Abstract
By selectively eliminating ubiquitin-conjugated proteins, the 26S proteasome plays a pivotal role in a large variety of cellular regulatory processes, particularly in the control of cell cycle transitions. Access of ubiquitinated substrates to the inner catalytic chamber within the 20S core particle is mediated by the 19S regulatory particle (RP), whose subunit composition in budding yeast has been recently elucidated. In this study, we have investigated the cell cycle defects resulting from conditional inactivation of one of these RP components, the essential non-ATPase Rpn3/Sun2 subunit. Using temperature-sensitive mutant alleles, we show that rpn3 mutations do not prevent the G1/S transition but cause a metaphase arrest, indicating that the essential Rpn3 function is limiting for mitosis. rpn3 mutants appear severely compromised in the ubiquitin-dependent proteolysis of several physiologically important proteasome substrates. Thus, RPN3 function is required for the degradation of the G1-phase cyclin Cln2 targeted by SCF; the S-phase cyclin Clb5, whose ubiquitination is likely to involve a combination of E3 (ubiquitin protein ligase) enzymes; and anaphase-promoting complex targets, such as the B-type cyclin Clb2 and the anaphase inhibitor Pds1. Our results indicate that the Pds1 degradation defect of the rpn3 mutants most likely accounts for the metaphase arrest phenotype observed. Surprisingly, but consistent with the lack of a G1 arrest phenotype in thermosensitive rpn3 strains, the Cdk inhibitor Sic1 exhibits a short half-life regardless of the RPN3 genotype. In striking contrast, Sic1 turnover is severely impaired by a temperature-sensitive mutation in RPN12/NIN1, encoding another essential RP subunit. While other interpretations are possible, these data strongly argue for the requirement of distinct RP subunits for efficient proteolysis of specific cell cycle regulators. The potential implications of these data are discussed in the context of possible Rpn3 function in multiubiquitin-protein conjugate recognition by the 19S proteasomal regulatory particle.
Selective degradation of short-lived proteins by the ubiquitin-proteasome machinery has increasingly emerged as a key regulatory mechanism for many essential cellular functions (25). Some of the best examples of this process involve instances of cell cycle control where the removal of negative regulatory factors by ubiquitin-mediated proteolysis provides the biochemical basis for ordered and unidirectional progression through successive cell cycle transitions (8, 24, 34, 42). Cell cycle progression is driven by a family of serine/threonine protein kinases known as cyclin-dependent kinases (Cdk’s) (46). Cdk activation requires association with positive regulatory subunits called cyclins, whose protein levels peak at different phases of the cell cycle. Additionally, Cdk’s are regulated by phosphorylation and binding of inhibitory proteins (39, 41). In budding yeast, there is only one major Cdk, Cdc28, whose sequential activation depends successively on three types of cyclin subunit, the G1 cyclins Cln1 to Cln3, the S-phase cyclins Clb5 and Clb6, and finally the mitotic cyclins Clb1 to Clb4 (40). Genetic studies of this organism have identified three main cell cycle transitions where the regulated proteolysis of key protein targets by different ubiquitin pathways becomes rate limiting (40). First, the Clb/Cdc28 kinase inhibitor Sic1 must be degraded for the S phase to be initiated (52). Next, after the cell has successfully duplicated its genome, proteins controlling chromatid cohesion, such as Pds1 in budding yeast and Cut2 in fission yeast, have to be rapidly destroyed for progression from metaphase to anaphase to occur (6, 16). Finally, inactivation of the mitotic form of Cdc28 kinase via the proteolysis of mitotic cyclins is essential for cytokinesis and exit from mitosis under some circumstances (59).
Polyubiquitination is a multistep process that ultimately results in the covalent attachment of a multiubiquitin chain to specific lysine residues of target proteins (30). The initial step consists of the activation of a ubiquitin monomer by ATP to a high-energy thiol ester intermediate in a reaction catalyzed by a ubiquitin-activating (E1) enzyme. Activated ubiquitin is then transferred to the reactive cysteine residue of a second protein, a ubiquitin-conjugating (Ubc or E2) enzyme. In a last step also catalyzed by the E2 enzyme but often with the help of a third component, a ubiquitin protein ligase (E3 enzyme), the ubiquitin moiety is attached to the target protein, forming an isopeptide bond between its activated C-terminal glycine residue and the ɛ-NH2 group of a lysine residue of the substrate. Generally, substrate specificity in the ubiquitination reaction is conferred by the E3 enzyme component, which recognizes structural determinants on the target protein. For example, the so-called “mitotic destruction box” (D box) initially identified at the N terminus of mitotic cyclins is a short sequence of nine residues able to confer M-phase-specific instability when fused to an otherwise stable polypeptide (21). Recognition of the D-box targeting signal by the ubiquitin machinery is mediated in an as-yet-unknown manner by a multisubunit complex E3 enzyme called anaphase-promoting complex (APC) or cyclosome (28, 33, 58). APC activity is subject to cell cycle regulation and, at least in certain organisms, correlates with the phosphorylation of some APC subunits. In budding yeast, D-box- and APC-dependent proteolysis of M-phase targets starts at anaphase and persists until late G1, until the activation of Cln/Cdc28 kinases (1). The temporally distinct degradation of the APC targets Pds1 and mitotic cyclins during mitosis likely involves additional factors, such as the evolutionarily conserved WD40 proteins Cdc20 and Hct1/Cdh1 (27).
The ubiquitination pathway that targets Sic1 for degradation differs from the one described above by the nature of both the E2 and the E3 enzymes that are used. In this case, the Ubc component is encoded by the CDC34 gene, while the E3 enzyme corresponds to another multicomponent ubiquitinating complex, termed SCFCdc4, named for the initials of its three constitutive subunits: Skp1, Cdc53 (also called cullin), and the F-box protein Cdc4 (2, 12, 56). The factor endowed with substrate specificity is the F-box-containing subunit Cdc4, which is one of a family of proteins that share the F-box structural motif (43). Cln1 and Cln2 ubiquitination also depends on Cdc34 (10) and on another F-box protein, Grr1 which, by analogy to Cdc4, functions in a different SCF complex with Cdc53 and Skp1, referred to as SCFGrr1 (4, 64). Unlike the APC, SCF complexes seem to be constitutively active once assembled, suggesting that the cell cycle-dependent degradation of Sic1 is regulated through a different mechanism. Although the multiple SCF target proteins do not seem to share any obvious consensus recognition motif, the degradation of many, if not all, is phosphorylation dependent (3, 9, 37, 56, 63, 65). For example, Cln-dependent phosphorylation of Sic1, which in turn allows its recognition by the SCFCdc4 ligase and subsequent degradation, provides a biochemical rationale for the cell cycle regulation of Sic1 proteolysis (50, 61).
Once polyubiquitinated, targeted proteins are recognized and degraded by the 26S proteasome, a ubiquitous multicomponent proteolytic enzyme. This barrel-shaped, self-compartmentalizing threonine protease of about 2 MDa consists of two functionally distinct subcomplexes, a 20S central core cylinder that has multicatalytic proteolytic activity and that is capped on both ends by 19S regulatory particles (5, 7). The Saccharomyces cerevisiae 20S proteasome is made of 14 distinct but related α-type and β-type subunits assembled in a highly compact particle as four stacked homotypic heptamer rings: α7β7β7α7. Because free access to the luminal catalytic chamber seems to be lacking in the yeast 20S proteasome, as suggested by crystallographic studies (22), it has been speculated that 19S regulatory particles act as gating devices in addition to their function in conferring both ATP and ubiquitin dependence (38). Far less is known about the ultrastructure and biochemical functions of the 19S complex. Recently, the presumably complete 19S complex subunit composition in budding yeast has been determined (19). In this study, we have adopted the new nomenclature recently proposed for the S. cerevisiae regulatory particle subunits (13). Among the 17 polypeptides identified, 6 are previously characterized members of the AAA family of ATPases and have been renamed Rpt1 to Rpt6 (for RP triple-A protein). Of the 11 remaining, non-ATPase (or Rpn) subunits, only two, Rpn9 and Rpn11, are newly identified components of this complex. Surprisingly, Rpn4/Son1, another regulatory subunit recently characterized (15), failed to copurify with 26S proteasome preparations in the same study, raising the possibility of additional, more loosely associated subunits. Besides Rpn11, which has some sequence homology with the catalytic domain of deubiquitinating enzymes (19), none of the other Rpn proteins shows obvious similarities to known enzymes. A crucial issue in this context concerns the identification of the proteasomal component(s) responsible for the recognition of ubiquitinated substrates (45). So far the only candidate specifically proposed is the highly conserved Rpn10 (also known as Mcb1 or Sun1) subunit, whose in vitro ubiquitin-binding properties are in good agreement with the expected specificity for such a function. However, whether it is the relevant factor in vivo is currently in question because it was demonstrated that RPN10 disruption did not cause lethality or strong proteolytic defects in yeast (14, 35, 62).
We initially isolated the RPN3/SUN2 gene by virtue of its strong dosage-dependent and CDC28 allele-specific genetic interaction with an M-phase-defective cdc28-1N mutant. Consistent with a functional interaction between the Cdc28 kinase and the 26S proteasome, we also established that the small Cdk-interacting protein Cks1 physically interacts with the proteasome to control the proteolysis of several M-phase targets (31). In agreement with recently published data (20, 35), the RPN3 gene product was then further characterized as an essential non-ATPase regulatory subunit of the 26S proteasome (31) with numerous structural homologs in other species (Fig. 1). More tellingly, some of these homologs have been shown to complement RPN3 null mutant strains, further supporting the notion that the essential function carried out by the regulatory subunit Rpn3 has been widely evolutionarily conserved (32, 35). Because the rpn3-54 mutant allele that we initially isolated is a null mutation (31) and because gradual titration of the presumably stable Rpn3 subunit by use of a conditional promoter yielded ambiguous results (35), we set out to generate thermosensitive RPN3 mutant alleles as a means of better assessing the role of Rpn3 in the ubiquitin-dependent proteolytic pathway. In this study, we report the phenotypic analysis of temperature-sensitive (ts) rpn3 mutants and discuss different possible models for Rpn3 function.
FIG. 1.
Sequence comparison of Rpn3 homologs. The following protein sequences (sizes and accession numbers) were aligned as indicated in Materials and Methods: S. cerevisiae Rpn3/Sun2 (523 amino acids [aa]; 603613), Homo sapiens p58/S3 (534 aa; D67025), Mus musculus P91A (529 aa; G387100), Drosophila melanogaster DoxA2 (510 aa; G157285), Caenorhabditis elegans open reading frame C30C11.2 product (504 aa; G156222), and S. pombe partial open reading frame SPBC 119.01 product (437 aa; g2959362). Identical amino acids are shown in black boxes, and similar residues are shaded in gray. Dashes indicate conserved residues. The bracket above the sequences indicates the PCI domain, as defined by Hofmann and Bucher (26), which is common to all Rpn3/Sun2 homologs shown here. D.m., H.s., and S.c. arrows mark the beginnings of the Drosophila, human, and budding yeast proteins, respectively, with complementing activity of a nullizygous rpn3 mutant (32, 35).
MATERIALS AND METHODS
Yeast strains, growth conditions, and synchronization.
The genotypes of the yeast strains used in the present study are given in Table 1. All YE strains have the BF264-15DU (MATa bar1 ade1 his2 leu2-3,112 trp1-1a ura3Δns) genetic background (48). Yeast cultures were grown in YEP medium (1% yeast extract, 2% Bacto Peptone, 0.005% adenine, 0.005% uracil) supplemented with 2% dextrose (YEPD) or raffinose (YEPR) as a carbon source. When plasmid selection was required, cells were grown in minimal medium supplemented with appropriate amino acids and/or bases (54). Induction experiments with the GAL1 promoter were performed by growing cells in YEPR before adding galactose (2% final concentration). Except for a few experiments (see Fig. 4B and C and Fig. 5C) in which 200 ng of mating pheromone per ml was used, synchronization experiments were done by arresting mid-log-phase cultures of bar1 cells in G1 with 50 ng of α-factor per ml for 2.5 h and, after a rapid wash, by returning the presynchronized cells to pheromone-free medium for various times.
TABLE 1.
Yeast strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| YE46 | MATa ade1 his2 leu2-3,112 trp1-1 ura3Δns | 48 |
| YE100 | MATa rpn3-4::TRP1 | This study |
| YE101 | MATa rpn3-7::TRP1 | This study |
| YE44 | MATa CLB2(HA)3::KANr | This study |
| YE102 | MATa CLB2(HA)3::KANrrpn3-4::TRP1 | This study |
| YE103 | MATa PDS1(HA)3::KANr | D. J. Clarke |
| YE104 | MATa PDS1(HA)3::KANrrpn3-4::TRP1 | YE100 × Y103 |
| YE106 | MATa LEU2::KANr::GAL1-CLB2(HA)3 | G. Mondésert |
| YE107 | MATa LEU2::KANr::GAL1-CLB2(HA)3 rpn3-4::TRP1 | YE100 × YE106 |
| YE108 | MATa LEU2::KANr::GAL1-PDS1(HA)3 | D. J. Clarke |
| YE109 | MATa LEU2::KANr::GAL1-PDS1(HA)3 rpn3-4::TRP1 | YE100 × YE108 |
| YE110 | MATa LEU2::GAL1:CLN2(HA)3 | This study |
| YE111 | MATa LEU2::GAL1:CLN2(HA)3 rpn3-4::TRP1 | This study |
| YE112 | MATa CLN2(HA)3::URA3 | This study |
| YE113 | MATa CLN2(HA)3::URA3 rpn3-4::TRP1 | This study |
| YE114 | MATa URA3::GAL1:SIC1(HA)1 | This study |
| YE115 | MATa URA3::GAL1:SIC1(HA)1 rpn3-4::TRP1 | This study |
| YE231 | MATα rpn3-54 YCpG2::GAL1:RPN3::URA3 | This study |
| YE416 | MATα rpn3-54 pds1::KANr YCpG2::GAL1:RPN3::URA3 | This study |
| YE417 | MATa CLN2(HA)3::URA3 rpn12-1::TRP1 | This study |
| YE471 | MATa LEU2::GAL1:CLN2(HA)3 cdc34-2 | This study |
| YE473 | MATa CLN2(HA)3::URA3 cdc34-2 | This study |
| RJD1021 | MATa Δsic1::HIS3 | R. J. Deshaies |
| YK109-NIN1 | MATa nin1-1 YCp19::NIN1::URA3::TRP1 | 36 |
| YK109-nin1-1 | MATa nin1-1 YCp19::URA3::TRP1 | 36 |
| Plasmids | ||
| YIpCLN2HAtr | CLN2(HA)3 URA3 (YIplac211) | C. Wittenberg and D. Stuart |
| YIpG2CLN2HA | GAL1:CLN2(HA)3 LEU2 (YIpG2) | C. Wittenberg and D. Stuart |
| pGAL-CLB5HA | GAL1:CLB5(HA)3 URA3 (YCplac33) | 17 |
| D347 | GAL1:SIC1(HA)1 URA3 (YIplac211) | E. Schwob |
| YCpG-SIC1 | GAL1:SIC1(HA)1 LEU2 (YCplac111) | This study |
| pCLB2(HA)3 | CLB2(HA)3 KANr (pKHA3) | This study |
| YCpRPN3 | RPN3 URA3 (YCp50) | This study |
| YCpGAL1-RPN3 | GAL1:RPN3::LEU2::URA3 (YIpG2) | This study |
| pCM250 | tetO2:CLN2(HA3)::URA3 (pCM188) | M. Aldea |
Plasmid derivatives are shown in parentheses at the end of the plasmid descriptions.
FIG. 4.
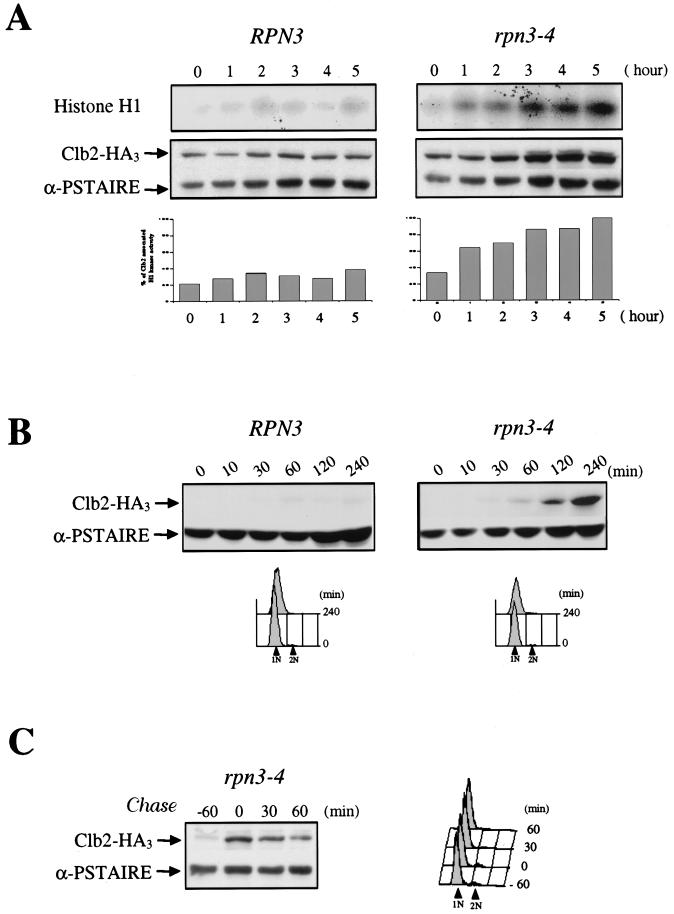
rpn3-4 mutants arrest with high levels of Clb2 and Clb2-associated histone H1 kinase activity and are defective in APC-dependent degradation of Clb2. (A) Wild-type YE44 (RPN3) cells and YE102 mutant (rpn3-4) cells carrying an HA-tagged CLB2 gene were grown and arrested as described in the legend to Fig. 3A. At hourly intervals, cell samples were recovered and Clb2-associated histone H1 kinase activity was determined by immunoprecipitation with an anti-HA antibody (upper panel; Histone H1). The same extracts were also analyzed by Western blotting with an anti-HA antibody to determine their Clb2 levels (lower panel; Clb2-HA3). Cdc28 immunodetected with an anti-PSTAIRE monoclonal antibody was used as a loading control (lower panel; α-PSTAIRE). Histograms represents quantification of the histone H1 kinase activity (percentage of Clb2-associated H1 kinase activity). (B) Accumulation of Clb2 in G1-arrested rpn3 mutant cells. YE106 (RPN3) cells and YE107 mutant (rpn3-4) cells harboring an HA-tagged CLB2 gene under the control of the inducible GAL1 promoter were grown to the early log phase in YEPR medium, arrested in G1 with α-factor for 2.5 h, and shifted to 37°C for an additional hour to inactivate the rpn3-4 gene product. At time zero, HA-tagged Clb2 expression was induced by adding galactose, and cell samples were collected at the indicated times for immunoblot analysis with an anti-HA antibody (Clb2-HA3). As in panel A, an immunoblot with an anti-PSTAIRE monoclonal antibody was used as a loading control (α-PSTAIRE). The effectiveness of the α-factor-induced G1 arrest in both experiments was assessed by FACScan analysis. 1N and 2N indicate cells with unreplicated and fully replicated nuclear DNA, respectively. (C) Clb2 is strongly stabilized in the rpn3 mutant. In an experiment similar to that shown in panel B, Clb2 expression was transiently induced with galactose for 60 min in G1-arrested YE107 (rpn3-4) cells and then repressed by transfer to prewarmed glucose-containing medium still in the presence of the pheromone. Cdc28 (α-PSTAIRE) is shown as a loading control.
FIG. 5.
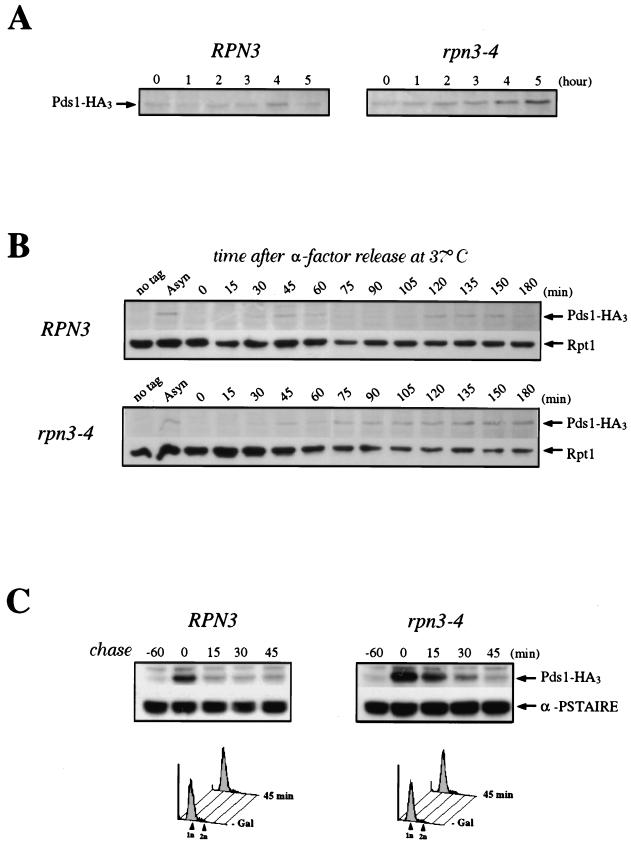
Role of Rpn3 in the proteolysis of Pds1. (A) Early-log-phase cultures of wild-type YE103 (RPN3) cells and YE104 mutant (rpn3-4) cells carrying an HA-tagged PDS1 gene were shifted from 25 to 37°C as described in the legend to Fig. 3A and analyzed by immunoblotting with an anti-HA antibody (Pds1-HA3) to monitor Pds1 levels at the indicated times. (B) Cell cycle regulation of Pds1 in the rpn3 mutant. Early-log-phase cultures of wild-type YE103 (RPN3) cells and YE104 mutant (rpn3-4) cells carrying an HA-tagged PDS1 gene were arrested in G1 with α-factor and then released into fresh medium lacking α-factor at 37°C. Samples for immunoblot analysis of HA-tagged Pds1 were withdrawn at the indicated times. Extracts from the wild-type strain with untagged PDS1 (no tag) were used as a control. A sample was also taken before the addition of α-factor as a source of asynchronous cells (Asyn). The 19S regulatory subunit Rpt1/Cim5 was immunodetected with specific antibodies as a loading control. (C) Reduced Pds1 turnover in the rpn3 mutant. Control YE108 (RPN3) cells and rpn3 mutant YE109 (rpn3-4) cells, both carrying an HA-tagged PDS1 gene under the control of the inducible GAL1 promoter, were grown in YEPR medium, arrested in G1, shifted to 37°C, and induced to express Pds1 for 60 min. Samples were collected before (lane −60) and after (lane 0) Pds1 induction and every 15 min after transfer of the cells to prewarmed α-factor-containing YEPD medium. A Cdc28 immunoblot is shown as a loading control (α-PSTAIRE). FACScan analysis of the first and last samples from this experiment is shown for each strain. (D) PDS1 deletion bypasses the metaphase arrest terminal phenotype of Rpn3-depleted cells. rpn3-54 (YE231) and rpn3-54 ΔPDS1 (YE416) cells, whose sole source of Rpn3 is expression from the GAL1 promoter, were inoculated into galactose-containing medium at 25°C. Rpn3 depletion was then induced at time zero by transferring both cultures to glucose-containing medium. At the indicated times, samples were collected for determination of their cellular DNA contents by flow cytometry. Cell samples were also recovered from the last (23-h) time point for phenotypic analysis of the arrested cells. DIC, differential interference contrast; DAPI, nuclear staining as observed with the DNA dye DAPI.
DNA manipulations and strain construction.
Yeast strains were constructed according to standard genetic procedures (53), except that transformations of yeast cells were carried out by a protocol described elsewhere (11). The ts rpn3 alleles were generated by random PCR mutagenesis under conditions described previously (60). Briefly, the RPN3 coding sequence was amplified by PCR with the following primers: 5′-CGGGATCCTGCTGTATTAAGCACA-3′ and 5′-ATGAATTCCTGTTATGAATGGTAG-3′ (the underlined sequences correspond to the BamHI and EcoRI restriction sites, respectively). The products of the PCR were digested with BamHI and EcoRI, gel purified, and ligated into the LEU2-marked centromeric plasmid YCplac111 linearized with the same restriction enzymes. After transformation and amplification into bacterial strain DH5α, the library of mutagenized RPN3 alleles was introduced into a strain carrying the rpn3-54 null allele and kept alive by the presence of an RPN3 gene on a URA3-marked centromeric plasmid (YCp50). LEU+ transformants were selected, and ts mutants were identified by replica plating colonies on 5-fluoro-orotic acid plates and screening for lack of growth at 37°C. Plasmids harboring ts alleles of RPN3 were recovered and retransformed into rpn3-54 mutant cells to confirm the plasmid dependence of the observed ts phenotype. Two of the ts rpn3 alleles, rpn3-4 and rpn3-7, were cloned into YIplac204 as 1.4-kb PstI-EcoRI fragments and introduced into the BF264-15DU genetic background by transforming cells with the resulting plasmids linearized with SacI to generate strains YE100 and YE101, respectively. Sequence analysis indicated that these two alleles are different with regard to their multiple point mutations. Hemagglutinin (HA) tagging of CLB2 (in YE44 and YE102) and CLN2 (in YE112, YE113, and YE473) at the chromosomal locus was done by transforming BF264-15DU (for YE44 and YE112), YE100 (for YE102 and YE113), and a cdc34-2 strain with pCLB2(HA)3 or YIpCLN2HAtr linearized with BglII and SpeI, respectively. The Gal1:SIC1(HA) construct, a kind gift from E. Schwob, was introduced at the URA3 locus of BF264-15DU and YE100 cells after linearization of D347 with EcoRV to generate strains YE114 and YE115, respectively. Integration of the GAL1:CLN2(HA)3 construct at the LEU2 locus was targeted by digestion with the BstEII restriction enzyme.
Cell biology methods.
Cellular DNA content was quantified by flow cytometry. Cells fixed in 70% ethanol were resuspended in phosphate-buffered saline (PBS) and incubated twice in a 1-mg/ml solution of boiled RNase A for at least 1 h at 37°C. After several washes in PBS, cells were stained with propidium iodide, briefly sonicated to disrupt cell aggregates, and analyzed with a fluorescence-activated cell sorter (FACScan; Becton Dickinson). The mitotic spindle status of arrested rpn3-4 cells was determined by indirect immunofluorescence according to standard protocols described elsewhere (47) and with a mouse monoclonal antibody to α-tubulin (Amersham, Les Ulis, France) and fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Nuclei were visualized with 0.5 μg of 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) per ml. Cells were observed with a Leica microscope equipped with epifluorescence and Nomarski optics.
Western blot and immunoprecipitation experiments.
Protein extracts for Western blot analysis were prepared essentially as described previously (57), with the exception that cells were lysed in PBS supplemented with a cocktail of protease inhibitors (5 μg each of leupeptin and pepstatin per ml, 50 μg of aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride). For the detection of Cln2 phosphoisoforms (see Fig. 9C), we used a slightly different protocol to prepare cell extracts. In brief, cells were broken at 4°C with glass beads in a lysis buffer that contained 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 0.1% NP-40, and 5 mM EDTA and that was supplemented with the same cocktail of protease inhibitors as that mentioned above plus 80 mM β-glycerophosphate and 5 mM NaF as phosphatase inhibitors. Also, proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 6.5% acrylamide gels (with a bisacrylamide/acrylamide ratio of 0.6/29.4). After determination of the protein concentration with the Bradford microassay and SDS-PAGE separation, proteins were transferred to Immobilon membranes (Millipore) and probed with the appropriate primary antibodies. The mouse monoclonal antibody 12CA5 was used at a 1:500 final dilution. A Sic1-specific antiserum kindly provided by M. Tyers was used to monitor the endogenous levels of the Cdk inhibitor. Cdc28 was detected with an anti-PSTAIRE monoclonal antibody. Anti-Cim5/Rpt1 rabbit antibodies were a kind gift from C. Mann. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories) were used at the dilutions recommended by the manufacturer and detected with the SuperSignal chemiluminescence system (Pierce).
FIG. 9.
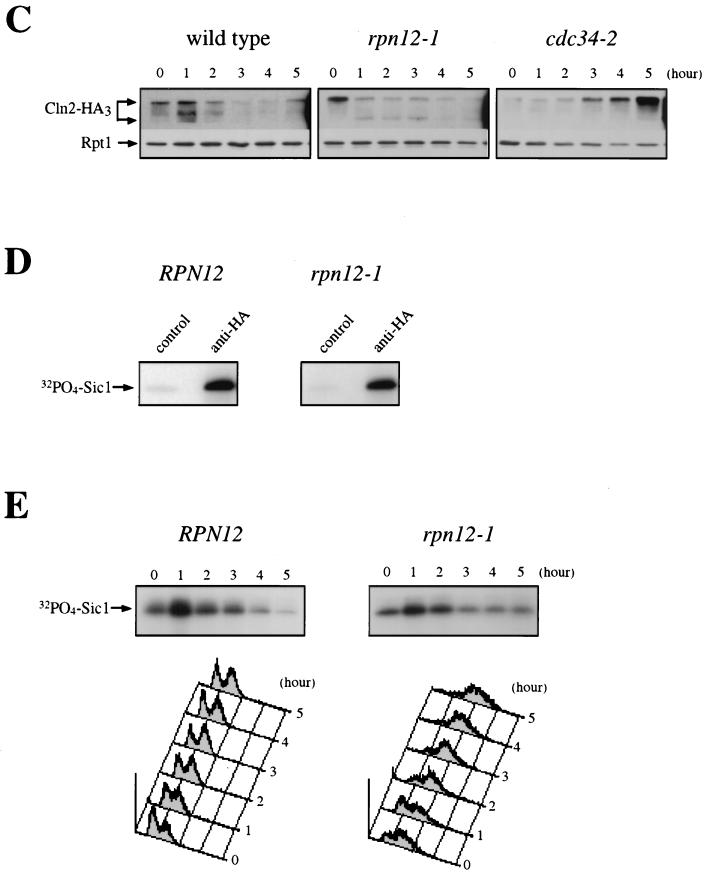
Sic1 regulation in an rpn12-1 mutant. (A) Hypersensitivity of the rpn12/nin1 mutant to elevated levels of Sic1. Control YK109-NIN1 (RPN12) cells and mutant YK109-nin1-1 (rpn12-1) cells were transformed with a centromeric plasmid carrying a SIC1 gene under the control of the GAL1 promoter. Two independent transformants of each strain were streaked on selective medium containing either dextrose or galactose as a carbon source. Plates were photographed after 3 days of incubation at 25°C. (B) Sic1 accumulation in the rpn12-1 mutant. Control YK109-NIN1 (RPN12) cells and mutant YK109-nin1-1 (rpn12-1) cells were grown to the early log phase at 25°C and shifted to 37°C. Samples taken before (lane 0) and at the indicated times after the shift to the restrictive temperature were probed by immunoblotting for Sic1 and Rpt1 (as an internal loading control) with antisera specific for these proteins. (C) Cln2 phosphoisoforms in the rpn12-1 mutant. Control YE112 (wild-type) cells, rpn12 mutant YE417 (rpn12-1) cells, and cdc34 mutant YE473 (cdc34-2) cells, all carrying an HA-tagged CLN2 allele, were grown at 25°C to the early log phase and shifted to 37°C. At the indicated times, samples were withdrawn for immunoblot analysis of Cln2 contents with an anti-HA-antibody (Cln2-HA3). Rpt1 was used as a loading control. (D) Asynchronous cultures of control YE112 (RPN12) cells and mutant YE417 (rpn12-1) cells, both carrying an HA-tagged CLN2 allele, were extracted and used for Cln2 immunoprecipitation with antibody 12CA5 directed against the HA epitope tag (anti-HA). As a control for the specificity of the HA epitope tag, the same cell lysates were immunoprecipitated with an irrelevant anti–glutathione S-transferase monoclonal antibody (control). Both immunoprecipitates were assayed for kinase activity with recombinant histidine-tagged Sic1 protein as a substrate. The incorporation of 32P-labeled phosphate into the Sic1 protein substrate (32PO4-Sic1) was monitored by autoradiography. (E) Control RPN12 (YE46) cells and rpn12-1 (YE413) cells harboring an HA-tagged CLN2 gene under the control of the tetracycline-repressible tetO2 promoter (pCM250) were grown to the early log phase at 25°C and shifted at time zero to 37°C. Samples were taken at hourly intervals for Cln2 immunoprecipitation with an anti-HA-antibody and for flow cytometric analysis of cellular DNA. Sic1 kinase activity present in Cln2 immunoprecipitates was assessed by the same in vitro assay as that used in panel D.
For some of the kinase assay experiments (see Fig. 4A and Fig. 9D and E), HA-tagged proteins were immunoprecipitated with monoclonal antibody 12CA5. The accumulation of Cln2-ubiquitin conjugates was assayed according to previously described methods (31) with purified anti-HA rabbit antibodies for Cln2 immunoprecipitation and a polyubiquitin-specific monoclonal antibody (kindly provided by E. Wayner, A. Kahana, and D. Gottschling) for immunoblot analysis. Immunoreactivity quantification was obtained by densimetric scanning and analysis of an appropriate film exposure with NIH Image 1.60 software.
Analysis of cyclin-associated kinase activity.
After immunoprecipitation of HA-tagged cyclins with HA-specific antibodies, the associated kinase activity was assayed with either a commercial stock of histone H1 (Sigma) as described previously (57) or purified Sic1 as a substrate. Recombinant histidine-tagged Sic1 protein was bacterially expressed from a pET15d-based vector (kindly provided by E. Schwob) in strain BL21-DE3 and purified on nitrilotriacetic acid-nickel columns (Qiagen) according to the manufacturer’s instructions. Kinase activity was visualized by autoradiography.
Protein sequence alignment.
Protein sequences shown in Fig. 1 were determined by use of the GenomeNet CLUSTALW Server (16a) and aligned with CLUSTAL W (1.7) software. Sequence boxing was obtained by use of BoxShade 3.21 software (29a).
RESULTS
Conditional loss of RPN3 function results in an M-phase arrest phenotype.
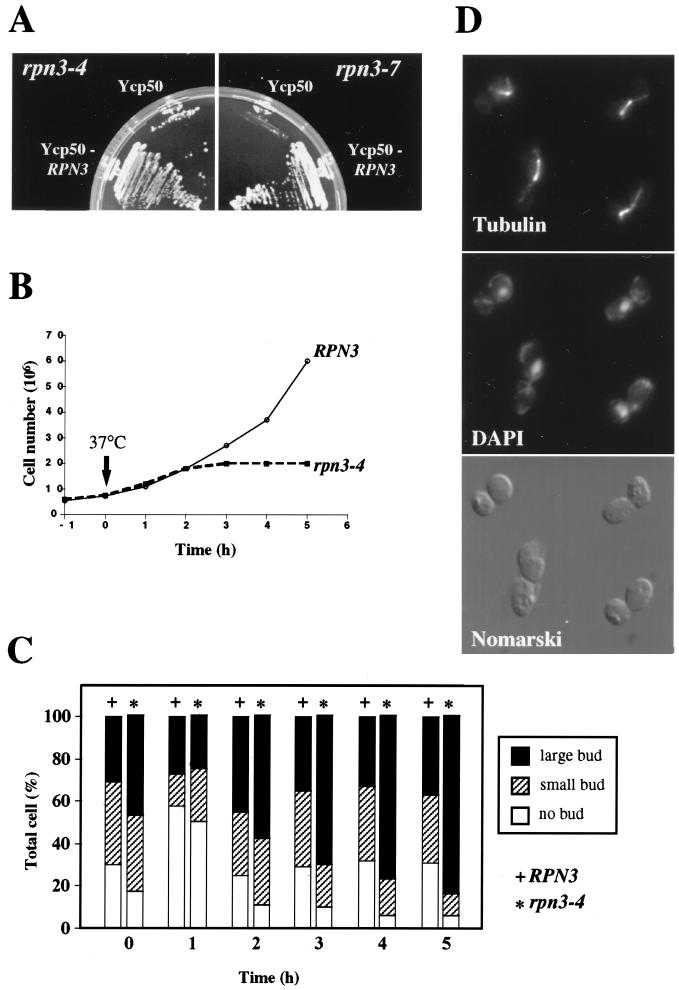
ts rpn3 mutants were generated by use of error-prone PCR mutagenesis and plasmid shuffling as described in Materials and Methods. The cell cycle data presented below were obtained independently with all 11 different mutant alleles tested. As shown in Fig. 2A, two of these chosen at random, rpn3-4 and rpn3-7, are unable to form colonies when incubated on plates at 37°C. This ts phenotype can be fully rescued by a plasmid-borne wild-type RPN3 gene, indicating that both mutations are recessive.
FIG. 2.
Thermosensitive phenotype of rpn3-4 and rpn3-7 mutants. (A) YE100 (rpn3-4) and YE101 (rpn3-7) mutant strains transformed with either an RPN3 gene-containing plasmid (YCp50::RPN3) or an empty vector (YCp50) were streaked on synthetic medium and incubated at 37°C for 2 days. (B) Wild-type YE46 (RPN3) cells and YE100 mutant (rpn3-4) cells were grown at 25°C to the early log phase and shifted at time zero (arrow) to 37°C. At hourly intervals, cell density was determined for both strains and plotted against time. (C) Wild-type YE46 (RPN3) cells and YE100 mutant (rpn3-4) cells were incubated as described for panel B and scored for bud morphology. Bars represent the percentages scored for each category. (D) Microtubule and nuclear phenotypes of rpn3-4 mutant cells at the restrictive temperature. YE100 (rpn3-4) cells were grown to the early log phase in YEPD medium at 25°C, shifted to 37°C, and incubated for 5 h prior to fixation and staining for mitotic spindles (tubulin) and nuclei (DAPI).
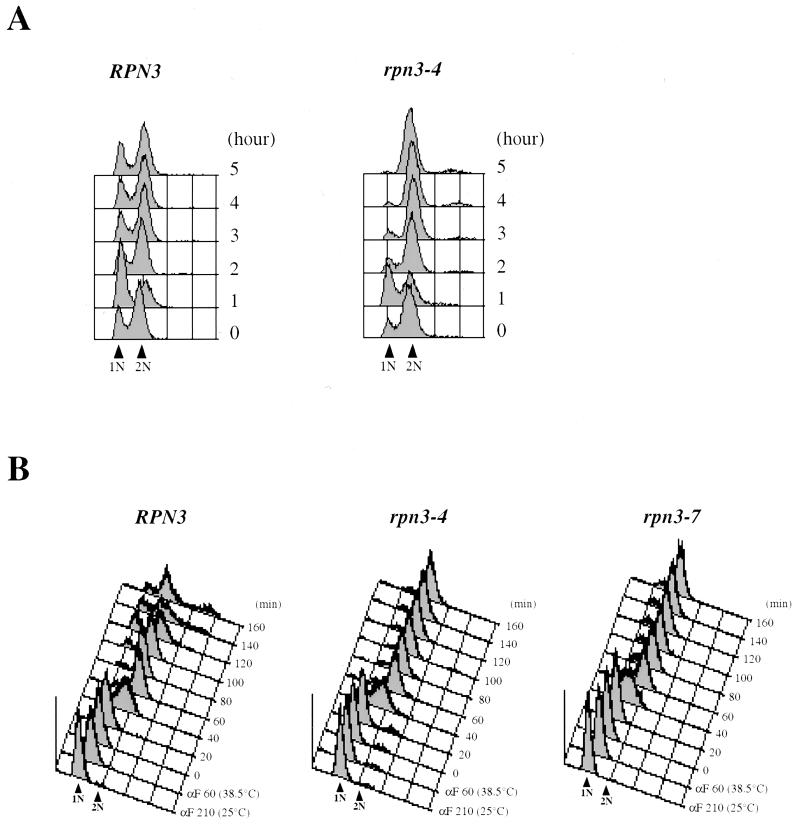
Temperature shift experiments revealed that both mutants rapidly cease cell division, as judged by cell numbers, which barely double following incubation at a restrictive temperature (Fig. 2B and data not shown). As illustrated for rpn3-4, all 11 mutants uniformly arrest as large-bud cells (Fig. 2C), with undivided nuclei and short bipolar spindles (Fig. 2D). The G2/M terminal phenotype suggested by these morphological criteria was confirmed by FACScan analysis (Fig. 3). We first observed that, even at the permissive temperature, early-log-phase cultures of rpn3-4 cells displayed a significantly higher peak of G2/M-phase cells (with fully replicated [2N] nuclear DNA) than wild-type control cultures grown under the same conditions (Fig. 3A). After 2 h of incubation at 37°C, the mutant had started accumulating with a 2N DNA content; ultimately, close to 100% of the cell population was found to arrest in this state by 5 h after the temperature shift. These data strongly suggest that Rpn3 exerts its limiting function primarily in mitosis.
FIG. 3.
Cell cycle arrest phenotype of rpn3 mutants. (A) Wild-type YE46 (RPN3) cells and YE100 mutant (rpn3-4) cells were grown at 25°C to the early log phase and shifted to 37°C. Cell samples were withdrawn before (0 h) and every hour after the temperature upshift and analyzed for nuclear DNA content by flow cytometry. 1N and 2N indicate cells with unreplicated and fully replicated nuclear DNA, respectively. (B) Nuclear DNA profile of wild-type and ts rpn3 cells presynchronized in G1 with α-factor (αF) and released to 38.5°C. YE46 (RPN3), YE100 (rpn3-4), and YE101 (rpn3-7) strains were grown in YEPD medium to the early log phase, incubated with α-factor for 3.5 h at 25°C [αF 210 (25°C)], shifted to 38.5°C for an additional hour [αF 60 (38.5°C)], and finally resuspended in medium preequilibrated to 38.5°C and without the pheromone (0 min). Upon release from the G1 arrest, cell aliquots were taken at 20-min intervals and processed for flow cytometric analysis of nuclear DNA content. 1N and 2N indicate cells with unreplicated and fully replicated nuclear DNA, respectively.
Rpn3 function is required for mitosis but not for the G1/S transition.
Rpn12/Nin1, another 19S proteasomal regulatory subunit, has been shown to be required for both the G1/S and G2/M transitions (36). Therefore, we were interested in determining whether Rpn3 also was required for S-phase entry. For this purpose, wild-type and mutant cells were presynchronized in G1 with the mating pheromone α-factor, released to a nonpermissive temperature, and analyzed for cell cycle progression by flow cytometry. We first performed this experiment at 37°C and found that both rpn3 mutants entered the S phase at the same time as wild-type cells, approximately 40 min after α-factor removal, and then uniformly arrested with 2N nuclear DNA content (data not shown). This result suggested that Rpn3 has no essential role at this stage of the cell cycle. However, because it could be argued that the lack of a G1 arrest phenotype for the rpn3 mutants might be due to the leakiness of the ts alleles that we used, we repeated this cell cycle experiment with more stringent nonpermissive temperature conditions, i.e., 38.5°C, which is the maximum temperature at which this particular genetic background can be propagated. As shown in Fig. 3B, cells presynchronized and arrested at 38.5°C recover from the G1 block and replicate their genomic DNA with similar kinetics, regardless of whether they have a wild-type or mutant RPN3 gene. However, in contrast to wild-type cells, which keep cycling upon prolonged incubation at this elevated temperature, both rpn3-4 and rpn3-7 cells do not progress past the M phase and accumulate with a 2N DNA content. When examined under the microscope for bud and mitotic spindle morphologies, the arrested cells exhibit a terminal phenotype identical to that described above for asynchronous cells: more than 95% of the cells arrest with a large bud and a short mitotic spindle (data not shown).
rpn3 mutant cells are defective in the proteolysis of two APC targets, Clb2 and Pds1.
Because the G2/M terminal phenotype of rpn3 mutants could stem from an inability to generate mitotic levels of Cdc28 kinase activity, we monitored both Clb2 protein levels and Clb2-associated Cdc28 kinase activity in mutant cells. Clb2 rapidly accumulates in rpn3-4 cells upon a temperature shift, and the steady-state accumulation of Clb2 is correlated with a dramatic increase in immunoprecipitable Cdc28 kinase activity throughout the duration of the shift to 37°C (Fig. 4A).
To check rpn3 mutants for a possible defect in the APC-dependent proteolysis of Clb2, we next examined the stability of this cyclin in early G1, i.e., under conditions where it is highly unstable due to the presence of active APC at that stage of the cell cycle. We first addressed the ability of Clb2 to accumulate in rpn3 cells. Control and rpn3-4 mutant strains arrested in G1 with α-factor and shifted to 37°C were induced to express HA-tagged Clb2 from the inducible GAL1 promoter. Consistent with its previously reported very short half-life in early G1, Clb2 remained undetectable in cells with a wild-type RPN3 gene. In striking contrast, Clb2 levels steadily accumulated in the mutant (Fig. 4B), reflecting a significant stabilization of the protein in the absence of Rpn3 function. Also in agreement with this interpretation, we observed that Clb2 that had accumulated in rpn3 cells decayed very slowly after its transient expression from the GAL1 promoter was repressed by glucose addition (Fig. 4C).
Pds1, an anaphase inhibitor in budding yeast, is another well-characterized mitotic target of the ubiquitin-proteasome pathway (6). Cells that are unable to degrade this APC substrate fail to execute the metaphase/anaphase transition. Since our ts rpn3 mutants uniformly arrest in a metaphase-like state, we wondered whether this phenotype could be correlated with an accumulation of Pds1 protein levels. Wild-type and rpn3-4 strains, both tagged with a triple HA epitope at their PDS1 chromosomal loci, were shifted to 37°C and harvested at hourly intervals for immunoblot analysis. As shown in Fig. 5A, Pds1 levels steadily increased over time in arrested rpn3-4 cells. We also monitored the cell cycle fluctuations of Pds1 in synchronous cultures following release from α-factor arrest. During the course of this experiment, wild-type cells underwent two successive rounds of cell division, as judged by cell counting and flow cytometry analysis (data not shown). This finding is also reflected by the Pds1 accumulation pattern, which showed two successive peaks of expression, at 45 and 135 min (Fig. 5B). Under these circumstances, rpn3-4 mutant cells similarly arrested in G1 with undetectable levels of Pds1 started expressing it with kinetics identical to those in control cells but then failed to degrade Pds1 and to initiate both spindle elongation and nuclear division (data not shown). Instead, the Pds1 concentration remained high for several hours, suggesting a strong defect in the M-phase-specific proteolysis of Pds1.
We addressed this issue more directly by comparing the rates of Pds1 degradation in wild-type and rpn3-4 cells arrested in G1 with α-factor. As for Clb2, we transiently expressed Pds1 from the inducible GAL1 promoter and measured protein levels by immunoblotting at 15, 30, and 45 min following repression with glucose. Pds1 was found to turn over very rapidly in wild-type cells, with an estimated half-life of less than 15 min. In the mutant background, however, Pds1 instability appeared significantly reduced, as the protein was still detectable 30 min after promoter shutoff (Fig. 5C). These results are consistent with the previous demonstration of Pds1 instability in G1 and suggest that Rpn3 function is important for Pds1 turnover in G1-arrested cells.
So far, the data presented here, both cytological and biochemical, all indicate that the failure to degrade Pds1 is likely to account for the metaphase arrest phenotype of ts rpn3 mutants. However, because some residual degradation activity was readily visible in the rpn3 mutant in our Pds1 instability assay, we wished to test this hypothesis more rigorously. If true, a simple prediction is that, as previously reported for APC subunit or APC activator mutants, such as cdc23-1 or cdc20-3 (55, 66), a PDS1 deletion, which is not lethal at 25°C, should bypass the metaphase/anaphase transition defect of rpn3 cells and cause rpn3 pds1 mutants to arrest in the telophase, i.e., when RPN3 function becomes necessary for mitotic cyclin destruction and thereby limiting for exit from mitosis. For this purpose, we compared the terminal arrest phenotype of cells lacking Rpn3 in either the presence or the absence of PDS1. Cells depleted of Rpn3 were obtained by shifting from galactose to glucose medium a conditional rpn3-54 strain whose sole source of the functional Rpn3 subunit is under the control of the GAL1 promoter (Fig. 5D). Both cell counting and flow cytometry analysis indicated that following their transfer to glucose medium, the conditional rpn3-54 cells went through three or four rounds of cell division before uniformly arresting with large buds, short mitotic spindles, 2N nuclear DNA contents, and undivided nuclei (Fig. 5D and data not shown). Similarly, rpn3-54 ΔPDS1 double mutants were observed to stop dividing after three or four generations upon a shift to glucose medium and to arrest as large-bud cells with 2N DNA. However, in striking contrast to the rpn3-54 single mutant, a significantly larger proportion of rpn3-54 ΔPDS1 cells terminally arrested with a telophase phenotype, as evidenced by the presence of segregated nuclei (Fig. 5D) and elongated mitotic spindles (data not shown) in more than 75% of the cells. These results therefore strongly support the conclusion that the metaphase arrest terminal phenotype of rpn3 mutants is due to their failure to degrade Pds1.
rpn3 mutants are also defective in the proteolysis of Cln2, an SCF-specific target.
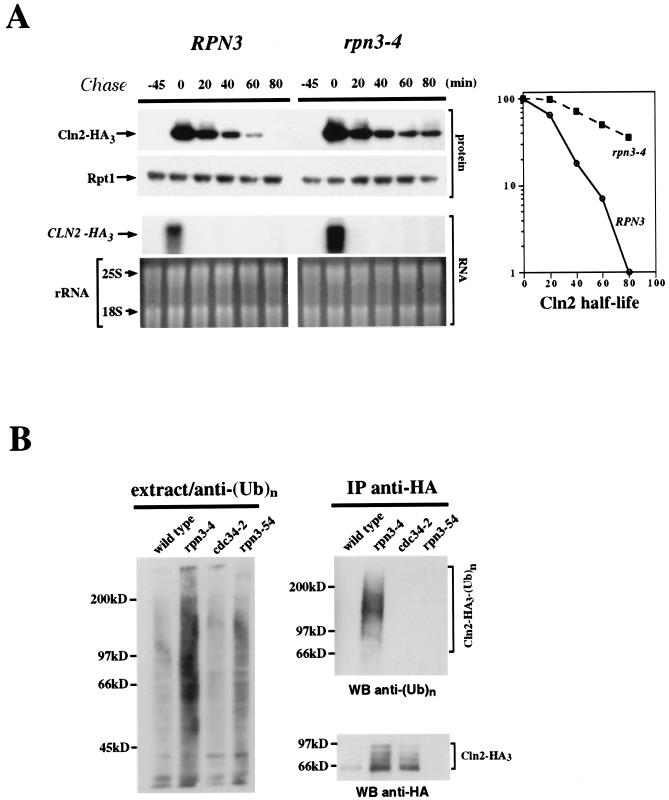
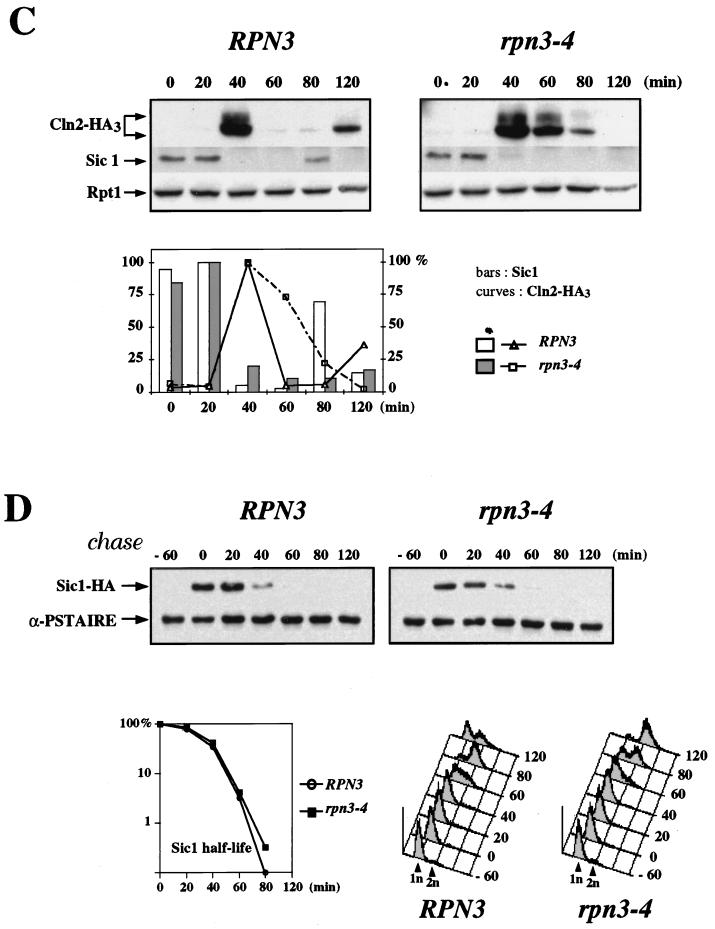
To determine whether rpn3 mutants are specifically affected in the proteolysis of M-phase targets of the APC pathway, we measured the stability of Cln2, a G1 cyclin whose degradation involves another type of E3-dependent ubiquitin pathway, namely, the SCFGrr1 complex. As for Clb2 and Pds1, we addressed this point by transient expression of HA-tagged Cln2 from the GAL1 promoter and repression with glucose (Fig. 6A). Although SCFGrr1 activity has not been reported to be tightly cell cycle regulated, we were concerned that possible differences in Cln2 stability might arise from cell cycle position effects. Accordingly, wild-type and mutant cells were synchronized in G1 with α-factor and shifted to 37°C before a 45-minute Cln2 induction. This step was followed by release from the block and the termination of Cln2 synthesis by incubation of the cells in glucose medium without the mating pheromone. rpn3-4 cells displayed a significantly reduced rate of Cln2 turnover. To rule out any possible interference of the rpn3 mutations with the shutoff or rapid decay of GAL-promoted mRNA, samples from different time points were also subjected to Northern blot analysis. As shown in Fig. 6A, CLN2-HA3 mRNA accumulated and decayed with the same kinetics, independent of the RPN3 genetic background.
FIG. 6.
Cln2 stabilization and ubiquitination in an rpn3 mutant. (A) Control YE110 (RPN3) cells and rpn3 mutant YE111 (rpn3-4) cells, both with an HA-tagged CLN2 gene fused to the GAL1 promoter, were grown in YEPR medium and arrested in G1 with α-factor. The cultures were then shifted to 37°C for an additional hour and induced to express Cln2. After 45 min, Cln2 induction was terminated by returning the cells to prewarmed pheromone-free glucose-containing medium. Aliquots were taken before (lane −45) and after (lane 0) Cln2 induction and every 20 min after glucose repression. HA-tagged Cln2 and Rpt1 (used as a loading control) were immunodetected with anti-HA and anti-Rpt1 antibodies, respectively (top panel; Cln2-HA3 and Rpt1). CLN2-HA3 mRNAs were detected by Northern blot analysis (middle panel). The bottom panel shows ethidium bromide staining of the corresponding gel as a control for equivalent loading of total RNA in the different lanes. Quantification of HA-tagged Cln2 immunoreactivity at different times as measured by densitometry is shown in the graph. (B) Cln2-ubiquitin conjugates accumulate in the rpn3 mutant. Wild-type (strain YE110), rpn3-4 (strain YE111), and cdc34-2 (strain YE471) cells expressing HA-tagged Cln2 under the control of the GAL1 promoter were arrested in G1 with α-factor and then shifted to 37°C to inactivate Rpn3 and Cdc34. Cln2 expression was induced for 30 min by the addition of galactose. As a control for untagged Cln2, we used rpn3-54 strain YE231 (no tag). HA-tagged Cln2 was immunoprecipitated (IP) with a rabbit serum directed against the HA epitope; half of the immunoprecipitate was probed for ubiquitin conjugates by immunoblotting with a monoclonal antibody to polyubiquitin (top right panel), while the other half was checked for HA-tagged Cln2 by Western blotting (WB) with the anti-HA epitope monoclonal antibody 12CA5 (bottom right panel). Cell lysates used for the immunoprecipitation were separated by SDS-PAGE and analyzed with the antipolyubiquitin [anti-(Ub)n] monoclonal antibody to detect total cellular polyubiquitinated proteins (left panel).
If, as suggested by the above biochemical data, the essential Rpn3 function is to promote the rapid turnover of ubiquitinated protein substrates, then rpn3 mutants should accumulate high levels of protein-ubiquitin conjugates. We addressed this point by analyzing mutant and wild-type cell lysates for Cln2-ubiquitin conjugates. Because protein modification by polyubiquitin chains is very efficiently reversed both in vivo and in vitro by deubiquitinating enzymes, we used a sensitive biochemical assay that enabled us to detect any significant increase over basal levels of Cln2-ubiquitin conjugates. We used ts cdc34-2 cells, which are defective in Cln2 ubiquitination, and a strain with untagged Cln2 as negative controls. Wild-type and rpn3 and cdc34-2 mutant cells were arrested in G1 with α-factor and shifted to 37°C, and the expression of HA-tagged Cln2 from the GAL1 promoter was induced for 30 min. HA-tagged Cln2 was immunoprecipitated and separated by SDS-PAGE, and Cln2-ubiquitin conjugates were detected by Western blotting with a ubiquitin-specific monoclonal antibody (Fig. 6B). As expected for mutations in a proteasome subunit, polyubiquitinated forms of Cln2 were found to accumulate to high levels in rpn3 mutant cells as opposed to wild-type cells. Consistent with an essential role of Cdc34 in the SCF-dependent degradation of Cln2, no Cln2-ubiquitin conjugates could be detected in the cdc34-2 cell lysate. Interestingly, a general increase in the levels of polyubiquitinated proteins was clearly apparent in the rpn3 mutants only, as evidenced by Western blot analysis of total proteins with ubiquitin-specific antibodies (Fig. 6B, left panel). This result suggests that many proteasome substrates depend on Rpn3 for their degradation by the ubiquitin-proteasome pathway.
Since the targeting of Cln2 to the SCFGrr1 ubiquitination pathway depends on the prior phosphorylation of this cyclin, our finding that the levels of Cln2-ubiquitin conjugates dramatically increased in rpn3 mutants strongly argues against the possibility of an indirect effect of rpn3 mutations on Cln2 stability due to a defect in Cln2 phosphorylation. Consistent with this idea, we observed a parallel accumulation of hyperphosphorylated Cln2 isoforms in immunoprecipitates from rpn3-4 cell lysates (Fig. 6B, bottom right panel).
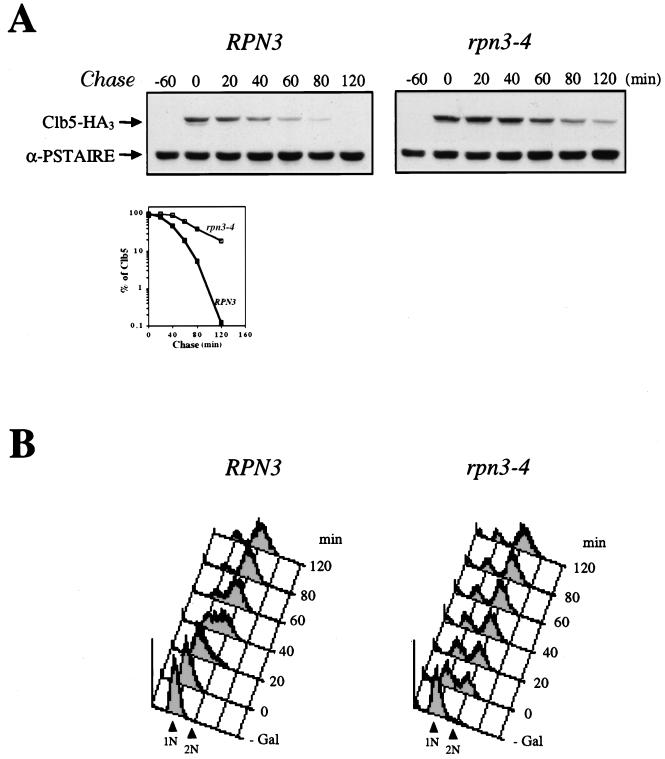
ts rpn3 mutants are impaired in Clb5 proteolysis.
Although less well characterized at the molecular level, the Clb5 destruction machinery has been proposed to rely on the ubiquitin proteolytic pathway (2, 17, 29, 53). As a means of investigating the possible involvement of Rpn3 function in Clb5 turnover, we analyzed the stability of this cyclin by using a GAL1:CLB5(HA)3 construct. For the same reasons as those mentioned above with regard to the exclusion of cell cycle position effects, wild-type and rpn3-4 mutant cells were synchronized in G1 with α-factor before the transient induction of HA-tagged Clb5 from the GAL1 promoter. Under these circumstances, Clb5 was found to be relatively short-lived in wild-type cells while strongly stabilized in the rpn3 mutant (Fig. 7A). This enhanced stability of Clb5 also correlated with a failure of the mutant to properly arrest in G1 in response to Clb5 overexpression, as evidenced by the large number of cells that had prematurely entered the S phase despite the continuous presence of the mating pheromone (Fig. 7B). From these biochemical and FACScan data, we conclude that RPN3 function also plays a critical role in the Clb5 destruction pathway.
FIG. 7.
Impaired degradation of Clb5 in a ts rpn3 mutant. (A) Wild-type YE46 (RPN3) cells and rpn3 mutant YE100 (rpn3-4) cells were transformed with a centromeric plasmid carrying an HA-tagged CLB5 gene fused to the GAL1 promoter (pGAL-CLB5HA). Cells were grown in selective medium with 2% raffinose to the early log phase, synchronized in G1 with a mating pheromone, and shifted to 37°C. After transient expression of Clb5 for 60 min by the addition of galactose, cells were transferred to prewarmed glucose-containing medium without the pheromone. Samples taken before (lane −60) or after (lane 0) Clb5 induction and at the indicated times following the termination of Clb5 expression were analyzed by SDS-PAGE and immunoblotting with an anti-HA antibody (Clb5-HA3). A Cdc28 immunoblot is shown as a loading control (α-PSTAIRE). Quantification of HA-tagged Clb5 immunoreactivity at the different times by densitometry is graphically represented. (B) G1 arrest failure of the rpn3 mutant upon ectopic expression of Clb5. Cell samples from the experiment shown in panel A were subjected to flow cytometric analysis. 1N and 2N indicate cells with unreplicated and fully replicated nuclear DNA, respectively.
Sic1 degradation is not impaired in rpn3 mutants.
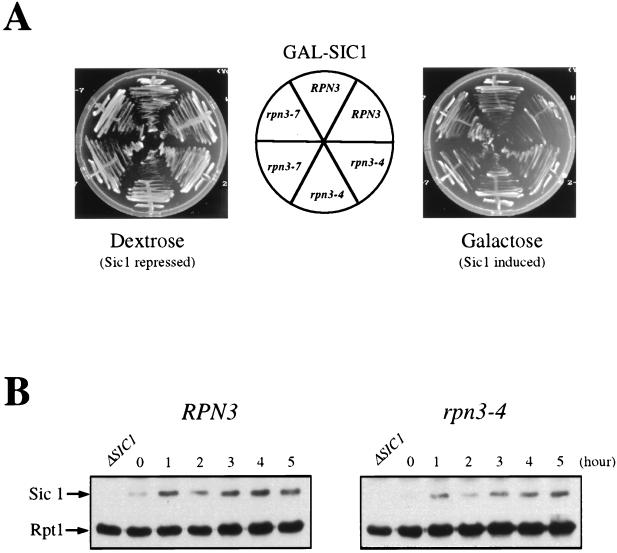
p40Sic1, which regulates the G1/S transition by inhibiting S-phase Clb5- and Clb6-associated Cdc28 kinase activities, is also targeted for destruction by the ubiquitin-proteasome pathway. The E3 enzyme responsible for the in vivo ubiquitination of Sic1 is SCFCdc4, a complex whose subunit composition differs from that of SCFGrr1 only by its distinct associated F-box protein, Cdc4. To determine whether Sic1 proteolysis also depends on Rpn3, we first assayed for possible genetic interactions between RPN3 and SIC1 by introducing a GAL-driven SIC1 gene construct into wild-type and rpn3 mutant cells. Figure 8A shows that Sic1 overexpression was well tolerated in wild-type cells and, likewise, did not cause any growth inhibition in either of two rpn3 mutants, as would be expected if Rpn3 were directly implicated in Sic1 turnover. It is noteworthy that identical results were obtained whether the plates were incubated at 25, 30, or 33°C (Fig. 8A and data not shown), further supporting the lack of interaction between these two genes.
FIG. 8.
Sic1 degradation in an rpn3 mutant. (A) rpn3 mutants tolerate high levels of Sic1. Wild-type YE46 (RPN3) cells and rpn3 mutant YE100 and YE101 (rpn3-4 and rpn3-7) cells were transformed with a centromeric plasmid carrying an HA-tagged SIC1 gene under the control of the GAL1 promoter (YCpG-SIC1). Two independent transformants of each strain were streaked on selective medium containing either dextrose or galactose as a carbon source. Plates were photographed after 3 days of incubation at 30°C. (B) Sic1 levels in an arrested rpn3 mutant. Wild-type YE46 (RPN3) cells and rpn3 mutant YE100 (rpn3-4) cells were grown to the early log phase at 25°C and shifted to 37°C. Cells taken before (lane 0) and at hourly intervals after the shift were subjected to immunoblotting analysis with anti-Sic1 and anti-Rpt1 antisera. An extract from a SIC1-disrupted strain (ΔSIC1) was run in parallel as a control for the specificity of the anti-Sic1 antiserum. (C) Sic1 protein levels in synchronized rpn3 mutant cells. Control YE112 (RPN3) cells and rpn3 mutant YE113 (rpn3-4) cells, both carrying an HA-tagged CLN2 allele, were grown at 25°C to the early log phase, synchronized in G1 with α-factor, and shifted to 37°C. After release from the G1 arrest at the restrictive temperature, samples were withdrawn at the indicated times and probed by immunoblotting for HA-tagged Cln2, Sic1, and Rpt1 (as a loading control) with an anti-HA antibody (Cln2-HA3), Sic1-specific antiserum, and Rpt1/Cim5-specific antiserum, respectively. A graphic representation of the Sic1 and HA-tagged Cln2 (Cln2-HA3) immunoreactivities obtained by densitometry is also presented. (D) Sic1 turnover in an rpn3 mutant. Control YE114 (RPN3) cells and rpn3 mutant YE115 (rpn3-4) cells, both with an integrated GAL1:SIC1(HA)1 construct, were grown at 25°C in YEPR medium to the early log phase, arrested in G1 with α-factor, and shifted to 37°C. Galactose was added for 60 min to induce Sic1 expression, and the cells were returned to prewarmed glucose-containing medium to shut off the GAL1 promoter. Samples withdrawn before (lane −60) and after (lane 0) Sic1 induction and at the indicated times following glucose repression were subjected to Western blot analysis with anti-HA (Sic1-HA) and anti-PSTAIRE (α-PSTAIRE) antibodies to monitor HA-tagged Sic1 and Cdc28 (as a loading control), respectively. Flow cytometric analysis of the corresponding samples is also presented for each strain. A graphic representation of the Sic1 half-life, as estimated by immunoblotting, is also presented.
Unlike the expression of Pds1 or Clb2, which rapidly accumulate in rpn3 mutant cells upon a shift to a nonpermissive temperature, Sic1 expression remained fairly constant under the same growth conditions (Fig. 8B). In a third set of experiments, we monitored the levels of Sic1 protein in synchronous cultures of cells that were arrested in G1 with α-factor and released to the restrictive temperature (Fig. 8C). We also checked for Cln2 variations in the same experiment by using wild-type and rpn3-4 strains carrying an HA-tagged CLN2 allele at the chromosomal locus. Sic1 was found to accumulate to wild-type levels both in α-factor-arrested cells and following the removal of the mating pheromone (Fig. 8C). Likewise, the kinetics of Sic1 destruction were superimposable when wild-type and mutant time courses were compared, with the bulk of endogenous Sic1 disappearing within 40 min of recovery from G1 arrest in both strains. Interestingly, this abrupt Sic1 degradation occurred concomitantly with S-phase entry, as judged by flow cytometry analysis (data not shown, but see Fig. 3B for an equivalent experiment). Probing the same immunoblots with an HA-specific monoclonal antibody for endogenous HA-tagged Cln2 revealed a clearly different picture. In this case, the Cln2 signal that appeared in both strains by 40 min after release persisted much longer in rpn3-4 mutant cells than in wild-type cells, consistent with our previous conclusion that Rpn3 is essential for Cln2 turnover. These results indicate that under conditions where Rpn3 function is severely compromised in the mutant, as judged by enhanced Cln2 stability, Sic1 destruction still takes place with wild-type kinetics.
To address more directly the issue of Sic1 turnover in the rpn3-4 mutant, we monitored the stability of the protein with the GAL1 promoter shutoff assay as described above for Cln2 and Clb5. Wild-type and rpn3-4 cells, both containing a GAL1:SIC1(HA)1 construct, were arrested in G1 with α-factor and shifted to 37°C, and Sic1 expression was induced by the addition of galactose. After 60 min, Sic1 synthesis was terminated by transferring the cells to prewarmed glucose medium in the absence of the pheromone. At regular intervals, cell samples were harvested and subjected to Western blot and FACScan analyses (Fig. 8D). As expected from the synchronization experiment described in the legend to Fig. 8C, we did not observe any significant stabilization of Sic1 in the mutant. Also in agreement with our conclusion that rpn3 mutations do not affect Sic1 turnover, flow cytometry analysis revealed that both wild-type and mutant cells entered the S phase with very similar if not identical kinetics, despite the transient Sic1 overexpression prior to the α-factor release. Taken together, these data strongly suggest that Rpn3 is dispensable for the ubiquitin-dependent proteolysis of Sic1.
Role of Rpn12/Nin1 in Sic1 regulation.
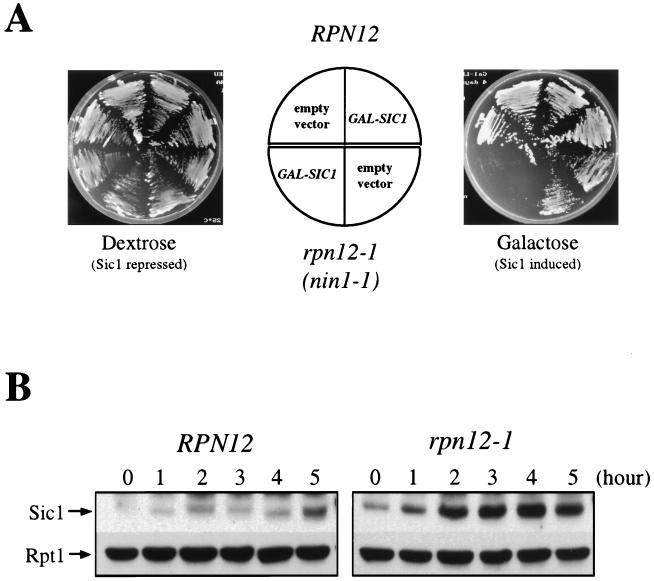
A conditional mutation in the 19S proteasome subunit Rpn12 has been previously reported to confer both G1/S and G2/M arrest phenotypes. Moreover, the rpn12 mutant was also shown to be severely impaired in sustaining wild-type levels of Cdc28 kinase activity upon incubation at the restrictive temperature (36). Because such a phenotype is fully consistent with a Sic1 proteolysis defect, we reasoned that overexpression of the Cdk inhibitor should strongly enhance the ts phenotype of the mutant. As illustrated in Fig. 9A, rpn12-1/nin1-1 mutant cells are hypersensitive to elevated levels of Sic1, whose overexpression from the GAL1 promoter is lethal even at the permissive temperature.
To lend further support for rpn12 cells having a severe defect in Sic1 degradation, we monitored the Sic1 steady-state levels in wild-type and rpn12-1 mutant cells before and after a shift of the temperature to 37°C (Fig. 9B). Consistent with the strong genetic interaction described above, we found that an early-log-phase culture of mutant cells expressed Sic1 to much higher levels than control cells. More significantly, Sic1 rapidly accumulated upon a temperature upshift in the mutant only. This steady increase in Sic1 protein levels in terminally arrested rpn12 mutant cells markedly contrasted with the periodic accumulation observed in wild-type cells.
We next examined whether the dramatic Sic1 accumulation seen in rpn12-1 cells could be an indirect effect due to the general defect in Cdc28 kinase activity, since there is compelling evidence that phosphorylation by Cln/Cdc28 complexes triggers rapid Sic1 degradation by targeting Sic1 to the SCFCdc4-dependent proteolytic pathway. As a first way to address this possibility, we undertook to analyze the electrophoretic pattern of Cln2 in rpn12-1 cells, the presence of slowly migrating hyperphosphorylated isoforms of Cln2 being a good marker of Cln/Cdc28 kinase activity. As a positive control, we used the cdc34-2 mutant, which is specifically impaired in Cln2 ubiquitination and therefore typically accumulates hyperphosphorylated isoforms of Cln2 (51). Immunoblot analysis of Cln2 under lysis conditions that preserve phosphoproteins (see Materials and Methods) enabled us to show that indeed a significant fraction of Cln2 exhibited a slow-migration profile in rpn12-1 cells (Fig. 9C). This finding was particularly apparent in an asynchronous culture of mutant cells, where most of the Cln2 protein comigrated with the upper band that accumulated in cdc34-2 cells, but was also apparent after several hours of incubation at 37°C. These results suggest that even under nonpermissive conditions, a sustained basal level of Cln/Cdc28 kinase activity is present in rpn12-1 cells.
To further examine Cln/Cdc28 kinase activity, we directly assayed HA-tagged Cln2 immunoprecipitates from wild-type and rpn12-1 cell lysates for associated kinase activity. For this experiment, we first used histone H1 as a substrate. However, because the assay appeared to be particularly unreliable, with Cln2 immunoprecipitates showing very low levels of kinase activity toward this substrate even when isolated from wild-type cells (data not shown), we next turned to a more sensitive assay with recombinant Sic1 protein as a more physiological substrate. As shown in Fig. 9D, significant amounts of Sic1 kinase activity were specifically recovered from asynchronous cultures of both wild-type and rpn12-1 cells when Cln2 was immunoprecipitated with anti-HA but not control antibodies. Taking advantage of this sensitive kinase assay, we next tested wild-type and rpn12-1 cell extracts for Cln/Cdc28 kinase activity following a shift to 37°C. Supporting the conclusion drawn from our data on the Cln2 electrophoretic pattern, we found that rpn12-1 cells were able to generate substantial levels of Cln2-dependent Sic1 kinase activity after several hours of incubation at the restrictive temperature (Fig. 9E). These results therefore suggest that phosphorylation by Cln/Cdc28 is unlikely to be rate limiting for Sic1 degradation in rpn12-1 cells and that the accumulation of Sic1 may be a direct consequence of a ts rpn12 mutation.
DISCUSSION
In this study, we have investigated the defects resulting from conditional inactivation of the 26S proteasome subunit Rpn3 in the budding yeast S. cerevisiae, with the ultimate goal of better understanding the function of this regulatory subunit in the control of cell cycle progression. The fact that 11 of 11 independent ts mutants terminally arrested with a large-bud-cell phenotype identical to that exhibited by spores with a disrupted RPN3 gene strongly argues for a complete loss of function of the mutant proteins. We have shown that ts rpn3 cells rapidly stop cell division after prolonged incubation at the restrictive temperature. Both cell counting and FACScan analysis indicated that most of these cells undergo one round of DNA replication before arresting with a uniform terminal phenotype that has all the hallmarks of a metaphase block, as evidenced by (i) their fully replicated genomic DNA content, (ii) their undivided nuclei and short bipolar spindles, and (iii) the presence of high levels of both Clb2 and Clb2-associated histone H1 kinase activity. Thus, one of the essential functions of RPN3 appears to be required during mitosis, as recently suggested by others (35). Also consistent with a mitotic function of Rpn3 is our finding that the half-life of Clb2 was dramatically increased in the mutants when assayed in G1, i.e., under conditions where Clb2 is normally highly unstable (1, 28). However, the observation that the rpn3 mutants uniformly arrested prior to anaphase implies that the proteolysis of the anaphase inhibitor Pds1 is also severely impaired in this genetic context. In agreement with this contention, rpn3-4 cells were found to terminally arrest with elevated levels of Pds1, and the stability of Pds1 ectopically expressed in G1 was significantly enhanced in the mutants. Furthermore, cells with a GAL-driven RPN3 gene as their sole source of Rpn3 protein have been found to terminally arrest with a large bud and an undivided nucleus when incubated in repressive glucose medium (35; this study), a phenotype that fully supports our conclusion that this proteasomal subunit actively participates in the destruction of Pds1 at the metaphase/anaphase transition. Finally, the best evidence that Pds1 is the limiting cell cycle target of Rpn3-dependent proteolysis is that a pds1 rpn3 double mutant no longer arrests at metaphase but instead progresses to telophase. However, we cannot eliminate the possibility that the metaphase arrest of rpn3 cells results indirectly from the stabilization of Pds1, for example, through activation of the replication or spindle checkpoint. Whether mutants affected in these checkpoint pathways similarly bypass the Rpn3-mediated metaphase arrest has not been investigated yet but should prove helpful in clarifying this issue.
RPN3 is also required for the proteolysis of non-APC substrates. This is the case for the G1 cyclin Cln2, whose targeting to the proteolytic machinery involves a different ubiquitin ligase, the SCFGrr1 complex (56, 64). The defect in Cln2 proteolysis of rpn3 mutants could be documented by both half-life measurements of Cln2 with the GAL-inducible system and a time course analysis of the endogenous protein upon α-factor release. Moreover, as described for mutants of the Cln2 ubiquitination pathway (2, 10), rpn3 mutant cells have been found to be very sensitive to high Cln2 levels (3). These results indicate that Rpn3 function is also necessary for normal Cln2 turnover and is not restricted to a single class of E3 enzyme-specific targets.
Because hyperactivation of the Cln2 pathway is known to strongly interfere with G1 arrest induced in response to the mating pheromone, we also addressed the issue of whether rpn3 mutations could confer some α-factor resistance owing to their Cln2-stabilizing effect. Surprisingly, halo assay experiments gave the opposite result, as cells mutated for RPN3 formed slightly larger halos in the presence of α-factor (3), thereby reflecting a modest but significant hypersensitivity of the mutants to the mating pheromone. In light of recent data demonstrating the involvement of the ubiquitin-dependent proteolytic machinery in regulating the level of Far1 (23), a Cln/Cdc28 kinase inhibitor required for mating pheromone arrest (44), a likely possibility to explain the pheromone sensitivity of rpn3 mutants is that Rpn3 is required for Far1 degradation. Further biochemical experiments are needed to test this hypothesis. Another physiologically important cell cycle regulator whose turnover was investigated in the present work is the S-phase cyclin Clb5. Consistent with a role of Rpn3 in the proteolysis of this cyclin, yeast cells harboring a ts rpn3 allele exhibit an extended Clb5 half-life. Moreover, transient expression of Clb5 from the GAL1 promoter in pheromone-arrested cells causes the mutant cells specifically to bypass G1 arrest and precociously enter the S phase. Thus, even though the Clb5 degradation pathway is still poorly understood, our data provide further arguments for an involvement of the ubiquitin-proteasome machinery.
Sic1 acts as a Clb-specific inhibitor of Cdc28 kinase activity, and its destruction after Start by the ubiquitin-dependent proteolytic machinery is a prerequisite for G1 cells to proceed into the S phase (52). Mutations in components of the ubiquitination pathway, such as the E2 enzyme Cdc34 or one of the E3 enzyme subunits, Cdc53, Skp1, or Cdc4, have all been shown to lead to a failure in Sic1 proteolysis and consequently to a G1 arrest phenotype (2, 50). Our data strongly suggest that Rpn3 plays no essential role in Sic1 proteolysis. Several observations support this conclusion. First, none of the ts rpn3 strains exhibit a G1 arrest phenotype, even under extreme restrictive-temperature conditions, such as those used for the experiment shown in Fig. 3B. Second, unlike what has been found with mutants of the Sic1 ubiquitination pathway, Sic1 overexpression fails to exacerbate the ts phenotype of rpn3 mutants at any of the temperatures tested. Third, under conditions where Cln2 proteolysis is found to be dramatically impaired in ts strains, Sic1 destruction takes place with wild-type kinetics. Fourth, using the same stability assay as the one used to document the increased half-life of Cln2 or Clb5 in rpn3 mutants, we consistently failed to detect any Sic1 stabilization. Although it still remains a formal possibility that all of the ts rpn3 alleles that we have isolated retain biological activity against Sic1, the fact that we have not been able to detect even a modest defect in Sic1 proteolysis argues against this notion. Nevertheless, additional experiments will be required to rule out this possibility.
The lack of a G1 arrest phenotype of rpn3 mutants strikingly contrasts with the dual cell cycle function of Rpn12/Nin1, which is required for both the G1/S and the G2/M transitions (36). Also remarkable is the inability of ts rpn12 mutants to maintain wild-type levels of Cdc28-dependent histone H1 kinase activity at a nonpermissive temperature, a finding which led the same authors to propose that Rpn12 is required for the degradation of some Cdc28-dependent kinase inhibitors, possibly including Sic1 (36). In favor of this proposal, we found that (i) Sic overexpression was highly toxic in rpn12 cells even at a permissive temperature and (ii) Sic1 rapidly accumulated in mutant cells upon a shift to 37°C. So far, our biochemical data indicate that rpn12-1 cells maintain significant levels of Cln2/Cdc28 kinase activity both in asynchronous cultures and upon a shift to a nonpermissive temperature. At first glance, this notion may appear contradictory to previous studies showing that most rpn12 mutant cells released from α-factor arrest fail to undergo bud emergence and to generate detectable Cdc28 kinase activity (36). One possibility to account for such a discrepancy is that SCF-dependent degradation of Far1 is strongly defective in this genetic background, thereby preventing pheromone-arrested cells from generating wild-type levels of Cln/Cdc28 kinase activity. Another explanation could stem from the fact that we used a different substrate in the kinase assay to monitor the activity of Cln2/Cdc28 complexes. Interestingly, we noted that in these kinase assay experiments, recombinant Sic1 protein incorporated significantly larger amounts of labeled phosphate than histone H1 when incubated with Cln2 immunoprecipitates. Although we cannot rule out completely the possibility of another indirect effect of the rpn12 mutation on Sic1 degradation, including a more subtle phosphorylation defect or the mislocalization of some critical component, our results suggest that Sic1 phosphorylation by Cln2/Cdc28 complexes is not rate limiting in an rpn12 mutant and that Sic1 accumulation in this context might be the direct consequence of a biochemical defect at the proteasome level. Further functional and biochemical studies in this direction should prove informative in this regard.
Like most of the other Rpn subunits, Rpn3 has no obvious sequence similarities to other proteins with known biochemical activities. Based on previous genetic studies, the essential region corresponds to the C-terminal half of the protein (32). This finding is perhaps not surprising in view of the sequence comparison with several structural and, in some cases, functional Rpn3 homologs from distantly related eukaryotes (Fig. 1); a significantly higher degree of homology is observed within the C-terminal portions of these Rpn3 homologs. Interestingly, it is also in this region that a new type of structural motif, called the PCI domain, has been recently identified (26). The occurrence of this motif in other Rpn subunits, such as Rpn5, Rpn6, Rpn7, and Rpn9, as well as in several unrelated proteins that all have the common feature of being part of large protein complexes, has led to the suggestion that PCI domains could provide a structural basis for assembly of proteins into highly ordered protein complexes. That the ts rpn3 alleles used in the present work confer defects in the assembly of 26S proteasomes is a possibility. Preliminary size exclusion chromatography experiments, however, tend to indicate that the rpn3 mutant phenotype does not result from a general failure of the mutants to assemble proteasomes of the correct molecular weight (data not shown).
The 19S regulatory complex is thought to provide the 20S core particle with a set of biochemical properties that are essential for its physiological activity. These include substrate recognition, substrate unfolding, energy dependence, and translocation of unfolded substrates into the central cavity of the catalytic core. How exactly the different 19S cap subunits contribute to these regulatory steps is still essentially unknown, as very few subunits have been functionally characterized at the biochemical level. Substrate recognition constitutes one of the first steps in the cascade of events that eventually leads to proteolysis once the polypeptide substrate has made its way to the inner cavity of the 20S catalytic core. It is believed to rely primarily on multiubiquitination of the protein substrates, which acts as a general but highly regulated proteasome targeting signal (for a recent review, see reference 45). Unfortunately, the molecular mechanisms by which ubiquitin-conjugated substrates are then recognized by the 26S proteasome are poorly understood, and the existence of an essential ubiquitin receptor within the 19S regulatory particle has yet to be confirmed. So far the only subunit known to have a high affinity for multiubiquitin chains in vitro is Rpn10 (also called Mcb1, S5a, or Sun1). However, Δrpn10 null mutants are viable and still able to degrade most of the model substrates of the ubiquitin-proteasome pathway. Therefore, other multiubiquitin receptors are likely to exist among the Rpn subunits. Whether Rpn3 function is involved in this process has not been addressed experimentally but certainly constitutes an attractive working model. However, if ubiquitin truly acts as a general targeting signal, such a model cannot easily accommodate our finding that rpn3 mutations have no effect on Sic1 degradation, unless Rpn3 serves as a receptor for a subset of ubiquitinated targets. It is worth mentioning that the proteasome also functions in a ubiquitin-independent degradation pathway that is used by several short-lived proteins, the prototype of which is ornithine decarboxylase (reference 45 and references therein). The strict dependence on 19S cap subunits in this proteolytic pathway strongly suggests that some of the regulatory particle subunits can serve as receptors for nonubiquitinated target proteins. Proof that Rpn3 is able to contribute to the ubiquitin-independent pathway will require further investigation, but this model appears to be another possible model for explaining the substrate specificity of this particular proteasomal subunit.
Alternatively, Rpn3 function may be required in the subsequent steps of 26S proteasome-mediated protein hydrolysis, namely, unfolding and translocation of the substrate, which presumably also involve the ATPase components of the 19S complex (5, 49). By analogy with their prokaryotic counterparts, it has been speculated that the six Rpt subunits could form a similar six-membered heteromeric ring ATPase that docks directly onto the α rings of the 20S cylinder (5). In excellent agreement with this hypothesis, a subcomplex of the 19S particle, named the base and comprising the six ATPases plus the three non-ATPase subunits Rpn1, Rpn2, and Rpn10, has been recently shown to correspond to a part of the regulatory particle most proximal to the 20S proteasome (20). Whether these proteasomal ATPases also require accessory proteins among the Rpn subunits to carry out their essential functions has yet to be addressed. Intriguingly, the rpn3 terminal phenotype reported here is highly reminiscent of that of rpt1/cim5 and rpt6/cim3 mutants (18). This finding could reflect a role of Rpn3 in modulating the activity of these ATPases. If true, one would predict that the three subunits are closely associated within the 19S particle. Although Rpn3 and both Rpt1 and Rpt6 clearly reside within two spatially distinct subcomplexes of the 19S cap (20), it remains possible that the three subunits function in the same biochemical pathway. Obviously, further structure-function analysis of Rpn3 should prove very helpful in discriminating among these various possibilities and in further defining the specific role of Rpn3 in the context of cell cycle control by the ubiquitin-proteasome pathway.
ACKNOWLEDGMENTS
We thank Ray Deshaies and Rati Verma for yeast strains; Doris Germain for pGAL-CLB5HA; Carl Mann for Rpt1/Cim5 antiserum; Etienne Schwob for the D347 construct and the pET15-Sic1 expression vector; Akio Toh-e for the NIN1 and nin1-1 yeast strains; Mike Tyers for Sic1 antibodies; E. Wayner, A. Kahana, and D. Gottschling for the ubiquitin monoclonal antibody; Spencer Brown, Danny Rouillard, and Marie Ange Deugnier for helpful advice on FACScan analysis; Oskar Smrzka for assistance with the sequence alignment programs and Northern experiments; Daniel Meur and Dominique Morineau for the artwork; and Duncan Clarke, Steve Haase, Guillaume Mondésert, Dave Stuart, Mark Watson, Curt Wittenberg, and Meira Wolff for the various DNA constructs and yeast strains used throughout this study. We express special thanks to Marie-Noëlle Simon and Peter Kaiser for stimulating discussions and critical review of the manuscript. Eric Bailly is particularly indebted to Michel Bornens for generosity in sharing laboratory space and for constant interest in this work as well as to all the members of the laboratory for their supporting encouragement and valuable discussions.
E.B. is supported by INSERM and acknowledges an international research fellowship from the Public Health Service Fogarty International Center and financial support from Michel Bornens. This work was supported by the Curie Institute, by CNRS, and by grants from the Association pour la Recherche sur le Cancer to E.B. and from NIH (grant GM38328) to S.I.R.
REFERENCES
- 1.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle: G2 cyclin proteolysis initiated at mitosis persists until activation of G1 cyclins in the next cell cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 2.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 3.Bailly, E. Unpublished results.
- 4.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister W, Walz J, Zhul F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Fix O, Peters J-M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 7.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:639–650. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 8.Deshaies R J. The self-destructive personality of a cell cycle in transition. Curr Opin Cell Biol. 1995;7:781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 9.Deshaies R J. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies R J, Chau V, Kirschner M W. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–314. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 12.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p and Cdc53/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 13.Finley D, Tanaka K, Mann C, Feldmann H, Hochstrasser M, Vierstra R, Johnston S, Hampton R, Haber J, McCusker J, Silver P, Frontali L, Thorsness P, Varshavsky A, Byers B, Madura K, Reed S I, Wolf D, Jentsch S, Sommer T, Baumeister W, Goldberg A, Fried V, Rubin D M, Glickman M H, Toh-e A. Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- 14.Fu H, Sadis S, Rubin D M, Glickman M, van Nocker S, Finley D, Vierstra R D. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoro M, Tanaka K, Yokosawa H, Toh-e A. Son1 is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- 16.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 16a.GenomeWet. 5 February 1998, revision date. [Online.] Sequence interpretation tools, Clustal W program, Institute of Medical Science, University of Tokyo, Tokyo, Japan. www.genome.ad.jp/SIT/SIT.html. [10 August 1999, last date accessed.]
- 17.Germain D, Hendley J, Futcher B. DNA damage inhibits proteolysis of the B-type cyclin Clb5 in S. cerevisiae. J Cell Sci. 1997;110:1813–1820. doi: 10.1242/jcs.110.15.1813. [DOI] [PubMed] [Google Scholar]
- 18.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 19.Glickman M H, Rubin D M, Fried V A, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 21.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of the 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 23.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 27.Hoyt M A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 28.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 29.Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- 29a.ISREC Bioinformatics Groupe. 17 July 1999, revision date. [Online.] Boxshade program, version 3.21. Institut Suisse de Recherche Experimentale sur le Cancer, Epalinges, Switzerland. www.isrec.isb-sib.ch. [10 August 1999, last date accessed.]
- 30.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser P, Moncollin V, Clarke D J, Watson M H, Bertolaet B L, Reed S I, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamura M, Kominami K, Takeuchi J, Toh-e A. A multicopy suppressor of nin1-1 of the yeast Saccharomyces cerevisiae is a counterpart of the Drosophila melanogaster diphenol oxidase A2 gene, Dox-A2. Mol Gen Genet. 1996;251:146–152. doi: 10.1007/BF02172912. [DOI] [PubMed] [Google Scholar]
- 33.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 34.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 35.Kominami K, Okura N, Kawamura M, Demartino G N, Slaughter C A, Shimbara N, Chung C H, Fujimuro M, Yokosawa H, Shimizu Y, Tanahashi N, Tanaka K, Toh-e A. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1-1. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kominami K, DeMartino G N, Moomaw C R, Slaughter C A, Shimbara N, Fujimoro M, Yokosawa H, Hisamatsu H, Tanahashi N, Shimizu Y, Tanaka K, Toh-e A. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995;14:3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 38.Larsen C N, Finley D. Protein translocation channels in the proteasome and other proteases. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]
- 39.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 40.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 41.Nigg E A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 42.Pagano M. Cell cycle regulation by the ubiquitin pathway. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 43.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 44.Peter M, Herskowitz I. Joining the complex: cyclin-dependent kinase inhibitory proteins and the cell cycle. Cell. 1994;79:181–184. doi: 10.1016/0092-8674(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 45.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 46.Pines J, Hunter T. Cyclin-dependent kinases: a new cell cycle motif? Trends Cell Biol. 1991;1:117–121. doi: 10.1016/0962-8924(91)90116-q. [DOI] [PubMed] [Google Scholar]
- 47.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 48.Reed S I, Hadwiger J A, Lorincz A. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci USA. 1985;82:4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin D M, Glickman M H, Larsen C N, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider B L, Yang Q-H, Futcher A B. Linkage of replication to Start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 51.Schneider B L, Patton E E, Lanker S, Mendenhall M D, Wittenberg C, Futcher B, Tyers M. Yeast G1 cyclins are unstable in G1 phase. Nature. 1998;395:86–89. doi: 10.1038/25774. [DOI] [PubMed] [Google Scholar]
- 52.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:239–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 53.Seufert W, Futcher B, Jentsch S. Role of ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 54.Sherman F, Fink G, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 55.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 57.Stueland C S, Lew D J, Cismowski M J, Reed S I. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sudakin V, Ganoth D, Dahan A, Heller A, Hershko J, Luca F C, Ruderman J V, Hershko A. The cyclosome, a large complex containing ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y, Reed S I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 61.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Nocker S, Sadis S, Rubin D M, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra R D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 64.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 65.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34CDC28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]