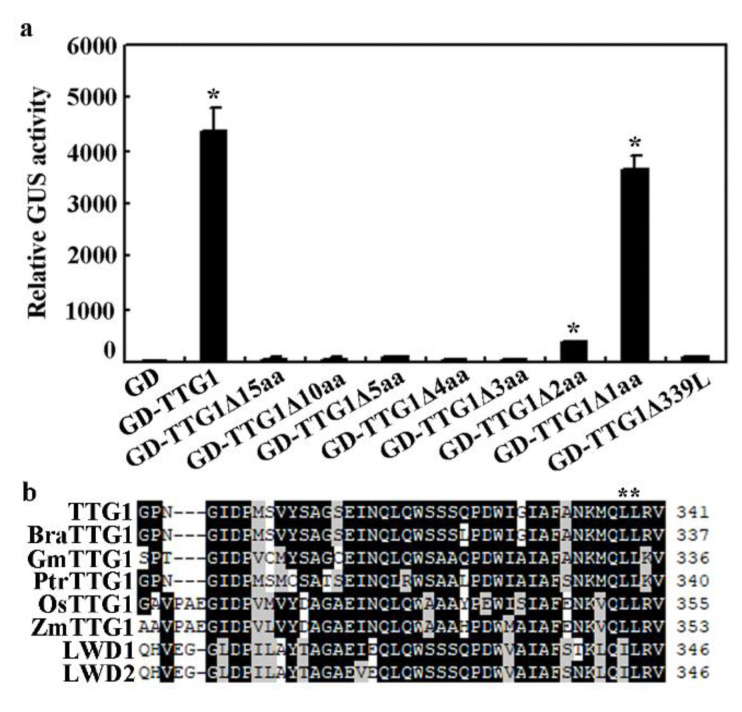
As we have previously shown that the bHLH transcription factor GL3 alone functions as an activator, whereas TTG1 alone is not able to activate reporter gene expression when recruited to the promoter region of the Gal:GUS reporter gene by a fused GD domain [40]. Therefore, we used GL3 as an example to examine if TTG1Δ15aa may be still able to interact with bHLH transcription factors. As shown in Figure 6a, the co-transfection of GD-TTG1 and GL3 activated the Gal4:GUS reporter gene expression, whereas the co-transfection of GD-TTG1Δ15aa and GL3 failed to do so, suggesting that TTG1Δ15aa is unable to interact with GL3. Assays with truncated TTG1 proteins indicate that the deletion of the last three C-terimnal amino acids or the 339L amino acid is sufficient to abolish the interaction of TTG1 with GL3 (Figure 6a, see Section 2.5 for details). Considering that amino acids in the C-terminus of TTG1 from different plants including Arabidopsis (TTG1), rapeseed (BraTTG1), soybean (GmTTG1), poplar (PtrTTG1), rice (OsTTG1), and corn (ZmTTG1) are highly conserved (Figure 6b), these results suggest that the C-terminus of TTG1 is critical for its functions.
Figure 6.
Interaction of full-length and truncated TTG1 with GL3. (a) Interaction of TTG1, TTG1Δ15aa, TTG1Δ10aa, TTG1Δ5aa, TTG1Δ4aa, TTG1Δ3aa, TTG1Δ2aa, TTG1Δ1aa, and TTG1Δ339L with GL3 in transfected protoplasts. Protoplasts were isolated from 3 to 4-week-old Col Arabidopsis rosette leaves. Plasmids of the effector genes GD-TTG1, GD-TTG1Δ15aa, GD-TTG1Δ10aa, GD-TTG1Δ5aa, GD-TTG1Δ4aa, GD-TTG1Δ3aa, GD-TTG1Δ2aa, GD-TTG1Δ1aa, or GD-TTG1Δ339L were co-transfected with the reporter gene Gal4:GUS and the effector gene HA-GL3 into protoplasts. The protoplasts were incubated in darkness for 20–22 h, and then GUS activity was assayed. Co-transfection of GD with Gal4:GUS and HA-GL3 was used as a control. Data represents the mean ± SD of three repeats. *, Significantly different from that in the GD control (Student’s t test, p < 0.0001). (b) Alignment of the C-terminal amino acids of TTG1, BraTTG1, GmTTG1, PtrTTG1, OsTTG1, ZmTTG1, AtLWD1, and AtLWD2 proteins. Identical amino acids are shaded in black, and similar in grey.
We then examined the expression of some TTG1 downstream genes, and we found that the expression of GL2 and CPC was decreased in the ttg1Δ15aa mutant, but largely recovered in the TTG1/ttg1Δ15aa transgenic plants (Figure 7).
Figure 7.
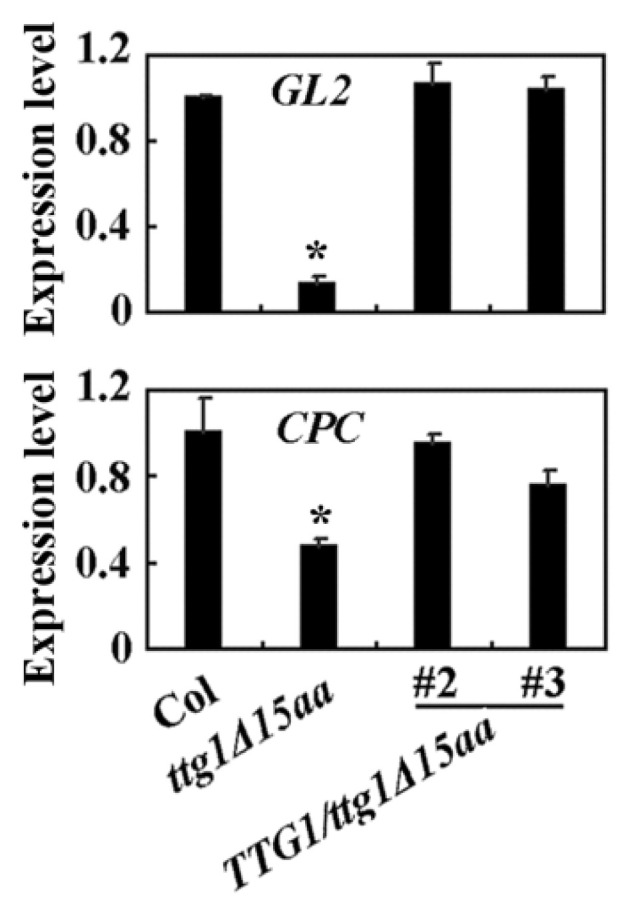
Expression of GL2 and CPC in the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. RNA was isolated from 10-day-old seedlings and qRT-PCR was used to examine the expression of GL2 and CPC. The expression of ACT2 was used as an inner control. The expression level of GL2 or CPC in the Col wild type was set as 1. Data represent the mean ± SD of three replicates. *, Significantly different from that in the Col wild type (Student’s t test, p < 0.01).
2.5. Deletion of the Last Three C-Terminalamino Acids Causes Loss-of-Function of TTG1
So far, more than 20 mutants of the TTG1 gene have been identified, and among them, only a few were caused by premature stop [2], with the ttg1-1 mutant having the least amino acids deleted at the C-terminus of TTG1 but still showing TTG1 loss-of-function mutant phenotypes [1,3]. In the ttg1-1 mutant, the mutation caused a premature stop at the position of the 317Q amino acid, resulting in the production of a truncated TTG1 protein with a deletion of the last 25 C-terminal amino acids [1]. In our case, however, only 15 amino acids were deleted, and it also caused TTG1 loss-of-function phenotypes in the mutant.
To further examine the minimum amino acids that may cause loss-of-function when deleted, we made truncated TTG1 constructs and examined their interaction with GL3 in transfected protoplasts. We found that the truncated TTG1 proteins with a deletion of the last three or more C-terminal amino acids, including TTG1Δ10aa, TTG1Δ5aa, TTG1Δ4aa, and TTG1Δ3aa, failed to interact with GL3 (Figure 6a). However, TTG1Δ2aa, truncated TTG1 with a deletion of the last two C-terminus amino acids, showed a weak interaction with GL3, whereas the interaction between TTG1Δ1aa, the truncated TTG1 with only the last one C-terminus amino acid deleted, and GL3 is largely unaffected (Figure 6a).
Amino acid alignment of the C-terminus of TTG1 from different plants showed that both of the third and fourth amino acids to the last are L residues and are conserved in all TTG1 proteins, but not LWD1 and LWD2, the two closely related proteins to TTG1 in Arabidopsis (Figure 6b). Previously research has also shown that ttg1-24, a mutant with an L339F amino acid substitution in TTG1, produced less trichomes and root hairs and reduced accumulation of proanthocyanidin [42]. In addition, TTG1L339F showed a weak interaction with GL3 [42]. We therefore examined if the deletion of one L amino acid may affect the functions of TTG1 by examining its interaction with GL3. As shown in Figure 6a, GD-TTG1Δ339L failed to interact with GL3 in the transfected protoplasts.
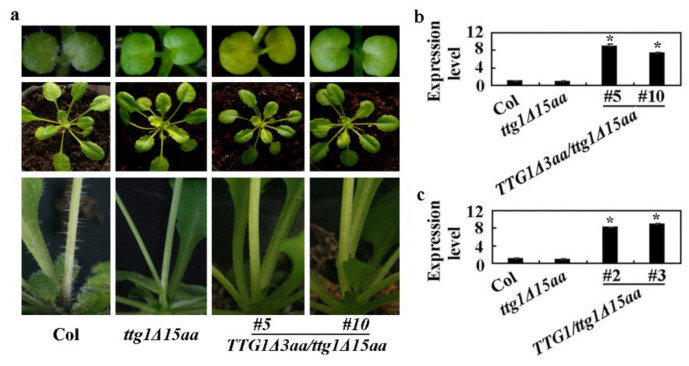
To further examine if the failure to interact with GL3 may indeed cause the loss-of-function of TTG1, we expressed TTG1Δ3aa under the control of TTG1′s native promoter in the ttg1Δ15aa mutant. We found that the transgenic plants still showed a glabrous phenotype (Figure 8a). The qRT-PCR results show that the expression level of TTG1Δ3aa in the TTG1Δ3aa/ttg1Δ15aa transgenic plants is elevated (Figure 8b) to a level similar to that of TTG1 in the TTG1/ttg1Δ15aa transgenic plants (Figure 8c).
Figure 8.
The expression of TTG1Δ3aa under the control of TTG1 native promoter failed to restore trichome formation in the ttg1Δ15aa mutant. (a) Trichome formation on leaves of 10-day-old seedlings (up panel), 4-week-old plants (middle panel), and stems of 5-week-old plants (lower panel) of the Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were germinated and grown in soil pots. Pictures were taken directly or under a Motic K microscope by using an EOS 1100D digital camera at indicated stages. (b) Expression of TTG1Δ3aa in Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. RNA was isolated from 10-day-old seedlings and qRT-PCR was used to examine the expression of TTG1Δ3aa. The expression of ACT2 was used as an inner control. The expression level of TTG1Δ3aa in the Col wild type was set as 1. Data represent the mean ± SD of three replicates. *, Significantly different from that in the Col wild type (Student’s t test, p < 0.01). (c) Expression of TTG1 in Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa transgenic plants. RNA was isolated from 10-day-old seedlings, and qRT-PCR was used to examine the expression of TTG1. The expression of ACT2 was used as an inner control. The expression level of TTG1 in the Col wild type was set as 1. Data represent the mean ± SD of three replicates. *, Significantly different from that in the Col wild type (Student’s t test, p < 0.01).
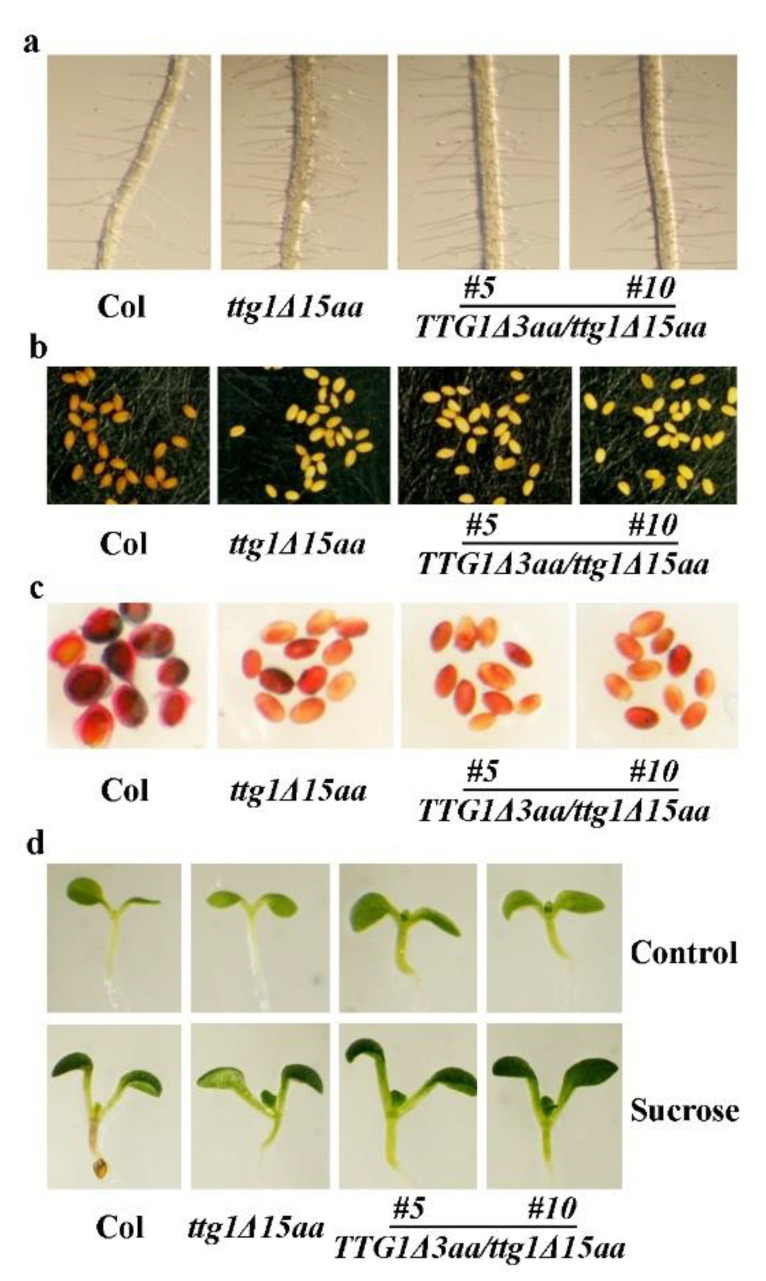
Root hair formation assays show that the TTG1Δ3aa/ttg1Δ15aa transgenic plant seedlings still produced more root hairs (Figure 9a), and a quantitative analysis indicates that the root hair density in the transgenic plant seedlings is similar to that in the ttg1Δ15aa mutant seedlings (Figure 4b). Furthermore, the TTG1Δ3aa/ttg1Δ15aa transgenic plants still produced yellow-colored seeds, as observed in the ttg1Δ15aa mutant (Figure 9b). The ruthenium red staining results show that the seed coats of the TTG1Δ3aa/ttg1Δ15aa transgenic plants are still not able to produce mucilage (Figure 9c), and the sucrose treatment suggests that the anthocyanin phenotypes observed in the ttg1Δ15aa mutant are also not restored in the TTG1Δ3aa/ttg1Δ15aa transgenic plants (Figure 9d).
Figure 9.
Expression of TTG1Δ3aa under the control of TTG1′s native promoter failed to restore root hair formation and secondary metabolism in the ttg1Δ15aa mutant. (a) Root hair formation in the Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were germinated and grown on half MS plates vertically. Pictures were taken from 7-day-old seedlings under a Motic K microscope by using an EOS 1100D digital camera. (b) Seed coat color in the Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. Pictures were taken under a Motic K microscope by using an EOS 1100D digital camera. (c) Mucilage production in the Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants. Seeds were stained with 0.01% (w/v) Ruthenium red for 2 h, and pictures were taken under a Motic K microscope by using an EOS 1100D digital camera. (d) Anthocyanin accumulation in seedlings of the Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants in response to sucrose. Seeds of Col wild type, the ttg1Δ15aa mutant, and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were germinated and grown on half MS plates with or without 3% sucrose. Pictures were taken from 6-day-old seedlings.
3. Discussion
TTG1 regulates the cell fate determination, flavonoid biosynthesis, and seed coat mucilage production in Arabidopsis via forming MBW activator complexes [2]. Experimental evidence in both yeast and plant cells indicate that TTG1 is able to interact with bHLH transcription factors [39,40]. However, the domain required for the interaction of TTG1 with the bHLH transcription factor remained largely unknown. So far, only the 25 C-terminal amino acids have been shown to affect the interaction of TTG1 with GL3 [39]. We provided evidence in this study that the C-terminus of TTG1 is required for the interaction of TTG1 with GL3 and may play a critical role for its functions in Arabidopsis.
First, we found that ttg1Δ15aa, a mutant with a premature stop, which is predicted to result in the production of a truncated TTG1 protein with its last 15 C-terminal amino acids deleted (Figure 2), is a TTG1 loss-of-function mutant. As the mutant shows a pleiotropic phenotype identical to the CRISPR/Cas9 gene-edited mutant ttg1-c1 (Figure 1), including glabrous leaves, increased root hairs, transparent testa seed coat, and reduced anthocyanin accumulation and seed coat mucilage production, typical phenotypes observed in the loss-of-function mutants of TTG1 [1,3,4,5,6]. In addition, the phenotypes observed in the ttg1Δ15aa mutant were recovered by the expression of TTG1 under the control of its native promoter (Figure 3, Figure 4 and Figure 5). Second, TTG1Δ15aa is unable to interact with the bHLH transcription factor GL3 in transfected protoplasts (Figure 6), and the expression levels of TTG1 downstream genes including GL2 and CPC were reduced in the ttg1Δ15aa mutant and were largely recovered in the TTG1/ttg1Δ15aa transgenic plants (Figure 7). Third, TTG1 with a deletion of the last three C-terminal amino acids or the 339L amino acid alone failed to interact with GL3 (Figure 6). Last but not least, the expression of TTG1Δ3aa, which encoded a TTG1 protein with a deletion of the last three C-terminal amino acids, in the ttg1Δ15aa mutant failed to recover its phenotypes (Figure 8 and Figure 9). It will be of interest to examine if the deletion of the last few amino acids may affect the binding of TTG1 to its target genes by using ChIP-seq or ChIP-PCR assays.
Even though both TTG1 and R2R3 MYB are able to interact with bHLH transcription factors, thereby forming MBW activator complexes [39,40], only the domains required for R2R3 MYB to interact with bHLH transcription factors have been well studied. The R3 domain has been identified as the interaction domains of R2R3 MYB proteins with bHLH transcription factors, the [D/E]L×2[R/K]×3L×6L×3R in R3 domain has been identified as an amino acid signature for this interaction [39,40,41], and the 92S in the R3 domain has also been found to be required for the interaction of R2R3 MYB transcription factor GL1 and bHLH proteins GL3/EGL3 [43]. As for TTG1, a previous study has only shown that the 25 C-terminal amino acids are required for the interaction of TTG1 with GL3 [39]. Our studies further narrow the region to the last three C-terminal amino acids, and we found that deletion of the 339L amino acid alone fully abolished the interaction of TTG1 with GL3 (Figure 6). Considering that the presence of the WD40 motifs is the only conserved feature of WD40 proteins [15] and WD40 proteins are able to provide a platform for interactions with other proteins [13,14,15], it will be of great interest to examine if the WD40 motifs in TTG1 may be required for its interaction with bHLH transcription factors.
As a matter of fact, a few amino acid substitution mutants of TTG1 have been found to affect the function of TTG1, including the ttg1-9 mutant with an S282F substitution [1,44], the ttg1-11 mutant with a G149R substitution [45], the ttg1-12 mutant with a G43R substitution [45], the ttg1-23 mutant with an S197F substitution [42], the ttg1-24 mutant with an L339F substitution [42], the urm23 mutant with a G302E substitution [46], and the ttg1 (Est) mutant with an S101F substitution [47]. Among them, amino acid substitution in the ttg1-9, ttg1-11, ttg1-23, and ttg1 (Est) mutant occurred in the WD40 motifs (Figure 2c), and all but ttg1-23 showed typical TTG1 loss-of-function mutant phenotypes [1,42,44,45,47], suggesting that WD40 motifs are important to the functions of TTG1. On the other hand, among the three mutants where amino acid substitution occurred outside the WD40 motifs, both ttg1-24 and urm23 have amino acid substitutions occurring at the C-terminus and showed weak phenotypes [42,46], but ttg1-12, a mutant with amino acid substitution occurred at the N-terminus, also showed typical TTG1 loss-of-function mutant phenotypes [45]. Examining the interactions of the TTG1 proteins with the above-mentioned amino acid substitutions with bHLH transcription factors may reveal addition domains/motifs required for the interaction of TTG1 and bHLH transcription factors.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col) wild type was used for protoplasts isolation, plant transformation, and served as controls for phenotypic analysis. The ttg1Δ15aa mutant was isolated from an EMS (ethyl methanesulfonate) mutant pool in the Col wild type background. The ttg1-c1 mutant was generated by using CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) gene editing in the Col wild type plants.
For trichome phenotypic analysis, protoplast isolation and/or plant transformation, seeds of the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants, the TTG1/ttg1Δ15aa and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were sown into soil pots directly and grew at 22 °C in a growth room with a photoperiod of 16 h light/8 h dark and light density at ~125 µmol m−2 s−1, as described previously [48].
For RNA isolation, anthocyanin biosynthesis, and/or root hair phenotypic analysis, seeds of the Col wild type, the ttg1Δ15aa, and the ttg1-c1 mutants, the TTG1/ttg1Δ15aa and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were bleach sterilized, washed five times with sterilized water, and then plated on plates containing 1% (w/v) sucrose and 1/2 Murashige & Skoog (MS) medium solidified with 0.6% (w/v) phytoagar (PlantMedia, Dublin, OH, USA). The plates were kept for 2 days in darkness at 4 °C and then transferred to a growth room, as described previously [49].
More than 15 plants/seedlings for each line were used for trichome and root hair phenotype analysis, and the experiments were repeated at least three times with similar results.
4.2. RNA Isolation and qRT-PCR
To examine the expression of trichome formation key regulator genes, or expression of TTG1 or TTG1Δ15aa, seedlings of 10-day-old Col wild type, the ttgΔ15aa, and the ttg1-c1 mutants, the TTG1/ttgΔ15aa and the TTG1Δ3aa/ttgΔ15aa transcription plants were collected, frozen in liquid nitrogen immediately after collection, and kept at −80 °C for RNA isolation.
Total RNA was isolated by using an EasyPure Plant RNA Kit (TransGene Biotech, Beijing, China), and 2 µg total RNA was used to synthesize cDNA by using the EazyScript First-Strand DNA Synthesis Super Mix (TransGene Biotech, Beijing, China) and following the manufacturer’s instructions. Synthesized cDNA was subjected to qRT-PCR analysis of the expression of TTG1, TTG1Δ3aa, and trichome formation core regulator genes including GL2 and CPC. The expression of ACTIN2 (ACT2) was used as an inner control for qRT-PCR. qRT-PCR was performed on an Applied Biosystems 7500 real time PCR System using SYBR Green/ROX Master Mix (TaKaRa Biomedicals, Dalian, China). Primers used for qRT-PCR examination of the expression of TTG1Δ3aa were 5′-CGAGCCAATCTCGGTTCTCA-3′ and 5′-CGAGACGTTTCGGCTCTACA-3′. Other primers used for qRT-PCR have been described previously [49].
4.3. Constructs
The reporter construct Gal4:GUS and the effector constructs GD (Gal4 DNA Binding Domain), GD-TTG1 and the HA-GL3 used for protoplasts transfection have been described previously [40,50].
To make GD-tagged TTG1Δ15aa, TTG1Δ10aa, TTG1Δ5aa, TTG1Δ4aa, TTG1Δ3aa, TTG1Δ2aa, TTG1Δ1aa, and TTGΔ339L constructs for protoplast transfection, the corresponding ORF sequences were amplified by PCR using GD-TTG1 plasmid DNA as a template and cloned in frame with an N-terminal GD tag under the control of the double 35S promoter into pUC19 vector [50,51]. The forward primer used to amplify all the truncated TTG1 genes and the reverse primer used to amplify TTG1Δ3aa are as described in 4.2, and the reverse primer used to amplify TTG1Δ15aa is 5′-CAAGAGCTCCTAATCAGGCTGCGAAGA-3′, for TTG1Δ10aa is 5′-CAAGAGCTCCTAAGCAATACCAATCCAATC-3′, for TTG1Δ5aa is 5′-CAAGAGCTCCTACATTTTGTTAGCAAAAGCAA-3′, for TTG1Δ4aa is 5′-CAAGAGCTCCTACTGCATTTTGTTAG-3′, for TTG1Δ2aa is 5′-CAAGAGCTCCTAAAGGAGCTGCATTTTGT-3′, for TTG1Δ1aa is 5′-CAAGAGCTCCTATCTAAGGAGCTGCATT-3′, and for TTGΔ339L is 5′-CAAGAGCTCTCAAACTCTGAGCTGCATTTT-3′.
To generate proTTG1:TTG1 and proTTG1:TTG1Δ3aa constructs, DNA fragments containing the 1062-bp upstream sequence and the CDS sequence of TTG1 or TTG1Δ3aa were PCR amplified by using DNA isolated from Col wild type seedlings as template and cloned before a nos terminator into the binary vector pPZP211. The forward primer used for PCR is 5′-CAAGTCGACGGATCAAGATCTTCATATTC-3′, and the reverse primers used for amplifying TTG1 and TTG1Δ3aa are as described in 4.2.
To generate gene editing CRISPR/Cas9 constructs of TTG1, the exon sequence corresponding the CDS sequence of TTG1 was scanned on CRISPRscan (http://www.crisprscan.org/, accessed on 2 September 2018) to identify potential target sequences, and selected target sequences were evaluated on Cas OFFinder (http://www.rgenome.net/cas-offinder/, accessed on 2 September 2018) to select sequence without potential off-targets. The specific target sequence selected for editing TTG1 was 5ʹ-GATGTAGAGCCGAAACGTCT(CGG)-3ʹ. The sequence was inserted into the pHDE vector to generate CRISPR/Cas9 construct by following the procedure described previously [52]. The primers used to generate the construct were 5ʹ-GATGTAGAGCCGAAACGTCTGTTTTAGAGCTAGAAATAGCAAGTTA-3ʹ and 5ʹ-AGACGTTTCGGCTCTACATCAATCACTACTTCGACTCTAGC-3ʹ.
4.4. Plant Transformation and Transgenic Plants Selection
About 5-week-old Arabidopsis Col wild type or ttg1Δ15aa mutant plants with several mature flowers on the main inflorescences were used for Agrobacterium tumefaciens GV3101 mediated plant transformation, and the plants were transformed by using the floral dip method [53].
The Col wild type Arabidopsis plants were transformed to generate CRISPR/Cas9 edited mutants for TTG1. T1 seeds from the transformed plants were collected and plated on plates containing solidified half MS medium with 100 µg/mL Carbenicillin and 30 µg/mL Hygromycin for T1 transgenic plants selection. Gene editing status in T1 transgenic plants identified was examined by amplifying and sequencing the genomic sequence of TTG1. T2 seeds collected from gene edited T1 plants were germinated and grew directly into soil pots and used to identify Cas9-free homozygous mutants.
The ttg1Δ15aa mutant plants were transformed with proTTG1:TTG1 and proTTG1:TTG1Δ3aa constructs, respectively, for phenotype rescue experiment. T1 seeds from the transformed plants were collected and plated on plates containing solidified half MS medium with 100 μg/mL Carbenicillin and 50 μg/mL Kanamycin for T1 transgenic plants selection. T2 seeds collected from identified T1 transgenic plants were plated on plates containing solidified half MS medium with 25 µg/mL kanamycin to select transgenic lines with 3:1 segregation. T3 seeds collected from individual plants of the lines with 3:1 segregation were plated on plates containing solidified half MS medium with 25 µg/mL kanamycin to select homozygous transgenic plants.
Expression of TTG1 and TTG1Δ3aa in the homozygous transgenic plants was examined by using RT-PCR. Confirmed homozygous overexpression transgenic plants, i.e., TTG1/ttg1Δ15aa lines #2 and #3, and TTG1Δ3aa/ttg1Δ15aa lines #5 and #10, were used for phenotypic analysis.
4.5. DNA Isolation and PCR
To examine the mutation occurring in the ttg1Δ15aa mutant, leaves of the ttg1Δ15aa mutant were collected, and DNA was isolated and used for PCR amplification of TTG1 genome sequence. PCR product obtained was sequenced, and the sequence obtained was aligned with the wild type TTG1 sequence.
To examine gene editing status of TTG1 in the CRISPR/Cas9 gene-edited plants, leaves of T1 transgenic plants were collected, DNA was isolated, and TTG1 genome sequence was amplified and sequenced. The sequences obtained were aligned with the wild type TTG1 sequence.
To obtain Cas9-free gene edited homozygous mutants, DNA isolated from T2 offspring of confirmed gene edited T1 plants was subjected to PCR amplification of the Cas9 fragments to identify Cas9-free plants, and TTG1 was amplified and sequenced to identify homozygous mutants. The primers used for amplifying Cas9 fragment have been described previously [54].
4.6. Plasmid DNA Isolation, Protoplast Isolation and Transfection
Plasmid DNA was isolated by using a GoldHiEndoFree Plasmid Maxi Kit (CWBIO, Beijing, China) following the manufacturer’s instructions and used for protoplast transfection.
Protoplasts were isolated from 80–90 rosette leaves collected from 3–4-week-old Col wild type plants and transfected with plasmids of the reporter and effector genes by following the procedure described previously [50].
To examine the possible interaction of TTG1Δ15aa, TTG1Δ10aa, TTG1Δ5aa, TTG1Δ4aa, TTG1Δ3aa, TTG1Δ2aa, TTG1Δ1aa, and TTG1Δ339L with GL3, plasmids of the effector genes GD-TTG1Δ15aa, GD-TTG1Δ10aa, GD-TTG1Δ5aa, GD-TTG1Δ4aa, GD-TTG1Δ3aa, GD-TTG1Δ2aa, GD-TTG1Δ1aa, or GD-TTG1Δ339L were co-transfected with plasmids of the reporter gene Gal4:GUS and the effector gene GL3 into protoplasts. Co-transfection of GD with Gal4:GUS and HA-GL3 were used as a control. The transfected protoplasts were incubated under darkness for 20–22 h at room temperature. GUS activities were measured by using a microplate reader (Synergy™ HT, BioTEK, Winooski, Vermont, USA). Transfection of each combination contains three biological replicates, and the experiments were repeated at least three times with similar results.
4.7. Mucilage Production Assays
Seeds of the Col wild type, the ttg1Δ15aa and the ttg1-c1 mutants, and the TTG1/ttg1Δ15aa and the TTG1Δ3aa/ttg1Δ15aa transgenic plants were stained with 0.01% (w/v) Ruthenium red and mounted, as described previously [55]. Mucilage was examined under a Motic K dissecting microscope (MOTIC, Xiamen, China), and pictures were taken by using a digital camera connected to the microscope. At least 30 seeds from each plant were used for the assays, and the experiments were repeated three times with similar results.
4.8. Anthocyanin Biosynthesis Assays
Anthocyanin biosynthesis in seedlings of the Col wild type, the ttg1Δ15aa mutant, and the TTG1/ttg1Δ15aa and TTG1Δ3aa/ttg1Δ15aa transgenic plants was assayed by using sucrose treatments, as described previously [43]. At least 30 seedlings from each plant were used for the assays, and the experiments were repeated three times with similar results.
4.9. Trichome and Root Hair Formation Assays
For trichome formation assays, seeds of Col wild type, the ttg1Δ15aa and the ttg1-1c mutants, and the TTG1/ttg1Δ15aa and TTG1Δ3aa/ttg1Δ15aa transgenic plants were germinated and grown in soil pots. Trichome formation was examined under a Motic K dissecting microscope, trichome numbers on the first pair, third, fourth, fifth, and sixth true leaves were counted, and pictures were taken at indicated growth stages by using a digital camera.
For root hair formation assays, seeds of Col wild type, the ttg1Δ15aa and the ttg1-c1 mutants, and the TTG1/ttg1Δ15aa and TTG1Δ3aa/ttg1Δ15aa transgenic plants were germinated on plates containing solidified half MS medium and grown vertically. Root hair formation was examined under a Motic K dissecting microscope, numbers of root hairs were counted, and pictures were taken by using a digital camera connected to the microscope.
5. Conclusions
Our results in this study indicate that the C-terminus is critical for TTG1’s functions in Arabidopsis, and the deletion of the last three C-terminal amino acids or the 339L amino acid alone is sufficient to abolish the interaction of TTG1 with GL3.
Acknowledgments
We thank all our lab members in both Linyi University and Northeast Normal University for their helpful discussion and suggestion.
Author Contributions
Conceptualization, S.W. and T.W.; investigation, Y.W., H.T., W.W., X.W., K.Z., S.H. and R.L.; data curation, Y.W., H.T. and S.W.; writing—original draft preparation, S.W. and Y.W.; writing—review and editing, S.W. and Y.W; project administration, S.W.; funding acquisition, S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32071938, and startup funding from Linyi University, grant number LYDX2019BS039.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data were obtained were presented in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Walker A.R., Davison P.A., Bolognesiwinfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian H., Wang S. TRANSPARENT TESTA GLABRA1, a key regulator in plants with multiple roles and multiple function mechanisms. Int. J. Mol. Sci. 2020;21:4881. doi: 10.3390/ijms21144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koornneef M. The complex syndrome of ttg mutants. Arabid Inf. Serv. 1981;18:45–51. [Google Scholar]
- 4.Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 5.Shirley B.W., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M., Goodman H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313X.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- 6.Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Chen M., Zhang B., Li C., Kulaveerasingam H., Chew F.T., Yu H. TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 2015;169:391–402. doi: 10.1104/pp.15.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C., Zhang B., Chen B., Ji L., Hao Y. Site-specifific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 2018;9:571. doi: 10.1038/s41467-018-03013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paffendorf B.A., Qassrawi R., Meys A.M., Trimborn L., Schrader A. TRANSPARENT TESTA GLABRA 1 participates in flowering time regulation in Arabidopsis thaliana. Peer J. 2020;8:e8303. doi: 10.7717/peerj.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan F., Leng B., Zhang H., Wang X., Han G., Wang B. A WD40-repeat protein from the Recretohalophyte Limonium bicolor enhances trichome formation and salt tolerance in Arabidopsis. Front. Plant Sci. 2019;10:1456. doi: 10.3389/fpls.2019.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong D., Li M., Dong Z., Ji H., Li X. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 2015;34:395–410. doi: 10.1007/s00299-014-1717-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Liu R., Chen L., Wang Y., Liang Y., Wu X., Li B., Wu J., Liang Y., Wang X., et al. Nicotiana tabacum TTG1 contributes to ParA1-induced signalling and cell death in leaf trichomes. J. Cell Sci. 2009;122:2673–2685. doi: 10.1242/jcs.049023. [DOI] [PubMed] [Google Scholar]
- 13.Van Nocker S., Ludwig P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics. 2003;4:50. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang Y., Huang X., Lu Z., Yao J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genomics. 2012;13:100. doi: 10.1186/1471-2164-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neer E.J., Schmidt C.J., Nambudripad R., Smith T.F. The ancient regulatory protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen S., Wang S. GLABRA2, a common regulator for epidermal cell fate determination and anthocyanin biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2019;20:4997. doi: 10.3390/ijms20204997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiefelbein J. Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 2003;6:74–78. doi: 10.1016/S136952660200002X. [DOI] [PubMed] [Google Scholar]
- 18.Lin Q., Aoyama T. Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: Update on the roles of the classic regulator. J. Integr. Plant Biol. 2012;54:729–737. doi: 10.1111/j.1744-7909.2012.01159.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Chen J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 2014;5:133. doi: 10.3389/fpls.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golz J.F., Allen P.J., Li S.F., Parish R.W., Jayawardana N., Bacic A., Doblin M.S. Layers of regulation—Insights into the role of transcription factors controlling mucilage production in the Arabidopsis thaliana seed coat. Plant Sci. 2018;272:179–192. doi: 10.1016/j.plantsci.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Pesch M., Hulskamp M. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 2004;14:422–427. doi: 10.1016/j.gde.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 24.Schiefelbein J., Huang L., Zheng X. Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front. Plant Sci. 2014;5:47. doi: 10.3389/fpls.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudry A., Heim M.A., Debreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 27.Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 29.Petroni K., Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive Organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Xu W., Grain D., Bobet S., Le Gourrierec J., Thévenin J., Kelemen Z., Lepiniec L., Dubos C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;202:132–144. doi: 10.1111/nph.12620. [DOI] [PubMed] [Google Scholar]
- 31.Deng Y., Lu S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017;36:1–34. doi: 10.1080/07352689.2017.1402852. [DOI] [Google Scholar]
- 32.Francoz E., Ranocha P., Burlat V., Dunand C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015;20:515–524. doi: 10.1016/j.tplants.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Voiniciuc C., Yang B., Schmidt M.H., Günl M., Usadel B. Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 2015;16:3452–3473. doi: 10.3390/ijms16023452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., Gonzalez A. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58:1431–1441. doi: 10.1093/pcp/pcx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson C.S., Kolevski B., Smyth D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Western T.L., Young D.S., Dean G.H., Tan W.L., Samuels A.L., Haughn G.W. MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 2004;134:296–306. doi: 10.1104/pp.103.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J.G. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008;8:81. doi: 10.1186/1471-2229-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J.C., Chezem W.R., Clay N.K. Ternary WD40 repeat-containing protein complexes: Evolution, composition and roles in plant immunity. Front. Plant Sci. 2015;6:1108. doi: 10.3389/fpls.2015.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payne C.T., Zhang F., Lloyd A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S., Chen J.G. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 2008;49:1792–1804. doi: 10.1093/pcp/pcn159. [DOI] [PubMed] [Google Scholar]
- 41.Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. Cell Mol. Biol. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 42.Long Y., Schiefelbein J. Novel TTG1 mutants modify root-hair pattern formation in Arabidopsis. Front. Plant Sci. 2020;11:383. doi: 10.3389/fpls.2020.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Wang X., Hu Q., Dai X., Tian H., Zheng K., Wang X., Mao T., Chen J.G., Wang S. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015;83:300–311. doi: 10.1111/tpj.12887. [DOI] [PubMed] [Google Scholar]
- 44.Larkin J.C., Oppenheimer D.G., Lloyd A.M., Paparozzi E.T., Marks M.D. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell. 1994;6:1065–1076. doi: 10.2307/3869885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin J.C., Walker J.D., Bolognesi-Winfifield A.C., Gray J.C., Walker A.R. Allele-specifific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics. 1999;151:1591–1604. doi: 10.1093/genetics/151.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development. 2009;136:1039–1048. doi: 10.1242/dev.030585. [DOI] [PubMed] [Google Scholar]
- 47.Bharti A.K., Khurana J.P. Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana(ecotype Estland) impaired in flavonoid biosynthetic pathway. Plant Sci. 2003;165:1321–1332. doi: 10.1016/S0168-9452(03)00344-3. [DOI] [Google Scholar]
- 48.Tian H., Wang X., Guo H., Cheng Y., Hou C., Chen J.G., Wang S. NTL8 regulates trichome formation in Arabidopsis by directly activating R3 MYB genes TRY and TCL1. Plant Physiol. 2017;174:2363–2375. doi: 10.1104/pp.17.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng K., Tian H., Hu Q., Guo H., Yang L., Cai L., Wang X., Liu B., Wang S. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci Rep. 2016;6:19254. doi: 10.1038/srep19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S., Tiwari S.B., Hagen G., Guilfoyle T.J. Auxin response factor7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell. 2005;17:1979–1993. doi: 10.1105/tpc.105.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiwari S.B., Wang X., Hagen G., Guilfoyle T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cel l. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao X., Chen J., Dai X., Zhang D., Zhao Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–1800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clough S.J., Bent A.F. Floral dip: A simplifified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen S., Zhang N., Zhang Q., Zhou G., Tian H., Hussain S., Ahmed S., Wang T., Wang S. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019;20:2695. doi: 10.3390/ijms20112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S., Barron C., Schiefelbein J., Chen J.G. Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 2010;185:387–400. doi: 10.1111/j.1469-8137.2009.03067.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data were obtained were presented in this article.