Abstract
Familial hypercholesterolemia (FH) is the most common genetic disorder of lipid metabolism, characterized by increased levels of total and LDL plasma cholesterol, which leads to premature atherosclerosis and coronary heart disease. FH phenotype has considerable genetic heterogeneity and phenotypic variability, depending on LDL receptor activity and lifestyle. To improve diagnosis and patient management, here, we characterized two single nucleotide missense substitutions at Methionine 1 of the human LDLR gene (c.1A>T/p.(Met1Leu) and c.1A>C/p.(Met1Leu)). We used a combination of Western blot, flow cytometry, and luciferase assays to determine the effects of both variants on the expression, activity, and synthesis of LDLR. Our data show that both variants can mediate translation initiation, although the expression of variant c.1A>T is very low. Both variants are in the translation initiation codon and codify for the same amino acid p.(Met1Leu), yet they lead to different levels of impairment on LDLR expression and activity, corroborating different efficiencies of the translation initiation at these non-canonical initiation codons. The functional data of these variants allowed for an improved American College of Medical Genetics (ACMG) classification for both variants, which can allow a more personalized choice of the lipid-lowering treatment and dyslipidemia management, ultimately improving patients’ prognosis.
Keywords: initiation codon, familial hypercholesterolemia, LDLR, functional characterization, ACMG classification
1. Introduction
Familial hypercholesterolemia (FH) (OMIM 143890) is an autosomal disorder of lipid metabolism characterized by increased plasma cholesterol levels and lipid accumulation in arteries and tendons, promoting premature atherosclerosis and coronary heart disease (CHD) [1]. FH is one of the most common genetic disorders with a frequency of around 1:250 in most populations, according to a recent meta-analysis [2]. The FH phenotype can be caused by pathogenic variants in the three FH-associated genes (APOB, LDLR, and PCSK9) or by pathogenic variants in several phenocopies or alternative molecular etiologies (ABCG5/8, APOE, LDLRAP1, and LIPA), or it can even be due to a polygenic cause [3,4,5,6]. Although the underlying genetics of the FH phenotype is complex, more than 90% of FH cases result from LDLR defects associated with an autosomal dominant inheritance pattern [3,4]. Up to date, there are more than 2300 LDLR variants exclusively identified in FH patients [7]. Another key feature of FH genetics is the phenotypic variability [3,8]; there is a diversity of phenotypes between groups sorted by causative gene and mutation type, and it is not uncommon to observe a wide range of lipid levels among carriers of an identical FH-causing mutation [3,8]. Moreover, the mutation type and the percentage of activity of LDLR variants can modulate the patients’ phenotype and response to lipid therapy [4,8]. LDLR variants can be primarily divided into null variants and defective variants, with complete or partial loss of function, respectively. Several studies showed an association between higher levels of LDL cholesterol and null variants compared with defective variants [9,10,11]. Additionally, the type of mutation can modulate response to statin therapy, as shown in Brazilian and Spanish cohorts, where null variants were associated with the lowest decrease of LDL levels upon treatment [12,13]. LDLR mutations can be functionally classified into five groups: class I—no protein synthesis (null variants); class II—partial or complete retention of the protein at the endoplasmic reticulum; class III—defective binding; class IV—defective internalization or endocytosis; class V—defective recycling [14]. Although the functional characterization is important for a correct patient diagnosis and management, less than 10% of all variants described in the LDLR gene have been functionally characterized. To improve diagnosis and patient management, here, we report two different missense variants (c.1A>T, p.(Met1Leu) and c.1A>C, p.(Met1Leu)) in the LDLR initiation codon, exhibiting distinct functional phenotypes.
2. Materials and Methods
2.1. Clinical Study
The Portuguese FH Study is a research project continuously running since 1999 with the aim of identifying the genetic cause of hypercholesterolemia in individuals with a clinical diagnosis of FH [15,16,17]. The study protocol and database have been approved by the National Institute of Health Ethics Committee and the National Data Protection Commission, respectively. Written informed consent was obtained from all participants before their inclusion in the study. Fasting blood samples were collected from all individuals at the time of their inclusion in the study. The biochemical characterization of lipids and lipoproteins was performed in a Cobas Integra 400 plus (Roche, Risch-Rotkreuz, Switzerland) by enzymatic colorimetric and immunoturbidimetric methods. All healthy individuals from whom the LDL used in the experiments was extracted followed the same exact procedure as any participant of the Portuguese FH Study.
2.2. Plasmid Constructs
For Western blot and cytometry assays, the two missense variants under study, NM_000527.5:c.1A>T and NM_000527.5:c.1A>C, were individually introduced into the human LDLR cDNA (NM_000527.5) in the mammalian expression vector pcDNA3 under control of an SV40 promoter and enhancer. For luminometry assays, the promoter and the 5′ untranslated region (5′UTR) of LDLR (−319 bp upstream from ATG) were cloned upstream the Firefly Luciferase (FLuc) coding region in the pGL4.10 reporter plasmid (Promega, Madison, WI, USA), obtaining the LDLR_pGL4-WT construct. Both plasmids were subjected to oligonucleotide site-directed mutagenesis using the NZYMutagenesis kit (NZYTech, Lisbon, Portugal), according to the manufacturer’s instructions. For every variant, the presence of the correct variant and the integrity of the region was confirmed by direct Sanger sequencing.
2.3. Cell Culture and Transfection
LDLR-deficient CHO-ldlA7 cells (Chinese hamster ovary cell line ldlA7), kindly provided by Dr. Monty Krieger, Massachusetts Institute of Technology, Cambridge, MA, were cultured in F-12 Nut Mix medium supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen, Waltham, MA, USA), 100 units/mL penicillin, and 100 μg/mL streptomycin, and cells were grown at 37 °C in a humidified incubator containing 5% CO2.
For Western blot and cytometry assays, cells were seeded in 6- or 24-well culture plates at 80% confluence (48 h) and were transfected with plasmids containing wild-type or mutant LDLR variants using Lipofectamine 2000 Transfection reagent (Invitrogen, Waltham, MA, USA) following manufacturer’s instructions. LDLR expression and LDL binding and internalization were assessed 24 h after transfection.
For luminometry assays, transient transfections were performed using Lipofectamine 2000 Transfection Reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions in 35-mm plates. Cells were co-transfected with 750 ng of the test DNA construct corresponding to the LDLR_pGL4-WT or its derivative plasmids and 500 ng of the pRL-TK plasmid (Promega, Madison, WI, USA), which encodes Renilla luciferase (RLuc) as an internal control, and then harvested after 24 h.
2.4. Western Blot Analysis
Protein expression was determined for whole-cell extracts and fractionated by electrophoresis using NuPAGE™ 4%–12% Bis-Tris gel (Invitrogen, Waltham, MA, USA) and NuPAGE™ MOPS SDS Running Buffer (Invitrogen, Waltham, MA, USA) for semi-quantitative immunoblotting. PVDF membranes were immunostained overnight at 4 °C with a specific rabbit polyclonal antibody to the human LDL receptor (Progen Biotechnik, Heidelberg, Germany) and a monoclonal anti-α-tubulin antibody (Boster Biological Technology, Pleasanton, CA, USA) for protein loading control. After incubation with appropriate secondary antibodies, proteins were detected by chemiluminescence using Pierce™ ECL Plus Western Blotting Substrate (Thermo Scientific, Waltham, MA, USA) on a ChemiDoc (BioRad, Hercules, CA, USA). Relative band intensities of mature (≈160 kDa) and precursor (≈120 kDa) forms of LDLR protein were quantified by densitometric analysis using Image Lab Software (BioRad, Hercules, CA, USA) and normalized to levels of α-tubulin.
2.5. LDL Isolation and Labeling
Plasma used for lipoprotein purification was collected from healthy individuals’ blood after 30 min centrifugation at 2465× g at room temperature (5810R, Eppendorf, Hamburg, Germany). For all samples, plasma density was adjusted to 1.21 g/mL with potassium bromide (KBr), and PBS buffer was added to obtain a clear separation between lipid fractions. LDL was isolated through ultracentrifugation carried out in a TST 41–14 rotor at 225,000× g for 19 h at 4 °C (Centrikon T-21X0, Kronton, Munich, Germany). The intermediate orange band corresponding to LDL was collected and stored at 4 °C.
LDL was fluorescently labeled with fluorescein isothiocyanate (FITC). LDL was loaded in a 0.1 M NaHCO3 (pH 9.0) pre-equilibrated Sephadex G-25 column and then mixed under constant agitation with 10 μL FITC (2 mg/mL in DMSO) per lipoprotein milliliter at room temperature for 2 h. After incubation, free FITC was removed by gel filtration on a Sephadex G-25 column equilibrated with PBS EDTA-free buffer. Lipoprotein quantification was determined by the Pierce BCA protein assay.
2.6. LDLR Expression and Activity (Lipoprotein Binding and Internalization) by FACS
LDLR cell surface expression was measured by FACS using mouse anti-human-LDLR (1:100; Progen Biotechnik, Heidelberg, Germany) as primary antibody and Alexa Fluor 488-conjugated goat anti-mouse IgG (1:200; Molecular Probes, Eugene, OR, USA) as the secondary antibody. Briefly, cells were incubated at 4 °C, overnight with the primary antibody after fixing (10 min in 4% paraformaldehyde) and blocking (1 h with PBS-5% FBS) steps. Next, cells were incubated for 1 h at room temperature with the secondary antibody. For each sample, the fluorescence of 15,000 events was acquired for data analysis, and measurements were performed in triplicate. All washes were done with PBS-1% BSA.
For quantification of LDLR activity, cells were seeded in 24-well culture plates and transfected, as previously described. Briefly, 24 h after transfection, cells were incubated for 3 h at 37 °C or 4 °C with 20 μg/mL FITC-LDL to determine LDL internalization or LDL-LDLR binding, respectively. After incubation, cells were washed three times, fixed for 10 min in 4% paraformaldehyde, and washed again three times. The amount of internalized LDL was determined by adding Trypan blue solution to a 0.2% final concentration (Sigma-Aldrich, Steinheim, Germany) to the samples. This dye quenches external fluorescence, eliminating the signal of non-internalized LDLR-LDL complexes, allowing the determination of the intensity of the remaining fluorescent particles inside the cells. Fluorescence intensities were measured by flow cytometry in a Facscalibur Flow cytometer. For each sample, the fluorescence of 15,000 events was acquired for data analysis, and measurements were performed in triplicate. All washes were done with PBS-1% BSA.
2.7. Luminometry Assay
Cell lysis was performed with Passive Lysis Buffer (Promega). The cell lysates were used to determine relative FLuc and RLuc activities using the Dual-Luciferase® Reporter Assay System (Promega), according to the manufacturer’s instructions, on a GloMax® 96 Microplate Luminometer (Promega). The collected data was expressed in arbitrary light units. Luciferase activity was obtained by normalizing FLuc to RLuc luminescence for each sample, and each value was derived from three independent experiments.
2.8. American College of Medical Genetics (ACMG) Classification and In Silico Analysis
Both variants were classified according to the Clinical Genome resource FH variant curation expert panel specifications for the ACMG/AMP Variant Interpretation Guidelines [18]. Additionally, the potential effects of the two variants were evaluated by 5 software tools: PROVEAN [19], SIFT [20], PolyPhen2 [21], MutationTaster [22], and REVEL [23].
2.9. Statistical Analysis
Results are expressed as mean ± standard deviation. The Student’s t-test was used for the estimation of statistical significance. Significance for statistical analysis was defined as a p < 0.001.
3. Results
3.1. In Silico and ACMG Classification
The bioinformatics prediction for the pathogenicity of the two variants displays conflicting classification between the different software but not among themselves (Table 1).
Table 1.
In silico results obtained by the different bioinformatics tools.
| Variant | PROVEAN | SIFT | Polyphen-2 | Mutation Tester 2 | REVEL |
|---|---|---|---|---|---|
| c.1A>T, p.(Met1Leu) | Neutral | Damaging | Benign | disease causing | 0.638 |
| c.1A>C, p.(Met1Leu) | Neutral | Damaging | Benign | disease causing | 0.638 |
Initially, variants c.1A>T, p.(Met1Leu), and c.1A>C, p.(Met1Leu) were classified as VUS (variant of uncertain significance) and Likely Pathogenic, respectively. Once the functional data were integrated into the algorithm, the classification has improved to Likely Pathogenic and Pathogenic (Table 2).
Table 2.
ACMG/AMP classification with and without the functional data.
| Variant | ACMG Classification without FS | Points without FS | ACMG Classification after FS * |
|---|---|---|---|
| c.1A>T, p.(Met1Leu) | VUS | PM2, PVS1_Moderate and PP4 | Likely pathogenic |
| c.1A>C, p.(Met1Leu) | Likely pathogenic | PM2, PVS1_Moderate PP1_Moderate, PP4, PS4_Supporting | Pathogenic |
FS—Functional study; VUS—variant of uncertain significance; * Classification with additional PS3 point.
3.2. Functional Profiling of Variants c.1A>T and c.1A>C
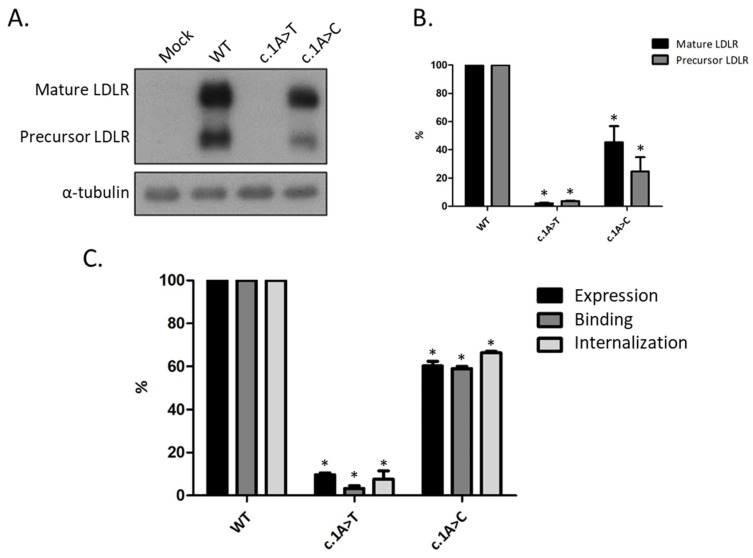
Aiming to understand the functional consequences of both variants, protein levels from each variant were analyzed by Western blot (Figure 1A), and relative levels of LDLR expression were determined by quantitative densitometry, using α-tubulin protein expression as an internal control (Figure 1B). This analysis revealed different mature and precursor LDLR expression levels between the two variants (Figure 1A). LDLR variant c.1A>T produces almost no protein, showing a similar protein profile to the mock control. Whereas in LDLR variant c.1A>C, both mature and precursor forms of LDLR were detected, being the expression lower compared to the wild-type (WT) receptor (Figure 1B).
Figure 1.
Expression and activity of LDLR in CHO-ldlA7 cells transfected with wild-type (WT), c.1A>T, p.(Met1Leu), or c.1A>C, p.(Met1Leu) variants. After 24 h of overexpression, cells were lysed, and protein extracts were analyzed (A) by Western blot, as previously described in Methods. (B) The relative band intensities of both mature (black bars) and precursor (grey bars) LDLR forms were calculated, respectively, as the ratio of 160 kDa or 120 kDa LDLR band intensity to that of α-tubulin. (C) Black bars—LDLR expression; Dark grey bars—LDL(FITC)-LDLR binding after 3 h incubation at 4 °C; Light grey bars—LDL(FITC) internalization after 3 h incubation at 37 °C. Geometric fluorescence intensity of 15,000 events was acquired in a Facscalibur Flow cytometer, as mentioned above. All values represent the mean of triplicate independent determinations (n = 3); error bars represent ± SD. * p < 0.001 compared to LDLR WT using a two-tailed Student’s t-test.
To quantify the expression and activity of both LDLR variants, CHO-ldlA7 cells were transiently transfected with WT LDLR, c.1A>T, p.(Met1Leu), or c.1A>C, p.(Met1Leu) constructs to determine the ability of the receptor to bind and internalize labeled LDL. Results for cell surface LDLR expression, LDL (FITC)-LDLR binding, and internalization are illustrated in Figure 1C. WT transfection was used for normalization of results, and it represents 100% of expression and activity. Both variants exhibit expression and activity levels below 70% of the WT LDLR; therefore, they are considered to affect LDLR function. Cut-offs for this limit were used from Chora et al. (2021) [18]. The two variants exhibit cell surface expression lower than WT LDLR, and consequently, binding and internalization levels are also diminished. Variant c.1A>T shows expression and activity levels below 10%, whereas variant c.1A>C has expression and activity levels around 60%.
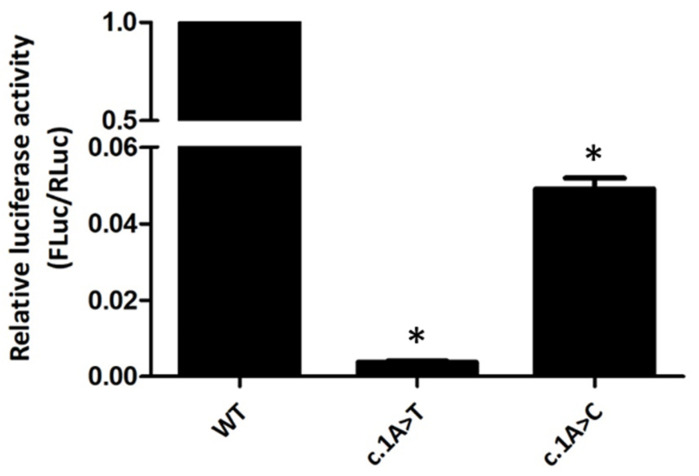
Additionally, luminometry assays were performed to check the effect of the two variants in protein synthesis (Figure 2). Relative luciferase activity of constructs pGL4-c.1A>T and pGL4-c.1A>C is 0.5% and 5% of the normal control, respectively. Thus, both c.1A>T and c.1A>C LDLR variants repress protein expression of the downstream reporter gene, although with different intensities.
Figure 2.
Relative luciferase activity of LDLR_pGL4 constructs with c.1A>T, p.(Met1Leu) or c.1A>C, p.(Met1Leu) variants. CHO-ldlA7 cells were transiently co-transfected with each one of the LDLR_pGL4 constructs (containing Firefly Luciferase—FLuc) and with the pRL-TK plasmid encoding the Renilla Luciferase (RLuc). Cells were lysed 24 h later, and the luciferase activity was measured by luminometry assays. FLuc activity values were normalized to RLuc activity to control for transfection efficiency. Relative luciferase activity of the LDLR_pGL4-WT (WT) construct was defined as one. All values represent the mean of triplicate independent determinations (n = 3); error bars represent ± SD. * p < 0.001 compared to LDLR WT using a two-tailed Student’s t-test.
3.3. Phenotype of Patients with Variant c.1A>C, p(Met1Leu)
Only the variant c.1A>C is identified in the Portuguese FH Study in four different families exhibiting an autosomal dominant inheritance pattern. A total of eight individuals carries this variant—four index cases and four relatives (Table 3). The eight carriers present untreated values of total and LDL cholesterol (LDL-C) ranging from 288 to 491 mg/dL and from 222 to 392 mg/dL, respectively, in adults. In children, total cholesterol and LDL-C range from 174 to 284 mg/dL and from 108 to 226 mg/dL, respectively.
Table 3.
Carriers of variant c.1A>C, p.(Met1Leu) found in the Portuguese FH Study.
| Family | Age at Referral | Sex | Total-C * [mg/dL] | LDL-C * [mg/dL] | HDL-C * [mg/dL] | TG * [mg/dL] | BMI (Kg/m2) | LLT |
|---|---|---|---|---|---|---|---|---|
| 1—Index | 22 | F | 439 | 297 | 115 | 135 | NK | No |
| 2—Index | 38 | M | 288 | 222 | 48 | 90 | NK | No |
| 2 | 9 | F | 174 | 108 | 52 | 53 | NK | No |
| 3—Index | 2 | M | 281 | 226 | 47 | 93 | 16.8 | No |
| 3 | 3 | M | 284 | 220 | 51 | 95 | NK | No |
| 3 | 32 | F | 491 | 392 | 77 | 110 | NK | No |
| 4—Index | 12 | M | 257 | 174 | 68 | 32 | 22.6 | Atorvastatin |
| 4 | 41 | F | 227 | 149 | 63 | 50 | NK | Atorvastatin |
* Values at referral for a genetic test. TG—triglycerides; BMI—body mass index; LLT—lipid-lowering therapy; NK—not known.
4. Discussion
Both LDLR variants (c.1A>T and c.1A>C) are predicted to change Methionine (AUG) to Leucine [UUG or CUG, respectively; p.(Met1Leu)]. Here, we investigated the functional consequences of these two LDLR variants. Our data show that both variants are able to mediate translation initiation, although the expression of variant c.1A>T is very low. Translation has been traditionally considered to initiate at the universal AUG start codon. However, due to the advent of the ribosome profiling technique [24], it is now clear that many non-AUG codons can function as non-canonical start codons [25]. Accordingly, here we show that the UUG and CUG Leucine codons are able to function as translation initiation codons for the LDLR protein. Our data also show that these two Leucine codons mediate translation initiation with different efficiencies, being the UUG codon a much less efficient initiator. Indeed, these results are in accordance with what has been described, showing that, in eukaryotes, the substitution of the initiating codon CUG for an alternate Leucine-encoding UUG codon drastically reduces protein expression [26]. Moreover, the CUG codon is the most prevalent non-AUG start codon found in upstream open reading frames [27]. Although the key eukaryotic initiation factor (eIF), eIF2, does not significantly bind other tRNAs besides Met-tRNAiMet [25], it is known that other eIFs can substitute eIF2 for initiation at non-AUG codons. eIF2A, for instance, can deliver Leu-tRNACUG and even Met-tRNAiMet to initiate translation at non-AUG start codons in a GTP-independent manner [28,29]. Accordingly, depletion of eIF2A does not significantly impair global protein synthesis, neither affects translation initiation at AUGs, but has a negative impact on initiation at CUG or UUG codons [28,29]. These pieces of evidence well support the contrasting in vitro phenotypes observed between the two variants regarding LDLR expression. As shown, the construct with the variant c.1A>T has residual LDLR expression and consequently very low LDL binding and internalization abilities. Nonetheless, the construct with the variant c.1A>C partially expresses LDLR, and the binding and internalization activities are coherently affected. Moreover, our results from the FLuc-based reporter constructs also support these data. However, the expression of these constructs is significantly lower than the one from the constructs with the LDLR open reading frame (ORF) (5% versus 60% and 0.5% versus 10% for c.1A>C and c.1A>T, respectively; Figure 1C and Figure 2). One possible explanation for this discrepancy could be the existence of RNA secondary structures downstream the start codons that could favor recognition of the non-canonical, UUG and CUG, codons on the pcDNA3 constructs (LDLR ORF) when compared to the pGL4 constructs (Luciferase ORF). This idea is supported by previous reports showing that recognition of non-canonical codons by the scanning ribosomes is favored by GC-rich downstream regions that potentiate the formation of hairpin-like structures. These secondary structures are thought to slow down the ribosomal scanning process, giving time for translation initiation to occur on sub-optimal codons [30,31]. Accordingly, the mRNA secondary structure obtained from mFOLD (http://www.unafold.org; accessed on 7 February 2021) using the first 100 nucleotides of the LDLR coding sequence is more stable than the one corresponding to the first 100 nucleotides of the Luciferase ORF.
Considering the functional data gathered on the two variants, variant c.1A>T is classified as an LDLR class I mutation, with no protein synthesis (null variant), whereas variant c.1A>C phenotype is similar to LDLR class II mutations, with partial retention of the immature protein at the endoplasmic reticulum [14]. These distinct phenotypes, a null and a defective allele type variant, highlight the importance of functionally characterizing and classifying LDLR variants. Carriers of a null allele variant present higher LDL values and higher rates of premature CHD than a defective allele carrier [10,32]. As such, functional characterization is important for cardiovascular risk stratification. Considering our in vitro results, a distinctive severity of FH phenotype in carriers of these variants is expected, with carriers of the variant c.1A>C exhibiting a milder phenotype compared to c.1A>T carriers.
According to ACMG/AMP guidelines for the classification of genetic variants, c.1A>T and c.1A>C are classified, respectively, as likely pathogenic and pathogenic variants, taking into consideration the functional study results [18]. However, functionally, they have different performances, as described above. ACMG classification classifies a variant as likely pathogenic or pathogenic or likely benign or benign but does not characterize the level to which the variant affects the protein, which can be determined only with functional studies.
Whereas the variant c.1A>T was first described in 1996 in a compound heterozygous (CH) patient from South Africa [33], variant c.1A>C was first identified in 2001 in a heterozygous patient from Spain (no lipid information provided) [34]. As expected for an FH homozygous or compound heterozygous individual, the CH patient carrying the variant c.1A>T exhibits extremely high levels of total and LDL cholesterol (LDL-C), 719 mg/dL and 669 mg/dL, respectively. Additionally, the variant c.1A>T is also identified in two first degree relatives of the CH patient with ages of 36 and 18 years old, presenting values of total and LDL-C of 363 mg/dL and 305 mg/dL and 305 mg/dL and 232 mg/dL, respectively. Unfortunately, no data on lipid-lowering treatment is provided for any of these individuals. To date, only the variant c.1A>C is identified in the Portuguese FH Study in four different families exhibiting an autosomal dominant inheritance pattern. As reported in the results, the eight carriers present a wide range of untreated values of total and LDL-C, showing the already mentioned heterogeneity in phenotypes presented by carriers of the same variant. There are several cholesterol modulators from age, body mass index, diet, physical exercise, and co-occurrence of other disorders and treatments that can account for this phenotypical heterogeneity. In this case, since the variants under study are in the LDLR translation initiation codon and originate a non-canonical initiator, we can speculate that some factors involved in non-canonical translation initiation may also be modulating it with different efficiencies among patients, leading to distinct phenotypes.
It is known that the type of mutation can modulate the response to statin therapy, where null variants have the lowest decrease of LDL levels upon treatment [12,13]. In adults, the lowest observed values of total and LDL-C (227 mg/dL and 149 mg/dL) correspond to an individual undergoing statin therapy, who before treatment had 400 mg/dL of total cholesterol (LDL-C values before treatment not known). The child of this individual also undergoing statin therapy shows a reduction of 23% and 30% of total and LDL-C, i.e., from 335 mg/dL and 250 mg/dL to 257 mg/dL and 174 mg/dL, respectively. This good response to treatment goes to the encounter of our results, to the extent that considering the mechanism of action of statins, theoretically, carriers of putative null variants should not greatly benefit from this therapy. However, from carriers of a variant with partial activity, such as c.1A>C, a good response to statin treatment is expected, as observed in these individuals. Both c.1A>C and c.1A>T variants exert their pathogenicity by means of low expression of LDLR; the second one is close to null expression. Although the expression is reduced, the activity of the synthesized receptors is normal, and so they play their role in the clearance of LDL from circulation. This way, the effect of statin therapy, which blocks cholesterol biosynthesis and promotes the hepatic expression of LDLR [35], should have a more beneficial impact on carriers of variant c.1A>C compared to c.1A>T carriers.
5. Conclusions
In conclusion, we have characterized 2 different LDLR missense variants in the translation initiation codon, which codify for the same amino acid p.(Met1Leu) but have a distinct impact on LDLR expression and activity. The two variants exhibit different functional behaviors: cytometry and luciferase assays show that variant c.1A>T/p.(Met1Leu) acts like a null variant with extremely low levels of protein expression and consequent null activity; conversely, variant c.1A>C/p.(Met1Leu) is able to initiate translation with a higher efficiency, which renders a significant LDLR activity to this variant. Our data gives new functional insight into these two variants, leading to an improved ACMG classification, which can be used in the choice of the lipid-lowering treatment and dyslipidemia management for a more personalized approach, which, in the end, can lead to a better patient prognosis. With this work, we have also contributed with evidence to the function of non-canonical initiation codons in translation initiation. However, additional experiments are required to fully unveil the underlying mechanism through which mRNA translation initiation is affected.
Acknowledgments
The authors acknowledge Fundação para a Ciência e a Tecnologia (Ph.D. scholarship no. SFRH/PD/BD/131427/2017 and SFRH/PD/BD/114392/2016).
Author Contributions
Conceptualization, L.R. and M.B.; methodology, R.G., R.F., A.C.A. and J.M.; validation, A.C.A. and J.M.; formal analysis, R.G. and R.F.; writing—original draft preparation, R.G. and R.F.; writing—review and editing, R.G., L.R. and M.B., supervision, L.R. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and the Portuguese FH Study was approved by the National Institute of Health Ethics Committee and the National Data Protection Commission on 21 September 2009, protocol code 3962/2009.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science (80-) 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Akioyamen L.E., Genest J., Shan S.D., Reel R.L., Albaum J.M., Chu A., Tu J.V. Estimating the prevalence of heterozygous familial hypercholesterolaemia: A systematic review and meta-analysis. BMJ Open. 2017;7:1–13. doi: 10.1136/bmjopen-2017-016461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berberich A.J., Hegele R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat. Rev. Cardiol. 2019;16:9–20. doi: 10.1038/s41569-018-0052-6. [DOI] [PubMed] [Google Scholar]
- 4.Bourbon M., Alves A.C., Sijbrands E.J. Low-density lipoprotein receptor mutational analysis in diagnosis of familial hypercholesterolemia. Curr. Opin. Lipidol. 2017;28:120–129. doi: 10.1097/MOL.0000000000000404. [DOI] [PubMed] [Google Scholar]
- 5.Mariano C., Alves A.C., Medeiros A.M., Chora J.R., Antunes M., Futema M., Humphries S.E., Bourbon M. The familial hypercholesterolaemia phenotype: Monogenic familial hypercholesterolaemia, polygenic hypercholesterolaemia and other causes. Clin. Genet. Clin. Genet. 2019;97:457–466. doi: 10.1111/cge.13697. [DOI] [PubMed] [Google Scholar]
- 6.Talmud P.J., Shah S., Whittall R., Futema M., Howard P., Cooper J.A., Harrison S.C., Li K., Drenos F., Karpe F., et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: A case-control study. Lancet. 2013;381:1293–1301. doi: 10.1016/S0140-6736(12)62127-8. [DOI] [PubMed] [Google Scholar]
- 7.Iacocca M.A., Chora J.R., Carrié A., Freiberger T., Leigh S.E., Defesche J.C., Kurtz C.L., DiStefano M.T., Santos R.D., Humphries S.E., et al. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 2018;39:1631–1640. doi: 10.1002/humu.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Taranto M.D., Giacobbe C., Fortunato G. Familial hypercholesterolemia: A complex genetic disease with variable phenotypes. Eur. J. Med. Genet. 2020;63:103831. doi: 10.1016/j.ejmg.2019.103831. [DOI] [PubMed] [Google Scholar]
- 9.Abul-Husn N.S., Manickam K., Jones L.K., Wright E.A., Hartzel D.N., Gonzaga-Jauregui C., O’Dushlaine C., Leader J.B., Kirchner H.L., Lindbuchler D.M., et al. Genetic identification of familial hypercholesterolemia within a single U.S. Health care system. Science (80-) 2016;354:aaf7000. doi: 10.1126/science.aaf7000. [DOI] [PubMed] [Google Scholar]
- 10.Bourbon M., Alves A.C., Alonso R., Mata N., Aguiar P., Padró T., Mata P. Mutational analysis and genotype-phenotype relation in familial hypercholesterolemia: The SAFEHEART registry. Atherosclerosis. 2017;262:8–13. doi: 10.1016/j.atherosclerosis.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Khera A.V., Won H., Peloso G.M., Lawson K.S., Bartz T.M., Deng X., van Leeuwen E.M., Natarajan P., Emdin C.A., Bick A.G., et al. Diagnostic Yield of Sequencing Familial Hypercholesterolemia Genes in Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez De Isla L., Alonso R., Watts G.F., Mata N., Saltijeral Cerezo A., Muñiz O., Fuentes F., Diaz-Diaz J.L., de Andrés R., Zambón D., et al. Attainment of LDL-cholesterol treatment goals in patients with familial hypercholesterolemia: 5-year SAFEHEART registry follow-up. J. Am. Coll. Cardiol. 2016;67:1278–1285. doi: 10.1016/j.jacc.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Santos P.C.J.L., Morgan A.C., Jannes C.E., Turolla L., Krieger J., Santos R.D., Pereira A.C. Presence and type of low density lipoprotein receptor (LDLR) mutation influences the lipid profile and response to lipid-lowering therapy in Brazilian patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2014;233:206–210. doi: 10.1016/j.atherosclerosis.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 14.Hobbs H.H., Brown M.S., Goldstein J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 15.Medeiros A.M., Alves A.C., Francisco V., Bourbon M. Update of the Portuguese Familial Hypercholesterolaemia Study. Atherosclerosis. 2010;212:553–558. doi: 10.1016/j.atherosclerosis.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Medeiros A.M., Alves A.C., Bourbon M. Mutational analysis of a cohort with clinical diagnosis of familial hypercholesterolemia: Considerations for genetic diagnosis improvement. Genet. Med. 2015;18:316–324. doi: 10.1038/gim.2015.71. [DOI] [PubMed] [Google Scholar]
- 17.Bourbon M., Rato Q. Estudo Português de hipercolesterolemia familiar: Apresentação do estudo e resultados preliminares. Rev. Port. Cardiol. 2006;25:999–1013. [PubMed] [Google Scholar]
- 18.Chora J.R., Iacocca M.A., Tichy L., Wand H., Kurtz C.L., Zimmermann H., Leon A., Williams M., Humphries S.E., Hooper A.J., et al. Title: The Clinical Genome Resource (ClinGen) Familial Hypercholesterolemia Variant Curation Expert Panel consensus guidelines for LDLR variant classification. medRxiv. 2021 doi: 10.1101/2021.03.17.21252755. [DOI] [PubMed] [Google Scholar]
- 19.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. [(accessed on 11 March 2021)];Nat. Methods. 2010 7:248–249. doi: 10.1038/nmeth0410-248. Available online: http://www.nature.com/articles/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 23.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-Wide Analysis in Vivo of Resolution Using Ribosome Profiling. [(accessed on 11 March 2021)];Science (80-) 2009 324:218–223. doi: 10.1126/science.1168978. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2746483&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearse M.G., Wilusz J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017;31:1717–1731. doi: 10.1101/gad.305250.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwab S.R., Li K.C., Kang C., Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science (80-) 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 27.Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starck S.R., Jiang V., Pavon-Eternod M., Prasad S., McCarthy B., Pan T., Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science (80-) 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 29.Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L. Translation from the 5’ untranslated region shapes the integrated stress response. Science (80-) 2016;351:aad3867. doi: 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guenther U.P., Weinberg D.E., Zubradt M.M., Tedeschi F.A., Stawicki B.N., Zagore L.L., Brar G.A., Licatalosi D.D., Bartel D.P., Weissman J.S., et al. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature. 2018;559:130–134. doi: 10.1038/s41586-018-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alves A.C., Alonso R., Diaz-Diaz J.L., Medeiros A.M., Jannes C.E., Merchan A., Vasques-Cardenas N.A., Cuevas A., Chacra A.P., Krieger J.E., et al. Phenotypical, clinical, and molecular aspects of adults and children with homozygous familial hypercholesterolemia in iberoamerica. Arterioscler. Thromb. Vasc. Biol. 2020;40:2508–2515. doi: 10.1161/ATVBAHA.120.313722. [DOI] [PubMed] [Google Scholar]
- 33.Langenhoven E., Warnich L., Thiart R., Rubinsztein D.C., Van Der Westhuyzen D.R., Marais A.D., Kotze M. Two novel point mutations causing receptor-negative familial hypercholesterolemia in a South African Indian homozygote. Atherosclerosis. 1996;125:111–119. doi: 10.1016/0021-9150(96)05871-6. [DOI] [PubMed] [Google Scholar]
- 34.Chaves F.J., Real J.T., García-García A.B., Civera M., Armengod M.E., Ascaso J.F., Carmena R. Genetic diagnosis of familial hypercholesterolemia in a South European outbreed population: Influence of low-density lipoprotein (LDL) receptor gene mutations on treatment response to simvastatin in total, LDL, and high-density lipoprotein cholesterol. J. Clin. Endocrinol. Metab. 2001;86:4926–4932. doi: 10.1210/jcem.86.10.7899. [DOI] [PubMed] [Google Scholar]
- 35.Mach F., Baigent C., Catapano A.L., Koskina K.C., Casula M., Badimon L., Chapman M.J., De Backer G.G., Delgado V., Ference B.A., et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Atherosclerosis. 2019;290:140–205. doi: 10.1016/j.atherosclerosis.2019.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.