Abstract
SF3b is a U2 snRNP-associated protein complex essential for spliceosome assembly. Although evidence that SF3b contains the spliceosomal proteins SAPs 49, 130, 145, and 155 has accumulated, a protein-mediated association between all of these proteins has yet to be directly demonstrated. Here we report the isolation of a cDNA encoding SAP 130, which completes the cloning of the putative SF3b complex proteins. Using antibodies to SAP 130 and other putative SF3b components, we showed that SAPs 130, 145, and 155 are present in a protein complex in nuclear extracts and that these proteins associate with one another in purified U2 snRNP. Moreover, SAPs 155 and 130 interact with each other (directly or indirectly) within this complex, and SAPs 49 and 145 are known to interact directly with each other. Thus, together with prior work, our studies indicate that SAPs 49, 130, 145, and 155 are indeed components of SF3b. The Saccharomyces cerevisiae homologs of SAPs 49 and 145 are encoded by essential genes. We show here that the S. cerevisiae homologs of SAPs 130 and 155 (scSAP 130/RSE1 and scSAP 155, respectively) are also essential. Recently, the SF3b proteins were found in purified U12 snRNP, which functionally substitutes for U2 snRNP in the minor spliceosome. This high level of conservation, together with the prior observation that the SF3b proteins interact with pre-mRNA very close to the branch site, suggest that the SF3b complex plays a critical role near or at the spliceosome catalytic core.
Many proteins essential for spliceosome assembly and splicing have been identified, and numerous human homologs of essential yeast splicing factors are now known (for reviews, see references 19, 20, 24, and 29). Among the best characterized of these are the components of U2 snRNP (for reviews, see references 19, 20, and 29). In mammals, functional 17S U2 snRNP can be assembled from 12S U2 snRNP and two essential splicing factors, SF3a and SF3b (5, 6). SF3a has been purified to homogeneity and contains three proteins (SF3a60, SF3a66 and SF3a120) (5, 6). SF3b has been purified through multiple chromatographic steps but has not been purified to homogeneity (5, 6). The components thought to constitute SF3b were identified by comparing purified 17S U2 snRNP and the spliceosomal complex A (for reviews see references 14 and 19). The abundant proteins common to both of these complexes are referred to as SF3b 53, 120, 150, and 160 in 17S U2 snRNP and SAPs 49, 130, 145 and 155, respectively, in the spliceosome (we use the latter nomenclature here) (2, 6, 19). Further evidence that at least two of these proteins are components of SF3b came from the observation that SAPs 49 and 145 interact directly with each other (7). In addition, SAPs 49, 145, and 155, as well as all three SF3a subunits, can be UV cross-linked to the region surrounding the branch site in the spliceosomal complex A (11, 12). Thus, these proteins are all located next to one another in functional spliceosomal complexes, consistent with the notion that they are present in a complex. Despite all of the circumstantial evidence that SAPs 49, 130, 145, and 155 correspond to SF3b, it remains to be established whether any or all of these proteins are indeed components of a single protein complex.
All of the mammalian SF3a components and three of the putative SF3b components (SAPs 49, 145, and 155) have been cloned (19). In addition, yeast counterparts of SF3a have been identified and shown to be essential for viability (for a review, see reference 19). In contrast to SF3a, none of the putative SF3b components were identified in the early genetic screens for yeast splicing factors. However, the likely Saccharomyces cerevisiae homologs of SAPs 145 and 155, scSAP 145 and scSAP 155, were identified in the GenBank database on the basis of their similarity to the corresponding mammalian proteins (7, 12, 26). One of these proteins, scSAP 145, was subsequently found to be the same as CUS1, a protein identified as a suppressor of a U2 snRNA mutation (27). scSAP 145 is essential for A complex assembly in yeast (27). scSAP 49/HSH49 was also identified in the database and shown to be essential for viability in yeast (15). Yeast SAPs 49 and 145, like their mammalian counterparts, interact directly with each other via protein-protein interactions and thus are presumed to be components of a yeast SF3b complex (10, 15). It is not yet known whether scSAP 155 is essential in yeast or whether a yeast counterpart of SAP 130 exists.
Here we report the isolation of a cDNA encoding SAP 130. Using antibodies to this protein, as well as antibodies to other putative SF3b components, we showed that SAPs 130, 145, and 155 are present in a protein complex and that SAPs 130 and 155 interact (directly or indirectly) with each other within this complex. Together with previous work, our data provide strong evidence that SAPs 49, 130, 145, and 155 are components of SF3b. We have also completed the description of the yeast SF3b counterparts by identifying the S. cerevisiae homolog of SAP 130 and showing that it and scSAP 155 are essential in yeast. Thus, together the data indicate that the spliceosomal proteins SAPs 49, 130, 145, and 155 are components of a highly conserved protein complex essential for splicing.
MATERIALS AND METHODS
Isolation of SAP 130 cDNA.
Spliceosomal complex A3′ was produced in large amounts by incubating biotinylated AD3′ENH pre-mRNA (which lacks a 5′ splice site but contains an exonic enhancer) in large-scale splicing reactions (11 ml) (3, 9). The A3′ complex was isolated by gel filtration followed by binding to avidin agarose. Total protein from the purified A3′ complex was then fractionated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and the SAP 130 band was excised from the Coomassie brilliant blue-stained gel. Two sequences (NVSEELDRTPPEVSK and KLEDIRTRYAF) were obtained by microsequencing (W. Lane, Microchemistry Facility, Harvard University). These sequences are encoded by a partial cDNA in the GenBank database (accession no. T92977). On the basis of this sequence, oligonucleotide probes were designed for screening human λ bacteriophage cDNA libraries. A partial cDNA encoding 718 amino acids from the carboxyl terminus of SAP 130 was isolated from this screen. To isolate the 5′ terminus of the SAP 130 cDNA, we used rapid amplification of cDNA ends and PCR amplification (Marathon Ready cDNA; Clontech). The full-length SAP 130 cDNA was created by joining the 5′ and the 3′ cDNA clones.
Western analysis and immunoprecipitation.
Rabbit polyclonal antibodies were raised against the SAP 130 carboxyl-terminal peptide sequence, VSKKLEDIRTRYAF (HRP Inc). The cap (4) and B" (13) monoclonal antibodies and SAP 155 (26), SAP 145 (23), and hPrp17 (31) rabbit polyclonal antibodies have been described. SF3a rabbit polyclonal antibodies will be described elsewhere (9a). For Western blots, spliceosomal complexes were isolated by gel filtration followed by biotin-avidin affinity purification (2, 21). Total proteins were fractionated on SDS–6% polyacrylamide gels and then immobilized on polyvinylidene difluoride membranes. Polyclonal antibodies were used at a 1/1,000 dilution, and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody was used at 1/5,000. Blots were blocked in 5% nonfat dry milk in phosphate-buffered saline containing 0.1% Tween 20. Proteins were detected with an ECL kit (Amersham). Immunoprecipitation of U2 snRNP with the cap or B" antibody was carried out by coupling the antibody to protein G-Sepharose and then rotating with nuclear extract for 4 h. After extensive washing with 250 mM NaCl–20 mM Tris (pH 7.8), proteins were eluted with SDS, RNase A, or 1 M urea as indicated below.
Immunoprecipitations of the RNase and urea eluates were carried out by rotating with the indicated antibodies, followed by washing with 250 mM NaCl–20 mM Tris (pH 7.8).
Disruption of scSAPs 130 and 155 in S. cerevisiae.
Disruption of scSAP 130 was carried out by using standard homologous recombination procedures (22). Briefly, ∼0.5 kb of the 5′ flanking region and ∼0.5 kb of the 3′ flanking region of the SAP 130 gene were amplified from total yeast genomic DNA by PCR with appropriate primers. These flanking regions were then cloned on either side of the LEU gene to generate the plasmid pscSAP 130Δ::LEU. This plasmid was digested with AlwNI on the 5′ end and StuI on the 3′ end of the SAP 130-flanking regions, and the DNA was used to transform a wild-type diploid yeast strain. For disruption of scSAP 155, 1.229 kb of the 5′ flanking region and 761 bp of the 3′ flanking region of the SAP 155 gene were amplified from total yeast genomic DNA by using PCR and appropriate primers. These flanking regions were then cloned on either side of the LEU gene to generate the plasmid pscSAP 155Δ::LEU. This plasmid was digested with ApaI on the 5′ end and XbaI on the 3′ end of the SAP 155 flanking regions, and the DNA was used to transform a wild-type diploid yeast strain. Transformants were selected on plates lacking leucine, and correct integration of the LEU gene into the SAP 130 locus was verified by Southern analysis. The transformants were sporulated and tetrads were dissected by standard procedures.
Nucleotide sequence accession number.
The accession number for the full-length SAP 130 cDNA is AJ001443.
RESULTS
Isolation of a human SAP 130 cDNA.
To isolate the cDNA encoding SAP 130, we purified spliceosomal complex A in large quantities (3). The proteins in this complex were fractionated by SDS-polyacrylamide gel electrophoresis, and peptide sequences were obtained from tryptic digestion products of SAP 130. These sequences were used to isolate a full-length cDNA (see Materials and Methods). The SAP 130 cDNA is predicted to encode a 1,217-amino-acid protein with a molecular mass of 130 kDa and an isoelectric point of 5.10 (Fig. 1). The initiator methionine is preceded by a well-conserved Kozak sequence, and the reading frame is closed upstream of this methionine, indicating that the cDNA encodes the full-length SAP 130 protein (18) (data not shown).
FIG. 1.
Predicted amino acid sequence of SAP 130 cDNA. The amino acid sequence predicted from the human SAP 130 cDNA sequence is shown. The two peptide sequences obtained from microsequencing are underlined once and twice, and the peptide used for making antibodies is boxed.
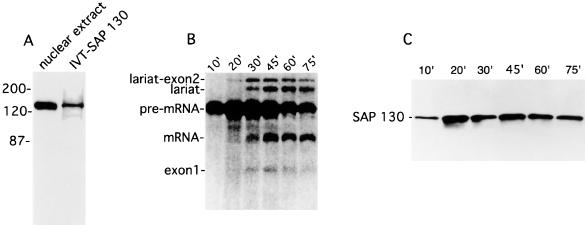
To further characterize the SAP 130 cDNA, we raised rabbit polyclonal antibodies to a C-terminal peptide (Fig. 1). This antibody is highly specific for SAP 130, as it detects only a single 130-kDa band in total HeLa cell nuclear extracts (Fig. 2A). The protein produced by in vitro translation of the SAP 130 cDNA comigrates with the protein detected on Western blots by SAP 130 antibodies (Fig. 2A). SAP 130 antibodies also detect the SAP 130 protein in affinity-purified spliceosomal complexes (Fig. 2B and C). This protein associates with spliceosomes by 10 min of assembly and remains bound throughout the splicing time course (Fig. 2B and C). The SAP 130 antibody also specifically immunoprecipitates the in vitro translation product of the SAP 130 cDNA (data not shown). Together, these observations indicate that we have isolated a full-length cDNA encoding the spliceosomal protein SAP 130. The SAP 130 antibodies do not immunoprecipitate the spliceosome or U2 snRNP from splicing extracts, indicating that the epitope (14 residues at the C terminus [Fig. 1]) is not accessible in these complexes (data not shown).
FIG. 2.
SAP 130 antibodies detect a single ∼130-kDa band in nuclear extract and spliceosomal complexes. (A) The in vitro translation product of the SAP 130 cDNA comigrates with a protein in the nuclear extract that is detected by SAP 130 antibodies. (B) 32P-labeled adenovirus major late pre-mRNA was incubated under splicing conditions for the times indicated and then total RNA was fractionated on a 15% denaturing gel. Splicing intermediate and products are indicated. (C) SAP 130 antibodies were used to probe a Western blot of purified spliceosomal complexes assembled for the indicated times under splicing conditions. IVT, in vitro translation.
SAPs 130, 145, and 155 exist in a protein complex in nuclear extracts and in U2 snRNP.
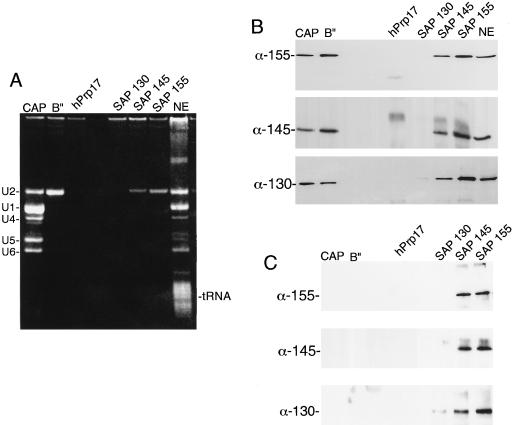
As mentioned in the introduction, SAPs 49, 130, 145, and 155 are components of 17S U2 snRNP and putative components of the essential splicing factor SF3b (5, 6). To determine whether these proteins associate with one another in splicing extracts and thus could correspond to the SF3b complex, we carried out coimmunoprecipitation assays. The antibodies used were directed against the snRNP-specific trimethyl cap, the U2 snRNP-specific protein B", SAPs 130, 145, and 155, and hPrp17 (see Materials and Methods). As shown in Fig. 3A, all of the spliceosomal snRNPs were immunoprecipitated by anticap antibodies, and U2 snRNP alone was immunoprecipitated by anti-B", anti-SAP 145, and anti-SAP 155. No snRNPs were immunoprecipitated by anti-SAP 130 (see above) or the negative control, anti-hPrp17.
FIG. 3.
SAPs 130, 145, and 155 are part of a protein complex in nuclear extracts. (A) The indicated antibodies were used for immunoprecipitations from total nuclear extracts (NE). Total RNA was fractionated on an 8% denaturing gel. The snRNAs and tRNA are indicated. (B) An aliquot of each immunoprecipitate was fractionated on an SDS–6% polyacrylamide gel, and then Western blots were probed with the indicated antibodies (α). (C) Same as panel B except that nuclear extract was incubated with 2 μl of RNase A (10 mg/ml) prior to the immunoprecipitations.
Western analysis showed that SAPs 155, 145, and 130 were present in all of the immunoprecipitates that contained U2 snRNA (Fig. 3B). Thus, as expected, these proteins are components of U2 snRNP. To determine whether these proteins exist in a protein complex independently of U2 snRNA, we preincubated nuclear extracts in RNase A, which completely digested the snRNAs (data not shown). Immunoprecipitations were then carried out with the antibodies described above. As shown in Fig. 3C, SAPs 155, 145, and 130 were no longer detected in the anticap or anti-B" immunoprecipitates. In contrast, all three proteins are detected in the anti-SAP 145 and anti-SAP 155 immunoprecipitates. These data indicate that SAPs 130, 145, and 155 exist in a protein complex independently of U2 snRNA.
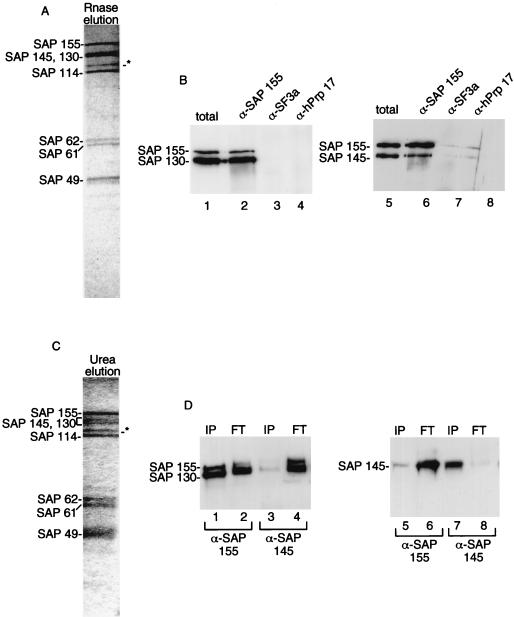
To determine whether SAPs 130, 145, and 155 interact with each other in a discrete protein complex within the purified U2 snRNP particle, we immunopurified 17S U2 snRNP from nuclear extracts using immobilized anti-B" antibody. The proteins were eluted from the antibody by incubation with RNase A (Fig. 4A). The major proteins in the eluate correspond to SF3a and the putative SF3b proteins (see below; also data not shown). When the eluate was used for immunoprecipitations with anti-SAP 155 (Fig. 4B, lanes 2 and 6) or anti-SAP 145 (data not shown) antibodies, SAPs 145, 155, and 130 were detected on Western blots of the immunoprecipitates. These three proteins were not detected when the eluate was immunoprecipitated with antibodies to SF3a (Fig. 4B, lanes 3 and 7) or the negative control antibody, anti-hPrp17 (Fig. 4B, lanes 4 and 8). Thus, SAPs 130, 145, and 155 are present in a specific protein complex which is associated with U2 snRNP. The protein complex does not associate with SF3a in the absence of U2 snRNA.
FIG. 4.
SAPs 130 and 155 interact with each other in the protein complex. (A) Antibodies to the U2 snRNP protein B" were used to immunoprecipitate U2 snRNP from nuclear extract. Total proteins were eluted with RNase A and fractionated on an SDS–9% polyacrylamide gel. The abundant SAPs are indicated and were identified on two-dimensional gels and on Western blots (panel B and data not shown). The asterisk designates a band of unknown identity. However, it is unlikely to be a component of SF3b since it does not coimmunoprecipitate with SF3b antibodies (data not shown). (B) The eluate from panel A was used for immunoprecipitations with the antibodies (α) indicated on the top of each lane, and Western blots were probed with the antibodies indicated to the left of each blot. (C) Same as panel A except that proteins were eluted with 1 M urea. (D) The urea eluate was used for immunoprecipitations with the SAP 155 and SAP 145 antibodies. The bound (IP) and unbound (FT) proteins were fractionated on an SDS–6% polyacrylamide gel, and Western blots were probed with the indicated antibodies.
To further define interactions among the SF3b proteins within U2 snRNP, we immunopurified U2 snRNP using the B" antibody and then eluted the proteins with 1 M urea. This eluate contained the set of proteins detected in the RNase eluate (compare Fig. 4A and C). When the urea eluate was used for immunoprecipitations with anti-SAP 155, we found that SAPs 155 and 130 coimmunoprecipitated (Fig. 4D, lane 1). In contrast, SAPs 130 and 155 were found in the flowthrough when anti-SAP 145 was used for the immunoprecipitation (Fig. 4D, lanes 3 and 4). Consistent with these data, SAP 145 was found in the flowthrough when anti-SAP 155 was used for immunoprecipitation (Fig. 4D, lanes 5 and 6) whereas SAP 145 was found in the immunoprecipitate when anti-SAP 145 antibodies were used (Fig. 4D, lanes 7 and 8). We conclude that SAPs 130 and 155 interact more tightly with each other than with SAP 145.
Together, the data presented above indicate that SAPs 130, 145, and 155 associate with each other via protein-protein interactions. These proteins are present as a protein complex in purified U2 snRNP, and SAPs 130 and 155 interact more tightly with each other than with SAP 145 within this complex. Further studies are needed to determine whether the interaction between SAPs 130 and 155 is direct or indirect (see Discussion).
scSAPs 130 and 155 are essential for viability in yeast.
Previously, the S. cerevisiae homologs of SAP 145/CUS1 (YMR240c) and SAP 49/HSH49 (YOR319w) were shown to be essential in yeast (15, 27). These and other data (see the introduction) indicate that yeast also contains the SF3b complex (15, 27). To obtain further evidence that this is the case, we asked whether yeast contains a SAP 130 homolog. The greatest similarity to the human SAP 130 gene in the database was an S. cerevisiae open reading frame that we designated scSAP 130. During the course of our study, Chen and coworkers (8) independently identified scSAP 130 as a protein, RSE1 (YML049c), in a screen of temperature-sensitive mutants defective in endoplasmic reticulum-to-golgi transport. Characterization of the temperature-sensitive RSE1 gene revealed that it is a splicing factor (8). Taken together, these data indicate that scSAP 130/RSE1 is the ortholog of SAP 130.
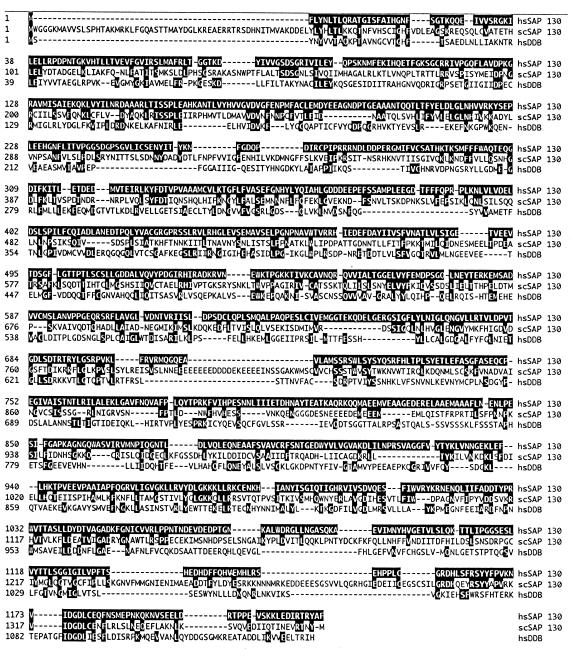
SAP 130 also resembles the large subunit of a human heterodimer, designated damage-specific DNA-binding protein (DDB), that has been implicated in DNA repair (8, 16, 17). Human SAP 130 is 27% identical and 40% similar to scSAP 130 and 20% identical and 29% similar to DDB127 (Fig. 5). The size of the three proteins is conserved, and the similarity is distributed throughout their lengths. Yeast SAP 130 is more related to human SAP 130 than to DDB (Fig. 5). Thus, it is likely that yeast SAP 130 is the functional homolog of human SAP 130 and not of DDB. Consistent with this conclusion, Chen and coworkers (8) showed that RSE1 mutants have normal DNA repair after treatment with UV light. Using antibodies to DDB, we detected this protein in nuclear extracts but not in either purified spliceosomes or the purified hnRNP complex H (data not shown). Thus, DDB does not appear to be a general spliceosomal protein. It is possible that the similarities between these proteins are due to a common function that remains to be identified.
FIG. 5.
Alignment of SAP 130 with scSAP 130 and human DDB. The alignment was done by the Clustal method (DNASTAR Inc.). Residues that are identical in hsSAP 130 and the other two proteins are shown in white on black. The overall identities of scSAP 130 and hsDDB to hsSAP 130 are 27 and 20%, respectively.
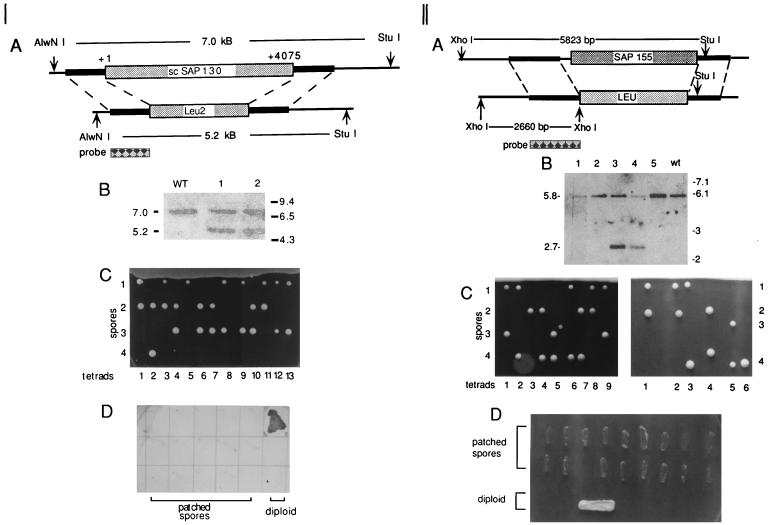
Recently, we identified the likely homolog of SAP 155 (YMR288w) in yeast (26). To determine whether scSAPs 130 and 155 are essential for viability in yeast, we constructed diploid yeast strains in which one copy of the genes was disrupted by replacement with the leu2 selectable marker (22). Southern analysis of total yeast genomic DNA confirmed the replacement of one allele by the Leu2 gene (Fig. 6A and B [panels I and II]). Tetrad analysis demonstrated a 2:2 segregation of nonviable and viable spores (Fig. 6C [panels I and II]), and the viable spores did not grow on plates lacking leucine (Fig. 6D [panels I and II]). Together, these data indicate that scSAP 130 and SAP 155 genes are both essential for viability in yeast.
FIG. 6.
The scSAP 130 and scSAP 155 genes are essential in S. cerevisiae. (Panel I) (A) Structure of scSAP 130 and scSAP 130Δ::LEU alleles. The sizes of AlwNI and StuI restriction fragments are indicated. The probe is shown below the region with which it hybridizes. (B) Southern analysis of total genomic DNA from a normal diploid strain (WT) and diploid strains transformed with the scSAP 130Δ::LEU allele (lanes 1 and 2). The sizes (in kilobases) of markers and restriction fragments are shown. (C) Tetrad analysis. The scSAP 130Δ::LEU strain was sporulated, and 13 individual tetrads were dissected. (D) Viable spores are Leu−. Fifteen viable spores were patched onto a plate lacking leucine. The diploid strain was patched as a control. (Panel II) (A) Structure of SAP 155 and scSAP 155Δ::LEU alleles. The sizes of the StuI and XhoI restriction fragments used to identify each allele are indicated. (B) Southern analysis of total genomic DNA from a normal diploid yeast strain (wt) and diploid strains transformed with the scSAP 155Δ::LEU allele (lanes 1 through 5). The sizes (in kilobases) of the markers and restriction fragments are shown. The differences in band intensities between lanes is most likely due to different levels of DNA loaded on the gel. (C) Tetrad analysis. The scSAP 155Δ::LEU strain was sporulated, and 15 individual tetrads were dissected. (D) Viable spores are Leu−. Eighteen viable spores were patched onto a plate lacking leucine. The diploid strain was patched as a control.
DISCUSSION
Here we report the isolation of a cDNA encoding the human spliceosomal protein SAP 130, which completes the cloning of the major U2 snRNP proteins in humans. Our data show that SAP 130, together with SAPs 145 and 155, exists in a protein complex in nuclear extracts. We do not have antibodies to the U2 snRNP protein SAP 49. However, this protein interacts very tightly with SAP 145 (7). Thus, the simplest interpretation of the data is that SAPs 49, 130, 145, and 155 are present in the same protein complex. Although functional studies are required to prove that this protein complex corresponds to the essential splicing factor SF3b, it is highly likely that this is the case. This contention is based on several observations. Functional 17S U2 snRNP is composed of 12S U2 snRNP together with SF3a and SF3b (5). Purified 17S U2 snRNP contains SAPs 61, 62, and 114, which are known to be the components of SF3a (5, 6). 17S U2 snRNP also contains SAPs 49, 130, 145, and 155 in equal stoichiometries (1). Our data show that these proteins are present in a protein complex within purified U2 snRNP. Thus, it is highly likely that this protein complex corresponds to the U2 snRNP-associated SF3b complex.
Previous work showed that purified 17S U2 snRNP contains two additional proteins (35 and 92 kDa) (1), but these proteins are present at lower levels than the other SF3a and SF3b proteins. Indeed, the 35- and 92-kDa proteins are not evident in our immunopurified U2 snRNP (Fig. 4A and C). Thus, whether they are essential components of U2 snRNP or SF3b remains to be determined.
Previous studies showed that SAPs 145 and 49 interact with each other through direct protein-protein interactions (7). Our data show that SAPs 130 and 155 interact with each other (directly or indirectly). However, it is not known how these two protein complexes (SAP 145-SAP 49 and SAP 155-SAP 130) associate. One possibility is that these complexes interact directly to form SF3b. However, we cannot rule out the alternative possibility that a protein not yet detected mediates the interaction, or that the 35- or 92-kDa protein mentioned above is involved. We note that we were unable to detect direct interactions between SAPs 155 and 130 or between SAP 155-SAP 130 and any of the other U2 snRNP proteins using far-Western analysis. Thus, further work is needed to determine how the SF3b proteins interact with each other.
Our study also provides new evidence that the counterpart of human SF3b is present in S. cerevisiae. Prior work identified scSAPs 145 and 49 and showed that they interact directly and that they are essential in yeast (12, 15, 27). We have shown here that scSAPs 130 and 155 are also essential in yeast, and recent work showed that scSAP 130 is a splicing factor (8). Studies with yeast showed that scSAP 145 suppresses lethal mutations in U2 snRNA near the region that base-pairs to the branch site (30). Likewise, in mammals, all of the SF3a and -b subunits interact with the 5′ end of U2 snRNA near the region that base-pairs with the branch point sequence (1). All of the mammalian SF3a and -b subunits, except SAP 130, also UV cross-link to pre-mRNA around that branch site prior to both catalytic steps I and II. One function for these interactions appears to be to anchor U2 snRNP tightly to the pre-mRNA (12). The SF3b proteins are also part of the U12 snRNP, which is the equivalent of U2 snRNP in the minor-abundance human spliceosome (28). This spliceosome functions to splice a rare class of introns (25). Thus, the high degree of conservation of SF3b, together with its positioning near the catalytic center of the spliceosome, indicates that this complex is likely to play key roles in the establishment and/or functioning of the catalytic center of the spliceosome for both steps of the splicing reaction. The availability of recombinant SF3b subunits in both yeast and mammals should now allow these possibilities to be tested directly.
REFERENCES
- 1.Behrens S E, Tyc K, Kastner B, Reichelt J, Luhrmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 3.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 4.Bochnig P, Reuter R, Bringmann P, Luhrmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987;168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 5.Brosi R, Groning K, Behrens S E, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 6.Brosi R, Hauri H P, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 7.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 8.Chen E J, Frand A R, Chitouras E, Kaiser C A. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol Cell Biol. 1998;18:7139–7146. doi: 10.1128/mcb.18.12.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiara M D, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 9a.Das, R., and R. Reed. Unpublished data.
- 10.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 11.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 13.Habets W J, Hoet M H, De Jong B A, Van der Kemp A, Van Venrooij W J. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- 14.Hodges P E, Beggs J D. RNA splicing. U2 fulfils a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 15.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 16.Kazantsev A, Mu D, Nichols A F, Zhao X, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc Natl Acad Sci USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keeney S, Chang G J, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 18.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 19.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 20.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 21.Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci USA. 1990;87:8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–230. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 23.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 25.Tarn W Y, Steitz J A. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 28.Will C, Schneider C, Reed R, Luhrmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- 29.Will C L, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 30.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Reed R. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]