Abstract
Potent induction of the gene coding for human prointerleukin 1β (il1b) normally requires a far-upstream inducible enhancer in addition to a minimal promoter located between positions −131 and +12. The transcription factor Spi-1 (also called PU.1) is necessary for expression and binds to the minimal promoter, thus providing an essential transcription activation domain (TAD). In contrast, infection by human cytomegalovirus (HCMV) can strongly activate il1b via the expression of immediate early (IE) viral proteins and eliminates the requirement for the upstream enhancer. Spi-1 has been circumstantially implicated as a host factor in this process. We report here the molecular basis for the direct involvement of Spi-1 in HCMV activation of il1b. Transfection of Spi-1-deficient HeLa cells demonstrated both the requirement of Spi-1 for IE activity and the need for a shorter promoter (−59 to +12) than that required in the absence of IE proteins. Furthermore, in contrast to normal, enhancer-dependent il1b expression, which absolutely requires both the Spi-1 winged helix-turn-helix (wHTH) DNA-binding domain and the majority of the Spi-1 TAD, il1b expression in the presence of IE proteins does not require the Spi-1 TAD, which plays a synergistic role. In addition, we demonstrate that a single IE protein, IE2, is critical for the induction of il1b. Protein-protein interaction experiments revealed that the wing motif within the Spi-1 wHTH domain directly recruits IE2. In turn, IE2 physically associates with the Spi-1 wing and requires the integrity of at least one region of IE2. Functional analysis demonstrates that both this region and a carboxy-terminal acidic TAD are required for IE2 function. Therefore, we propose a protein-tethered transactivation mechanism in which the il1b promoter-bound Spi-1 wHTH tethers IE2, which provides a TAD, resulting in the transactivation of il1b.
The clinical importance of human cytomegalovirus (HCMV) infection has increased over the past several decades, in particular as immunosuppressive posttransplantation therapies and AIDS and other immunosuppressive states have become significant medical concerns. Although a majority of the infected population remains subclinically infected, an active infection occurring in a debilitated individual usually leads to mortality. Monocytes and tissue macrophages play prominent roles in host response to HCMV infection and are also an important reservoir during latent infection. After infecting the host, HCMV stimulates the production of various monocyte/macrophage inflammatory cytokines, such as interleukin 1β (IL-1β), IL-2, tumor necrosis factor alpha, IL-6, IL-8, and beta interferon (10, 12, 17, 18).
A markedly increased level of IL-1β observed during the active phase of HCMV infection indicates a significant role for this mediator in the interaction between the host defense mechanism and disease progression (25). We have previously reported that the HCMV immediate early (IE) gene products upregulate the expression of the human prointerleukin 1β gene (il1b) in infected monocytes as well as in transient-transfection assays (12, 25). Interestingly, in contrast to normal expression of il1b, which requires both a far-upstream enhancer and a promoter element (30, 48), transactivation of il1b by HCMV IE gene products eliminates the need for the essential upstream enhancer located between positions −3134 and −2729 and requires only the il1b promoter element located at positions −131 to +12 (11). Within the il1b promoter, there are two binding sites for the myelomonocyte- and B-cell-specific ETS transcription factor Spi-1 (also called PU.1) which have been demonstrated to be critical for basal il1b promoter activity (30) (sites A and B in Fig. 1). Spi-1, a winged helix-turn-helix (wHTH) transcription factor, plays a pivotal role in the development of hematopoietic cell lineages and has also been implicated as an essential host factor in viral transactivation of various cellular genes (17, 24, 26, 28, 54). Studies have suggested direct interactions between Spi-1 and viral proteins, such as the human T-cell leukemia virus type 1 Tax protein (54) and the Epstein-Barr virus nuclear protein-2 (EBNA-2) (26). In the case of il1b transactivation by HCMV, mutation of the Spi-1-binding site located at −50 to −39 results in a dramatic loss of IE protein-dependent transactivation, suggesting a possible involvement of Spi-1 (24). However, this report did not demonstrate a direct role for Spi-1 protein in activation by IE protein because the Spi-1-binding site is also required in the absence of viral protein (30).
FIG. 1.
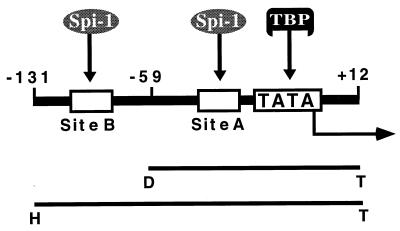
Schematic representation of the il1b promoter showing two Spi-1-binding sites, one located adjacent to the transcription start site and the other located further upstream. The TATA sequence (TATAAAA) and the TBP are also illustrated. Two fragments of the il1b promoter, HT (−131 to +12) and DT (−59 to +12), used in the transfection experiments in this study are shown as bars (see Kominato et al. [30] for a detailed description of these sequences).
Due to the fact that there are three predominant HCMV IE proteins, IE1(p72), IE2(p55), and IE2(p86), reported to activate the transcription of cellular genes by considerably different mechanisms (20, 37), it is important to identify the roles of individual IE proteins in il1b transactivation. It has been shown by many studies that IE1 and IE2, either independently or synergistically, transactivate various promoters. These promoters include those regulating cellular expression of the gene for the 70,000-molecular-weight heat shock protein (hsp70), c-myc, c-fos, and c-jun, as well as the promoter for the human immunodeficiency virus (HIV) long terminal repeat (LTR) (3, 4, 21, 31, 55). IE1(p72) has been described as a transactivator for the HCMV major IE promoter (6) and for some cellular genes via NF-κB (17, 32, 44, 57). IE2(p86) transactivates the HCMV UL 112/113 early promoter and represses the major IE promoter via direct DNA binding (34, 46). It is also a potent transactivator for many cellular genes (14, 33, 36, 41, 47, 56). In contrast to that by IE1, which has not been reported to bind any DNA sequence, transactivation by IE2 appears to involve both direct DNA binding and interaction with various cellular proteins (14). Recent studies have revealed physical associations of IE2 with a number of transcription factors, including TATA-binding protein (TBP), TFIIB (5), Sp1, Tef-1, Rb (19), p53, c-Jun, JunB (47), and p300/CREB-binding protein (45). The function of IE2(p55), an alternatively spliced version of IE2, is unclear, but it does not appear to directly support transactivation (40).
The activation of gene transcription has been widely demonstrated to involve numerous protein-protein interactions. Many of these involve various members of the ETS transcription factor family, including Spi-1 (1, 23, 30, 51, 58). Our study was aimed at determining whether Spi-1 protein is involved in a physical interaction with HCMV IE proteins in enhancer-independent il1b transactivation. Using transient transfection of Spi-1-deficient HeLa cells, we directly demonstrate a requirement for Spi-1. Furthermore, we show that the wHTH DNA-binding domain of Spi-1 is sufficient to support significant transactivation of il1b by HCMV IE proteins. A protein-protein interaction assay was used to demonstrate that IE2, but not IE1, interacts with Spi-1 and to map specific regions of IE2 and Spi-1 which are essential for a physical association between these two proteins. Functional analysis indicates that, in addition to a centrally located region of IE2 required for strong interaction with the Spi-1 wHTH wing motif, a carboxy-terminal region is required and an amino-proximal region is supportive for il1b transactivation. These two regions have previously been reported to serve as an acidic activation domain (40) and a TBP-binding domain (52), respectively. Based on this evidence, we propose a protein-tethered transactivation (PTT) mechanism of il1b by IE2 in which the Spi-1 wHTH directly binds to the il1b promoter and tethers IE2 protein, which is unable to directly bind the promoter, providing a multifunctional TAD required for gene expression.
MATERIALS AND METHODS
Cell culture.
HeLa cells (strain S3) were cultured as previously described (30). The cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum and 0.5% penicillin-streptomycin. Every three days, cells were split 1:10 by adding cold phosphate-buffered saline (PBS) containing 25 mM EDTA to detach the cells.
Reporter constructs and expression vectors.
Plasmids 3MHT and 3MDT, previously described (48), contain the il1b promoter sequence −131 to +12 and the sequence −59 to +12 (Fig. 1), respectively, ligated to chloramphenicol acetyltransferase (CAT) reporter plasmid pA10CAT3M (3M).
Plasmid pGL3B-DT contains the il1b promoter sequence −55 to +12 ligated to the luciferase reporter pGL3-Basic vector (Promega) at XbaI and BglII sites.
The sense and antisense Spi-1 pRc/CMV expression vectors contain the entire wild-type Spi-1 cDNA as previously reported (15). The series of Spi-1 mutant constructs (see Fig. 3a) with deletions in the PEST, PN, and NN regions were constructed by shuttling the cDNAs (43), gifts from Richard Maki (The Scripps Research Institute, La Jolla, Calif.), into pRc/CMV expression vectors. The Spi-1 Δ100 expression vector for the truncated Spi-1 protein, lacking the first 100 amino acids of Spi-1 but containing amino acids 101 to 272, was constructed by digesting the Spi-1 cDNA with NcoI followed by ligation into pRc/RSV (Invitrogen) in the presence of a small oligonucleotide complementary to the overhangs generated by NcoI and HindIII and was provided by Jack Hensold (Case Western Reserve University, Cleveland, Ohio). The Spi-1 Δ8/32 pECE expression vector, which lacks amino acids 8 to 32 and was provided by Richard Maki, had an additional 50-bp T7 promoter sequence located upstream of the cDNA. The Spi-1 ETS domain (wHTH) cDNA corresponding to amino acids 161 to 272 was PCR amplified with the following primers: 5′ TTG CAA GCT TCC GCC ATG CTT CTG CAC GGG GAG ACA G 3′ and 5′ TTG CTC TAG ATC AGT GGG GCG GGA GGC G 3′. The overhangs were generated with HindIII and XbaI and ligated into the pRc/CMV expression vector.
FIG. 3.
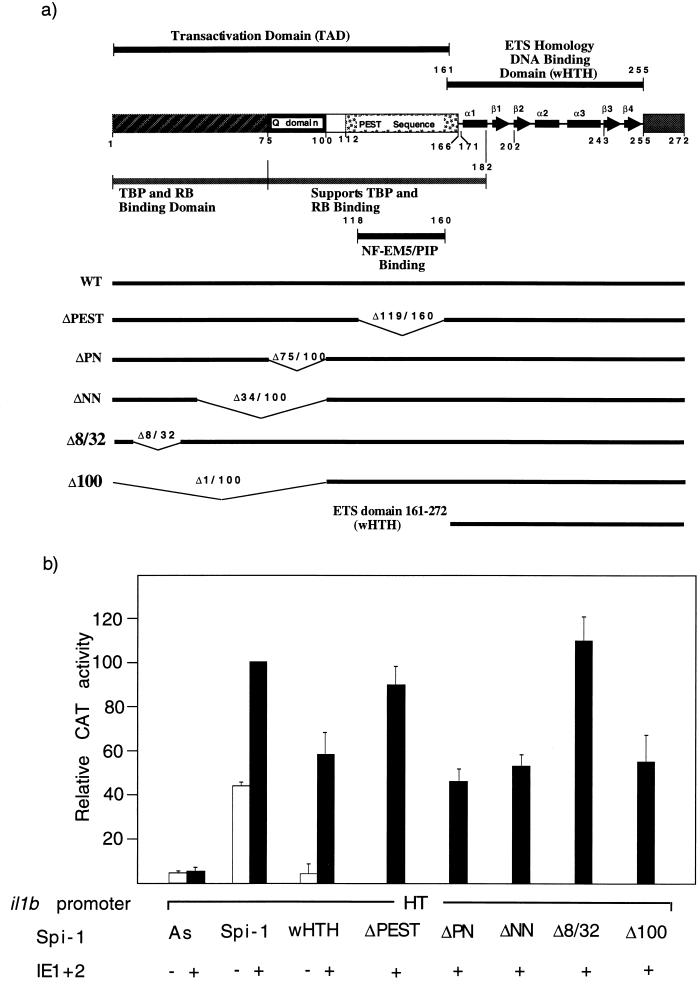
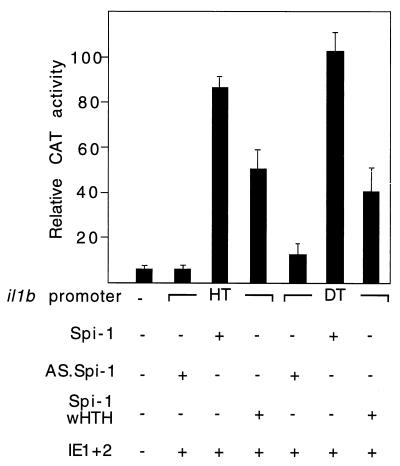
Transactivation of il1b by HCMV IE protein is differentially mediated by discrete domains of the transcription factor Spi-1. (a) Schematic representation of Spi-1 illustrating the ETS homology DNA-binding domain (wHTH) and TAD. The previously identified functional regions are also shown, including the TBP/Rb-binding region, Q domain, and PEST sequence containing the NF-EM5/PIP-binding region. Various Spi-1 expression vectors containing different regions of Spi-1 are shown by horizontal bars. Numbers indicate the amino acid sequences. (b) The Spi-1 wHTH DNA-binding domain alone can support IE transactivation of il1b. The Q domain is important for maximal transactivation. CAT activities observed for the il1b promoter from positions −131 to +12 (HT) are shown. The antisense Spi-1 (As), wild-type Spi-1 (WT), and various mutated Spi-1 expression vectors (as shown in panel a) were each cotransfected with the il1b promoter (HT) CAT reporter along with the HCMV IE1-IE2 expression vector into HeLa cells. Twenty micrograms of the CAT vector and 10 μg of each expression vector were used for each analysis. The results are expressed as average percentages of the activity observed for the wild-type promoter and the Spi-1 expression vector after stimulation with 50 ng of PMA per ml. Open bars correspond to transfections in the absence of the IE1-IE2 expression vector. Error bars indicate the deviations in results from a minimum of three repetitions.
The HCMV IE expression vectors pEQ273(IE1), pEQ326(IE2), and pEQ276(IE1+2), gifts from Adam Geballe (Fred Hutchinson Cancer Research Center, Seattle, Wash.), contain the genomic HCMV IE DNA inserted into the pGEM1 vector. The expression vector pEQ336 contains only the HCMV IE promoter without any IE protein coding sequence. The vector pEQ273(IE1) contains genomic DNA coding for the IE1(p72) protein. The vector pEQ326(IE2) contains genomic DNA for IE2 protein. The vector pEQ276(IE1+2) contains the entire genomic DNA sequence for both IE1 and IE2 via alternative splicing.
The HCMV IE protein expression vectors IE1(p72), IE2(p86), and IE2(p55), as well as the mutated IE2 expression vectors IE2 ΔMX, IE2 ΔSN, IE2 ΔSX519, IE2 ΔXN, and IE2NheI were gifts from Deborah Spector (University of California, San Diego) and contained individual IE cDNAs (see Fig. 6a and b) in the pSG5 (Stratagene) expression vector.
FIG. 6.
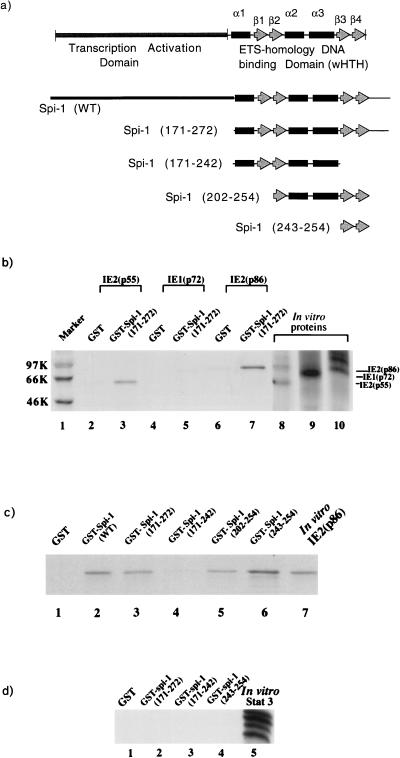
Physical interaction of Spi-1 with IE2. (a) Schematic representation of various GST–Spi-1 fusion constructs. The GST–wild-type Spi-1 fusion construct is indicated as Spi-1 (WT). Other deletion constructs are shown with numbers of amino acids deleted (in parentheses) relative to the amino acid sequence of Spi-1. Quantitation of expressed GST fusion proteins was determined by Coomassie brilliant blue staining (data not shown). (b to d) For all experiments, in vitro-translated 35S-labeled proteins (IE proteins in panels b and c or Stat 3 protein in panel d) were incubated with glutathione-Sepharose beads bound to either GST or GST–Spi-1 fusion proteins (as indicated). After incubation, the glutathione-Sepharose beads were washed extensively and bound proteins were resolved by SDS-PAGE. (b) IE2(p55) and IE2(p86) physically interacted with the Spi-1 DNA-binding domain (wHTH) (lanes 3 and 7, respectively). IE proteins bound weakly to the GST control (lanes 2, 4, and 6). Lanes 8, 9, and 10 indicate the mobilities of these radiolabeled IE proteins as well as the efficiency of in vitro translation. Molecular weight markers (in thousands [K]) are indicated at the left. (c) IE2 directly bound to Spi-1 through a portion (antiparallel β3 and β4) of the wing motif (lane 6). Lane 7 indicates the mobility of the in vitro-35S-labeled IE2(p86) protein on SDS-polyacrylamide gel. In lane 1, the radiolabeled IE2 was incubated with glutathione-Sepharose bead-linked GST alone. In lanes 2 to 6, the radiolabeled IE2 was incubated with immobilized GST–wild-type Spi-1 or various GST–Spi-1 deletion mutation fusion proteins. (d) Stat 3 is unable to bind Spi-1 wHTH. Lane 5 shows the mobility of the Stat 3 protein on SDS-polyacrylamide gel.
The Stat 3 expression vector contains Stat 3 cDNA in the pRc/CMV expression plasmid, a gift from Z. Zhong and J. Darnell, Jr. (Rockefeller University, New York, N.Y.).
Transfections and reporter gene assays.
The CaPO4 transfection and CAT assays were performed as previously described (48). CAT assays were carried out by liquid scintillation (50). In this kinetic method, which is not affected by the saturation limitation of endpoint methods such as conventional thin-layer chromatography, HeLa cell lysates (60 μg of total protein) were adjusted to a final volume of 150 μl with TT buffer (0.25 M Tris-Cl [pH 7.8], 0.5% Triton X-100], and 100 μl of 2.5 mM chloramphenicol (Sigma) solution containing 0.25 μCi of [3H]acetyl-coenzyme A (NEN) was added. The CAT activities were evaluated by calculating the slopes within a linear range of the kinetic response generated by 10 cycles of scintillation counting (total reaction time of approximately 6 h). The first-order slopes were derived by a polynomial curve fit generating a coefficient of correlation which was routinely very close to unity, and only those fits generating r values between 0.998 and 1.000 were used.
For luciferase reporters, liposomal transfection was performed by using DOTAP transfection reagent (Boehringer Mannheim GmbH) according to the manufacturer’s instructions. Briefly, HeLa cells (5 × 104) were plated in 16-mm-diameter culture plates 24 h before transfection. Immediately before transfection, cells (60 to 80% confluence) were washed with 500 μl of DMEM three times and incubated in 1 ml of DMEM containing 10% fetal bovine serum and penicillin-streptomycin for 3 h. Plasmid DNA (2 μg of total DNA [1 μg of the reporter and 0.5 μg of each expression vector]) was gently mixed with DOTAP (1 μg of DNA per 7.5 μl of DOTAP) and HEPES buffer (20 mM, pH 7.4, cell culture grade) in a sterile reaction tube (total volume, 120 μl). The transfection mixture was incubated for 20 min at room temperature before being added to the cells. The plates were incubated for 5 h and washed with 500 μl of DMEM. After 40 h of incubation, cells were harvested and lysed in 150 μl of cell culture lysis reagent (Promega). The lysates were assayed for protein concentrations by using Coomassie protein assay reagent (Pierce) and processed for the luciferase assay by using a Promega luciferase assay kit according to the instructions of the manufacturer. Fifty microliters of the lysate was mixed with 100 μl of luciferase assay reagent (Promega), and the luciferase activity was measured in a luminometer (AutoLumat LB 953; EG&G Berthold). The values obtained were normalized to the protein concentration in each cell lysate.
Human THP-1 cells were transfected by the DEAE-dextran method as described previously (48). Briefly, 107 cells (per 100-mm-diameter plate) were transfected with 10 μg of plasmid. After transfection, cells were treated with lipopolysaccharide (LPS) (10 ng/ml) for 16 h prior to CAT assays.
Whole-cell extraction and Western blotting.
HeLa cells were plated on 100-mm-diameter plates and incubated for 24 h in order to allow growth to 60 to 80% confluence prior to transfection. Transfections were performed with DOTAP reagent as described above. After 40 h of incubation at 37°C with 5% CO2, cells were washed with cold PBS once and detached with cold PBS containing 25 mM EDTA. The cells were collected by centrifugation and resuspended in 200 μl of cold lysis buffer (20 mM HEPES [pH 7.9], 1 mM MgCl2, 10 mM KCl, 300 mM NaCl, 0.1% Triton X-100, 20% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM NaF, 1 mM ZnCl2, 1 mM Na orthovanadate, 1 protease inhibitor cocktail tablet [Boehringer Mannheim]/50 ml). After incubation on ice for 10 min, the lysates were cleared by centrifugation for 10 min at 12,000 × g at 4°C. Protein concentrations were measured by the Bradford method (Bio-Rad). Ten micrograms of whole-cell extract for each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. The membranes were incubated with a 1:1,000 dilution of mouse anti-IE2 antibody (Godwin Institute of Cancer Research, Inc.), followed by incubation with a 1:2,000 dilution of conjugated horseradish peroxidase-labeled anti-mouse antibody. Horseradish peroxidase activity was detected with the enhanced-chemiluminescence system (Amersham).
GST fusion vectors.
The glutathione S-transferase (GST)–wild-type Spi-1 fusion vector (WT) containing the Spi-1 coding sequence was provided by D. G. Tenen (Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Mass.).
GST fused with various motifs of Spi-1 fusion vectors, specifically, GST–Spi-1 (171-272), GST–Spi-1 (171-242), and GST–Spi-1 (202-254) (the amino acid coding region of each construct is indicated by the numbers after GST–Spi-1), which contain different motifs of the Spi-1 DNA-binding domain as shown in Fig. 6a, were constructed by PCR amplification by combining the following primers, as required: 5′ wHTH, 5′ ATG GGA TCC AAG ATT CGC CTG TAC 3′; 5′ β2 motif, 5′ ATC GGA TCC CAG TTC TCG TCC AAG C 3′; 3′ wHTH, 5′ GAT GAA TTC ATC AGT GGG GCG GGA G 3′; 3′ β4 motif, 5′ ATG AAT TCA GAA CTG GTA GGT GAG C 3′; and 3′ α3 motif, 5′ GAT GAA TTC CTC GCC TGT CTT GCC GTA GT 3′. Each PCR was performed in a total volume of 100 μl composed of 10 μl of 10× Pfu polymerase buffer (Stratagene), 0.8 μl of deoxynucleoside triphosphates (25 mM each deoxynucleoside triphosphate), 1.0 μl of the DNA template (100 ng/μl), 2.5 μl of each primer (100 ng/μl), 2.0 μl of the cloned Pfu DNA polymerase (2.5 U/μl; Stratagene), and 81.2 μl of distilled water. The sample was overlaid with 70 μl of mineral oil, and amplification was performed with a program consisting of 94°C for 1 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 45 s, and a final cycle of 72°C for 10 min. The PCR fragments with overhangs generated with BamHI and EcoRI were purified with a Mermaid Kit (Bio 101, Inc.) and ligated into the pGEX-2T (Pharmacia) GST expression plasmid. GST–Spi-1 (243-254) was constructed with synthetic oligonucleotide sequences 5′ GAT CCG TGA AGA AAG TCA AGA AGA AGC TCA CCT ACC AGT TCG 3′ and 5′ AAT TCG AAC TGG TAG GTG AGC TTC TTC TTG ACT TTC TTC ACG 3′, annealed, and ligated into the pGEX-2T expression vector (Pharmacia) at the BamHI and EcoRI restriction sites.
Expression and purification of GST fusion protein.
Overnight culture of Escherichia coli DH5α containing one of the GST fusion constructs was diluted 1:10 in Luria broth with ampicillin (100 μg/ml) and grown for 1 h with shaking at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and cultures were incubated at 37°C for 3 h with shaking. Bacteria (20 ml) were pelleted and suspended in 0.6 ml of cold NETN buffer (20 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0], 200 mM NaCl, 0.5% NP-40, 0.2 mM PMSF, 1 mM dithiothreitol, 1% Triton X-100, 1 protease inhibitor cocktail tablet [Boehringer Mannheim]/50 ml). The cells were sonicated three times for 15 s on ice and then centrifuged at 10,000 × g for 10 min. The protein concentrations of the lysates were measured with a Bio-Rad protein assay kit. Glutathione-Sepharose beads (Pharmacia) were equilibrated with NETN buffer. Equivalent amounts of GST fusion proteins (as determined by Bio-Rad and confirmed by Coomassie blue staining) were bound to 50 μl of glutathione-Sepharose beads by incubation in a total volume of 500 μl of NETN buffer for 1 h at 4°C. The beads were washed three times in NETN buffer.
Protein-protein interaction assay.
The GST fusion protein beads were incubated with 35S-labeled, in vitro-translated protein at 4°C for 1 h. The beads were washed five times in NETN buffer, and the bound proteins were eluted by boiling the beads for 5 min in SDS-PAGE loading buffer (50 mM Tris [pH 6.8], 30% glycerol, 0.4% SDS, 0.1% bromphenol blue) containing β-mercaptoethanol. Proteins were separated by SDS-PAGE (10% polyacrylamide gel), which was then soaked in Amplify fluorographic reagent (Amersham Life Science) and exposed to Kodak X-Omat film.
In vitro-translated proteins.
Coupled in vitro transcription-translation was carried out to obtain recombinant proteins by using the TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions in the presence of [35S]methionine. Briefly, each reaction mixture was composed of 40 μl of TNT T7 Quick Master Mix, 2 μl of [35S]methionine (1,000 Ci/mmol) at 10 mCi/ml, 1 μg of the DNA template, and nuclease-free water to a final volume of 50 μl. The reaction mixture was incubated at 30°C for 90 min. The translated proteins were analyzed by SDS-PAGE and stored at −80°C.
GST one-hybrid DNA-binding assays.
A modification of a technique previously reported by Chittenden et al. (7) was used as a sensitive and versatile method to detect protein interaction with DNA. Equivalent amounts of GST fusion proteins (as determined by SDS-PAGE and Coomassie blue staining) were bound to 50 μl of glutathione-Sepharose beads as described above. After being washed with NETN buffer three times, the beads were resuspended in 200 μl of DNA probe-binding buffer [20 mM HEPES (pH 8.0), 1 mM EDTA, 50 mM NaCl, 3 mM MgCl2, 20 μg of poly(dA-dT)/reaction mixture, 1 mM dithiothreitol, 0.2 mM PMSF] and incubated for 5 min at 4°C. The beads were then incubated for 20 min at 4°C with either 200,000 cpm of a 32P-labeled Spi-1 DNA probe derived from the il1b promoter (sense, GCC TCC TAC TTC TGC TTT TGA AAG CTA TAA AA; antisense, CTG TTT TTA TAG CTT TCA AAA GCA GAA GTA GG) or 200,000 cpm of a 32P-labeled IE2 DNA probe (cis repression sequence [CRS]) derived from the major IE promoter (34, 46) (sense, 5′ AGC TTG AGG TCT ATA TAA GCA GAG CTC GTT TAG TGA ACC GTC AGG ATC 3′; antisense, GAT CCT GAC GGT TCA CTA AAC GAG CTC TGC TTA TAT AGA CCT CAA GCT 3′). The beads were washed twice with the binding buffer, and their radioactivity was counted. Specific binding was assessed by retention of the protein-DNA complex on the glutathione-Sepharose beads.
GST two-hybrid interaction assay (GHIA).
In vitro-translated Spi-1 wHTH was incubated with either GST-IE2 or GST control proteins bound to glutathione-Sepharose beads. The optimal condition required to obtain a GST-IE2–Spi-1 wHTH was the same as that described above for the protein-protein binding assay. After extensive washing, binding of the GST-IE2–Spi-1 wHTH bead complexes with radiolabeled oligonucleotides containing either the strong β-globin B1-A Spi-1-binding site (5′ ACC TTC CTA TCA GAA AAA AAG GGG AAG CGA TTC T 3′) or this site with a specific mutation (underlined) that results in a loss of Spi-1 binding (5′ ACC TTC CTA TCA GAA AAA CCC GGG AAG CGA TTC T 3′) (15, 30) was carried out in 200 μl of probe binding buffer at 4°C for 20 min. The probe-binding buffer [20 mM HEPES (pH 8.0), 1 mM EDTA (pH 8.0), 50 mM NaCl, 3 mM MgCl2, 0.2 μg of poly(dI-dC) per ml, 1 mM dithiothreitol, 0.2 mM PMSF] was modeled after that previously reported to be suitable for the binding of Spi-1 to its recognition DNA sequence (15). Gentle washing with a probe elution buffer (same as the probe binding buffer but containing 0.5% NP-40) was performed. Special attention was paid to probe washing, especially during the evaluation of ternary-complex formation by Spi-1, IE2, and DNA. Although the complex formed quite readily, association with the DNA probe was not maintained by the GST–IE2–Spi-1 wHTH complex as well as was that with the same probe by the GST–Spi-1 wHTH complex (approximately 10-fold less) (data not shown). This result suggests that the protein-protein interaction between IE2 and the Spi-1 wHTH decays much more rapidly than the interaction between the Spi-1 wHTH complex and the DNA probe. The amount of radioactive probe bound to the GST-IE2–Spi-1 wHTH was determined by Cerenkov counting of the Sepharose beads in a scintillation counter.
RESULTS
Strong transactivation of the il1b promoter by IE proteins requires a single Spi-1-binding site.
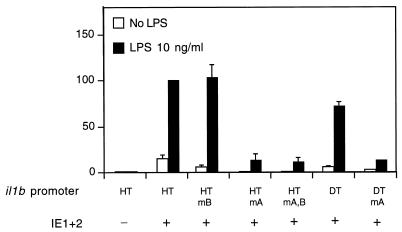
Consistent with our previous report (24), we found that, in transfected THP-1 monocytes, IE proteins upregulate il1b expression in the absence of the il1b enhancer (Fig. 2). This expression did not require any additional exogenous stimuli, but in combination with LPS, IE proteins supported a stronger (approximately 70-fold) total induction. When il1b promoter CAT reporters (HT and DT) carrying single point mutations in Spi-1-binding site A (Fig. 2) were cotransfected with a genomic IE1-IE2 expression vector [pEQ276(IE1+2)], a dramatic loss of IE protein-dependent activation was observed. The upstream Spi-1-binding site B, which we previously reported to be essential for enhancer-dependent il1b expression in the absence of IE protein (30), was not required for IE protein-dependent expression (Fig. 2). Mutation at both Spi-1 sites resulted in IE protein-dependent activity comparable to that observed for the mutation at site A alone. The il1b promoter fragment containing only site A (DT fragment) demonstrated IE protein-dependent activity comparable to that of the longer sequence (HT fragment). These results indicate that Spi-1-binding site A, but not site B, is critical for IE protein-dependent induction of il1b.
FIG. 2.
The il1b promoter is strongly transactivated by IE proteins in human THP-1 monocytes. Shown are the CAT activities of the il1b promoter CAT reporters (HT and DT) containing point mutations at the distal (B) and proximal (A) Spi-1-binding sites, respectively, and cotransfected with a genomic IE1-IE2 expression vector [pEQ276(IE1+2)] which expresses both IE1 and IE2. The results are expressed as average percentages of the relative CAT activity observed for the wild-type HT promoter when it was cotransfected with the IE1-IE2 expression vector. Open bars correspond to unstimulated cells, whereas filled bars correspond to cells treated with 10 ng of LPS per ml. Standard deviations are indicated for a minimum of three repetitions. mA, mutation at site A; mB, mutation at site B; mA,B, mutations at both sites.
Enhancer-independent il1b transactivation by HCMV IE gene products requires only the Spi-1 DNA-binding domain (wHTH).
We have previously reported that potent il1b transcription is controlled by two independent elements, an upstream inducible enhancer and a cell-type-specific promoter (48). This requires the binding of transcription factor Spi-1 to two sites within the promoter (30) (Fig. 1). The HCMV IE proteins support strong il1b transcription in the absence of an enhancer. Mutation of the Spi-1-binding site located adjacent to the transcription start site has indicated a significant role for this transcription factor in both the presence (24) and absence of HCMV IE protein (30). However, the direct involvement of Spi-1 in the HCMV transactivation of il1b has not been demonstrated. In order to address this, we transiently transfected Spi-1-deficient HeLa cells (16), which do not express il1b, even when they are induced by agents such as phorbol myristate acetate (PMA) (8). We have previously demonstrated that transfection of Spi-1 into HeLa cells can support strong transactivation of an il1b promoter sequence in the presence of an abbreviated simian virus 40 enhancer (30) but not in the presence of the il1b far-upstream enhancer, as was previously reported for monocytes (48).
An enhancerless CAT reporter vector containing the il1b promoter sequence −131 to +12 (fragment HT in Fig. 1) was cotransfected into HeLa cells along with a genomic IE1-IE2 vector that expresses both HCMV IE1 and IE2. Transfections were executed in the presence of various Spi-1 expression vectors such as antisense Spi-1, wild-type Spi-1, and several mutated Spi-1 expression vectors containing cDNAs coding for deleted protein regions (Fig. 3a). As shown in Fig. 3b, the antisense Spi-1 expression vector failed to support IE protein-activated il1b promoter activity. In the absence of both an enhancer and the IE proteins, wild-type Spi-1 induced a 9-fold increase in CAT activity over that of the control and an additional 2.5-fold increase in activity when it was supplemented with IE proteins. This confirms the absolute requirement for Spi-1 in il1b transactivation by HCMV IE proteins. Surprisingly, an expression vector coding only for the Spi-1 wHTH, which is inactive in the absence of IE, was able to support significant il1b promoter activity when it was cotransfected with the IE protein expression vector.
The PEST sequence and the TBP-binding domain of Spi-1 have been previously shown to be differentially critical for the expression of various Spi-1-dependent genes (2, 27, 30, 43). However, for enhancer-dependent activity of il1b in the absence of IE protein, both the TBP and Q domains are required (30). Figure 3 demonstrates that with IE proteins, deletion of either the PEST sequence (ΔPEST vector) or amino acids 8 to 32 (Δ8/32 vector) within the TBP-binding domain of Spi-1 did not affect the transactivation activities of IE proteins, indicating the dispensability of both of these regions in il1b transactivation by IE proteins. Other Spi-1 deletion constructs, namely, those with deletions of the PN sequence, the NN sequence, and the first 100 amino acids of Spi-1 (ΔPN, ΔNN, and Δ100 vectors, respectively), all lacked the Q domain and resulted in a 50% loss of CAT activity when levels were compared to that of the wild-type Spi-1 expression vector. This result is similar to what was observed in the absence of IE protein (30), which suggests that the Spi-1 Q domain is essential for maximal il1b transactivation both in the presence and absence of IE proteins. Based on these data, we conclude that HCMV IE proteins use an unique transcriptional activation pathway which partially circumvents the requirement for both an enhancer and the Spi-1 TAD. Furthermore, the Spi-1 DNA-binding module (wHTH) is sufficient to support significant il1b transactivation by HCMV IE proteins.
The −59-to-+12 il1b promoter sequence functions as an IE protein-dependent and enhancer-independent promoter.
We have previously reported that in the presence of IE proteins the il1b promoter from −131 to +12 (region HT in Fig. 1) is able to support transactivation in the absence of the normally required far-upstream enhancer. Within this promoter sequence, there are two binding sites for Spi-1 (Fig. 1). The proximal Spi-1-binding site is located between −50 and −39 and has been shown to be crucial for il1b transactivation in both the presence and absence of HCMV IE proteins. However, the requirement for the more upstream Spi-1-binding site located between −115 and −97 has been determined only in the absence of IE protein (30).
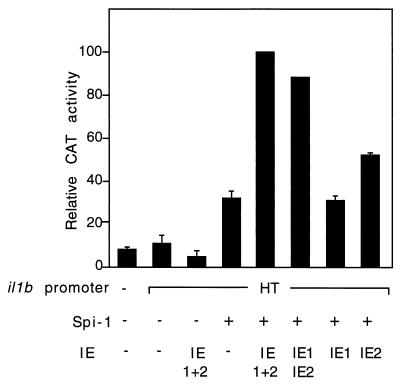
Transient transfections were executed with HeLa cells. An enhancerless CAT reporter containing either the il1b promoter sequence −131 to +12 (fragment HT; which contains both of the Spi-1-binding sites) or the shorter sequence −59 to +12 (fragment DT; which contains only the TATA-box-proximal Spi-1 site) was cotransfected with the IE1-IE2 expression vector. The results shown in Fig. 4 argue that in the presence of Spi-1, the −59-to-+12 il1b promoter (DT) can function as effectively as the −131-to-+12 promoter (HT) in IE protein (enhancer-independent) transactivation of il1b, resulting in about a 20-fold-greater activity than that of the control. The activities of IE protein-induced HT and DT promoter fragments in the presence of the Spi-1 wHTH expression vector are also indistinguishable from each other and are about 50% of the activity mediated by the full-length Spi-1 expression vector. Therefore, the il1b promoter sequence from −59 to +12 serves as a minimal IE protein-dependent (enhancer-independent) promoter requiring only one Spi-1-binding site.
FIG. 4.
The −59-to-+12 il1b promoter (DT) containing only one Spi-1-binding site adjacent to the transcription start site can function as an IE-activated promoter via an enhancer-independent pathway. The upstream Spi-1-binding site is not essential for il1b transactivation by HCMV IE proteins. The il1b promoter CAT reporters were transiently cotransfected with an IE1-IE2 expression vector into HeLa cells in the presence of 50 ng of PMA per ml. The il1b promoter CAT reporter (20 μg) and IE1-IE2 expression vector (IE1+2) (10 μg) were used for each analysis. Ten micrograms of the Spi-1, antisense Spi-1 (AS.Spi-1), or Spi-1 DNA-binding domain (wHTH) expression vector was cotransfected for each analysis. The results are shown as average percentages of the CAT activity of the wild type. Error bars indicate the deviations in results from a minimum of three repetitions.
Both IE1 and IE2 synergistically transactivate il1b.
To determine the role of individual IE proteins in il1b transactivation, HeLa cells were cotransfected with an il1b promoter CAT reporter and Spi-1 expression vectors along with one of three different IE protein genomic expression vectors: pEQ273(IE1), pEQ326(IE2), and pEQ276(IE1+2). Whole-cell extracts from the transfected HeLa cells were analyzed by SDS-PAGE and Western blotting with an antibody directed against both IE1 and IE2 in order to confirm the expression of IE proteins in HeLa cells (data not shown). Figure 5 reveals that in the presence of the Spi-1 expression vector, IE2 alone significantly induced il1b promoter CAT activity and that IE1 did not. Synergy was observed when both the IE1 and IE2 expression vectors were cotransfected along with the Spi-1 expression vector, suggesting that IE1 augments the IE2 function in il1b transactivation. This observation is in agreement with several reports that IE1 and IE2 synergistically transactivate a variety of genes (21, 37, 38).
FIG. 5.
IE1 and IE2 synergistically transactivate il1b expression. Shown is CAT activity of the enhancerless il1b promoter (HT) CAT vector (20 μg) cotransfected with 10 μg of the Spi-1 or antisense Spi-1 (AS.Spi-1) expression vector into PMA-treated HeLa cells, in the presence of an individual IE expression vector [pEQ273(IE1) or pEQ326(IE2)] or a genomic IE1-IE2 expression vector [pEQ276(IE1+2)] expressing both IE1 and IE2. The total amount of transfected DNA was kept constant by the addition of the parental vector [pEQ336]. The CAT data were normalized to the average activity elicited by the IE1-IE2-activated HT CAT construct in the presence of the Spi-1 expression vector. Error bars represent the deviations in results from a minimum of three repetitions.
IE2, but not IE1, physically associates directly with the Spi-1 wHTH DNA-binding domain.
A number of studies have shown that HCMV IE proteins function as powerful transactivators for various genes by associating with numerous cellular transcription factors. It has also been reported that the DNA-binding module (wHTH) of Spi-1 and other ETS factors interact with a variety of transcription factors, resulting in a cooperative gene activation (reviewed in reference 1). Together with our functional data demonstrating that HCMV requires only the Spi-1 wHTH for il1b transactivation, we hypothesized that il1b transactivation by HCMV is mediated by a direct association between IE proteins and the Spi-1 wHTH.
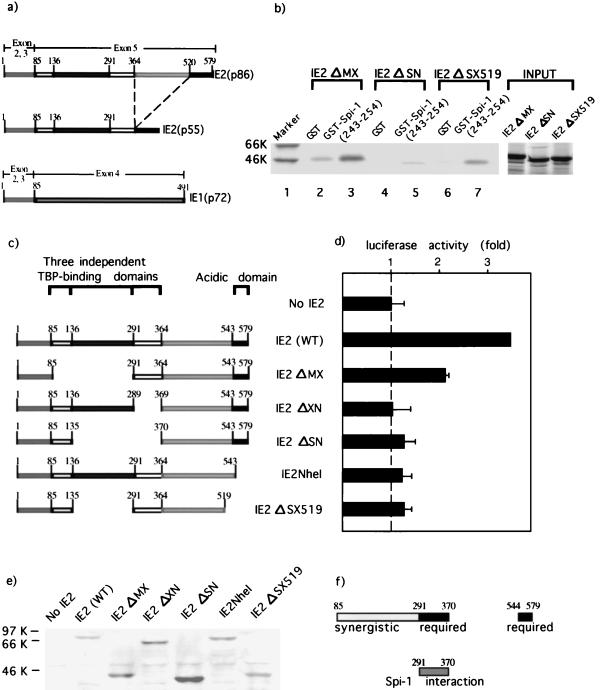
To test our hypothesis, GST pulldown protein-binding assays were performed. Since there are three major HCMV IE gene products, IE1(p72), IE2(p55), and IE2(p86), which possess considerably different functions, we evaluated the binding activities of individual IE proteins. GST fusion proteins containing the Spi-1 wHTH (amino acids 171 to 272) (Fig. 6a) or a GST control was bound to glutathione-Sepharose beads. In vitro-translated 35S-labeled IE1(p72), IE2(p55), or IE2(p86) was then added. The beads were washed, and bound proteins were analyzed by boiling followed by SDS-PAGE. Figure 6b reveals that both IE2(p55) and IE2(p86) bound strongly to the GST–Spi-1 wHTH (amino acids 171 to 272) (lanes 3 and 7, respectively) but only weakly bound to the GST control (lanes 2 and 6). In contrast, the interaction of IE1(p72) with GST-Spi-1 wHTH (lane 5) was much weaker than those of IE2 proteins, although the level of the in vitro-translated IE1 was significantly higher than that of IE2 (lanes 8 to 10).
The strong binding of IE2 to the Spi-1 wHTH is in agreement with the functional activity demonstrated in this study in that, although IE1 and IE2 synergistically transactivate il1b expression, IE2 provides a stronger transactivation effect than IE1. Furthermore, the results from the protein-protein interaction assays support our hypothesis that HCMV transactivates il1b expression via a physical association of IE2 with the Spi-1 wHTH.
IE2 associates with Spi-1 via the wing of the wHTH.
The X-ray crystal structure of the Spi-1 DNA-binding domain reveals that the wHTH consists of three α-helices containing a classical HTH along with a four-stranded antiparallel β-sheet wing (29). It has been reported that Spi-1 binds to the major groove of DNA through the HTH and that the wing provides additional interactions with the DNA backbone (29). To further localize a specific region within the Spi-1 wHTH responsible for the recruitment of IE2(p86), GST pulldown binding assays were performed with a series of GST fusion constructs carrying various cDNA motifs of Spi-1 (Fig. 6a). In vitro-translated 35S-labeled IE2(p86) was incubated with either the GST control or GST fused to various subregions of Spi-1 (Fig. 6a). Figure 6c shows that IE2 binds to GST–Spi-1 (lane 2) as well as to the GST–Spi-1 wHTH (amino acids 171 to 272) (lane 3) but not to GST alone (lane 1). The GST fusion constructs containing amino acids 202 to 254 (lane 5) and 243 to 254 (lane 6) were also capable of binding to IE2(p86) protein. The data demonstrate that all GST constructs containing amino acids 243 to 254 (antiparallel β-strands β3 and β4) of the Spi-1 wHTH are able to physically interact with IE2, suggesting that the minimal structure important for the association of Spi-1 with IE2 is the portion of the wing corresponding to the antiparallel β3-β4 strand pair. This is confirmed by the observation that the GST–Spi-1 wHTH fusion construct lacking this structure (amino acids 171 to 242) (lane 4) does not efficiently bind IE2.
To verify that protein-protein interaction directed by the wing is not due to nonspecific binding of this motif, in vitro-translated 35S-labeled Stat 3 protein was incubated with GST–Spi-1 (171-272), –Spi-1 (171-242), or –Spi-1 β3-β4 (243-254). The radiolabeled Stat 3 protein did not bind to any part of Spi-1 (Fig. 6d). This result supports the argument that the association of the Spi-1 wing with IE2 is specific.
The sequence between amino acids 291 and 370 of IE2 is necessary for interaction with Spi-1.
As demonstrated above, the wing is a minimal region within the Spi-1 wHTH essential for interaction with IE2 protein. In order to map a specific region of IE2 responsible for interaction with Spi-1, the sequences of IE1(p72), IE2(p55), and IE2(p86) were examined, revealing that all have a common amino-terminal 85 amino acids. IE2(p55) and IE2(p86) are identical, except that IE2(p55) lacks amino acids 364 to 520 of IE2(p86) (Fig. 7a). The GST pulldown protein-binding assay showed that IE2(p55) and IE2(p86) bind strongly to the Spi-1 DNA-binding domain but that IE1(p72) binds weakly (Fig. 6b). Therefore, amino acids 1 to 85 are probably not responsible for the association with Spi-1, since all three proteins contain this region. IE2(p55) lacks amino acids 364 to 520 of the IE2(p86) (Fig. 7a) and is still able to interact with Spi-1 as effectively as IE2(p86), suggesting that a sequence essential for Spi-1 binding is not located within the sequence from amino acids 364 to 520 but is rather within the sequence from amino acids 85 to 364.
FIG. 7.
Mapping the region of IE2(p86) responsible for cooperativity with Spi-1 in il1b transactivation. (a) Schematic representation of the HCMV IE gene products, IE2(p86), IE2(p55), and IE1(p72). The exons from which each IE protein arises and their amino acids are indicated. (b) The region between amino acids 291 and 370 of IE2 is essential for the interaction with Spi-1. GST or GST–Spi-1 β3-β4 (243-254) fusion proteins linked to glutathione-Sepharose beads were incubated with equal amounts of the in vitro-translated 35S-labeled mutated IE2 proteins (as shown in Fig. 6c). After extensive washing, the bound proteins were resolved by SDS-PAGE. All the IE2 deletion mutants bound weakly to the GST control (lanes 2, 4, and 6). Molecular weight markers (in thousands [K]) are indicated at the left. (c) Schematic representation of mutated IE2 expression vectors. All vectors are as described by Sommer et al. (52). The wild-type IE2 (WT) and the deletion mutants are illustrated with numbers representing the positions of important amino acid residues. Also shown are the locations of three previously reported TBP-binding domains (52) and an acidic activation domain (40). (d) Functional data support the significance of the IE2 interaction motif (amino acids 291 to 370), the sequence between amino acids 544 and 579, and a region within amino acids 85 to 291 of IE2 protein in the transcriptional activation of il1b. The il1b promoter luciferase reporter (pGL3B-DT) was cotransfected with Spi-1 and mutated IE2 expression vectors (as shown in Fig. 6c) into HeLa cells. The total amount of transfected DNA was kept constant by the addition of the parental vector. Shown are average relative luciferase activities. A broken line indicates the activity level in the absence of IE2. Error bars indicate the deviations in results from a minimum of three repetitions. (e) Whole-cell extracts from the transfected HeLa cells were subjected to Western blot analysis with a monoclonal antibody recognizing the IE2 protein in order to quantitate mutated IE2 protein expression. (f) Summary of the results from the protein-protein binding assay and transactivation study. Two regions of IE2 required for il1b transactivation are shown as black bars. A region providing synergistic activation is shown as a gray bar. The likely Spi-1 interaction motif of IE2 is illustrated as a dark-gray bar. The numbers indicate amino acid sequences of IE2.
GST pulldown protein-binding assays were employed in order to identify the region of IE2 responsible for the interaction with Spi-1. GST or GST–Spi-1 (243-254) (β3 and β4 of the wing) was bound to glutathione-Sepharose beads. In vitro-35S-labeled mutated IE2(p86) proteins carrying various deletions as shown in Fig. 7c were tested for binding activity. Figure 7b demonstrates that IE2 ΔMX and IE2 ΔSX519 interacted with GST–Spi-1 (243-254) (lanes 3 and 7, respectively). IE2 ΔSN possesses significantly weaker binding activity than IE2 ΔMX and IE ΔSX519. IE2ΔSN does not contain amino acids 136 to 370. Nevertheless, the deletion of amino acids 136 to 291 did not result in loss of binding activity, as shown by the interaction of IE2 ΔMX and IE2 ΔSX519 with GST–Spi-1 (243-254). Thus, the data suggest that the association of IE2 with Spi-1 is strongly dependent upon amino acids 291 to 370.
Transactivation of il1b by IE2 is dependent upon two different IE2 domains.
To investigate the biological role of amino acids 291 to 370, which are critical for Spi-1–IE2 interaction, we performed transient transfections in HeLa cells. Expression vectors carrying various IE2(p86) deletions (Fig. 7c) were cotransfected along with a Spi-1 expression vector and a reporter vector containing the il1b promoter sequence from −55 to +12 (pGL3B-DT). In the presence of Spi-1 (Fig. 7d), IE2 ΔMX activated il1b transcription to a level approximately 50% of that induced by wild-type IE2. This result suggests that a region between amino acids 85 and 291, although not absolutely required, plays a synergistic role in activation of il1b expression. The mutated IE2 vectors which lack the critical Spi-1 interaction sequence (IE2 ΔXN and IE2 ΔSN) failed to transactivate il1b beyond the level generated by Spi-1 alone, supporting the argument that il1b transactivation by HCMV IE2 is dependent upon a strong physical association between Spi-1 and the critical amino acids 291 to 370 of IE2. Interestingly, in the presence of Spi-1, IE2NheI and IE2 ΔSX519, lacking the acidic amino acids between 544 and 579, were unable to transactivate il1b. Our results argue that both of these two regions of IE2 (amino acids 291 to 370 and 544 to 579) are necessary for transcriptional activation by IE2 and that a third region between amino acids 85 and 291 provides a synergistic activity (Fig. 7f).
IE2 and the Spi-1 wHTH DNA-binding domain form a ternary complex with DNA that requires a protein-protein interaction.
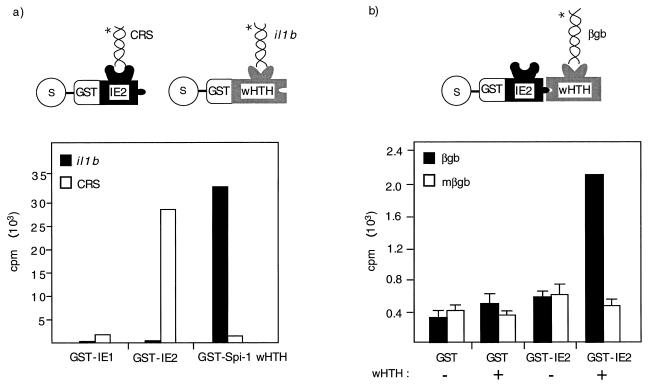
In order to measure IE2 binding activity to the il1b promoter, in vitro DNA-binding assays were performed. A GST-protein fusion technique was used because of its ability to detect weak complexes possessing high decay rates. As shown in Fig. 8a, GST-IE2 protein strongly bound to one of its cognate binding sites found in the HCMV genome (CRS element) but was unable to bind the il1b promoter. In contrast, GST–Spi-1 wHTH bound to the il1b promoter but not to the CRS. The binding of IE2 and Spi-1 to their respective cognate probes was stable and withstood extensive washing, suggesting a low rate of probe dissociation. Because the in vitro binding data argue that IE2 protein does not directly recognize the il1b promoter but that it is capable of binding to Spi-1 wHTH in vitro and transactivating il1b transcription in vivo, we hypothesized that IE2 is recruited to the il1b promoter via association with the DNA-binding domain of Spi-1.
FIG. 8.
An in vitro ternary complex involving IE2–Spi-1 and DNA requires protein-protein interaction. (a) IE2 protein does not bind to the il1b promoter. DNA-binding assays were performed as described in Materials and Methods. GST fusion proteins were incubated with either a radiolabeled Spi-1 DNA probe (il1b) or an IE2 DNA probe (CRS). Following two rounds of washing, specific DNA-binding activities were determined by measuring the radioactive counts per minute from the Sepharose beads. Reactions with GST-IE1 were performed simultaneously as negative controls. (b) The Spi-1 wHTH is required for the interaction between IE2 and DNA. The GHIA used in vitro-translated Spi-1 wHTH, which was incubated with an equal amount of either GST-IE2 or the GST control bound to glutathione-Sepharose beads. The in vitro-translated control protein was made by the same method as was used for the Spi-1 wHTH but in the absence of the DNA template. After incubation and extensive washing, the beads were incubated with either a wild-type or a mutated Spi-1 radiolabeled DNA probe (15). Following two rounds of washing, the amount of bound radioactive probe was determined from the Sepharose beads. Error bars indicate the deviations in results from a minimum of three repetitions. Cartoons explain the details of the experiment in which the natures of individual proteins and radiolabeled DNAs ( ) are indicated. βgb, β-globin-binding site; mβgb, mutated β-globin-binding site.
) are indicated. βgb, β-globin-binding site; mβgb, mutated β-globin-binding site.
To support this hypothesis, we used a GHIA to evaluate the existence of an IE2-wHTH-DNA ternary complex. For GHIA, a GST-protein fusion was used to detect the formation of a ternary complex involving a second unlabeled protein and a radiolabeled DNA probe (Fig. 8b). Experiments with the il1b probe were inconclusive and suggested that under the experimental conditions used, the association of Spi-1 and the il1b probe may not have been sufficiently strong to allow a stable complex to be observed. Therefore, we attempted to detect a complex using the β-globin B1-A-binding site (16), which has at least a fivefold-higher affinity for Spi-1 than did the il1b probe (30). Our results show that the radiolabeled DNA probe containing the β-globin binding site can be tethered to GST-IE2 only in the presence of the Spi-1 wHTH, demonstrating the existence of an IE2-wHTH-DNA ternary complex. In contrast, a radiolabeled mutated DNA probe containing a specific mutation previously reported (15, 30) not to support Spi-1 binding resulted in the loss of DNA association with GST-IE2. It should be noted that the ternary complex, unlike the direct binding of either IE2 or the Spi-1 wHTH to its cognate probe, could not withstand extensive washing (not shown). This resulted in approximately a 10-fold higher level of bound probe for direct binding as compared with protein-protein-mediated binding (compare the counts per minute bound in Fig. 8a and b). Therefore, the ternary complex probably has a high decay rate that may be due to a tenuous protein-protein interaction.
DISCUSSION
In the present study, we have investigated the molecular mechanism by which HCMV IE proteins transactivate il1b via the transcription factor Spi-1. It has been previously demonstrated that the Spi-1 binding site located at positions −50 to −39 of the il1b promoter sequence is crucial for both enhancerless IE-dependent (24) and normal enhancer-dependent (30) il1b transcription. In addition, many studies have reported that other cellular genes coding for the IL-1 receptor antagonist, IL-6, and IL-8, which possess a potential Spi-1-binding site, are activated by HCMV (28). A recent report of tumor necrosis factor alpha (tnfa) gene transactivation has also suggested an essential role for Spi-1 in HCMV IE activation (17). Based on these lines of evidence, Spi-1 appears to play a central role as a host factor required for HCMV transactivation. Here, we show that IE-dependent il1b promoter activity absolutely depends upon Spi-1. Moreover, we discovered that HCMV IE proteins circumvent the normal enhancer-dependent il1b transcription pathway and upregulate il1b by a mechanism that eliminates the requirement not only for an enhancer but also, to a significant degree, for the Spi-1 TAD. This TAD is required for enhancer-dependent transcription in the absence of HCMV IE (30). In the presence of IE, only the Spi-1 wHTH DNA-binding domain is required to support significant il1b expression. This observation is distinct from those made with some other systems in which additional sequences not essential for DNA binding (extra-wHTH) mediated transcription. Examples include PIP/NF-EM5, which binds to the phosphorylated PEST sequence of Spi-1 (42), and FOG, which interacts with the non-DNA-binding amino (N) zinc finger of GATA-1 (53).
The il1b promoter from −131 to +12 supports both normal enhancer-dependent and IE protein-mediated transcription (11, 24, 30). Within this promoter, there are two Spi-1-binding sites, one located adjacent to the transcription start site and the other located further upstream (Fig. 1), raising the question of whether the functions of these two sites are different for il1b transactivation by HCMV IE proteins. In the absence of IE proteins, a shorter −59-to-+12 il1b promoter is inactive (30). We now demonstrate that, in the presence of IE proteins, the −59-to-+12 promoter can function as effectively as the −131-to-+12 promoter, indicating that the presence of the upstream Spi-1-binding site, which is normally required, is not imperative for activation by IE. In other words, this finding demonstrates that the IE proteins specifically target the proximal Spi-1-binding site in order to activate the expression of il1b.
Our study shows that IE2 protein plays a significant role in il1b transactivation. Since IE2 is able to transactivate il1b, even though there is no IE consensus binding site (34, 46) in the il1b promoter and we cannot detect any in vitro binding, it is not unreasonable to speculate that IE2 activates il1b transcription by, at least in part, directly interacting with Spi-1 bound to il1b. Spi-1 has been reported to bind various viral proteins in the activation of cellular genes. An interaction between Spi-1 and EBNA-2 has been shown to play an important role in latent membrane protein 1 (LMP1) gene activation (26). Furthermore, we recently showed that il1b transactivation by human T-cell leukemia virus type 1 Tax protein likely involves the cooperation of Tax with Spi-1 and with NF-IL6 (C/EBPβ) (54). None of these reports identified a specific region of Spi-1 responsible for the interaction with viral proteins. The findings in our present study have confirmed a direct binding of Spi-1 with IE2 protein and indicate that a specific region between amino acids 291 and 370 of IE2 is important for a strong physical association with a portion (antiparallel β3 and β4 strands) of the Spi-1 wHTH wing. Our in vitro experiments further reveal that IE2 is able to form a Spi-1 wHTH-dependent ternary complex on DNA. These data appear to support a mechanism for IE2-dependent transcriptional activation mediated by direct interaction with il1b promoter-bound Spi-1. Moreover, we find that both IE1 and IE2 synergistically upregulate il1b expression, although direct IE1 interaction with Spi-1 could not be detected. The mechanism of synergy between the two IE proteins is not clearly understood. It has been demonstrated that activation by individual IE proteins is dependent upon both the promoter-regulatory region of the cellular gene and the cell type (39). A report that IE1 does not directly interact with IE2 (14) argues that the cooperative activation of il1b is unlikely to occur via physical association between IE1 and IE2. Since IE1 has been shown to transactivate a variety of genes, including that coding for IE2 (38), it is possible that IE1 induces the expression of a coactivator, which in turn is able to directly bind IE2 and upregulate il1b transcription.
The wing of the wHTH proteins, including Spi-1, all other ETS factors, and various non-ETS wHTH proteins such as heat shock factor (HSF), is a small β-sheet consisting of three or four antiparallel β-strands and their interconnecting loops, which extend to one side of the triple α-helical bundle containing the HTH (9, 29). We have recently determined that GATA-1 and GATA-2 are also capable of interacting with Spi-1 via the wing (59). In addition, a recent crystal structure report reveals that HSF multimerization on DNA occurs via wing-mediated protein-protein interaction (35). Therefore, the wHTH wing may represent a site for protein-protein interaction among many other diverse factors that have been reported to functionally cooperate in transcriptional activation.
In accordance with our observation that IE2 transactivation of il1b requires only the Spi-1 wHTH DNA-binding domain but not an enhancer or the Spi-1 TAD, we hypothesize that IE2 functions by providing a TAD normally supplied by the Spi-1 TAD in cooperation with critical factors that bind to the il1b upstream enhancer. This speculation is supported by our functional data which indicate that, in addition to the putative interaction motif in IE2 (amino acids 291 to 370), amino acids 544 to 579, which contain a potent independently acting acidic TAD (40), are also necessary for activation. In addition, the amino acid sequence between 85 and 291 of IE2, containing two previously reported TBP-binding domains (Fig. 7c), appears to be important for maximal il1b transactivation. IE2 has been reported to bind several transcription factors, including TAFs, and has been demonstrated to perform a TAF-like function as a component of TFIID (36). It is likely that IE2 interacts with the wing of the il1b promoter-bound Spi-1 and either recruits TBP via its TBP-binding domain (amino acids 85 to 291) or tethers other transcriptional activators to the transcription start site.
A well-characterized mechanism of transcriptional activation in which a viral protein, herpes simplex virus (HSV) VP16, interacts with the DNA-binding domain of a cellular factor (POUs domain of Oct-1) in order to mediate transcription has been reported previously (22). In this case, VP16 is required to bind a host cell factor (HCF) before recruitment to DNA via interaction with the Oct-1 POUs domain. However, the mechanism of IE protein transactivation of il1b appears to be different in that IE protein is able to directly associate through the Spi-1 DNA-binding domain without the requirement of an additional factor and form an in vitro ternary complex with DNA. This possibility is strongly suggested by the functional requirement (Fig. 7d) for the same region that is critical for the strong in vitro interaction between Spi-1 and IE2 (Fig. 7b). Additionally, IE does not appear to interact with the il1b promoter sequence whereas VP16 in the HCF-VP16-POU complex has been shown to recognize DNA.
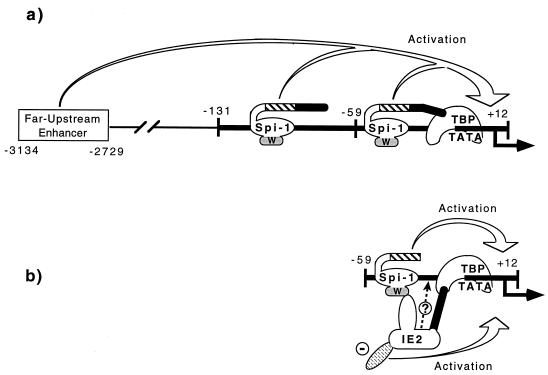
In conclusion, our present study has demonstrated a mechanism, which we refer to as PTT, by which HCMV IE2 transactivates il1b expression. In PTT, a DNA-binding transcription factor can activate an apparently irrelevant target gene which is devoid of a cognate recognition site by means of a protein-protein interaction. With IE2 and the il1b gene, the Spi-1 wHTH provides both DNA-binding activity and a protein recruitment site for IE2 (Fig. 9b). Although the il1b promoter does not appear to directly bind IE2, the transactivation of il1b by IE2 can occur because an interaction between the two proteins, IE2 and Spi-1, is possible. The Spi-1 wHTH provides DNA-binding activity, while the IE2 protein provides the activation function. The PTT model does not exclude the possibility that association between the tethered factor and a weak nonspecific site does occur following the initial formation of the protein-protein interaction. However, the lack of any observable in vitro cooperative binding to DNA with Spi-1 suggests that this is not the case for IE2. Consequently, the functional cooperativity observed between PIP/NF-EM5 and Spi-1 (13) may also be considered to be an example of PTT. However, in this case the tethered PIP factor is incapable of binding DNA until the Spi-1 interaction unmasks the latent DNA-binding activity that is structurally repressed in the full-length protein. This interaction is also distinct in that it results in cooperative binding in vitro and also requires the phosphorylated PEST region, which is not required either for IE2 function or for protein-protein interaction (fig. 3 and 6).
FIG. 9.
Proposed model of PTT-mediated transcriptional activation of il1b by HCMV IE2. (a) Potent il1b transcription normally requires both a far-upstream enhancer and the −131-to-+12 il1b promoter which contains two essential Spi-1-binding sites. Two Spi-1 TAD subregions are also required. One of these is a glutamine-rich (Q) domain (hatched areas), and the other directly binds TBP (filled areas). The Spi-1 wHTH DNA-binding domain is shown as an oval, with the wing indicated by “w”. (b) IE2 replaces the functions of the Spi-1 TBP-binding domain and the il1b enhancer. Furthermore, only a minimal il1b promoter located between −59 and +12 is sufficient to support IE-activated il1b expression. IE2 is tethered to the il1b promoter via a direct interaction with the Spi-1 wing and possibly interacts with TBP. Also, an acidic domain of IE2 (shown by a minus sign) provides an activation effect eliminating the absolute requirement for the Spi-1 TAD.
Some recently reported examples demonstrating a PTT-like mechanism include the upregulation of macrophage colony-stimulating factor (M-CSF) receptor promoter via an interaction of Spi-1 with c-Jun (2) and the transcriptional activation of the HIV-1 LTR mediated by cooperative interaction of ETS-1 with USF-1 (49). In the case of M-CSF receptor transactivation, c-Jun does not directly bind to the M-CSF receptor promoter but is tethered to this promoter via interaction with the Spi-1 wHTH DNA-binding domain, providing transcriptional activation via its TAD (2). In this report, in addition to the Spi-1 wHTH DNA-binding domain, the Spi-1 TAD was also shown to be critical for the function of the c-Jun–Spi-1 complex. This is in contrast to the mechanism of il1b transactivation by IE2 reported here, in which the Spi-1 TAD is not required for significant IE2–Spi-1 function. A more related phenomenon has been demonstrated for the transactivation of the HIV LTR, which requires the interaction between Ets-1 and USF-1. Binding of USF-1 to its recognition site on the distal enhancer of the HIV-1 LTR and the presence of the Ets-1 TAD are essential for the activation of HIV-1 LTR transcription in the absence of the Ets-1 DNA-binding site. The process of PTT suggests that DNA-binding transcription factors may play significant roles in the expression of genes which do not contain strong and cognate DNA target sites. Consequently, PTT may be a general phenomenon which can play a major role in gene expression.
ACKNOWLEDGMENTS
We are deeply indepted to Deborah Spector and Chuck Clark for the cDNA wild-type and mutated IE expression constructs and informative discussions. We also thank James Alwine and Adam Geballe, who provided GST-IE constructs and genomic HCMV IE expression vectors, respectively. Richard Maki, Dan Tenen, and Jack Hensold kindly made additional constructs available. Deborah Galson and James Listman provided helpful discussions, and Changmin Chen is acknowledged for technical assistance.
This work was supported by NIH grant CA 68544.
REFERENCES
- 1.Bassuk A G, Leiden J M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 2.Behre G, Whitmarsh A J, Coghlan M P, Hoang T, Carpenter C L, Zhang D E, Davis R J, Tenen D G. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J Biol Chem. 1999;274:4939–4946. doi: 10.1074/jbc.274.8.4939. [DOI] [PubMed] [Google Scholar]
- 3.Biegalke B J, Geballe A P. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology. 1991;183:381–385. doi: 10.1016/0042-6822(91)90151-z. [DOI] [PubMed] [Google Scholar]
- 4.Boldogh I, AbuBakar S, Albrecht T. Activation of protooncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 5.Caswell R, Hagemeier C, Chiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 6.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chittenden T, Livingston D M, Kaelin W G., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- 8.Clark B D, Fenton M J, Rey H L, Webb A C, Auron P E. Characterization of cis and trans acting elements involved in human proIL-1 beta gene expression. In: Powanda M C, Oppenheim J J, Kluger M J, Denarello C, editors. Monokines and other non-lymphocytic cytokines. New York, N.Y: Alan R. Liss; 1988. pp. 47–53. [Google Scholar]
- 9.Clark K L, Halay E D, Lai E, Burley S K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 10.Craigen J L, Yong K L, Jordan N J, MacCormac L P, Westwick J, Akbar A N, Grundy J E. Human cytomegalovirus infection up-regulates interleukin-8 gene expression and stimulates neutrophil transendothelial migration. Immunology. 1997;92:138–145. doi: 10.1046/j.1365-2567.1997.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump J W, Geist L J, Auron P E, Webb A C, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus require only proximal promoter elements to upregulate expression of interleukin-1 beta. Am J Respir Cell Mol Biol. 1992;6:674–677. doi: 10.1165/ajrcmb/6.6.674. [DOI] [PubMed] [Google Scholar]
- 12.Dudding L, Haskill S, Clark B D, Auron P E, Sporn S, Huang E S. Cytomegalovirus infection stimulates expression of monocyte-associated mediator genes. J Immunol. 1989;143:3343–3352. [PubMed] [Google Scholar]
- 13.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 14.Furnari B A, Poma E, Kowalik T F, Huong S M, Huang E S. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galson D L, Hensold J O, Bishop T R, Schalling M, D’Andrea A D, Jones C, Auron P E, Housman D E. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galson D L, Housman D E. Detection of two tissue-specific DNA-binding proteins with affinity for sites in the mouse β-globin intervening sequence 2. Mol Cell Biol. 1988;8:381–392. doi: 10.1128/mcb.8.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geist L J, Hopkins H A, Dai L Y, He B, Monick M M, Hunninghake G W. Cytomegalovirus modulates transcription factors necessary for the activation of the tumor necrosis factor-alpha promoter. Am J Respir Cell Mol Biol. 1997;16:31–37. doi: 10.1165/ajrcmb.16.1.8998076. [DOI] [PubMed] [Google Scholar]
- 18.Geist L J, Monick M M, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the interleukin-2 and interleukin-2 receptor genes. Am J Respir Cell Mol Biol. 1991;5:292–296. doi: 10.1165/ajrcmb/5.3.292. [DOI] [PubMed] [Google Scholar]
- 19.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagemeier C, Walker S M, Sissons P J, Sinclair J H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently transactivate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol. 1992;73:2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 22.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 23.Hohaus S, Petrovick M S, Voso M T, Sun Z, Zhang D E, Tenen D G. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol. 1995;15:5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunninghake G W, Monks B G, Geist L J, Monick M M, Monroy M A, Stinski M F, Webb A C, Dayer J M, Auron P E, Fenton M J. The functional importance of a cap site-proximal region of the human prointerleukin 1 beta gene is defined by viral protein trans-activation. Mol Cell Biol. 1992;12:3439–3448. doi: 10.1128/mcb.12.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwamoto G K, Monick M M, Clark B D, Auron P E, Stinski M F, Hunninghake G W. Modulation of interleukin 1 beta gene expression by the immediate early genes of human cytomegalovirus. J Clin Investig. 1990;85:1853–1857. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemsz M J, Maki R A. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kline J N, Geist L J, Monick M M, Stinski M F, Hunninghake G W. Regulation of expression of the IL-1 receptor antagonist (IL-1ra) gene by products of the human cytomegalovirus immediate early genes. J Immunol. 1994;152:2351–2357. [PubMed] [Google Scholar]
- 29.Kodandapani R, Pio F, Ni C Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain–DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 30.Kominato Y, Galson D, Waterman W R, Webb A C, Auron P E. Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;15:59–68. [PMC free article] [PubMed] [Google Scholar]
- 31.Koval V, Jault F M, Pal P G, Moreno T N, Aiken C, Trono D, Spector S A, Spector D H. Differential effects of human cytomegalovirus on integrated and unintegrated human immunodeficiency virus sequences. J Virol. 1995;69:1645–1651. doi: 10.1128/jvi.69.3.1645-1651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalik T F, Wing B, Haskill J S, Azizkhan J C, Baldwin A S, Jr, Huang E S. Multiple mechanisms are implicated in the regulation of NF-kappa B activity during human cytomegalovirus infection. Proc Natl Acad Sci USA. 1993;90:1107–1111. doi: 10.1073/pnas.90.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Littlefield O, Nelson H C. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- 36.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monick M M, Geist L J, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol. 1992;7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 40.Pizzorno M C, Mullen M A, Chang Y N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pizzorno M C, O’Hare P, Sha L, LaFemina R L, Hayward G S. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pongubala J M, Atchison M L. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc Natl Acad Sci USA. 1997;94:127–132. doi: 10.1073/pnas.94.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pongubala J M, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambucetti L C, Cherrington J M, Wilkinson G W, Mocarski E S. NK-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz R, Helmich B, Spector D H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz R, Sommer M H, Scully A, Spector D H. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J Virol. 1994;68:5613–5622. doi: 10.1128/jvi.68.9.5613-5622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirakawa F, Saito K, Bonagura C A, Galson D L, Fenton M J, Webb A C, Auron P E. The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13:1332–1344. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieweke M H, Tekotte H, Jarosch U, Graf T. Cooperative interaction of ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J. 1998;17:1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eukaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 51.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 52.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 54.Tsukada J, Misago M, Serino Y, Ogawa R, Murakami S, Nakanishi M, Tonai S, Kominato Y, Morimoto I, Auron P E, Eto S. Human T-cell leukemia virus type I Tax transactivates the promoter of human prointerleukin-1beta gene through association with two transcription factors, nuclear factor-interleukin-6 and Spi-1. Blood. 1997;90:3142–3153. [PubMed] [Google Scholar]
- 55.Walker S, Hagemeier C, Sissons J G, Sinclair J H. A 10-base-pair element of the human immunodeficiency virus type 1 long terminal repeat (LTR) is an absolute requirement for transactivation by the human cytomegalovirus 72-kilodalton IE1 protein but can be compensated for by other LTR regions in transactivation by the 80-kilodalton IE2 protein. J Virol. 1992;66:1543–1550. doi: 10.1128/jvi.66.3.1543-1550.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo Y D, Chiou C-J, Choi K S, Yi Y, Michelson S, Kim S, Hayward G S, Kim S-J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yurochko A D, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang D E, Hetherington C J, Chen H M, Tenen D G. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P, Behre G, Pan J, Iwama A, Wara-aswapati N, Radomska H S, Auron P E, Tenen D G, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]