Abstract
The role of protein kinase A in regulating transcription of the cholinergic gene locus, which contains both the vesicular acetylcholine transporter gene and the choline acetyltransferase gene, was investigated in PC12 cells and a protein kinase A-deficient PC12 mutant, A126.1B2, in which transcription of the gene is reduced. The site of action of protein kinase A was localized to a neuron-restrictive silencer element/repressor element 1 (NRSE/RE-1) sequence within the cholinergic gene. Neuron-restrictive silencer factor (NRSF)/RE-1-silencing transcription factor (REST), the transcription factor which binds to NRSE/RE-1, was expressed at similar levels in both PC12 and A126.1B2 cells. Although nuclear extracts containing NRSF/REST from A126.1B2 exhibited binding to NRSE/RE-1, nuclear extracts from PC12 cells did not. The NRSF/REST isoform REST4 was expressed in PC12 cells but not in A126.1B2. REST4 inhibited binding of NRSF/REST to NRSE/RE-1 as determined by gel mobility shift assays. Coimmunoprecipitation was used to demonstrate interaction between NRSF/REST and REST4. Expression of recombinant REST4 in A126.1B2 was sufficient to transcriptionally activate the cholinergic gene locus. Thus, in PC12 cells, protein kinase A promotes the production of REST4, which inhibits repression of the cholinergic gene locus by NRSF/REST.
The cholinergic gene locus contains the genes for both the enzyme choline acetyltransferase (ChAT; EC 2.3.1.6) and the vesicular acetylcholine transporter (VAChT) (1, 2, 5). ChAT is the enzyme responsible for the biosynthesis of the neurotransmitter acetylcholine, while VAChT is the vesicle membrane transporter that translocates cytoplasmic acetylcholine to the interior of synaptic vesicles. ChAT and VAChT are both required for cholinergic neurotransmission.
Previous reports showed that the ChAT gene contains three 5′ noncoding exons: exon 1, named the R exon; exon 2, named the N exon; and exon 3, named the M exon. The three exons result in multiple 5′ mRNA species, which are produced from different promoters and by alternative mRNA splicing (14, 18, 30). The VAChT gene lies uniquely within the first intron of the ChAT gene, between the R and N exons, and has the same transcriptional orientation. Multiple VAChT mRNA species with different 5′ noncoding exons have also been reported, and some of these share the R exon with ChAT mRNA species. This organization of the ChAT and VAChT genes is suggested to lead to coordinate regulation of the two genes at the transcriptional level (2, 3, 9, 19, 25). Although the mechanism of transcriptional regulation controlling the cholinergic gene locus is poorly understood, a neuron-restrictive silencer element/repressor element 1, (NRSE/RE-1) sequence is implicated in silencing the cholinergic gene locus in nonneuronal cells (16). The NRSE/RE-1, which comprises ∼23 nucleotides, is found in a number of neuron-specific genes, including the type II sodium channel (7), synapsin I (15), SCG10 (20), Ng-CAM (13), and the m4 muscarinic receptor (17), to name but a few.
The transcription factor that binds to the NRSE sequence, the neuron-restrictive silencer factor (NRSF) or RE-1-silencing transcription factor (REST), is a 210-kDa glycoprotein containing nine zinc finger domains (7). It was reported that NRSF/REST bound to the NRSE and repressed the expression of neuron-specific genes in nonneuronal cell lines (24). It was also found that NRSF/REST could act as a silencer of neuron-specific gene expression in undifferentiated neuronal progenitor cells (6, 7, 24). Recently, it was reported that several NRSF/REST splice variants were expressed in mature neurons of adult brain, albeit at low levels (21). The neuron-specific isoforms have an insertion of either 16 nucleotides (REST4) or 28 nucleotides (REST5) in the region of the gene encoding the spacer between zinc fingers 5 and 6. These splice variants lead to truncated proteins containing only five zinc fingers. The physiological function of these splice variants has been, until now, unclear.
Our previous studies using PC12 cells and protein kinase A (PKA)-deficient mutant PC12 cells suggested that PKA signaling pathways regulate the transcription of the cholinergic gene locus. The site of interaction was proposed to reside in the 5′ noncoding region of the cholinergic gene locus upstream of the VAChT gene (12, 25). To study further the role of PKA in regulating the cholinergic gene locus, we focused on the role of this kinase in regulating NRSF/REST activity. The results suggest that in PC12 cells, PKA controls the expression of the neuron-specific isoform REST4, produced by alternative splicing of NRSF/REST. REST4 appears to regulate cholinergic gene expression by forming a complex with NRSF/REST, thereby preventing the binding of the latter to the NRSE/RE-1 sequence.
MATERIALS AND METHODS
Materials.
RNase inhibitor and DNase I were purchased from Boehringer Mannheim (Indianapolis, Ind.). Deoxyribonucleotides, Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), Geneticin (G418), horse serum, and Superscript II reverse transcriptase were obtained from Gibco BRL (Gaithersburg, Md.). Oligonucleotide primers were synthesized with a Beckman Oligo1000 DNA synthesizer or purchased from commercial sources. [γ-32P]ATP, [α-32P]dCTP, and [3H]acetyl coenzyme A were from ICN Pharmaceuticals (Irvine, Calif.). Taq bead hot-start polymerase was purchased from Promega (Madison, Wis.). The bicinchoninic acid (BCA) protein assay kit was obtained from Pierce (Rockford, Ill.). poly(dI-dC) · poly(dI-dC) and random hexamers [pd(N)6] were obtained from Pharmacia Biotech (Piscataway, N.J.). The ECL+Plus Western blotting detection system and Hybond-P membranes were purchased from Amersham Life Science Inc. (Arlington Heights, Ill.). SuperFect transfection reagent, plasmid kits, and RNeasy MINI kits (Total RNA system) were obtained from Qiagen Inc. (Valencia, Calif.). The GalactoLight system was purchased from Tropix Inc. (Bedford, Mass.). The pcDNA3 vector was obtained from Invitrogen, while pXP2 was obtained from the American Type Culture Collection. All other reagents were from Sigma Chemical Co. (St. Louis, Mo.) and were of the highest quality available.
Cell lines and cell culture.
The cell lines used in this study were a wild-type parental PC12 cell line and a PC12 mutant cell line, A126.1B2, generated by using nitrosoguanidine (29). The A126.1B2 cell line has an overall reduced level of cytosolic PKA activity and is devoid of nuclear PKA activity (4). Cells were routinely grown in DMEM supplemented with 10% heat-inactivated FBS and 5% heat-inactivated horse serum at 37°C in a humidified atmosphere of 90% air and 10% CO2. The medium was changed every 3 days, and cells were harvested and subcultured every 6 days. HEK293 cells were cultured in DMEM containing 10% heat-inactivated FBS.
Antisera.
A monoclonal antibody (MAb) to mouse NRSF/REST, 12C11-1, was generated as previously described (6). A rabbit antiserum against the catalytic subunit of PKA (23) was a generous gift of Y. Fukami, Kobe University, Kobe, Japan. A MAb against the myc epitope, a goat anti-myc epitope immunoglobulin G (IgG) coupled to agarose, a goat anti-FLAG epitope IgG coupled to agarose, and normal goat IgG coupled to agarose were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.).
Electrophoretic mobility shift assays.
Nuclear extracts from cells were prepared as described by Lonnerberg et al. (16) except for the inclusion of additional protease inhibitors: 20 mM leupeptin, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, 1 μg of pepstatin/ml, and 18 μg of aprotinin/ml. DNA fragments were end labeled with [γ-32P]ATP by using T4 polynucleotide kinase. For binding, 10 μg of nuclear protein was preincubated on ice, with or without a 100-fold excess of unlabeled competitor DNA, for 10 min in 20 μl of a solution containing 20 mM HEPES (pH 7.6), 0.1% Nonidet P-40, 10% glycerol, 1 mM dithiothreitol, 2.5 mM MgCl2, 250 mM KCl, and 2 μg of poly(dI-dC) · poly(dI-dC). Labeled oligonucleotide (100 fmol) was mixed with nuclear protein, and the mixture was incubated for 10 min at 25°C. For supershift assays, 10 μg of nuclear protein was preincubated on ice with or without MAb 12C11-1 for 30 min before addition of the labeled probe. The reaction mixture was loaded onto a 6% nondenaturing polyacrylamide gel with 0.25× Tris-borate-EDTA (TBE) buffer. The ChAT NRSE/RE-1 and control oligonucleotides described by Lonnerberg et al. (16) were used in these studies.
Western blot analysis.
Nuclear extract (100 μg) was solubilized in Laemmli sample buffer. After separation on a reducing sodium dodecyl sulfate (SDS)-polyacrylamide gel (7%), proteins were transferred onto a Hybond-P membrane as described previously (28). The membrane was then incubated with anti-NRSF MAb (12C11-1), followed by horseradish peroxidase-labeled goat anti-mouse antiserum, and visualized by using an ECL+Plus detection kit as per the manufacturer’s instructions.
RNA preparations and RT-PCR.
Total RNA was prepared from cell lines by using RNeasy kits. The recovery of RNA was quantified spectrophotometrically. RNA was digested with DNase I to eliminate contaminating endogenous DNA. Single-stranded cDNAs were synthesized with Superscript II reverse transcriptase, using random hexamers [pd(N)6] as primers. Each PCR cycle consisted of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a 5-min extension at 72°C. PCR products were separated in a 1.5% agarose gel with TBE buffer; this was followed by ethidium bromide staining. Primers used were as follows: rNRSF-S, GACAGGTTCACAACGGGCC; rNRSF-AS, CCCTTCGGCACTTCGCCGCT; p2-S, CTACATGGCACACCTGAAGCACCAC; pR4-AS, GGCTTCTCACCCAACTAGATCACACT; Actin-S, ATTCCTATGTGGGCGACGAG; and Actin-AS, TGGATAGCAACGTACATGGC.
For ChAT and VAChT, single-stranded cDNAs were synthesized by using specific primers for VAChT or ChAT (25). PCR was performed with 1/10 volume of reverse transcription (RT) mixture in a 50-μl reaction volume that included deoxynucleoside triphosphates (10 nmol each), appropriate sense and antisense primers (10 pmol), [α-32P]dCTP (1.67 pmol), and Taq polymerase in a buffer supplied by the manufacturer. After an initial denaturation at 94°C for 5 min, each PCR cycle included 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with variable numbers of cycles. After completion of the desired number of PCR cycles, a 5-min extension at 70°C was performed. PCR products were electrophoresed on a 5% polyacrylamide gel with TBE followed by autoradiography.
Enzyme activity assays.
ChAT activity was assayed by a modification of the method of Fonnum as described previously (11). The protein concentrations of cell extracts were determined with a BCA protein assay kit (Pierce). Cyclic AMP (cAMP)-dependent PKA activity was measured as described previously (25).
Construction and expression of PKA catalytic subunit.
The Chinese hamster ovary cell PKA catalytic subunit β (Catβ) or a mutated form of Catβ (CatβM) (kindly provided by R. Maurer) was cloned into the multiple cloning site of the pcDNA3 mammalian expression vector. Transfections were carried out with SuperFect transfection reagent according to the manufacturer’s protocol. Briefly, 104 A126.1B2 cells on a 100-mm-diameter plate were first washed twice with DMEM. Ten micrograms of pcDNA3-Catβ or pcDNA3-CatβM and 50 μl of SuperFect transfection reagent were mixed in 300 μl of serum-free DMEM, and the cells were incubated in this mixture for 3 h. Subsequently, the medium was discarded, the cells were washed with phosphate-buffered saline, and fresh medium was added. After 3 days, the medium was changed to one containing 800 μg of G418/ml, and after 3 weeks, clonal cell lines were isolated by using cloning wells.
Reporter gene assays.
Fragments of the human cholinergic gene were inserted into the multiple cloning site of the pXP2 vector. Thus, pXP2NX is a derivative of pXP2 containing a 1,156-bp NheI-XhoI fragment of the human cholinergic gene upstream of the VAChT gene. pXP2EX is a similar construct but contains a 2,200-bp 5′ extension of the NheI-XhoI fragment. After transfection of these constructs into cells by using SuperFect reagent, cell lysates were prepared, and luciferase activity in these lysates was measured by a luminescence assay (25). β-Galactosidase activity was measured by using the GalactoLight system according to the manufacturer’s procedure. Luminescence was detected by using an Opticomp I luminometer (GEM Biomedical, Inc.)
Expression of NRSF/REST and REST4.
The mouse full-length NRSF cDNA or that of the truncated isoform REST4 was inserted into the pCS2+MT expression vector, which added five myc epitope tags to its N terminus (22). The inserts were cloned from an NcoI to an XbaI site by using primers that added these sites to the ends of the cDNA. The truncated form of REST, REST4, was prepared by PCR amplification with a forward primer (ATGGCCACCCAGGTGATG) and a reverse primer (TCACCCAACTAGATCACACTCTGAGTGAGTACGCATGTG) including REST4-specific sequences. The PCR product, amplified with Pfu polymerase, was gel purified and subcloned into the SmaI site of pBluescriptII KS(+); this was followed by digestion with PstI and XbaI. The fragment (PstI-XbaI) of pCS2+MTNRSF was removed and replaced with the PstI-XbaI fragment of pBluescript-REST4. A second REST4 construct was prepared in which the five N-terminal myc epitope tags were replaced with a FLAG epitope tag to yield pCS2+FLAGREST4. The sequences of PCR products were confirmed by DNA sequencing. Constructs were transfected into HEK293 cells by using SuperFect transfection reagent in accordance with the instructions in the manufacturer’s manual. After 72 h, cells were harvested and nuclear extracts were prepared as described above.
Immunoprecipitation.
HEK293 cells were transfected, using SuperFect transfection reagent, with either pCS2+MTNRSF or FLAG epitope-tagged REST4, pCS2+ FLAGREST4. The cells were cultured for 3 days after transfection, and then collected, and nuclear extracts were prepared as described above. Each nuclear extract was preincubated with normal goat IgG coupled to agarose at 4°C for 2 h, and the supernatant was obtained by centrifugation. The supernatant was subjected to immunoprecipitation with goat anti-myc IgG conjugated to agarose or goat anti-FLAG IgG conjugated to agarose, with gentle rotation at 4°C for 16 h. The immunoprecipitate was collected by centrifugation, solubilized with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, resolved by SDS-PAGE, and subjected to Western blot analysis with the NRSF MAb 12C11-1.
RESULTS
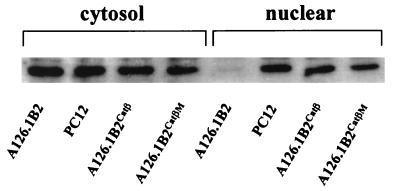
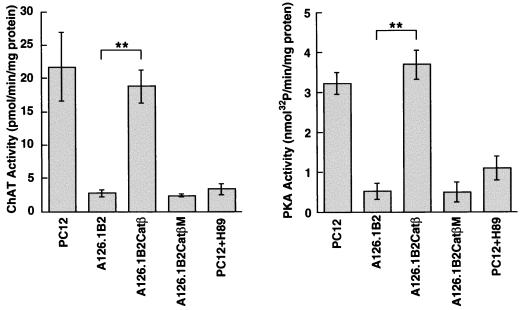
Previous studies have shown that the level of transcription of the cholinergic gene locus, comprising the ChAT gene and the VAChT gene, is greatly reduced in two mutant PC12 cell lines (12, 25), A123.7 (8, 10) and A126.1B2 (29), which both exhibit reduced PKA activity. We tested the effect of introducing the catalytic subunit of PKA (Catβ) into the mutant PC12 cell line A126.1B2. This cell line, which was generated by nitrosoguanidine mutagenesis, fails to accumulate Catβ in the nucleus and, although exhibiting normal levels of Catβ in the cytosol, has reduced cytosolic PKA activity (4). We have generated A126.1B2 cell lines stably transfected with Catβ (A126.1B2Catβ) or a mutated inactive CatβM (A126.1B2CatβM). In agreement with the findings of Cassano et al. (4), we found that the parental and A126.1B2 cell lines contained similar cytosolic levels of the catalytic subunit of PKA; however, only the parental cell line contained the nuclear catalytic subunit. The nuclear PKA catalytic subunit was expressed at comparable levels in A126.1B2Catβ and A126.1B2CatβM (Fig. 1). The specific activity of PKA in the A126.1B2Catβ cell line, measured with a synthetic peptide substrate, was increased from 0.6 to 3.6 nmol of 32P incorporated/min/mg of protein, while the specific activity of ChAT was increased from 2.7 to 18.8 pmol of acetylcholine formed/min/mg of protein (Fig. 2). There was no change in the specific activity of PKA or ChAT in A126.1B2CatβM. The PKA inhibitor H89 reduced both PKA activity and ChAT activity in the parental cell line. The ability to increase ChAT activity in the A126.1B2 cell line to parental levels clearly demonstrates that the ChAT gene is intact in this cell line. Furthermore, these results demonstrate that PKA alone is sufficient to restore ChAT activity.
FIG. 1.
Expression of the PKA catalytic subunit in the cytosolic and nuclear fractions of PC12 cell lines. Nuclear and cytosolic extracts (10 μg) from the indicated cell lines were subjected to SDS-PAGE followed by Western blot analysis with a rabbit anti-PKA catalytic subunit antibody and then detection by enhanced chemiluminescence.
FIG. 2.
PKA activity and ChAT activity in control cells and cells transfected with the PKA catalytic β subunit. A126.1B2 cells stably transfected with either the plasmid pcDNA3-Catβ, containing the catalytic subunit of PKA (A126.1B2Catβ), or the plasmid pcDNA3-CatβM, containing an inactive catalytic subunit of PKA (A126.1B2CatβM), were grown in DMEM containing G418. The medium was changed every 3 days, and cells were subcultured every 6 days. After 3 weeks, cells were harvested and lysates were prepared. Also shown are results obtained with untreated wild-type PC12 cells and wild-type PC12 cells treated for 3 days with 10 μM H89. ChAT and PKA activity were assayed as described in Materials and Methods. Data are means ± standard errors of the means (n = 6). ∗∗, P < 0.01 versus the respective control value (Student’s t test).
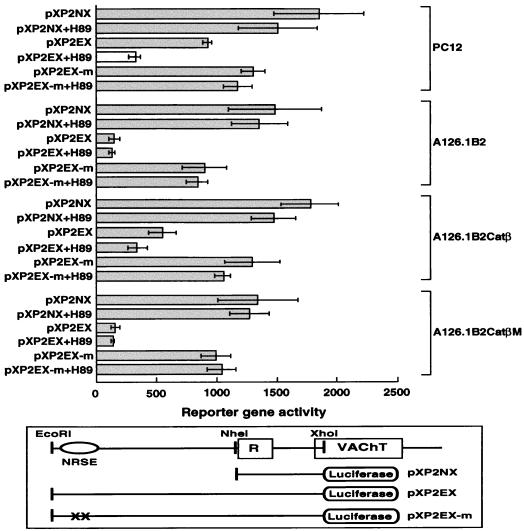
We next investigated whether PKA might regulate transcription of the ChAT gene through elements in the 5′ upstream region of the gene. We tested reporter gene constructs which contained fragments of the human cholinergic gene and luciferase as a reporter gene: a 1,156-bp NheI-XhoI fragment referred to as pXP2NX and a 3,356-bp EcoRI-XhoI fragment referred to as pXP2EX. The construct pXP2NX contains a basal VAChT promoter from the cholinergic locus (31), while pXP2EX contains an additional 2,200 bp of upstream sequence including the NRSE/RE-1. The NRSE/RE-1 is the silencer element that mediates negative regulation of a number of neuronal genes. We also used a construct, pXP2EX-m, in which the NRSE/RE-1 was inactivated by mutagenesis. Figure 3 shows the results of a typical transient-transfection experiment with these constructs. In the parental PC12 cell line, expression of the reporter gene from pXP2EX was somewhat reduced compared to that from pXP2NX, and this effect was eliminated by mutation of the NRSE. The PKA inhibitor H89 had no significant effect on the basal promoter in pXP2NX but inhibited expression of the pXP2EX construct. In contrast, H89 had no effect on the pXP2EX-m construct, which contains the inactive mutant NRSE. These results suggest that the NRSE/RE-1 is minimally active in the parental PC12 cell line and that inhibition of PKA decreases transcription of the gene dependent on the region of the gene containing the NRSE. In the PKA-deficient PC12 cell line A126.1B2, the reporter gene activity of the extended fragment pXP2EX containing the NRSE was greatly reduced. Reporter gene activity could be restored by mutation of the NRSE, while addition of a PKA inhibitor had no effect on the pXP2EX-m construct. The expression pattern seen in A126.1B2Catβ recapitulated that of the parental cell line, while the expression pattern in A126.1B2CatβM was the same as that seen in A126.1B2. These results provide evidence that PKA can increase transcription of the cholinergic gene locus by acting through the NRSE/RE-1 repressor element.
FIG. 3.
Transient-transfection assays using the 5′ noncoding region of the human ChAT gene driving the luciferase reporter gene. A schematic showing the 5′ noncoding region of the human cholinergic gene and the constructs generated from this region is shown on the bottom. The position of the VAChT gene is indicated, while R represents the first exon of the cholinergic gene. The positions of restriction sites for EcoRI, NheI, and XhoI are shown. Constructs were named on the basis of the restriction fragment inserted into the pXP2 vector, with N corresponding to the NheI site, X corresponding to the XhoI site, and E corresponding to the EcoRI site. PXP2EX-m contains a mutated inactive NRSE. H89 is a PKA inhibitor. Constructs were transiently transfected into the indicated cell lines as described in Materials and Methods. After 48 h, cells were harvested, and extracts prepared from them were subsequently assayed for luciferase activity. Luciferase activity was normalized to β-galactosidase activity.
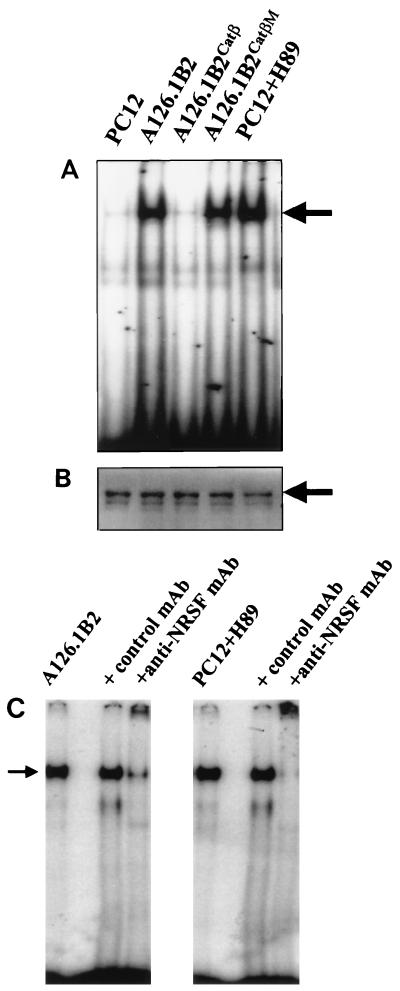
Preliminary evidence previously obtained suggested that PKA might regulate the activity of NRSF/REST (25). Thus, we analyzed NRSF/REST expression in nuclear extracts prepared from wild-type PC12, A126.1B2, A126.1B2Catβ, or A126.1B2CatβM cells by Western blot analysis with an anti-NRSF MAb, using the ECL+Plus detection system. As shown in Fig. 4A, comparable levels of NRSF/REST immunoreactive protein were detected in all of the PC12 cell lines, although the level of NRSF/REST present in PC12 cells was considerably lower than that observed in nonneuronal cell lines such as NIH 3T3 and HeLa. Since the levels of NRSF/REST were approximately the same in all of the PC12 cell lines examined, the effect of PKA cannot be attributed to regulation of NRSF/REST gene transcription, NRSF/REST mRNA stability, or NRSF/REST protein turnover.
FIG. 4.
NRSE binding activity in PC12 and A126.1B2 cell lines. (A) Electrophoretic mobility shift assay. Nuclear cell extracts prepared from the indicated cell lines were analyzed for binding to the cholinergic NRSE/RE-1 by electrophoretic mobility shift assays. (B) Western blot analysis of NRSF. Nuclear extracts (100 μg) from each cell line were subjected to SDS-PAGE followed by Western blotting with an anti-NRSF MAb and detection with the ECL+Plus detection system. PC12 cells were treated with 10 μM H89 as for Fig. 2. (C) Supershifting of NRSE/RE-1 gel shift band. Electrophoretic mobility shift assays were conducted as described for panel B except that the nuclear extract was preincubated with 2 μg of anti-NRSF MAb or 2 μg of a control MAb prepared against mouse ChAT where indicated.
The DNA binding activity of NRSF/REST in nuclear extracts from the various cell lines was analyzed by determining the ability to bind to an oligonucleotide containing the human ChAT NRSE in an electrophoretic mobility shift assay. A gel shift band was barely detectable with a nuclear extract from wild-type PC12 cells (Fig. 4B), consistent with the lack of repression of the cholinergic gene in this cell line. However, a prominent gel shift band was detected for the A126.1B2 cell line, in which transcription of the cholinergic gene is repressed. The gel shift band was also detected in nuclear extracts from A126.1B2CatβM, but not in nuclear extracts from A126.1B2Catβ. Binding was competed by an excess of cold NRSE oligonucleotide but not by an unrelated sequence (data not shown). Furthermore, the gel shift band could be supershifted with an anti-NRSF antibody (Fig. 4C). Thus, although NRSF/REST is expressed in both wild-type and mutant PC12 cells, only nuclear extracts derived from PKA-deficient PC12 cells show significant binding to the NRSE/RE-1. This finding is consistent with the observation that PKA-deficient PC12 cells exhibit repressed levels of ChAT activity and of ChAT and VAChT mRNAs.
We examined the possibility that PKA directly phosphorylates NRSF/REST by incubating recombinant NRSF with the catalytic subunit of PKA in the presence of [γ-32P]ATP and the phosphatase inhibitor sodium vanadate. Analysis of the products by radioautography following SDS-PAGE failed to reveal the presence of 32P-labeled NRSF. The same results were obtained when we tested REST4 (see below) as a substrate for PKA. In this experiment, the catalytic subunit of PKA was shown to be active by its ability to phosphorylate the synthetic peptide substrate Leu-Arg-Arg-Ala-Ser-Leu-Gly.
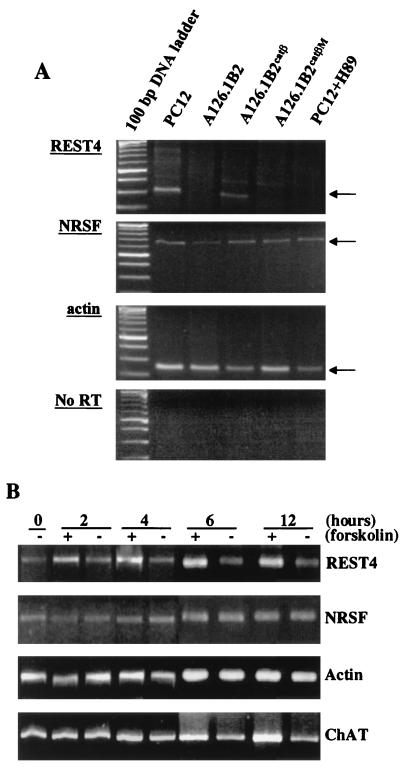
We thus looked for alternative mechanisms that might lead to the regulation of NRSF/REST activity by PKA. Recently, it was reported that a multiple splicing pattern of NRSF/REST mRNA occurred (21), leading to novel isoforms. Two of these splice variants, referred to as REST4 and REST5, were found to be neuron-specific NRSF/REST truncated isoforms expressed in adult brain neurons (21). We used RT-PCR to detect the presence of these isoforms in the wild-type PC12, A126.1B2, A126.1B2CatβM, and A126.1B2Catβ cell lines. Total RNA was prepared from these cell lines and transcribed with random hexamers; this was followed by PCR amplification with oligonucleotide primers specific for the neuron-specific NRSF sequences.
As shown in Fig. 5A, a PCR product was detected in all cell lines by using primer pairs specific for NRSF/REST. On the other hand, a PCR product for the neuron-specific isoform REST4 was detected in PC12 and A126.1B2Catβ but not in A126.1B2 or A126.1B2CatβM. The PKA inhibitor H89 blocked the expression of REST4 in PC12 cells (Fig. 5A). REST4 mRNA was induced within 4 h of treatment of PC12 cells with 10 μM forskolin, an adenylate cyclase activator, and appeared to reach a maximal level by 6 h (Fig. 5B). ChAT mRNA was also induced by forskolin, but an increase was just detectable at 6 h and a maximum was reached at 12 h (Fig. 5B). In contrast, NRSF/REST mRNA was unaffected by forskolin.
FIG. 5.
Analysis of NRSF/REST and REST4 mRNA expression in PC12 and A126.1B2 cell lines and the effect of forskolin. (A) NRSF/REST and REST4 mRNA levels in PC12 cell lines. PCR was performed with primer pairs specific for NRSF/REST, REST4, and actin as described in Materials and Methods. The bottom pattern is a control in which the primer pairs for REST4 were used without RT of RNA. PC12 cells were treated with H89 as described in the legend to Fig. 2. (B) Effect of forskolin on NRSF/REST, REST4, and ChAT mRNAs. PC12 cells were untreated (−) or were treated with 10 μM forskolin (+) for the time periods indicated. PCR was performed as described in Materials and Methods.
We attempted to estimate the relative levels of NRSF/REST and REST4 in PC12 cells. A low level of NRSF/REST was detectable by Western blot analysis, as shown in Fig. 4A; however, REST4 was difficult to detect, and its level appeared to be at or just below our detection limits. Thus, there is more endogenous NRSF/REST than REST4, although as noted REST4 mRNA levels can be increased by agents that lead to the activation of PKA. The presence of excess NRSF/REST relative to REST4 is supported by the data in Fig. 3, which show that expression of a reporter gene containing the NRSE/RE-1 is partially repressed in PC12 cells.
No PCR product was detected in any cell line when primer pairs specific for the isomer REST5 were used. These data suggest that PKA activity, presumably nuclear PKA activity, is required for the production of the REST4 isoform. Furthermore, the presence or absence of REST4 mRNA, but not NRSF/REST, correlates with transcription of the cholinergic gene.
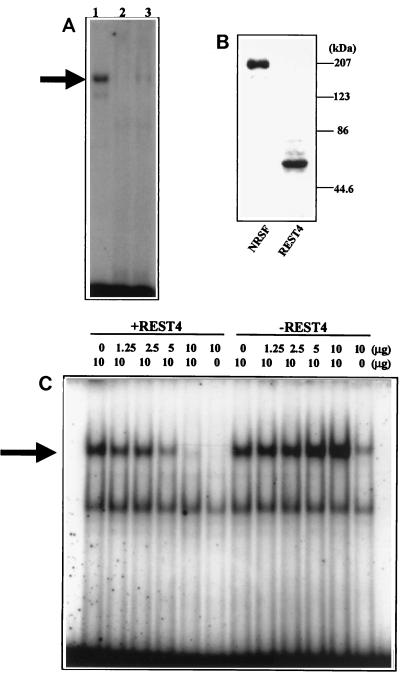
To study whether the neuron-specific isoform REST4 binds to the NRSE sequence, we performed electrophoretic mobility shift assays using recombinant NRSF/REST and REST4 expressed in HEK293 cells. As shown in Fig. 6A, the expected gel shift band was detected with nuclear extracts containing NRSF/REST (lane 1). No gel shift band was detected with nuclear extracts containing REST4 (lane 2). However, when a nuclear extract containing REST4 was mixed with a nuclear extract containing full-length NRSF/REST, the gel shift band was eliminated (lane 3). No such effect was seen with nuclear extracts from control HEK293 cells. Western blotting verified the presence of NRSF/REST or REST4 in nuclear extracts. Increasing the amount of REST4 progressively diminished the binding of NRSF/REST to the NRSE/RE-1 sequence (Fig. 6C). In contrast, a slight increase in the intensity of the gel shift band was seen with increasing amounts of nuclear extract from vector-transfected HEK293 cells. This slight increase presumably reflects the endogenous NRSF/REST.
FIG. 6.
REST4 inhibits binding of NRSF/REST to the NRSE element of the cholinergic gene. (A) Electrophoretic mobility shift assays performed with nuclear extracts from HEK293 cells expressing NRSF/REST (lane 1), REST4 (lane 2), and a 1:1 mixture of the two extracts (lane 3). (B) Western blot analysis of HEK293 cells transfected with NRSF/REST or REST4. The positions of molecular mass markers are shown on the right. (C) Electrophoretic mobility shift assays performed with 10 μg of a nuclear extract from HEK293 cells expressing NRSF and increasing amounts (0 to 10 μg) of a HEK293 nuclear extract expressing REST4 (+REST4) or control plasmid (−REST4). The arrow indicates the gel shift band produced by the binding of NRSF/REST to the NRSE from the cholinergic gene. The faster-moving gel shift band is a nonspecific band seen in all experiments.
Since REST4 interferes with the binding of NRSF/REST to the NRSE, we looked for a direct interaction between REST4 and NRSF/REST. As shown in Fig. 7, mixing of an N-terminally myc-tagged NRSF/REST with an N-terminally FLAG-tagged REST4 led to the immunoprecipitation of both proteins with either an anti-myc antiserum or an anti-FLAG antiserum.
FIG. 7.
REST4 forms a hetero-oligomer with NRSF/REST. Nuclear extracts were prepared from HEK293 cells expressing either FLAG-REST4 or myc-NRSF/REST. The individual extracts or a mixture of the two were subjected to immunoprecipitation (IP) with either an immobilized goat IgG to the FLAG epitope (FLAG), an immobilized goat IgG to the myc epitope (Myc), or immobilized nonimmune goat IgG (NI). The resulting samples were subjected to SDS-PAGE and probed with MAb 12C11-1, which is directed against the N-terminal region of NRSF/REST and thus recognizes both NRSF/REST and REST4. The bands corresponding to REST4 and NRSF/REST are indicated. The positions of molecular mass markers are shown on the left.
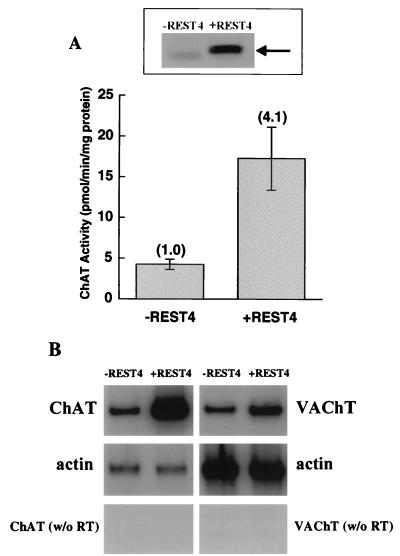
To determine whether expression of REST4, in the absence of nuclear PKA, is sufficient to transcriptionally activate the ChAT gene, we transiently transfected the A126.1B2 cell line with recombinant REST4. As shown in Fig. 8A, this led to an approximately fourfold increase in ChAT activity in this cell line, with a concomitant increase in ChAT mRNA (Fig. 8B). VAChT mRNA levels also increased, but to a lesser extent (Fig. 8B).
FIG. 8.
ChAT activity and ChAT and VAChT mRNAs in A126.1B2 cells transfected with REST4. (A) A126.1B2 was transfected with pCS2+MT-REST4 (+REST4) or vector alone (−REST4), using SuperFect transfection reagent. After 72 h, cell extracts were prepared and assayed for REST4 expression by Western blot analysis (top) or for ChAT activity (bottom) as described in Materials and Methods. (B) PCR analysis for ChAT mRNA (30 cycles) and VAChT mRNA (35 cycles) of A126.1B2 treated as described above.
DISCUSSION
Utilizing mutant PC12 cell lines which have reduced levels of PKA activity, we previously found that basal transcription of the ChAT and VAChT genes is regulated coordinately by PKA (25). In this study, we have shown that reintroduction of PKA activity into one of these PC12 cell lines, A126.1B2, can restore ChAT activity.
Studies conducted with reporter gene constructs further demonstrate that the effect of PKA is at the transcriptional level and that the action of PKA requires an intact NRSE/RE-1 sequence. This element, which is though to be responsible for silencing the activity of a number of neuronal genes in nonneuronal cells, has been shown to be active in the cholinergic gene locus (16). The observation that the concentrations of the transcription factor that binds to the NRSE/RE-1, NRSF/REST, are essentially the same in the parental and PKA mutant PC12 cell lines indicates that PKA does not affect the levels of this transcription factor. On the other hand, electrophoretic mobility shift assays indicate that the NRSF/REST in the PKA-deficient cell line can bind to the NRSE/RE-1 sequence. In contrast, nuclear extracts derived from the wild-type PC12 cell line exhibited barely detectable binding to the NRSE/RE-1 sequence despite the fact that these extracts contain the same levels of NRSF/REST immunoreactive protein. Restoration of PKA activity in the PKA-deficient cell line resulted in a loss of the ability of the NRSF/REST to bind to the NRSE/RE-1.
The finding of an active NRSF/REST in the PKA-deficient cell line can thus account for the low ChAT activity in these cells, caused by repression of the cholinergic gene locus by this transcription factor. The fact that the catalytic subunit of PKA can lead to an apparently inactive NRSF/REST and concomitantly increase ChAT gene expression indicates that the actions of PKA can be solely explained by a PKA signaling pathway regulating NRSF function. Labeling experiments suggest that PKA does not directly phosphorylate NRSF/REST.
In contrast to the finding that full-length NRSF/REST is expressed in both the parental and the PKA mutant PC12 cell lines, the alternatively spliced REST4 isoform is expressed only in the parental cell line. REST4 is formed by the utilization of an alternative exon between exons V and VI of the NRSF/REST gene, which introduces a stop codon (21). Thus, REST4 contains only five of the nine zinc finger domains found in NRSF/REST.
Expression of the PKA catalytic subunit in the mutant PC12 cell line induced expression of REST4, and the direct introduction of REST4, in the absence of PKA, mimicked the action of PKA on the cholinergic gene. In other words, the expression of exogenous REST4 was sufficient to rescue the mutation in PKA and restore expression of ChAT, bypassing the requirement for PKA activity.
The effect of PKA can be accounted for by its regulation of the splicing of the NRSF/REST gene to produce the REST4 isoform. In this regard, the defect in the mutant PC12 cell line A126.1B2 is an absence of nuclear PKA activity. This localization is consistent with a role for PKA in regulating NRSF/REST splicing, although we do not yet know whether regulation is direct or indirect. Treatment of PC12 cells with forskolin, which increases intracellular cAMP levels by activating adenylate cyclase, induced REST4 mRNA within 4 h and ChAT mRNA within 6 to 12 h. The kinetics of forskolin induction is consistent with PKA first increasing REST4 levels, which in turn leads to increased levels of ChAT mRNA.
We have shown that although REST4 itself does not bind to the NRSE, it acts as a dominant negative by inhibiting the binding of NRSF/REST to the NRSE sequence. Dominant-negative forms of NRSF/REST, generated by both N- and C-terminal truncation of NRSF/REST such that there are eight DNA binding zinc finger domains, have been generated and utilized (6, 7). Palm et al. (21) found that REST2-5trunc acted as a transcriptional repressor of a reporter gene in Neuro-2A cells, which do not contain detectable NRSF/REST. Relatively weak repression of the reporter gene was seen in C6 cells, which contain relatively high levels of endogenous NRSF/REST. They concluded that the truncated forms of REST produce repression independent of the activity of endogenous NRSF/REST and that the repressor activity of the truncated forms resides in an N-terminal repressor (26, 27). It was suggested (21) that the repressor activity of truncated forms of REST such as REST4 might result from their formation of a complex with essential components of the transcription machinery.
Our data show that REST4 acts as a dominant negative. The mechanism by which REST4 acts as a dominant negative can be accounted for, at least in part, by the physical interaction, either direct or indirect, between REST4 and NRSF/REST. Consistent with the results shown in Fig. 4B and C and 6C, the complex containing REST4 and NRSF/REST cannot bind to the NRSE.
The finding that REST4 can act as a dominant negative, coupled with its expression in neuronal cells, can account for the presence of NRSF/REST in neuronal cells without its repressing neuronal gene expression. Palm et al. (21) reported the presence of NRSF/REST in adult neurons, albeit at rather low levels relative to its expression in nonneuronal cells. Both Palm et al. (21) and Thiel et al. (27) suggested that the inability of NRSF/REST to repress gene expression in neurons could be explained by its low concentration. Although this is one possible reason, we propose a second mechanism: that REST4, which is found coexpressed with NRSF/REST in neurons, modulates NRSF/REST as a transcriptional repressor of neuronal genes in neurons. Thus, the relative levels of NRSF/REST and REST4 determine the extent of transcriptional repression. Although NRSF/REST levels appear to be constant in PC12 cells, we found that the levels of REST4 mRNA respond to effectors of cAMP levels and PKA activity. Thus, at least in the PC12 cell line that we studied, the cholinergic gene was not maximally active since there was an excess of NRSF/REST relative to REST4. The transcriptional activity of the cholinergic gene can thus be regulated by basal levels of PKA, and the levels of REST4 can be increased or decreased by effectors of PKA. This mechanism, rather than acting on a cAMP response element, can explain the effects of cAMP on cholinergic gene expression in PC12 cells. Since REST4 is only expressed in neurons (21), its action is limited to these cells. Whether this mechanism can be extended to cholinergic and other neurons in vivo remains to be determined.
In summary, our results suggest that REST4 itself does not act as a transcriptional repressor but rather regulates the repressor activity of NRSF/REST. We propose that this mechanism be called the antisilencer mechanism of gene regulation.
ACKNOWLEDGMENTS
This research was supported in part by grants from the National Institute on Aging (AG05893 and AG05144). A.J.P. is a recipient of an NIH predoctoral training grant.
REFERENCES
- 1.Bejanin S, Cervini R, Mallet J, Berrard S. A unique gene organization for two cholinergic markers, choline acetyltransferase and a putative vesicular transporter of acetylcholine. J Biol Chem. 1994;269:21944–21947. [PubMed] [Google Scholar]
- 2.Berrard S, Varoqui H, Cervini R, Israel M, Mallet J, Diebler M F. Coregulation of two embedded gene products, choline acetyltransferase and the vesicular acetylcholine transporter. J Neurochem. 1995;65:939–942. doi: 10.1046/j.1471-4159.1995.65020939.x. [DOI] [PubMed] [Google Scholar]
- 3.Berse B, Blusztajn J K. Coordinated up-regulation of choline acetyltransferase and vesicular acetylcholine transporter gene expression by the retinoic acid receptor α, cAMP, and leukemia inhibitory factor/ciliary neurotrophic factor signaling pathways in a murine septal cell line. J Biol Chem. 1995;270:22101–22104. doi: 10.1074/jbc.270.38.22101. [DOI] [PubMed] [Google Scholar]
- 4.Cassano S, Gallo A, Buccigrossi V, Porcellini A, Cerillo R, Gottesman M E, Avvedimento E V. Membrane localization of cAMP-dependent protein kinase amplifies cAMP signaling to the nucleus in PC12 cells. J Biol Chem. 1996;271:29870–29875. doi: 10.1074/jbc.271.47.29870. [DOI] [PubMed] [Google Scholar]
- 5.Cervini R, Houhou L, Pradat P-F, Bejanin S, Mallet J, Berrard S. Specific vesicular acetylcholine transporter promoters lie within the first intron of the rat choline acetyltransferase gene. J Biol Chem. 1995;270:24654–24657. doi: 10.1074/jbc.270.42.24654. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z-F, Paquette A J, Anderson D J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 7.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 8.Correll L A, Woodford T A, Corbin J D, Mellon P L, McKnight G S. Functional characterization of cAMP-binding mutations in type I protein kinase. J Biol Chem. 1989;264:16672–16678. [PubMed] [Google Scholar]
- 9.Erickson J D, Varoqui H, Schafer M K, Modi W, Diebler M F, Weihe E, Rand J, Eiden L E, Bonner T I, Usdin T B. Functional identification of a vesicular acetylcholine transporter and its expression from a “cholinergic” gene locus. J Biol Chem. 1994;269:21929–21932. [PubMed] [Google Scholar]
- 10.Ginty D D, Glowacka D, DeFranco C, Wagner J A. Nerve growth factor-induced neuronal differentiation after dominant repression of both type I and type II cAMP-dependent protein kinase activities. J Biol Chem. 1991;266:15325–15333. [PubMed] [Google Scholar]
- 11.Hersh L B. Induction of choline acetyltransferase in the neuroblastoma × glioma cell line NG108-15. Neurochem Res. 1992;17:1063–1067. doi: 10.1007/BF00967282. [DOI] [PubMed] [Google Scholar]
- 12.Inoue H, Li Y P, Wagner J A, Hersh L B. Expression of the choline acetyltransferase gene depends on protein kinase A activity. J Neurochem. 1995;64:985–990. doi: 10.1046/j.1471-4159.1995.64030985.x. [DOI] [PubMed] [Google Scholar]
- 13.Kallunki P, Jenkinson S, Edelman G M, Jones F S. Silencer elements modulate the expression of the gene for the neuron-glia cell adhesion molecule, Ng-CAM. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 14.Kengaku M, Misawa H, Deguchi T. Multiple mRNA species of choline acetyltransferase from rat spinal cord. Mol Brain Res. 1993;18:71–76. doi: 10.1016/0169-328x(93)90174-n. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci USA. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonnerberg P, Schoenherr C J, Anderson D J, Ibanez C F. Cell type-specific regulation of choline acetyltransferase gene expression. Role of the neuron-restrictive silencer element and cholinergic-specific enhancer sequences. J Biol Chem. 1996;271:33358–33365. doi: 10.1074/jbc.271.52.33358. [DOI] [PubMed] [Google Scholar]
- 17.Mieda M, Haga T, Saffen D W. Expression of the rat m4 muscarinic acetylcholine receptor gene is regulated by the neuron-restrictive silencer element/repressor element 1. J Biol Chem. 1997;272:5854–5860. doi: 10.1074/jbc.272.9.5854. [DOI] [PubMed] [Google Scholar]
- 18.Misawa H, Ishii K, Deguchi T. Gene expression of mouse choline acetyltransferase. Alternative splicing and identification of a highly active promoter region. J Biol Chem. 1992;267:20392–20399. [PubMed] [Google Scholar]
- 19.Misawa H, Takahashi R, Deguchi T. Coordinate expression of vesicular acetylcholine transporter and choline acetyltransferase in sympathetic superior cervical neurons. Neuroreport. 1995;6:965–968. doi: 10.1097/00001756-199505090-00004. [DOI] [PubMed] [Google Scholar]
- 20.Mori N, Schoenherr C J, Vandenbergh D J, Anderson D J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 21.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth M B, Zahler A M, Stolk J A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahara S, Sato K, Kaise H, Mori K, Sato A, Aoto M, Tokmakov A A, Fukami Y. Biochemical evidence for the interaction of regulatory subunit of cAMP-dependent protein kinase with IDA (inter-DFG-APE) region of catalytic subunit. FEBS Lett. 1996;384:138–142. doi: 10.1016/0014-5793(96)00302-x. [DOI] [PubMed] [Google Scholar]
- 24.Schoenherr C J, Anderson D J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 25.Shimojo M, Wu D, Hersh L B. The cholinergic gene locus is coordinately regulated by protein kinase A II in PC12 cells. J Neurochem. 1998;71:1118–1126. doi: 10.1046/j.1471-4159.1998.71031118.x. [DOI] [PubMed] [Google Scholar]
- 26.Tapia-Ramirez J, Eggen B J, Peral-Rubio M J, Toledo-Aral J J, Mandel G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc Natl Acad Sci USA. 1997;94:1177–1182. doi: 10.1073/pnas.94.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiel G, Lietz M, Cramer M. Biological activity and modular structure of RE-1-silencing transcription factor (REST), a repressor of neuronal genes. J Biol Chem. 1998;273:26891–26899. doi: 10.1074/jbc.273.41.26891. [DOI] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Buskirk R, Corcoran T, Wagner J A. Clonal variants of PC12 pheochromocytoma cells with defects in cAMP-dependent protein kinases induce ornithine decarboxylase in response to nerve growth factor but not to adenosine agonists. Mol Cell Biol. 1985;5:1984–1992. doi: 10.1128/mcb.5.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu D, Hersh L B. Choline acetyltransferase: celebrating its fiftieth year. J Neurochem. 1994;62:1653–1663. doi: 10.1046/j.1471-4159.1994.62051653.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, Y., and L. B. Hersh. Unpublished data.