Abstract
This study describes a potential new function of hnRNP U as an RNA polymerase (Pol II) elongation inhibitor. We demonstrated that a subfraction of human hnRNP U is associated with the Pol II holoenzyme in vivo and as such recruited to the promoter as part of the preinitiation complex. hnRNP U, however, appears to dissociate from the Pol II complex at the early stage of transcription and is therefore absent from the elongating Pol II complex. When tested in the human immunodeficiency virus type 1 transcription system, hnRNP U inhibits elongation rather than initiation of transcription by Pol II. This inhibition requires the carboxy-terminal domain (CTD) of Pol II. We showed that hnRNP U can bind TFIIH in vivo under certain conditions and inhibit TFIIH-mediated CTD phosphorylation in vitro. We find that the middle domain of hnRNP U is sufficient to mediate its Pol II association and its inhibition of TFIIH-mediated phosphorylation and Pol II elongation. The abilities of hnRNP U to inhibit TFIIH-mediated CTD phosphorylation and its Pol II association are necessary for hnRNP U to mediate the repression of Pol II elongation. Based on these observations, we suggest that a subfraction of hnRNP U, as a component of the Pol II holoenzyme, may downregulate TFIIH-mediated CTD phosphorylation in the basal transcription machinery and repress Pol II elongation. With such functions, hnRNP U might provide one of the mechanisms by which the CTD is maintained in an unphosphorylated state in the Pol II holoenzyme.
Transcription of a variety of cellular and viral genes is regulated, at least in part, at the level of elongation. Prior to the activation of these genes, RNA polymerase II (Pol II) initiates but pauses after synthesizing a short transcript. Transcription activation for these genes appears to be achieved by stimulating phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of Pol II (12, 31). The CTD, which consists of multiple repeats of the heptad sequence (YSPTSPS) ranging from 26 repeats in yeast to 52 in mammals, is essential in vivo, and transcription from many promoters is sensitive to CTD truncation (12, 18, 36, 55). Its role in elongation control has been further suggested by the observation that the CTD in paused Pol II is partially or unphosphorylated, but that in elongating Pol II is hyperphosphorylated (35).
One of the kinases that are thought to phosphorylate CTD in vivo for Pol II elongation is TFIIH, a complex of nine subunits (13, 43). TFIIH binds tightly to nonphosphorylated Pol II as a component of holoenzyme and dissociates from Pol II after transcription of 30 to 50 bp (53). It was suggested that the phosphorylation of CTD by TFIIH kinase may be important for promoter clearance for a certain promoter (2). However, recent studies indicated that the TFIIH-associated kinase is important to stimulate transcription elongation (1, 9, 10, 17, 38, 52). Although the exact role of TFIIH-mediated CTD phosphorylation in elongation control is not clear, strong evidence for the involvement of TFIIH in elongation comes from experiments that used antibodies against subunits of TFIIH in the Xenopus oocyte system. When anti-TFIIH antibodies against individual subunits were injected (52), transcription was inhibited due to the failure of elongation, suggesting an essential role of TFIIH in Pol II elongation.
TFIIH is present in the Pol II holoenzyme, where it is thought to regulate CTD phosphorylation at an early stage of transcription prior to the recruitment of other CTD kinases, such as P-TEFb, that are important for productive elongation (10, 56). Despite the presence of TFIIH, the CTD in Pol II holoenzyme remains unphosphorylated (28, 37). As the hyperphosphorylated Pol II (Pol IIO) cannot enter into a preinitiation complex (PIC), the unphosphorylated Pol II (Pol IIA) is thought to be the initiation-competent form in vivo (26, 27). One possibility is that a putative negative regulator that inhibits CTD phosphorylation may be present in the Pol II holoenzyme complex. In support of this view, previous in vitro transcription studies indicated that abortive elongation is an inherent property of the PICs derived from crude nuclear extract (24, 30, 49). In contrast, the reconstituted PICs with purified Pol II and general transcription factors can generate full-length transcripts efficiently, suggesting that elongation inhibitors may be associated with the Pol II holoenzyme.
The results in this study suggest that a ubiquitous nuclear protein, hnRNP U (heterogeneous nuclear ribonucleoprotein U) (14), may be one such elongation inhibitor in the Pol II holoenzyme. hnRNP U (120 kDa, 806 amino acids) is known as an RNA- and a scaffold/matrix attachment region DNA-binding protein. Approximately 50% of total hnRNP U is present in the nuclear matrix and 20% is tightly associated with chromatin, whereas half of hnRNP U in the remaining 30% soluble fraction is found in the hnRNP particles (16, 19, 20). hnRNP U is thought to participate in pre-mRNA processing together with other hnRNP proteins and/or to play a role in the higher-order organization of chromatin. Although most abundant proteins of the nuclear matrix are hnRNP proteins (33) and pre-mRNA is tightly associated with nuclear substructures, the RNA-binding RGG domain at its C terminus is dispensable for its interaction with nuclear matrix or chromatin. Rather, the N-terminal domain was found to be important for these interactions, while the RGG domain was shown to mediate interactions with other hnRNP proteins to form hnRNP particles.
This study suggests an unexpected function of hnRNP U as an inhibitor of Pol II elongation. We found that a fraction of hnRNP U is associated with the Pol II holoenzyme and is recruited to the promoter as part of a PIC. It appears that hnRNP U, via its middle domain, suppresses the CTD phosphorylation by TFIIH in the basal transcription machinery and inhibits Pol II elongation. This study suggests that the hnRNP U-mediated inhibition of TFIIH might be one of the mechanisms by which the CTD is maintained in an unphosphorylated state in the PIC. The involvement of hnRNP proteins in transcription regulation is not unprecedented. For example, hnRNP K has been shown to function as a transcription activator for c-myc gene expression (34, 48). The new role of hnRNP U as a Pol II elongation inhibitor underscores the diversity of potential roles of this class of proteins in a cell.
MATERIALS AND METHODS
Plasmid construction.
The cytomegalovirus (CMV) expression vectors for wild-type hnRNP U [HN(WT)] or various deletion mutants were constructed by inserting the full-length or corresponding deletion fragments in frame to a hemagglutinin epitope (HA) tag and the nuclear localization signal derived from simian virus 40 (SV40) T antigen. To construct various expression vectors in this study, the pCG-CMV expression vector was digested with XbaI and BamHI and ligated with a synthetic linker containing XbaI, KpnI, NotI, SalI, and BamHI sites. The nuclear localization signal derived from the SV40 T antigen was inserted into upstream NotI site (pCMV-1). For HA fusion proteins, the PCR fragment containing HA sequences was inserted into the XbaI/KpnI sites in pCMV-1 (pCMV-HA), and the corresponding hnRNP U fragments (NotI/SalI) were inserted in frame into pCMV-HA. All constructs made by PCR were verified by DNA sequencing.
Depletion of hnRNP U from HeLa nuclear extract.
HeLa nuclear extract (5 to 7 μl or 60 μg of protein; typical amount used for one in vitro transcription reaction) was incubated with heparin-agarose (1/2 volume of the nuclear extract; Pharmacia) at 4°C for 1 h. The supernatant was collected and incubated with the hnRNP U antibody-protein A/G-Sepharose complex at 4°C for 1 h. For the anti-hnRNP U-Sepharose complex, 2 μl of the antibody was bound to 5 μl of protein A/G-Sepharose for 1 h at 4°C, and the immobilized beads were washed three times with buffer A (10 mM Tris, 10 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT]) containing 0.5% Triton X-100. Although a considerable amount of hnRNP U was removed from nuclear extract after the heparin-agarose step, immunodepletion with anti-hnRNP U was necessary to remove the remaining hnRNP U protein.
Immunopurification of HA-HN from HeLa nuclear extract.
HeLa cells (107) were transfected with 20 μg of the expression vector for various HA-tagged hnRNP U (HA-HN) proteins. After 24 h of transfection, nuclear extract was prepared as described previously (22, 23). Prior to the addition of antibody, the nuclear extract was precleared by incubation with 1/10 volume of protein A/G-Sepharose. To 100 μl of nuclear extract containing 500 μg of protein, 15 μl of anti-HA antibody was added, and the mixture was incubated for 1 h at 4°C. The nuclear extract-antibody mixture was spun for 5 min at 14,000 rpm to remove any protein aggregates. Protein A/G-Sepharose was added (15 μl), and the mixture was incubated for 1 h at 4°C to precipitate the immune complexes. After five washes with buffer A, the bound protein was eluted with excess amount (5 μg) of HA peptide (Boehringer Mannheim).
Production of Strep-U in Schizosaccharomyces pombe.
Modified S. pombe expression vector pESP-1-Strep (Stratagene), where the original glutathione-S-transferase (GST) tag was replaced with the streptococcal epitope (Strep) tag (WSHPQFEK), has been described elsewhere (4). After creation of NotI site in frame to the Strep tag, a NotI/BamHI fragment containing the hnRNP U cDNA was removed from the pCMV/hnRNP U construct and inserted into the NotI/BamHI-digested pESP-1-Strep vector. The transformed S. pombe cells were grown in the presence of 20 μM thiamine to repress the nmt1 promoter until expression was desired. Cells were lysed at 4°C in sorbitol buffer containing 0.5% Triton X-100 and protease inhibitors (Boehringer minitablets) by using a French press. Strep-U was purified by using a StrepTactin-Sepharose column (Genosys, The Woodlands, Tex.). Protein concentration was measured by the Bradford method (Bio-Rad).
In vitro transcription.
For in vitro transcription reactions in Fig. 1 and 6, the linearized human immunodeficiency virus type 1 long terminal repeat (HIV-1 LTR)-chloramphenicol acetyltransferase (CAT) template (150 ng) was incubated for 30 min at 4°C with 60 μg of HeLa nuclear extract in a total volume of 25 μl containing transcription buffer (25 mM HEPES-KOH [pH 8.4], 7.5 mM MgCl2, 4 mM DTT, 65 mM KCl, 10.5% glycerol). After PIC formation, a nucleoside triphosphate (NTP) mix (0.5 mM each ATP, GTP, CTP, and UTP) was added and the transcription reaction was performed for an additional 45 min at 30°C. RNA transcripts were purified and processed for hybridization in the presence of the probes. For Fig. 5B, a thymidine kinase (TK)-CAT expression plasmid (in which a point mutation was introduced to the EcoRI site in the CAT coding region to destroy the EcoRI site) was linearized with BamHI and biotinylated by using the Klenow enzyme in the presence of 10 μM biotin-16-dUTP (Boehringer Mannheim). To remove the biotin residue at the 3′ end, the template was cleaved with SphI. PICs were formed by incubating the 5′-biotinylated template (500 ng) with 50 μg of HeLa nuclear extract in transcription buffer lacking NTPs for 30 min at 4°C. After formation of PICs, the templates were isolated by magnetic centrifugation following addition of streptavidin-coated magnetic beads (Promega) and washed with transcription buffer as described in a previous study (53). Prior to immunoprecipitation, the immobilized templates containing the PIC were digested with 100 U of EcoRI for 10 min at 30°C to cleave the TK promoter at position −80 and the magnetic beads were removed. After EcoRI digestion, the transcription complexes were immunoprecipitated and immunoblotted as indicated. For transcription initiation and elongation, PICs were formed as described above and subsequently incubated in the presence of various combinations of nucleotides such as ATP alone, ATP and CTP, or all four NTPs. Transcription reactions were performed for 20 min at 30°C. The templates were digested with EcoRI and washed in transcription buffer as described above, and the transcription complexes were immunoprecipitated. To monitor CTD phosphorylation in the reaction containing all four NTPs, the in vitro transcription reaction was carried out in the presence of 80 μCi of [γ-32P]ATP.
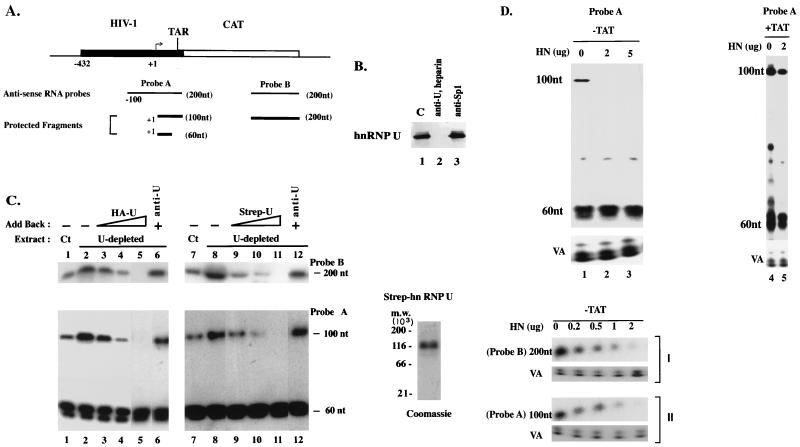
FIG. 1.
hnRNP U inhibits processive transcription by the HIV-1 LTR promoter. (A) RNA probes used for RNase protection assay. Probe A (200 nt) contains positions −100 to +80 of the HIV-1 LTR and the first 20 bp of the CAT gene. Probe B contains the C-terminal 200 nt of the CAT gene. (B) The presence of hnRNP U in nuclear extracts was examined by immunoblotting with anti-hnRNP U. hnRNP U depletion in lane 2 was done as described in Materials and Methods. In lane 3, Sp1 was depleted with anti-Sp1. C, control (undepleted extract). (C) In vitro transcription reactions were performed with the HIV-1 LTR (−432 to +80) promoter and the hnRNP U-depleted HeLa nuclear extract (U-depleted) with or without addition of the HA-RN protein (HA-U; HeLa) or Strep-U (S. pombe) protein. Ct, control (undepleted extract). The recombinant Strep-U was purified to near homogeneity and detected by colloidal blue staining (Novex) on an SDS–6% polyacrylamide gel (Coomassie). In lanes 3 to 5 and 9 to 11, increasing amounts of the recombinant hnRNP U proteins containing 20, 40, and 100 ng of hnRNP U (as judged by immunoblotting with anti-hnRNP U [anti-U]) were added back. In lanes 6 and 12, the HA-U or Strep-U preparation containing 100 ng of hnRNP U (the same amount as used in lanes 5 and 11) was preincubated with anti-hnRNP U (2 μl) and added back. RNA transcripts were hybridized to antisense RNA probes A and B (A) and analyzed by RNase protection assays. The consistent level of the 60-nt transcripts served as an internal control. Negative controls from reactions performed without the template or without NTP, or performed in the presence of α-amanitin or a nonspecific probe (−128 to +100 region of the human TSH promoter), yielded no protected band (data not shown). For the RNase protection assays using probe A, relative levels of the long transcripts are indicated as percentages of the total protected RNA (short transcript + long transcript) (lanes 1 to 6, 4, 15, 12, 2, 0, 14; lanes 7 to 12, 3, 11, 5, 0.5, 0, and 13). (D) HeLa cells (5 × 106) were transfected with the HIV-1 LTR-CAT reporter (10 μg) and expression vector for hnRNP U as indicated. HN, hnRNP U. In lanes 4 and 5, the expression vector for Tat (pCMV/Tat) (20 ng) was cotransfected (Tat expression in transfected cells with only 20 ng of expression vector was not measurable by immunoblotting). Nuclear and cytoplasmic RNAs were extracted separately. The results shown were obtained by RNase protection assay using nuclear RNAs. Cytoplasmic RNAs showed identical results (data not shown). Pol III-driven transcripts from adenovirus VA1 (pSPVA1) were processed as previously described (52). A negative control containing RNA samples obtained from untransfected HeLa cells did not show any band (data not shown). Relative levels of the nonprocessive transcript as a percentage of the total protected RNA are indicated (lanes 1 to 5, 2.5, 0, 0, 26, and 24).
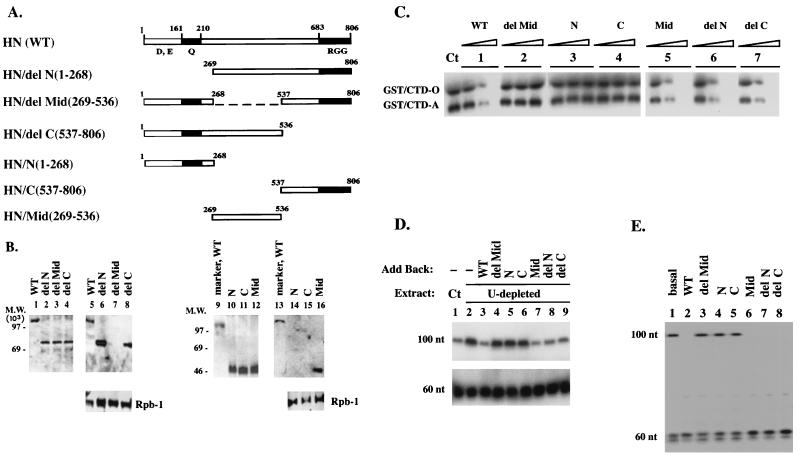
FIG. 6.
The middle domain of hnRNP U is sufficient to mediate its Pol II holoenzyme association and its inhibition of the TFIIH kinase and Pol II elongation. (A) HN(WT) and various HN deletion mutants. Modular structure of hnRNP U. Acidic (D, E), glutamine-rich (Q), and RNA-binding RGG domains are indicated. CMV expression vectors were constructed to contain the HA tag and the nuclear localization signal derived from the SV40 T antigen at the N termini of various hnRNP U fragments. (B) The middle domain of hnRNP U mediates its association with Pol II holoenzyme. HeLa cells (5 × 106 cells) were transfected with 10 μg of each expression vector. The nuclear extracts were immunoprecipitated with anti-Rap74. The bound proteins were released and reprecipitated with anti-HA antibody, and the resulting complexes were released in the presence of an excess amount of the HA peptide. Lanes 1 to 4 and 10 to 12, expression of each HA-HN protein in transfected cells probed with anti-HA; lanes 5 to 8 and 14 to 16, HA-HN proteins associated with the holoenzyme. In lanes 9 and 13, HA-HN(WT) was used as a marker. (C) The middle domain is essential for the inhibition of TFIIH-mediated CTD phosphorylation. Ct, control. Increasing amounts of each HA-HN protein were added to the kinase reaction as indicated. Similar amounts of each HA-HN protein measured by immunoblotting with anti-HA (data not shown) were used for comparison. (D) The middle domain of hnRNP U functions as a Pol II elongation block in vitro. In vitro transcription assays were performed as described for Fig. 1C. Similar amounts of HA-HN proteins as determined by immunoblotting (data not shown) were added back to the transcription reaction, and the transcripts were analyzed by RNase protection assay using probe A (Fig. 1A). Ct, control. (E) Exogenously expressed HA-HN proteins containing the middle domain inhibit elongation in vivo. HeLa cells were transfected with the HIV-1 LTR reporter (10 μg) and the expression vector for HA-HN proteins (2 μg) as described for Fig. 1D. RNase protection assays were performed with probe A (Fig. 1A), using the cytoplasmic RNAs.
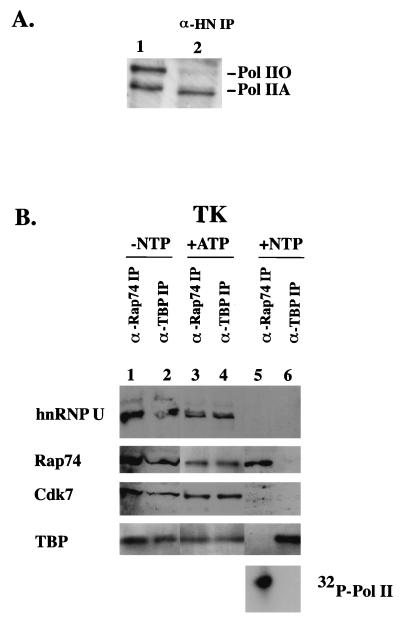
FIG. 5.
HnRNP U is recruited to the promoter as a part of the PIC and dissociates from the elongating Pol II complex. (A) Anti-hnRNP U coimmunoprecipitates only the IIA form of Pol II in vivo. HeLa whole-cell lysate (lane 1) and the anti-hnRNP U immunoprecipitate (α-HN IP; lane 2) were immunoblotted with anti-CTD antibody 8WG16. (B) The association of hnRNP U with Pol II was monitored in different stages of transcription in vitro as described in Materials and Methods. Transcription reactions were performed with the 5′-biotinylated TK-CAT template and HeLa nuclear extract. Transcription complexes formed from reactions incubated with DNA but without NTPs (lanes 1 and 2), with ATP (lanes 3 and 4), and with all four NTPs (lanes 5 and 6) were immunoprecipitated and immunoblotted as indicated. The hyperphosphorylation of Pol II in the reaction containing NTPs (lanes 5 and 6) was monitored in the presence of [γ32P]ATP.
RNase protection assay.
The RNase T1 protection assay was performed as described by the manufacturer (Ambion), using a range of input sample RNA amounts (1 to 20 μg) from transfected cells and a constant amount (5 × 104 cpm) of probe. The linear increase in intensity of the protected fragment was seen with increasing amounts of RNA, indicating that the assay was performed under conditions of probe excess. Typically, 5 to 10 μg of RNA was hybridized to 5 × 104 cpm of probe for the assay reported here. To generate the riboprobe, the linearized probe-containing plasmids were used as templates for T7 RNA polymerase.
Purification of Pol II holoenzyme.
Pol II holoenzyme shown in Fig. 3A and B was purified as described previously (28). HeLa nuclear extract from 109 cells was precleared with protein A/G-Sepharose and incubated with anti-Rap74 (250 μg; Santa Cruz)-bound protein A/G-Sepharose at 4°C overnight, washed extensively (50 mM HEPES-KOH [pH 7.8], 150 mM NaCl, 0.5% NP-40, 1% bovine serum albumin), and eluted with an excess amount of blocking peptide (Santa Cruz). The eluate containing approximately 100 μg of protein was fractionated on a gel filtration Sepharose CL-4B column (10-ml column, equilibrated in buffer containing 20 mM HEPES-KOH, 0.5 mM EDTA, 5 mM DTT, 0.01% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, and 1 μg each of aprotinin, leupeptin, and pepstatin per ml). Since the peak of Pol II holoenzyme activity was shown to coelute with the peak of the blue dextran marker (2,000 kDa), fractions were collected as described previously (28). For immunoblotting, 30 μl of each 300-μl fraction was analyzed. All antibodies used for immunoblotting were purchased from Santa Cruz except the anti-CTD monoclonal antibody 8WG16 (QED Bioscience). Anti-hnRNP U and anti-hnRNP A1 were gifts from G. Dreyfuss and D. Levens. Holoenzyme immunopurification with anti-hnRNP U (Fig. 3C) was performed as follows. Per assay, 1 μl of monoclonal anti-hnRNP U was incubated with 20 μl of protein A/G-Sepharose beads for 4 h at 4°C. The beads were extensively washed in phosphate-buffered saline containing 0.1% Triton X-100 and incubated with nuclear extracts containing 200 μg of protein for 1 h at 4°C. The beads were then washed three times with 400 μl of washing buffer.
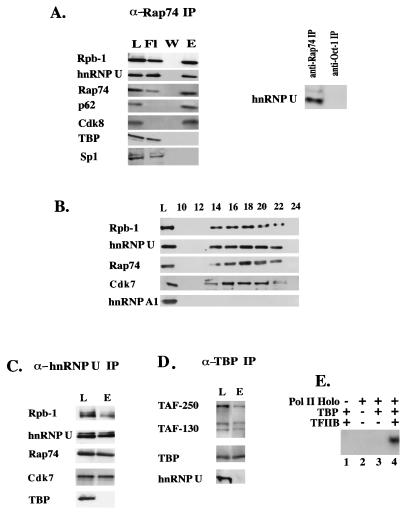
FIG. 3.
hnRNP U is coimmunoprecipitated with Pol II holoenzyme. (A) The anti-Rap74 antibody immunoprecipitates hnRNP U from HeLa cell nuclear extract. Western blots of load (L), flowthrough (Fl), second wash (W), and eluate (E) with various antibodies are shown. The largest subunit of Pol II (Rpb-1) was detected with anti-CTD monoclonal antibody 8WG16 (QED Bioscience). (B) Fractionation of the anti-Rap74 IP eluate by gel filtration (Sepharose CL-4B) as described previously (28). (C) HeLa nuclear extract was immunoprecipitated with anti-hnRNP U. The eluate corresponding to 20 μl of nuclear extract was used for Western blotting. L, load; E, eluate. (D) The TFIID complex does not contain hnRNP U. L, load; E, eluate. (E) The Pol II-containing complex in panel C obtained by immunoprecipitation with anti-hnRNP U supports transcription in vitro. A linear G-free cassette template of the HIV-1 LTR (150 ng) was incubated with immobilized beads containing anti-hnRNP U immunoprecipitates (Pol II Holo) from 200 μg of HeLa nuclear extract and the mixture of ATP, CTP, and [α-32P]UTP. The holoenzyme preparation was or was not supplemented with recombinant TBP (40 ng) and TFIIB (40 ng) (Promega) as indicated. The runoff transcript (390 nt) was resolved on a 6% polyacrylamide-urea gel.
Immunoprecipitation with whole-cell lysate.
Cells were solubilized with 0.5% Triton X-100 and 0.5% sodium deoxycholate (50 mM HEPES-KOH [pH 7.8], 0.1 M NaCl, 10 mM EDTA, 5 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin per ml, 10 μg of leupeptin per ml) at 4°C for 15 min. The suspension was passed through a needle repetitively and centrifuged briefly. The supernatant was used for immunoblot and immunoprecipitation. For immunoprecipitation in Fig. 5A, a volume of lysate equivalent to 106 cells was incubated with 2 μl of anti-hnRNP U antibody for 1 h at room temperature. After incubation with protein A/G-Sepharose beads for 2 h, the immunoprecipitates were washed and solubilized in sodium dodecyl sulfate (SDS) sample buffer. Pol II was detected by immunoblotting with anti-CTD monoclonal antibody 8WG.
TFIIH, Cdk8, and PITARLE (Cdk9) preparation.
Anti-p62, anti-Cdk8, and anti-Cdk9 antibodies (2 μl of each; Santa Cruz) were incubated with HeLa nuclear extract (50 μg) precleared with protein-Sepharose beads for 1 h on ice. The complexes were immunoprecipitated by the addition of protein A/G-Sepharose beads and then washed five times in buffer containing 10 mM HEPES (pH 7.5), 10 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, and 0.1% NP-40; the beads containing the immunoprecipitates (IPs) were used for the protein phosphorylation assay. Western blotting confirmed the presence of the specific kinase in each preparation (data not shown). To test whether other kinase activities are coimmunoprecipitated, the kinases were eluted from the beads and used for the CTD phosphorylation reaction in the presence or absence of the antibody specific to each kinase. CTD phosphorylation was blocked by the antibodies specific to these kinases (for TFIIH as shown in Fig. 4A; for Cdk8 and Cdk9, data not shown).
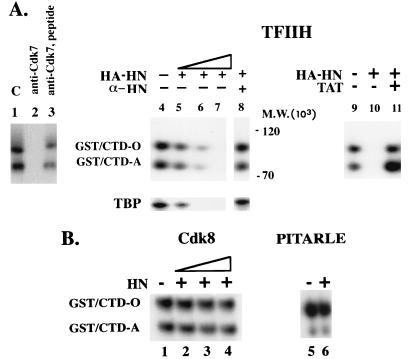
FIG. 4.
hnRNP U inhibits the CTD phosphorylation mediated by TFIIH-associated kinase. (A) hnRNP U inhibits kinase activities of TFIIH. For the TFIIH preparation in each lane in panel A, anti-p62 IP beads obtained by incubating 2 μl of anti-p62 antibody with 50 μg of HeLa nuclear extract were used. Lanes 1 to 3, TFIIH-mediated CTD phosphorylation. Lane 1, control (phosphorylation reaction was performed in kinase buffer in the presence of [γ-32P]ATP). Lane 2, CTD phosphorylation is blocked by anti-Cdk7 antibody. Lane 3, Cdk7 antigenic peptide releases the anti-Cdk7 blocking. Lanes 4 to 7, effect of HA-HN on TFIIH-mediated phosphorylation of GST-CTD (200 ng) or TBP (200 ng; Promega). HA-HN protein was immunopurified from transfected HeLa cells. HA-HN preparations containing 5, 25, and 50 ng of hnRNP U, as judged by immunoblotting with anti-hnRNP U, were used for lanes 4 to 7, respectively. Lane 8, HA-HN (50-ng equivalent [the same amount as used for lane 7]) was pretreated with anti-hnRNP U antibody (α-HN; 2 μl) for 2 h at room temperature and added to the kinase reaction. Lanes 10 and 11, the amount of HA-HN was same as that used in lane 7 (50-ng equivalent). Lane 11, effect of GST-Tat (25 ng) on hnRNP U-mediated inhibition of CTD phosphorylation. (B) HA-HN does not inhibit Cdk8 and PITALRE (Cdk9) kinases. Cdk8 and PITARLE (Cdk9) were prepared essentially as described above for the TFIIH preparation. The amounts of HA-HN used in lanes 2 to 4 are same as those in lanes 5 to 7 in panel A. Lane 6, the amount of HA-HN that completely inhibited the TFIIH-mediated phosphorylation was used. HnRNP U did not inhibit the activities of Cdk8 or PITALRE (Cdk9) even when increasing amounts of substrate (HA-HN) or enzyme preparations (anti-Cdk8 IP and anti-Cdk9 IP) were tested (data not shown).
Protein phosphorylation assay.
Protein phosphorylation assays were performed as described in other studies (10, 21, 32, 38). Briefly, reactions (20 μl) were performed in kinase buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM MnCl2, 40 mM KCl, 5 mM MgCl2, 2% glycerol, 0.25 mg of bovine serum albumin per ml, 1 mM DTT, and 0.3% NP-40. Portions (from 5 μl of nuclear extract) of anti-p62 IP, anti-Cdk8 IP, or anti-Cdk9 IP beads were incubated with 20 μCi of [γ-32P]ATP, 10 μM unlabeled ATP, 1 μg each of aprotinin, leupeptin, and pepstatin per ml, and a GST-CTD protein (200 ng) or recombinant TATA-binding protein (TBP; 200 ng; Promega) at 30°C for 1 h. The kinase reactions were terminated by addition of 5 μl of 5× SDS loading buffer. The GST-CTD fusion protein was expressed as described elsewhere (39). Various HA-HN proteins were immunopurified with anti-HA antibody 12C5A from HeLa nuclear extracts transfected with the expression vectors for various fusion proteins (20 μg of each CMV expression vector/107 cells) and eluted from the beads in the presence of excess amount of HA peptide. A range of HA-HN proteins (5 to 50 ng, as judged by immunoblotting) was used for the CTD phosphorylation assay.
RESULTS
hnRNP U as a potential basal repressor.
Several studies have indicated that a number of nonhistone chromosomal- or nuclear matrix proteins that had been originally described as structural proteins have transcription activities. As a preliminary screen to identify repressors among this group of proteins, we measured CAT activities in cells transfected with expression vectors for several nuclear matrix proteins (hnRNP U, lamin B, topoisomerase II, hnRNP A1, and HMG I/Y) and found that hnRNP U inhibits the expression of the reporter genes tested (Table 1). Cotransfection of the expression vector for human hnRNP U with the reporter constructs containing Pol II promoters (HIV-1 LTR, TK, SV40, growth hormone [GH], and thyrotropin β [TSH]) resulted in repression in basal expression. The hnRNP U-mediated repression was released by the promoter-specific activators when tested in the previously described transcription activation system (22, 23) (Tat for the HIV-1 LTR promoter, Pit-1 for the GH promoter, Pit-1/AP-1 for the TSH promoter) (data not shown).
TABLE 1.
hnRNP U represses basal expression of the reporter genesa
| Reporter promoter | Relative CAT activity (avg ± SD)
|
||||||
|---|---|---|---|---|---|---|---|
| hnRNP U | Lamin B | Topo II | hnRNP A1 | HMG I(Y) | Sp1 | Oct-1 | |
| HIV-1 (−432 to +80) | 0.2 ± 0.05 | 0.9 ± 0.15 | 0.9 ± 0.1 | 1.2 ± 0.2 | 3.1 ± 0.1 | 2.5 ± 0.02 | 1.2 ± 0.05 |
| TK (105 to +55) | 0.3 ± 0.02 | 1.0 ± 0.1 | 1.2 ± 0.02 | 1.5 ± 0.07 | 1.4 ± 0.2 | 3.1 ± 0.1 | 3.7 ± 0.1 |
| SV40 (−225 to +50) | 0.3 ± 0.01 | 0.9 ± 0.03 | 1.1 ± 0.1 | 1.3 ± 0.05 | 1.0 ± 0.05 | 2.7 ± 0.04 | 3.9 ± 0.01 |
| GH (−250 to +58) | 0.1 ± 0.01 | 1.1 ± 0.01 | 1.0 ± 0.05 | 1.0 ± 0.1 | 0.5 ± 0.1 | 1.2 ± 0.01 | 1.5 ± 0.04 |
| TSH (−128 to +8) | 0.12 ± 0.05 | 1.0 ± 0.05 | 1.0 ± 0.02 | 1.4 ± 0.12 | 0.4 ± 0.01 | 1.0 ± 0.1 | 0.7 ± 0.05 |
Basal-level CAT activity derived from each reporter construct in the absence of transfected expression plasmid was set as 1.0 and used to calculate relative CAT activity in the presence of transfected expression plasmid. The data were obtained from cotransfection experiments with the indicated CAT reporter constructs (10 μg) and CMV expression plasmids (2 μg). Reporter activities of the HIV-1 LTR, TK, and SV40 constructs were measured in HeLa cells, while those of the GH and TSH constructs were measured in the pituitary cell line GH3 cells as described in other studies (22, 23). Averages and standard deviations were determined from three independent transfections.
hnRNP U inhibits Pol II elongation.
One reason for the repression shown in Table 1 might be the inhibition of basal transcription by hnRNP U. The effect of hnRNP U on transcription was examined in the HIV-1 LTR transcription system as a model. For the in vitro transcription assays shown in Fig. 1C, HeLa nuclear extract was depleted of hnRNP U, and the HA-HN immunopurified from transfected HeLa cells (HA-U in Fig. 1C) or the highly purified recombinant Strep-U (Fig. 1C) was added back. Conventional immunodepletion with anti-hnRNP U resulted in coimmunoprecipitation of Pol II holoenzyme (see Fig. 3C), and the depleted extract by this method did not support transcription efficiently (data not shown). For this reason, we depleted hnRNP U by incubating the nuclear extract with heparin-agarose prior to immunodepletion with anti-hnRNP U by taking advantage of the ability of hnRNP U to bind heparin with high affinity (40). Since heparin-agarose chromatography is frequently used in purification of transcription complexes, it was likely that the combination of heparin-agarose and anti-hnRNP U would successfully remove hnRNP U from nuclear extract without compromising the transcription activities of the nuclear extract. In Fig. 2B, immunoblot analysis indicated that hnRNP U was removed to levels below detection (lanes 1 to 3) following a combination treatment of heparin-agarose plus immunodepletion, whereas Pol II holoenzyme components such as Rpb-1 (Pol II), Rap74 (TFIIF), and p62 (TFIIH) or some other component of hnRNP particles such as hnRNP A1 (40) that had been reported to be resistant to the heparin treatment were retained in the nuclear extract (data not shown).
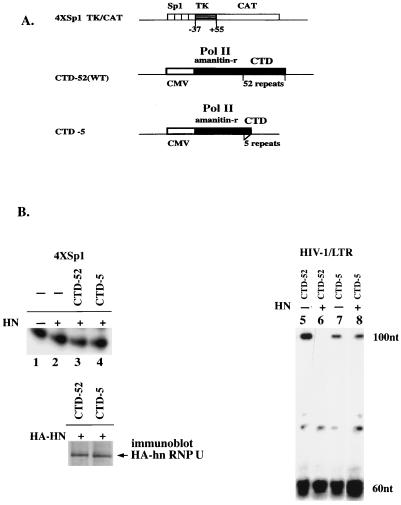
FIG. 2.
hnRNP U-mediated block of elongation requires the CTD of Pol II. (A) The 4×Sp1 reporter and expression vectors for α-amanitin-resistant mutant of the Pol II largest subunit with CTD-52 and CTD-5. (B) Role of CTD in hnRNP U-mediated inhibition of Pol II elongation. HeLa cells (5 × 106 cells) were transfected with the 4×Sp1 or HIV-1 LTR reporter (10 μg) with or without expression vectors for hnRNP U (HN) (2 μg) and α-amanitin-resistant mutants of Pol II (CTD-52 and CTD-5; 10 μg of each). α-Amanitin (2.5 μg/ml) was added after 12 to 18 h of transfection, and the cells were incubated for an additional 48 h. Cytoplasmic RNAs were obtained and analyzed by RNase protection assay. For RNAs transcribed from the 4×Sp1 promoter (lanes 1 to 4), the 200-nt probe containing two Sp1 sites, TK promoter (−37 to +55), and the first 45 bp of the CAT gene was used to protect the 100-nt fragment. The levels of exogenously expressed hnRNP U in the α-amanitin-treated cells were monitored by measuring the transfected HA-HN level in cells treated in the same manner as those in lanes 3 and 4 (immunoblot HA-hn RNP U). For the RNase protection assays in lanes 5 to 8 probe A (Fig. 1A) was used. Relative levels of the nonprocessive transcripts as percentages of the total protected RNA are indicated (lanes 5 to 8, 6, 0, 2, and 1.5).
When the HIV-1 promoter is transcribed, two types of transcription complexes are observed: a processive type that results in full-length transcripts and the nonprocessive type that yields short transcripts consisting of the stable TAR RNA stem-loop (11). In RNase protection assays with two antisense probes (Fig. 1A), transcription in the absence of Tat, using the undepleted HeLa nuclear extract (Fig. 1C, lanes 1 and 7), gave rise to short (∼60-nucleotide [nt]) transcripts and elongated transcripts that protected a 100-nt fragment with probe A or a 200-nt fragment with probe B. The hnRNP U-depleted extract stimulated processive transcription (100 and 200 nt) but not nonprocessive transcription (60 nt) (Fig. 1C, lanes 2 and 8), while the extract depleted of hnRNP A1 by anti-hnRNP A1 (data not shown) contained the same levels of transcription as with the undepleted extract (lanes 1 and 7). Since it is not possible to produce a heparin-agarose-treated control extract without removing hnRNP U, it was not clear whether the observed stimulation in lanes 2 and 8 was due at least in part to the removal of an unknown inhibitor(s). However, add-back of increasing amounts of HA-HN (lanes 3 to 5) or Strep-U (lanes 9 to 11) to the hnRNP U-depleted extract resulted in repression of the processive transcription, whereas similar experiments with the HA-hnRNP A1 or Strep-hnRNP A1 did not show such an effect (data not shown). These results suggested that hnRNP U can repress Pol II elongation in vitro. In further support of this view, preincubation of the recombinant hnRNP U proteins with anti-hnRNP U (lanes 6 and 12) but not with control antibodies such as anti-hnRNP A1 or -Oct-1 (data not shown) abolished the repressive effect, indicating that hnRNP U, but not the contaminating proteins in the recombinant preparations, is responsible for the repressor activities.
To date, we have not been able to express a full-length recombinant hnRNP U in bacteria or in the in vitro translation system. Although obtained from two different sources, both HA-HN and Strep-U used for Fig. 1C contained hnRNP U, as confirmed by immunoblotting (data not shown), and their transcription activities were indistinguishable in our in vitro transcription assays. However, yeast cells expressing Strep-U were difficult to grow, and the yeast recombinant hnRNP U was very unstable. For these reasons, we chose to use the HA-HN proteins from HeLa cells for further assays.
A similar effect on Pol II processivity was seen in vivo (Fig. 1D). In the absence of Tat expression, overexpression of hnRNP U had no effect on nonprocessive transcription (60 nt) (Fig. 1D, lanes 1 to 3), but it inhibited processive transcription (100 and 200 nt) in a dose-dependent manner (lanes 1 to 3, panels I and II). Tat-activated processive transcription (100 nt; compare lanes 5 and 2) occurred in the presence of overexpressed hnRNP U. This release of the hnRNP U-mediated repression by Tat activation was consistently observed even in the presence of increasing amounts of transfected hnRNP U expression vector (up to 10 to 20 μg; data not shown). Because a direct interaction between Tat and hnRNP U was not observed in the in vitro GST pull-down assay (21a), it is unlikely that the effect of Tat shown in lane 5 resulted from blocking or titrating hnRNP U with Tat. The hnRNP U effect was specific for Pol II, since Pol III-dependent VA1 transcription was not affected (Fig. 1D, lower panels).
The hnRNP U-mediated block to elongation requires the CTD of Pol II.
One of the mechanisms by which hnRNP U may block elongation is to inhibit CTD phosphorylation. Previous in vivo studies have shown that transcription from many promoters is sensitive to CTD truncation. However, transcription activation by Sp1 does not depend on CTD (54, 55), and transcription from an enhancerless promoter such as 4×Sp1-TK/CAT (hereafter called 4×Sp1) (Fig. 2A), consisting of a TATA box and four Sp1-binding sites, has been shown to be CTD independent in mammalian cells, including HeLa cells (7, 18). If hnRNP U blocks Pol II elongation by inhibiting CTD phosphorylation, CTD-independent transcription such as that from the 4×Sp1 promoter may not be affected by hnRNP U. To assess the CTD requirement in hnRNP U-mediated transcription repression, we used an approach developed by Gerber et al. (18), which relies on the efficient expression of α-amanitin-resistant mutants of the large subunit of Pol II with different numbers of CTD repeats (Fig. 2A). α-Amanitin treatment of cells transfected with these constructs results in inhibition of endogenous Pol II such that subsequent transcription depends on the exogenously expressed resistant mutant. To confirm that transcription from 4×Sp1 promoter in HeLa cells is CTD independent (7, 18), we transfected cells with the 4×Sp1 reporter and an expression vector for an α-amanitin-resistant mutant with either 52 (wild-type; CTD-52) or 5 (CTD-5) repeats in the CTD (Fig. 2A). The effect of the residual endogenous Pol II activity that might have escaped α-amanitin inhibition was assessed as described previously (7) by including a control transfection with the 4×Sp1 reporter and pUC19 without α-amanitin-resistant mutants. Transcription from the 4×Sp1 promoter was not affected by CTD truncation, and the transcription signal was absent in control cells cotransfected with pUC19 (data not shown).
We then tested whether hnRNP U overexpression would affect transcription from the 4×Sp1 promoter. Results of RNase protection assays indicate that hnRNP U does not inhibit transcription from this promoter (Fig. 2B, lanes 1 and 2). Similar results were obtained for cells transfected with increasing amounts of hnRNP U expression vector (up to 10 μg), suggesting that this resistance is not due to an insufficient amount of exogenously expressed hnRNP U (data not shown). CTD truncation and the overexpression of hnRNP U also did not affect transcription from the 4×Sp1 promoter when cells were treated with α-amanitin following transfection with the reporter and expression vectors for Pol II mutants and hnRNP U (lanes 3 and 4). To rule out the possibility that the absence of repression in elongation in the CTD-5-expressing cells is due to an insufficient amount of the exogenously expressed hnRNP U, the levels of hnRNP U expression were monitored in cells transfected with the expression vector for HA-HN (pCMV/HA-hnRNP U) and treated in the same manner as those in lanes 3 and 4. The results of immunoblotting with anti-HA (Fig. 2B) indicated that the CTD-52- and CTD-5-expressing cells expressed similar levels of HA-HN, in agreement with a previous report that transcription from the CMV promoter is not sensitive to CTD truncation (5). Overall, the results in Fig. 2B suggest that hnRNP U does not inhibit CTD-independent transcription from the 4×Sp1 promoter.
The CTD requirement in hnRNP U-mediated repression was further examined in cells transfected with the HIV-1 LTR reporter and the α-amanitin-resistant mutants of Pol II (Fig. 2B, lanes 5 to 8). As shown in other studies (7, 36), the level of processive transcription (100 nt), but not nonprocessive transcription (60 nt), was reduced upon CTD truncation (compare lanes 5 and 7). When hnRNP U was overexpressed in the presence of the wild-type CTD-52 (lane 6), only processive transcription (100 nt) was inhibited (compare lanes 5 and 6), consistent with the results in Fig. 1D. When the CTD-5 construct was used (lane 8), however, the low level of processive transcription (100 nt) was resistant to hnRNP U as was nonprocessive transcription (60 nt) (compare lanes 7 and 8), indicating that CTD-dependent transcription of the HIV-1 LTR is sensitive to hnRNP U. The simplest hypothesis suggested by the results in Fig. 2 is that the hnRNP U-mediated block to elongation may require the CTD of Pol II.
hnRNP U copurifies with Pol II holoenzyme in vivo.
If hnRNP U functions as a transcription repressor, because hnRNP U is not a sequence-specific DNA-binding protein, it may be recruited to the promoter through protein-protein interactions. The possible association of hnRNP U with Pol II holoenzyme or TFIID in vivo has been examined. The Pol II holoenzyme was immunoprecipitated with anti-Rap74 (TFIIF) antibody (Fig. 3A) followed by gel filtration as described by Maldonado et al. (28) (Fig. 3B). Western blotting of the eluate from the anti-Rap74 IP (Fig. 3A) showed retention of Pol II (Rpb-1), hnRNP U, TFIIF (Rap74), TFIIH (Cdk7), and Cdk8. As previously reported (9, 28), however, transcription factors such as TBP, Sp1 (Fig. 3A), and TFIIB or Oct-1 (data not shown) were not detected. Similarly, unlike the eluates from the anti-Rap74 IP, those from the anti-Oct-1 IP (Fig. 3A) or anti-Sp1 IP (data not shown) did not contain hnRNP U, suggesting a possible interaction of hnRNP U with the Pol II holoenzyme complex. The addition of DNase or RNase to the immunoprecipitation reaction did not affect the results, indicating that hnRNP U was not artificially bound to the holoenzyme by contaminating DNA or RNA (data not shown). Fractionation of the anti-Rap74 IP eluate by gel filtration showed that hnRNP U copurifies with Pol II, TFIIF (Rap74), and TFIIH (Cdk7) with an apparent molecular mass of greater than 2 MDa (Fig. 3B). hnRNP A1, another component of hnRNP particles, was not detected, however. This rules out the possibility that the presence of hnRNP U in the fractions containing holoenzyme is due to the contaminating hnRNP particles comigrating with the holoenzyme.
The possible association of hnRNP U with Pol II holoenzyme was further suggested by the observation that eluates from the anti-hnRNP U IP contained Pol II, TFIIF, Cdk7, and hnRNP U but not TBP (Fig. 3C). In contrast, the TFIID complex immunoprecipitated with anti-TBP did not contain hnRNP U but did contain known components of TFIID such as TAF-250, TAF-130, and TBP (Fig. 3D). To determine if the Pol II-containing complex immunopurified with anti-hnRNP U in Fig. 3C was competent for transcription, the anti-hnRNP U IP immobilized on washed beads was added to the transcription reaction (Fig. 3E). Specific transcription depended on the addition of recombinant TBP and TFIIB (lane 4) and was sensitive to α-amanitin (data not shown). In contrast, no transcription was observed with the IPs generated by anti-hnRNP A1 (data not shown). These results suggest that hnRNP U may be recruited to promoters through its association with Pol II holoenzyme. How hnRNP U is incorporated into holoenzyme and what fraction of holoenzyme is associated with hnRNP U in vivo remain unknown.
hnRNP U inhibits the CTD phosphorylation by TFIIH in vitro.
The results in Fig. 2 suggest that hnRNP U can inhibit CTD phosphorylation. One of the cellular targets of hnRNP U action could be a CTD kinase. Among a large number of kinases capable of phosphorylating the CTD in vitro, the targets for hnRNP U may be those in the PICs such as TFIIH-Cdk7 or -Cdk8 (13, 21, 25). Alternatively, hnRNP U may target CTD kinases that are not associated with the Pol II complex such as PITALRE (Cdk9), a catalytic subunit in the elongation factor P-TEFb complex that has been shown to play a role in productive elongation (29, 31, 56). These three CTD kinases have been widely postulated to play a role in CTD phosphorylation and elongation in vivo.
The effect of hnRNP U on CTD phosphorylation by these kinases was assayed on GST-CTD by using HA-HN (Fig. 4). The immunopurified kinase preparations used in these experiments did not contain any contaminating hnRNP U (data not shown). All three kinases phosphorylated GST-CTD (Fig. 4A, lanes 1 and 4, and B, lanes 1 and 5), as reported in other studies (10, 21, 38, 56). This phosphorylation was blocked when each kinase was pretreated with the corresponding antibody and the antibody-specific blocking was released in the presence of the antigenic peptide (for TFIIH, Fig. 4A, lanes 2 and 3; for other kinases, data not shown), indicating that contaminating kinases were not responsible for the phosphorylation shown. When HA-HN was added to the TFIIH-mediated phosphorylation reaction, 32P incorporation into both hypo- and hyperphosphorylated forms (GST/CTD-A and GST/CTD-O) was inhibited in a dose-dependent manner (lanes 4 to 7). As with CTD, hnRNP U inhibited phosphorylation of TBP, another substrate of TFIIH (lower panel, lanes 4 to 7). Preincubation of the immunopurified HA-hnRNP U with anti-hnRNP U (lane 8) but not with control antibodies such as anti-hnRNP A1 and anti-Oct-1 (data not shown) effectively neutralized the inhibition, indicating that contaminating kinase inhibitor activities or phosphatases in the HA-HN preparation are not responsible for the observed inhibition. Furthermore, if HA-HN was added to the reaction 1 h after the start of the kinase reaction, TFIIH-mediated phosphorylation was not inhibited (data not shown). In contrast to the effect on TFIIH, Cdk8 and PITARLE (Cdk9) activities (Fig. 4B) were not affected by the amount of HA-HN that completely inhibited the TFIIH-mediated reaction. These results indicate that hnRNP U specifically inhibits TFIIH-associated kinase and that this inhibition is not due to phosphatase activities. We then tested whether a transactivator such as Tat that has been reported to bind the Cdk7 subunit of TFIIH to activate TFIIH (10) might be able to release the inhibitory effect of hnRNP U. Interestingly, the inhibitory effect of hnRNP U on the TFIIH-mediated phosphorylation in vitro was neutralized by Tat (Fig. 4A, lanes 9 to 11). Further biochemical studies are required to elucidate the mechanism for this neutralization by Tat whether Tat might change conformation of TFIIH or compete with hnRNP U for binding to the TFIIH complex.
The hnRNP U-mediated inhibition in CTD phosphorylation in Fig. 4A can be attributed to many different reasons. One possibility is that hnRNP U, by binding to the CTD, sterically hinders its phosphorylation. However, a direct interaction between CTD and hnRNP U was not detected in GST pull-down assays (data not shown). Further, the result in Fig. 4B that hnRNP U did not inhibit the Cdk8- or PITARLE (Cdk9)-mediated CTD phosphorylation makes this possibility unlikely. This result also rules out the possibility that hnRNP U inhibits the kinase reaction by binding ATP nonspecifically. The second possibility is that hnRNP U competes with CTD as a substrate for TFIIH kinase. This is unlikely because 32P incorporation into hnRNP U (>120 kDa) was not detected in the in vitro phosphorylation reaction (Fig. 4A). The third possibility is that hnRNP U disrupts the assembly of TFIIH. If this is the case, however, it is unlikely that Tat would neutralize the effect of hnRNP U (Fig. 4A, lanes 9 to 11). Moreover, the observation that TBP phosphorylation by TFIIH was also inhibited by hnRNP U (Fig. 4A, lanes 4 to 7) suggests that hnRNP U inhibits TFIIH activity rather than sterically hindering CTD phosphorylation sites on TFIIH. The fourth possibility is that hnRNP U interacts with TFIIH (see Fig. 7) and possesses TFIIH-specific kinase inhibitor activities.
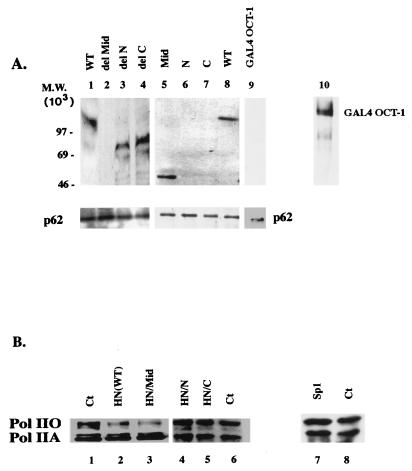
FIG. 7.
HnRNP U can bind TFIIH and inhibit CTD phosphorylation in vivo. (A) Coimmunoprecipitation of endogenous TFIIH complex with HA-HN proteins (lanes 1 to 8). HeLa cells were transfected with 10 μg of each expression vector, and the nuclear extract was processed as described in the text. Retention of HA-HN proteins in the TFIIH complex was monitored by immunoblotting with anti-HA. For the negative control, cells were transfected with the vector for GAL4–Oct-1, and the TFIIH complex in the GAL4–Oct-1 containing nuclear extract was isolated as described above. GAL4–Oct-1 was abundantly expressed in cells, as detected by immunoblotting with anti-GAL4 (lane 10). In the TFIIH complex, however, GAL4–Oct-1 was not present (lane 9). (B) HnRNP U inhibits CTD phosphorylation in vivo. HeLa cells (5 × 106) were transiently transfected with 15 μg of expression vectors as indicated. Whole-cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-CTD 8WG16. Ct, control (whole-cell lysate from untransfected cells).
hnRNP U is recruited to the promoter in the form of a PIC and released from elongating Pol II.
Pol II exists in two forms in cells, IIA and IIO. Although hnRNP U is a component of Pol II holoenzyme (Fig. 3) and the CTDs in the holoenzyme remain hypophosphorylated, these results do not necessarily indicate that hnRNP U is exclusively associated with Pol IIA in vivo. Because Pol IIO is predominantly associated with elongating complexes and discarded with chromatin during nuclear extract preparation (47), it was difficult to detect in nuclear extract. As reported in other studies (37), however, both IIO and IIA forms were detected in whole-cell lysate obtained from HeLa cells (Fig. 5A, lane 1). When the lysate was immunoprecipitated with anti-hnRNP U and immunoblotted with anti-CTD antibody (lane 2), only Pol IIA was detected, indicating that hnRNP U mainly associates with the nonprocessive form of Pol II in vivo.
If Pol IIO is derived from Pol IIA as currently thought, these results imply that hnRNP U dissociates from Pol IIA during or after CTD phosphorylation. To test this hypothesis, the association of hnRNP U with Pol II was monitored during different stages of transcription in vitro on the immobilized TK (−105 to +55) template. The HIV-1 LTR template was not used, because HIV-1 transcription results in a mixture of paused (IIA) and elongating (IIO) complexes that are difficult to isolate separately. A PIC was formed on the 5′-biotinylated TK template with HeLa nuclear extract (without NTP), and the template was immobilized by binding to streptavidin-coated magnetic beads. The DNA templates containing the transcription complexes were released from magnetic beads by restriction enzyme digestion, and the resulting transcription complexes were immunoprecipitated with anti-Rap74 or anti-TBP antibody (Fig. 5B, lanes 1 and 2). Components of the holoenzyme and TFIID including TFIIH (Cdk7), TFIIF (Rap74), TBP, and hnRNP U were present in both IPs, indicating that hnRNP U is recruited to the promoter with holoenzyme (Fig. 3) and incorporated into the PIC. When the transcription reaction was performed with ATP alone (lanes 3 and 4) or with ATP and CTP, which would allow formation of the first phosphodiester bond from the TK promoter (data not shown), hnRNP U and Cdk7 were still present in the IPs with anti-Rap74 and anti-TBP antibodies. When NTP was added to allow transcription elongation (Fig. 5B, lanes 5 and 6), the anti-TBP antibody could no longer coimmunoprecipitate Pol II with TBP (TFIID) and the anti-Rap74 could not immunoprecipitate TBP (TFIID) with Pol II, indicating that Pol II had left the initiation complex. When the hyperphosphorylation of Pol II was monitored with [γ-32P]ATP in the reaction containing all NTPs, the anti-Rap74 IP (lane 5, bottom) but not the anti-TBP IP (lane 6, bottom) contained Pol IIO, further confirming that Pol II was released from the initiation complex. At this stage of transcription, hnRNP U and Cdk7 were not detected in the anti-TBP- and anti-Rap74 IPs (lanes 5 and 6). These results suggest that hnRNP U and TFIIH dissociate from the initiation complex prior to productive elongation and that the release of hnRNP U and TFIIH requires transcription.
The middle domain of hnRNP U is sufficient to mediate its Pol II holoenzyme association and its inhibition of the TFIIH kinase and Pol II elongation.
HN(WT) has a modular structure, as indicated in Fig. 6A. The N-terminal domain (acidic and glutamine rich) is important for interaction with nuclear matrix and chromatin, while the RGG box-containing C-terminal domain is important for interaction with other hnRNP proteins to form hnRNP particles. To determine which domain of hnRNP U is essential for the different properties of hnRNP U described in this study (Pol II holoenzyme association, inhibition of the TFIIH kinase, and elongation), HA-HN proteins indicated in Fig. 6A were expressed in HeLa cells. In Fig. 6B (lanes 1 to 4 and 10 to 12), all HA-HN proteins were expressed efficiently in transfected cells. Pol II holoenzyme was isolated from cells expressing each HA-HN protein by immunoprecipitation with anti-Rap74. Because these preparations contain the mixture of Pol II holoenzyme complexes from transfected and untransfected cells, to enrich the population with the complexes containing HA-HN proteins, the anti-Rap 74 IPs were released and reprecipitated with anti-HA antibody. The Pol II holoenzymes released from the anti-HA IPs were tested for the presence of HA-HN proteins. As shown in Fig. 6B (lanes 5 to 8 and 14 to 16), only the HA-HN proteins containing the middle domain [HN(WT), HN/del N, HN/del C, and HN/Mid] were present in Pol II holoenzyme (lanes 5 to 8), indicating that the middle domain mediates the Pol II holoenzyme association. The presence of Rpb-1 (the largest subunit of Pol II) in the anti-Rap74 IPs was confirmed by immunoblotting with anti-CTD antibody (lower panels, lanes 5 to 8 and 14 to 16). As expected, in the final Pol II holoenzyme preparation released from the anti-HA IPs, Rpb-1 was present only in the complexes containing the HA-HN proteins with the middle domain (data not shown).
We then tested which domain is important for the hnRNP U-mediated inhibition of TFIIH kinase activity. When the immunopurified HA-HN proteins were added to the TFIIH kinase reaction as described in Fig. 4, hnRNP U with the middle domain deletion (HN/del Mid) and the truncated protein containing the N or C terminus only (HN/N or HN/C) showed no inhibition (Fig. 6C, lanes 2 to 4), whereas the middle domain-containing mutants (HN/Mid, HN/del N, and HN/del C) all showed inhibition (lanes 5 to 7), indicating that the middle domain inhibits the TFIIH kinase.
The middle domain was also sufficient for the inhibition of Pol II elongation from the HIV-1 LTR in vitro (Fig. 6D) and in vivo (Fig. 6E). When various HA-HN proteins were added back to the in vitro transcription reaction using the HeLa nuclear extract depleted of hnRNP U (Fig. 6D), the middle domain-containing mutants (lanes 7 to 9), but not the mutants lacking the middle domain (lanes 4 to 6), restored elongation inhibition. Inhibition of elongation was also dependent on the middle domain in cells transfected with the expression vectors for the various HA-HN proteins (Fig. 6E).
These results indicated that the middle domain of hnRNP U is sufficient for interaction with Pol II holoenzyme and for inhibition of TFIIH kinase and Pol II elongation, a function that has not been described previously for any protein.
hnRNP U can interact with TFIIH and inhibit CTD phosphorylation in vivo.
Next, we tested if HA-HN could copurify with the endogeneous TFIIH in vivo. To obtain TFIIH that contains HA-HN proteins, the nuclear extract used in Fig. 6 was first immunoprecipitated with anti-p62, and the released TFIIH complexes from the anti-p62 IPs were reprecipitated with anti-HA. Immunoblotting of the resulting TFIIH complexes released from the anti-HA IP showed that HA-HN(WT) and only HA-HN proteins containing the middle domain were retained (lanes 1 to 8). These results indicated that hnRNP U may interact with TFIIH directly or indirectly in vivo and that the middle domain is sufficient to mediate this interaction (Fig. 7A). When the anti-p62 IP (first immunoprecipitation) was subjected to immunoblotting, p62, but not Rap74 or Rpb-1 (data not shown), was detected in all lanes, confirming that TFIIH, not Pol II holoenzyme, was immunoprecipitated. To confirm the specificity of the interaction of hnRNP U with TFIIH, we transfected HeLa cells with the expression vector for GAL4–Oct-1 (GAL4 DNA-binding protein fused to the N terminus of Oct-1) and immunopurified TFIIH from GAL4–Oct-1-containing nuclear extract in a manner similar to that described above. GAL4–Oct-1 was efficiently expressed in cells, as detected by immunoblotting with anti-GAL4 (lane 10), but was not detected in the TFIIH preparation after the first immunoprecipitation with anti-p62 (lane 9). As expected, GAL4–Oct-1 was not detected when the anti-p62 IP was reprecipitated with anti-GAL4 (data not shown). Consistently, the same pattern of binding was observed when the HA-HN proteins were incubated with the anti-p62-purified TFIIH complex in vitro (data not shown). To generate a stable TFIIH complex associated with hnRNP U in vivo, however, it may be necessary to use different purification schemes for the TFIIH complex such as biochemical purifications or immunopurification with antibodies against a different subunit of TFIIH. To date, hnRNP U has not been detected in the immunopurified TFIIH complex with anti-p62, possibly because of a transient interaction between hnRNP U and TFIIH in vivo.
To test whether the inhibition of TFIIH kinase activity by hnRNP U and the ability of hnRNP U to associate with Pol II holoenzyme and TFIIH are correlated to hypophosphorylation of the CTD in vivo, HeLa cells were transfected with expression vectors for HA-HN(WT) and HA-HN/Mid, and the levels of Pol IIA and Pol IIO in whole-cell lysates were compared with those in nontransfected cells by immunoblotting (Fig. 7B). As expected, the Pol IIO form was substantially decreased in lysates from cells expressing HA-HN proteins, which can inhibit TFIIH kinase activity and associate with Pol II, HA-HN(WT), and HA-HN/Mid (lanes 2 and 3). In contrast, cells expressing HA-HN/N, HA-HN/C, or Sp1 (lanes 4 to 8) showed no difference from the control (lanes 5 and 8).
Overall, this study shows correlative evidence linking hnRNP U-mediated inhibition in CTD phosphorylation by TFIIH to the hnRNP U-mediated repression in Pol II elongation. Together, these results suggest that a subfraction of hnRNP U is recruited to the Pol II holoenzyme, where it appears to inhibit CTD phosphorylation by downregulating TFIIH and may thereby repress Pol II elongation. Although it remains unknown how hnRNP U might regulate TFIIH, our preliminary results suggest that hnRNP U specifically interacts with Cdk7 but not with other subunits of TFIIH in vitro (21a). Detailed mutational analyses of the middle domain of hnRNP U and the Cdk7 subunit are under way to begin to understand the possible mechanisms.
DISCUSSION
This study reports a new role for hnRNP U: downregulation of CTD phosphorylation and inhibition of Pol II elongation. We showed that a fraction of hnRNP U is associated with the Pol II holoenzyme in vivo and is recruited to the promoter as part of a PIC. hnRNP U appears to dissociate from the Pol II complex at the early stage of transcription and is therefore absent from the elongating complex. The results shown in this study suggest that hnRNP U, as a component of the Pol II holoenzyme, may inhibit TFIIH-mediated CTD phosphorylation and repress Pol II elongation. Although tested with a limited number of promoters, these findings may have wider applications for Pol II transcription. Overall, this study identifies, for the first time, an elongation inhibitor associated with the Pol II holoenzyme.
HnRNP U confers a negative elongation potential to the basal transcription machinery.
Although the role of the TFIIH-mediated CTD phosphorylation in transcription has not been clearly defined, previous studies suggest that it may be important for the processivity of Pol II. For example, the long-transcript production but not the short-transcript production from the HIV-1 LTR requires CTD and TFIIH (10). Together with the results of antibody injection experiments (52) where antibodies against each subunit of TFIIH injected into Xenopus oocytes selectively repressed long-transcript production from the coinjected HIV-1 LTR promoter, these findings indicated that long-transcript production from the HIV-1 LTR requires TFIIH. Considering that TFIIH is released from the transcription complex after a synthesis of short transcripts (53), it is difficult to explain how TFIIH selectively affects the long-transcript production that occurs after the release of TFIIH. One possibility is that the CTD phosphorylation by TFIIH does not have a major role in the early stage of transcription but is necessary to establish an elongation-competent form of Pol II (12). Indeed, several studies proposed that the early stage CTD phosphorylation by TFIIH is coupled to the functional transition to productive elongation (29, 56). In this manner, TFIIH would be able to influence the rate of processive elongation even after its release from the transcription complex, and accordingly, hnRNP U would be able to regulate Pol II processivity by regulating TFIIH. If hnRNP U controls Pol II elongation by inhibiting TFIIH kinase activities, it likely would not inhibit short-transcript production from the HIV-1 LTR or transcription from the 4×Sp1 promoter, that does not require CTD or the kinase activities of TFIIH.
The hnRNP U-mediated inhibition of TFIIH may be one of the mechanisms by which the CTD remains unphosphorylated in the Pol II holoenzyme despite its presence in the complex. Inhibition of TFIIH is likely critical for transcription initiation, as the hyperphosphorylated Pol IIO cannot enter into a PIC. However, the removal of hnRNP U does not affect initiation but elongation of the HIV-1 transcripts, suggesting that CTD phosphorylation at the early stage of transcription may require at least two different signals, one to derepress TFIIH by inactivating its inhibitors such as hnRNP U and the other to activate TFIIH. Transcription activators may remove hnRNP U and/or provide signals to inactivate hnRNP U. It remains to be determined whether a transcription factor such as Tat, retinoic acid receptor alpha, p53, or VP16 that binds TFIIH or Cdk7 (3, 9, 10, 17, 41, 50) would affect the interaction of hnRNP U with TFIIH in vivo. In the presence of an activating signal, the negative effect of hnRNP U on TFIIH would be neutralized, leading to derepression and activation of TFIIH to stimulate CTD phosphorylation in the basal transcription machinery. During the early stage of CTD phosphorylation, both hnRNP U and TFIIH appear to dissociate from Pol II and are most likely recycled for the next round of transcription. As a phosphoprotein that can be heavily phosphorylated in vivo (14), hnRNP U may be able to respond to various phosphorylation signals during the transcription cycle.
The importance of TFIIH in Pol II elongation.
It has been difficult to establish which of the CTD kinases plays a role in the in vivo phosphorylation of the CTD and subsequent elongation control, because doing so requires examination of Pol II phosphorylation in cells where specific CTD kinases have been inactivated. Given that CTD has multiple phosphorylation sites, the extent of phosphorylation may be differentially regulated according to the stages of transcription by distinct CTD kinases. Previous studies suggested that in vitro, TFIIH possesses a level of CTD kinase activities similar to that of P-TEFb, which is important for transition to productive elongation (32). Nevertheless, it is thought that the level of CTD phosphorylation by TFIIH in the Pol II complex in vivo is lower than that in the hyperphosphorylated CTD in the elongating complex (56). One of the functions of hnRNP U in the transcription machinery may be to control the level of CTD phosphorylation by modulating TFIIH kinase activities during the early stage of transcription. Such a tight regulation on TFIIH may be critical for productive elongation. If PITARLE (Cdk9), which is thought to be recruited to the promoter during the CTD hyperphosphorylation, is required for productive elongation as some studies have proposed (29, 31, 56), the results in this study suggest that TFIIH that phosphorylates the CTD before the recruitment of PITARLE (Cdk9) is necessary, although possibly not sufficient, for productive elongation. As has been proposed in other studies (29, 56), the TFIIH-mediated phosphorylation of CTD in the basal transcription machinery may be a prerequisite for the productive elongation that may involve other CTD kinases.
The hnRNP U-mediated block of elongation is a feature of higher organisms.
To date, an hnRNP U homologue has not been found in yeast or Drosophila, which may reflect fundamental differences in transcription activation and elongation mechanisms between mammals and lower eukaryotes. For example, transcription activation in yeast is achieved primarily by acidic activators, whereas a variety of activators are used in higher organisms (45). Yeast Pol II cannot substitute human Pol II in the reconstituted in vitro transcription (8), although yeast and human TBPs are interchangeable (6). Further, Pol II from higher eukaryotes shows frequent pausing and arrest in the in vitro transcription analyses, while yeast Pol II in vitro exhibits close to in vivo elongation rates (30, 44, 46). Mammalian Pol II might use additional mechanisms to regulate CTD phosphorylation and elongation due to the additional number of repeats in its CTD. hnRNP U-mediated elongation control may represent one of those mechanisms.
Potential diverse biological roles of hnRNP U.
The new role of hnRNP U described in this study, as a Pol II elongation inhibitor, underscores the diversity of roles that this class of proteins may play in a cell. Involvement of hnRNP proteins in transcription regulation is not unprecedented. Previous studies (34, 48) reported that hnRNP K is recruited to the c-myc gene promoter through a protein-protein interaction with TFIID to activate this gene (34). Further, a recent study reported that hnRNP U interacts with a ligand-dependent transcription factor, glucocorticoid receptor, suggesting a possible role of hnRNP U in the transcription of genes regulated by steroid hormones (15).
The modular structure of hnRNP U appears to be essential for its possible diverse roles. hnRNP U associates with the nuclear matrix or chromatin through its N terminus, while it forms hnRNP particles through its RGG box in its C terminus to participate in pre-mRNA processing with other hnRNP proteins. This study identifies yet another domain, the middle domain, through which hnRNP U interacts with the basal transcription machinery and inhibits Pol II elongation. That this may be mediated through the inhibition of the TFIIH-mediated CTD phosphorylation suggests that hnRNP U may have other important functions yet to be discovered. For example, the catalytic subunit of TFIIH, Cdk7, is also present in free Cdk-activating kinase, which has been proposed to regulate cell cycle progression (42, 51). These observations raise the interesting question of whether hnRNP U is able to regulate the function of free Cdk-activating kinase and as a result could be connected to cell-cycle regulation. Further studies about the mechanism by which hnRNP U regulates transcription and/or chromatin structure may shed more light on a variety of cellular processes.
ACKNOWLEDGMENTS
The cDNA clone for human hnRNP U and anti-hnRNP U monoclonal antibody 3G6 were generous gifts from G. Dreyfuss. We thank J. L. Corden, W. S. Dynan, K. T. Jeang, S. Y. Kim, D. Levens, R. Roeder, and J. McKlasky for sharing plasmids and antibodies and P. Hsieh and S. Simons for reading the manuscript.
REFERENCES
- 1.Akhtar A, Faye G, Bentley D L. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- 2.Akoulitchev S, Makel T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 3.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown A L, Lee C-H, Schwarz J K, Mitiku N, Piwnica-Worms H, Chung J H. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buermeyer A B, Thompson N E, Strasheim L A, Burgess R R, Farnham P J. The HIP1 initiator element plays a role in determining the in vitro requirement of the dihydrofolate reductase gene promoter for the C-terminal domain of RNA polymerase II. Mol Cell Biol. 1992;12:2250–2259. doi: 10.1128/mcb.12.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavallini B, Huet J, Plassat J L, Sentenac A, Egly J-M, Chambon P. A yeast activity can substitute for the HeLa TATA box factor. Nature. 1988;334:77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- 7.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B P, Strubin M, Ponticelli A S, Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991;65:341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- 9.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen B R. Regulation of HIV gene expression. AIDS. 1995;9(Suppl. A):19–32. [PubMed] [Google Scholar]
- 12.Dahmus M E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 13.Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:506–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 14.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 15.Eggert M, Michel J, Schneider S, Bornfleth H, Baniahmad A, Fackelmayer F O, Schmidt S, Renkawitz R. The glucocorticoid receptor is associated with the RNA-binding nuclear matrix protein hnRNP U. J Biol Chem. 1997;272:28471–29478. doi: 10.1074/jbc.272.45.28471. [DOI] [PubMed] [Google Scholar]
- 16.Fackelmayer F O, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP U/SAF-A, a nuclear matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 19.Gohring F, Fackelmayer F O. The scaffold/matrix attachment region binding protein hnRNP U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry. 1997;36:8276–8283. doi: 10.1021/bi970480f. [DOI] [PubMed] [Google Scholar]
- 20.Gohring F, Schwab B L, Nicotera P, Leist M, Fackelmayer F O. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold M O, Tassan J-P, Nigg E A, Rice A P, Herrmann C H. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Kim, M. Unpublished observations.
- 22.Kim M K, McClaskey J H, Bodenner D L, Weintraub B D. An AP-1-like factor and the pituitary-specific factor Pit-1 are both necessary to mediate hormonal induction of human thyrotropin beta gene expression. J Biol Chem. 1993;268:23366–23373. [PubMed] [Google Scholar]
- 23.Kim M K, Lesoon-Wood L A, Weintraub B D, Chung J H. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Mol Cell Biol. 1996;16:4366–4377. doi: 10.1128/mcb.16.8.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 25.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Zawel L, Fischer L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 28.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Inostroza J A, Rickett P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 29.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall N F, Price D H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 32.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 33.Mattern K A, Humbel B M, Muijsers A O, De Jong L, Van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J Cell Biochem. 1996;62:275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 34.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto H, Sheline C T, Corden J, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 38.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 39.Peterson S R, Dvir A, Anderson C W, Dynan W S. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 40.Pinol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 41.Rochette-Egly C, Adam S, Rossignol M, Egly J-M, Chambon P. Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by Cdk7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 42.Rossignol M, Kolb-Cheynel I, Egly J-M. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 44.Stargell L A, Struhl K. Transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:3111–3115. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 45.Struhl K. Transcriptional enhancement by acidic activators. Biochim Biophys Acta. 1996;1288:O15–O17. doi: 10.1016/s0304-419x(96)00029-7. [DOI] [PubMed] [Google Scholar]
- 46.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 47.Svejstrup J Q, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg R D. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 49.Xie Z, Price D. Drosophila factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J Biol Chem. 1997;272:31902–31907. doi: 10.1074/jbc.272.50.31902. [DOI] [PubMed] [Google Scholar]
- 50.Yankulov K, Yamashita K, Roy R, Egly J-M, Bentley D. The transcriptional elongation inhibitor 5,6-dichlor-a-β-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 51.Yankulov K Y, Bentley D L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yankulov K Y, Pandes M, McCracken S, Bouchard D, Bentley D. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zawel L, Kumar P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 54.Zehring W A, Greenleaf A L. The carboxyl-terminal repeat domain of RNA polymerase II is not required for transcription factor Sp1 to function in vitro. J Biol Chem. 1990;265:8351–8353. [PubMed] [Google Scholar]
- 55.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]